Abstract
Inulin is a well-known fructose-based prebiotic which has been shown to stimulate the growth of bifidobacteria, a bacterial group generally considered beneficial for intestinal health. In the present study, we analyzed inulin-associated shifts in the total bacterial community of wild-type mice and mice carrying a genetically inactivated adenomatous polyposis coli tumor suppressor gene by using DNA-based approaches independent of bacterial culturability. Mice were fed a high-fat, nonfiber diet with or without inulin inclusion at a 10% (wt/wt) concentration. Cecal contents were analyzed after 0, 3, and 9 weeks on the experimental diets. Inulin inclusion significantly affected the total bacterial community structure of the cecum as determined by both a nonselective percent-guanine-plus-cytosine-based profiling analysis and a more specific 16S ribosomal DNA sequence analysis. The shifts included stimulation of bifidobacteria and suppression of clostridia, but sequence comparison revealed that the major shifts were within previously unknown bacterial taxa. Concomitantly, significantly higher bacterial densities, determined by flow cytometry, were observed with the inulin-amended diet, and the metabolism of the cecal bacterial community was altered, as indicated by higher levels of residual short-chain fatty acids, particularly lactic acid. With regard to all of the microbiological parameters measured, the wild-type mice and mice carrying a genetically inactivated adenomatous polyposis coli tumor suppressor gene were essentially identical. Studies of the implications of pre- and probiotics may need to be expanded to include careful analysis of their effects on the entire microbial community, rather than just a few well-known species. Further studies are needed to increase our understanding of the possible roles of currently unknown gastrointestinal bacteria in health and disease.
The bacterial community of the gastrointestinal (GI) tract is metabolically perhaps the most active conglomerate of cells in an animal's body. Thus, disorders in the microbial community may affect the intestinal health and disease of the host. Indeed, it is possible that some diseases, the causes of which are currently unknown, may stem from such disorders in the microbial community.
The role of GI tract bacteria with regard to the aberrant growth of epithelial cells in the intestinal tract is unclear. In several studies comparing germ-free and conventional mice or rats, microflora have been found to play an important role in the development of intestinal tumors. For example, microflora reduced the incidence of spontaneous polyps in the small intestine of BALB/c mice (38), whereas adenocarcinomas in T-cell receptor β and p53 double-knockout mice developed only in the presence of bacteria (27). In studies with mice carrying a genetically inactivated adenomatous polyposis coli tumor suppressor gene (ApcMin/+), conventionally raised mice exhibited increased numbers of adenomas in the middle section of the small intestine while germ-free mice did not (11). Based on reports like these, GI microflora and/or microbial metabolites appear to be among several, as yet unidentified, factors controlling epithelial cell growth and development.
Currently, a number of products are being developed and sold to improve GI health through modulation of the microflora. Prebiotics, carbohydrates bypassing the small intestine, are often especially intended to stimulate the growth of bifidobacteria due to their presumed beneficial role on GI health (19). Inulin, a fructo-oligomer isolated from chicory root, has been shown to stimulate the growth of bifidobacteria in several well-designed in vitro studies using either pure bacterial cultures (16, 28) or undefined inocula of GI origin (41, 50). Inulin and/or other fructo-oligosaccharides have been demonstrated to exert similar effects in vivo both in rodent models (7, 10) and in studies with human volunteers (5, 15, 30). Inulin has also been examined for its role in carcinogenesis. There are studies showing that inulin can reduce the formation of preneoplastic aberrant crypt foci in rats (42, 45). However, another study with ApcMin/+ mice suggested that inulin might increase the incidence of intestinal adenomas (34).
In studies exploring the effects of diets, prebiotics, or probiotics on GI tract microflora, a number of bacterial groups have become established as being of interest and thus are the ones regularly analyzed. Some of these are taxonomically homogenous groups such as bifidobacteria, whereas some are taxonomically heterogeneous bacteria grouped only for their capability to grow under the same set of selected culture conditions (e.g., total anaerobes or gram-positive cocci). The taxonomy of several bacterial genera resident in the GI tract is unclear and is currently undergoing a lengthy process of reorganization (e.g., the genera Clostridium, Eubacterium and Ruminococcus). Another equally important factor that limits the ability to study these microbe-animal interactions is that there are few methods available for the analysis of the total GI tract bacterial community. In many environments inhabited by complex bacterial communities such as the animal GI tract, only a fraction of the species can be effectively monitored by culture-based methods (2, 13, 25, 26, 36). Indeed, this may have led to a historical bias wherein the dietary responses of only a small fraction of total bacteria in the GI tract have been effectively studied and potentially important microbes have been missed altogether.
In the present study we monitored shifts in the total cecal bacterial community of mice fed a typical Western type diet (high-fat, nonfiber) with or without inulin amendment. Bacterial shifts in the ApcMin/+ mice model in response to inulin were of particular interest since such changes might have a role in the previously observed aberrant growth of the intestinal mucosa (34). For analysis of the total microbial community of the cecum, the chromosomal DNA of the total intestinal bacterial community (enterome) was subjected to a nonselective DNA-based analysis, percent-guanine-plus-cytosine (%G+C)-based profiling (24). This method has previously been shown to depict the total bacterial community of the GI tract in a highly reproducible manner (3, 4). In order to define the taxonomic units of bacteria affected by the inulin amendment, we used 16S ribosomal DNA (rDNA) sequencing in combination with the %G+C profiling. This study, based on methods independent of culturability, indicated that inulin significantly affected the cecal bacterial community.
MATERIALS AND METHODS
Animals.
Male C57BL/6J ApcMin/+ mice (designated hereafter ApcMin/+ mice) and wild-type female C57BL/6J Apc+/+ mice (designated hereafter wild-type mice) were obtained from the Jackson Laboratory (Bar Harbor, Maine). They were mated, and the genotypes of the offspring were determined by using allele-specific PCR (9). The animals were reared in plastic cages in a temperature- and humidity-controlled animal facility with a 12-h light-dark cycle. The weanlings were fed standard rodent pellets (Altromin 1314; Chr. Petersen A/S, Ringsted, Denmark) (hereafter referred to as starter diet) until the age of 6 weeks. They were then assigned to the two treatment groups (control diet or inulin diet). The animals had free access to feed and water throughout the experiment. Animals were sacrificed at the ages of 6, 9, and 15 weeks, at which time they had been on the experimental diets for 0, 3, or 9 weeks, respectively. The number of animals per time point and treatment was 8 to 12, including both females and males. The Laboratory Animal Ethics Committee of the Faculty of Agriculture and Forestry, University of Helsinki, approved the study protocol.
Diet.
The diets were semisynthetic and based on the recommendations of the American Institute of Nutrition (AIN-93 [43]). The control diet was a high-fat, nonfiber diet, with casein as the protein source and dextrose as the source of carbohydrate (39). The dietary fat was a mixture of butter, sunflower oil, and rapeseed oil, providing all essential fatty acids and giving a fatty acid profile similar to that found in Western-type diets. The diet was supplemented with a vitamin and mineral mixture and choline chloride to provide all essential nutrients (39). This regimen is hereafter referred to as the control diet. The inulin diet was prepared by supplementing the control diet with 10% (wt/wt) inulin (Raftiline; Orafti, Tienen, Belgium). This regimen is hereafter referred to as the inulin diet. The diets were prepared weekly and stored at 4°C.
Sampling.
Mice were sacrificed by CO2 asphyxiation, and the entire intestine of each mouse was removed. The cecum was opened longitudinally, and the pH was measured in situ. The contents of the ceca were collected into plastic vials and stored at −70°C prior to analysis. Digesta from the ceca of individual animals within each group were combined randomly to produce two pools of digesta for each experimental group.
Bacterial recovery.
Bacteria in cecal digesta were recovered after extensive washing by diluting the samples 1:30 with 50 mM sodium phosphate buffer (pH 8), shaking on a reciprocating horizontal platform shaker at 200 rpm for 10 min, and centrifuging at 30,000 × g for 30 min at room temperature. The supernatant was discarded, and the pellet was washed three more times as described above. Prior to the last pelleting of bacteria, a subsample from each suspension was withdrawn for counting of total bacterial numbers. After the fifth centrifugation, the pellet was resuspended in 3.1 ml of 50 mM Tris (pH 8) buffer with 50 mM EDTA.
Recovery of enteromic DNA.
Bacterial suspensions were subjected to freeze-thaw cycles and lysed according to a previously described protocol (3), which combines physical (bead-beating), chemical (sodium dodecyl sulfate), and enzymatic (lysozyme and proteinase K) steps. This protocol has been shown to lyse more than 99% of the bacteria in chicken cecum samples (3). The recovered total bacterial DNA was further purified by using Qiagen Genomic-tip 100/G according to the manufacturer's instructions (Qiagen Sciences, Germantown, Md.).
Analysis of the cecal enterome by %G+C profiling.
Fifty micrograms of DNA from each enterome sample was subjected to cesium chloride-bisbenzimidazole gradient analysis (3). This approach fractionates the DNA of the component populations of the bacterial community based on their characteristic %G+C content through differential density, which is imposed by the AT-dependent DNA-binding dye, bisbenzimidazole (24). Determination of the %G+C content represented by each gradient fraction was accomplished by regression analysis (r2 > 0.99) of data obtained from gradients containing standard DNA samples of known %G+C content (Clostridium perfringens, Escherichia coli, and Micrococcus lysodeikticus). The %G+C profile of the total bacterial community was divided into twelve 5% increments spanning from 20 to 80% G+C, which represents the total range of G+C contents observed. The proportion of microbes within each increment was obtained by integrating the corresponding area for each analyzed sample.
16S rDNA sequence analysis.
DNA samples from cesium chloride gradients with a defined %G+C were desalted by using PD-10 Sephadex columns (Pharmacia, Uppsala, Sweden), and partial 16S rDNA genes of the organisms present in each sample were amplified by using PCR and the universal primers 536f (5′-CAGC(AC)GCCGCGGTAAT(AT)C-3′) and 907r (5′-CCCCGTCAATTCCTTTGAGTTT-3′) as described in detail previously (4, 26). These primers are based on highly conserved regions of the 16S and 18S rDNA genes of all organisms (hence, universal) and are thus suitable for this DNA sequence-based study. Amplified bacterial PCR products (∼400 bp) were subsequently cloned into the EcoRV site of the pT7Blue-3 vector using the Perfectly Blunt Cloning Kit (Novagen, Madison, Wis.). Plasmid DNA was purified from randomly picked clones, and insert size was confirmed by restriction fragment analysis using EcoRI. Confirmed clones were subjected to sequence analysis (MWG Biotech, High Point, N.C.) and sequence comparison by using the Ribosomal Database Project II (31) at http://rdp.cme.msu.edu/html as described previously (4, 26).
All of the sequences included in the phylogenetic tree analysis were first aligned by using Bionumerics 2.0 software (Applied Maths, Kortrijk, Belgium). Phylogenetic trees were generated by the maximum-parsimony method with UPGMA cluster analysis and by the neighbor-joining method based on the multiple alignment similarity matrix. Each phylogram was bootstrapped 100 times to assess the degree of support for the phylogenetic branching indicated by the optimal tree for each method.
Direct determination of bacterial numbers.
For the determination of total bacterial numbers, subsamples withdrawn during the bacterial recovery process were fixed with 37% formaldehyde to obtain a final concentration of 4%. The fixed bacterial suspensions were diluted as appropriate, and the cells were stained with a fluorescent, nucleic acid binding dye, SYTO24 according to the manufacturer's instructions (Molecular Probes, Leiden, The Netherlands). The stained samples were analyzed by using a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, N.J.) with parameters adjusted for bacterial counting. The bacteria were quantified by using BD TrueCount tubes (Becton Dickinson) with a preset number of fluorescent beads included as an internal standard. The number of events counted per sample was between 2,000 and 8,000.
SCFA analysis.
Samples of cecal digesta were analyzed for short chain fatty acid (SCFAs) by gas chromatography using an internal standard method as described previously (26).
Statistical analysis.
The Student's t test was used to detect statistically significant differences of pair-wise combinations of variables (P ≤ 0.05), e.g., between the groups of mice on the control diet versus the inulin diet.
Principal-component analysis was carried out to visualize the most important factors (diet, age, genetic background) affecting the microbiological parameters (4). For a review of principal-component analysis, see reference 1. The results of the analysis were summarized in a figure with unitless axes where every point represents data from one pooled sample. Points that cluster together in that graphical representation share similar overall data patterns.
The probability that the abundance of a certain phylotype in the total community was affected by inulin was calculated for each phylotype. When a total of N sequences were analyzed from a sample, the observed number of sequences nx, representing a certain phylotype x, follows multinomial distribution if the proportion of organism x is px. The likelihood of a change between control and inulin diet was calculated by multiplying the probability of observing nx,con sequences in the control treatment and the probability of observing nx,inu sequences in the inulin treatment and summing these probabilities over all proportions px,con and over such proportions px,inu that px,inu is less than px,con.
Nucleotide sequence accession numbers.
All sequences included in these analyses have been deposited in GenBank under accession numbers AY122340 through AY122355.
RESULTS
Inulin stimulates the growth of cecal bacteria.
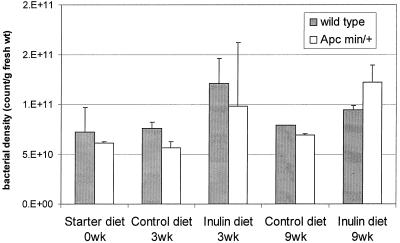
Mice with two different genetic backgrounds were fed standard pellets from weaning until the age of 6 weeks and then transferred to two experimental diets, control diet and inulin diet. The effects of the control and inulin diets on the density of the cecal bacterial community were measured by direct flow cytometric enumeration, which counts all bacterial cells, dead or live, provided that they have intracellular DNA. Inulin increased the total bacterial densities by an average of 55% (inulin versus control diet, P = 0.0007) with absolute numbers increasing from 7.0 × 1010 to 1.1 × 1011 bacteria per g of fresh cecal digesta (Fig. 1). It is worth noting that although the starter diet contained fiber, it did not give rise to bacterial densities higher than those of the nonfiber control diet.
FIG. 1.
Effects of diet on total bacterial densities in the ceca of wild-type and ApcMin/+ mice. Bacterial densities in the ceca of wild-type and ApcMin/+ mice were determined by flow cytometry before the animals were transferred to the experimental diets at the age of 6 weeks (Starter diet 0 wk) and again after 3 and 9 weeks on the experimental diets (Control diet and Inulin diet). Bacterial densities are shown in shaded columns for wild-type mice and in white columns for ApcMin/+ mice. Each column represents two pools of 8 to 12 animals. Error bars show ± standard errors of the means.
Inulin increases the residual concentration of short-chain fatty acids.
To evaluate the overall metabolic activity of the bacterial community in the cecum, the pH of digesta and the levels of major products of bacterial metabolism, SCFAs, were determined. The pH in the ceca of mice fed the inulin diet from week 6 onward did not significantly differ from the pH of those fed only starter diet (starter versus inulin diet mice, P > 0.05; Table 1), whereas, the cecal pH of animals fed the control diet after the starter diet was significantly higher than the pH of the animals on the starter diet (starter versus control diet, P < 0.0001). Animals kept on the control diet for 3 weeks showed an average cecal pH that was 0.8 units higher than that of mice on the starter diet (Table 1).
TABLE 1.
Effects of diet on cecal pH in wild-type and ApcMin/+ mice
| Diet | Age (wks) | Cecal pH ± SE (n)
|
|
|---|---|---|---|
| Wild type | ApcMin/+ | ||
| Starter | 6 | 7.1 ± 0.12 (11) | 6.7 ± 0.08 (9) |
| Control | 9 | 7.7 ± 0.12 (8) | 7.6 ± 0.04 (11) |
| Inulin | 9 | 6.9 ± 0.11 (9) | 7.2 ± 0.09 (9) |
| Control | 15 | 7.6 ± 0.05 (8) | 7.5 ± 0.03 (8) |
| Inulin | 15 | 7.3 ± 0.05 (8) | 6.9 ± 0.13 (9) |
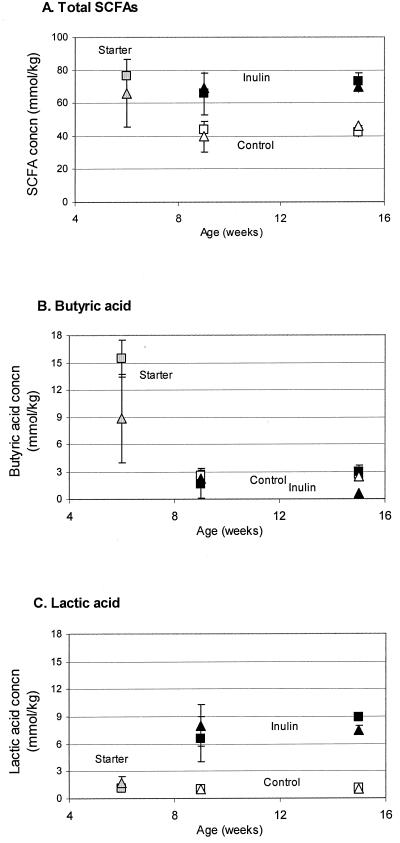
SCFA analysis showed that the total residual acid concentration in the cecum was at its highest level, approximately 70 mM, after mice had been fed the starter diet for 6 weeks (Fig. 2A). Inulin in the diet maintained the concentration of total SCFAs at about the same level (starter versus inulin diet, P > 0.05), suggesting that the overall bacterial fermentation activity was unchanged. However, the mice on the control diet had a level of total cecal SCFAs 40% lower than that of mice fed the inulin diet (control versus inulin diet, P = 0.0005). Wild-type and ApcMin/+ mice displayed almost identical responses to the diets with regard to SCFA (Fig. 2A; wild-type versus ApcMin/+ mice on all diets, P > 0.05).
FIG. 2.
Effects of diet on cecal concentrations of short-chain fatty acids. Residual concentrations of short-chain fatty acids in the ceca of wild type and ApcMin/+ mice were analyzed by gas chromatography following the sampling scheme described in the legend of Fig. 1. (A) Concentration of total short-chain fatty acids; (B) concentration of butyric acid; (C) concentration of lactic acid. Colors of the symbols indicate different diets (gray, starter diet; white, control diet; black, inulin diet), and the shapes of symbols indicate the genetic backgrounds of the mice (triangle, wild type; square, ApcMin/+). Each data point represents two pools of 8 to 12 animals. Error bars show ± standard errors of the means.
The relative proportions of different SCFAs changed in response to dietary treatment, suggesting that differences occurred in the metabolism of the cecal bacterial community. The residual butyric acid concentration was 9 and 15 mM (13 and 20% of the total SCFAs) in the ceca of wild-type and ApcMin/+ mice on the starter diet, respectively (Fig. 2B). When the animals were fed the Western-type diet, butyric acid levels dropped to <3 mM for both diets (starter versus control and starter versus inulin diet, P < 0.0001) in both mouse types. As a result, the proportion of butyric acid dropped to 6 and 3% of the total SCFAs in the ceca of both mouse types on the control and inulin diets, respectively. Absolute butyric acid levels were similar (control versus inulin diet, P > 0.05), whereas the relative proportion of butyric acid was lower in the group of inulin mice than in the control group (control versus inulin diet, P = 0.01).
While the residual concentration of lactic acid was about 1 mM in the ceca of all animals on the starter and control diets, inulin in the diet significantly increased the concentration of lactic acid, from 1 to 8 mM (control versus inulin diet, P < 0.0001; Fig. 2C). Consequently, lactic acid constituted about 11% of the total SCFAs in the inulin treatment, meaning that it became as abundant as propionic acid in the cecum. Overall, the genetic background (wild type versus ApcMin/+) and sampling time (9 versus 15 weeks) had no significant effect on the residual concentration of SCFAs in the cecum.
The composition of the cecal bacterial community is modulated by diet.
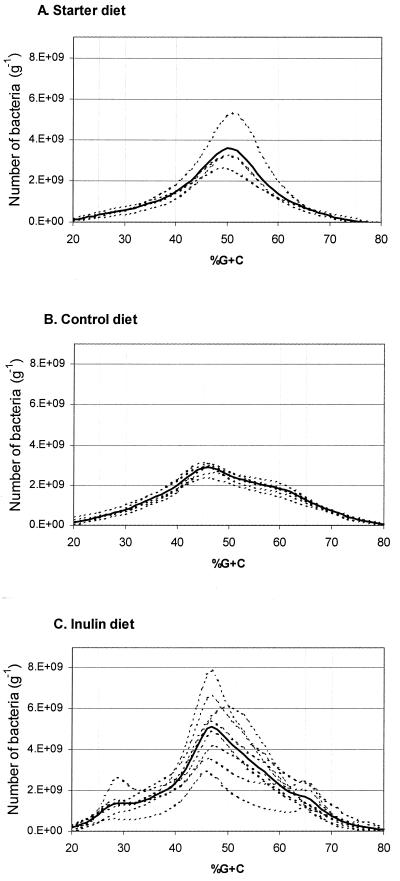
Total bacteria were recovered by differential centrifugation from two pools of digesta per treatment, and their DNA was purified. These enterome samples were then subjected to %G+C profiling. To better illustrate the shifts in the respective bacterial communities, the integral of each profile was corrected for the bacterial density in that sample of digesta (see Fig. 1). After this transformation, the DNA profiles indicate the absolute abundance (numbers of bacteria) and not just the relative distribution of bacteria with different G+C percentages in their genome (Fig. 3).
FIG. 3.
Percent G+C profiles of cecal bacterial communities in mice on different diets. (A) Four cecal bacterial profiles each representing pools of digesta from 8 to 12 animals on the starter diet. (B) Six cecal bacterial profiles each representing pools of digesta from 8 to 12 animals on the control diet. (C) Eight cecal bacterial profiles each representing pools of digesta from 8 to 12 animals on the inulin diet. Dotted lines, profiles of individual pools; solid lines, average profile on a specified diet.
Statistical analysis showed that the genetic background and age of the mice had no effect on the %G+C profiles of the cecal bacterial communities (wild type versus ApcMin/+, P > 0.05) for any 5% G+C increment, time point, or diet (data not shown), and therefore in Fig. 3 the profiles were combined based on diet.
All four bacterial community profiles analyzed from animals on the starter diet (two pooled samples each from wild-type and ApcMin/+ mice) were similar and revealed that bacteria with G+C content of approximately 50% dominated the cecal digesta. No additional peaks or shoulders were detected (Fig. 3A). Mice on the control diet showed a shift in bacterial community composition in all of the profiles analyzed. Ideally, there would be two pooled samples for each genetic background and each time point for a total of eight. However, due to the small amount of digesta and low bacterial densities in animals on the control diet, we did not recover enough DNA for two of the samples (the 9-week time point of ApcMin/+ mice and the 15-week time point of wild-type mice) to run duplicate %G+C profiles. The major peak of the profiles was at 45% G+C, and a sizeable shoulder in the profiles was observed at 62% G+C (Fig. 3B). The cecal bacterial community of mice on the inulin diet was noticeably different from those of mice on the other diets. The major peak appeared at about 47% G+C (Fig. 3C). In addition, evident peaks and shoulders were seen at 28 and 65% G+C. Even though the variability in cecal bacterial community structure in the pooled samples from the inulin group was substantially higher than that of the animals on the other diets, all of the profiles shared these features characteristic of the inulin diet (Fig. 3C).
Adaptation of the microflora to a new diet regimen seemed to have been completed after 3 weeks, since %G+C profiles from 9-week-old mice did not significantly differ from the profiles of 15-week-old animals (P > 0.05 for any 5% G+C increment) for either treatment.
16S rDNA sequencing reveals abundant populations of unknown phylotypes.
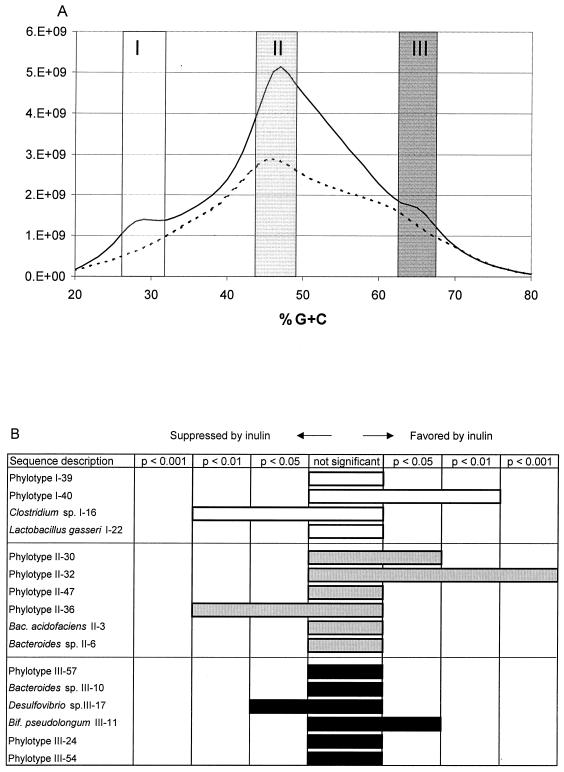
Since the major bacterial community shifts associated with inulin in the diet were localized to three regions of the %G+C profiles centered around 28, 47, and 65% G+C, we concentrated our efforts on studying these areas in more detail. Since age and genetic background had little or no effect on any of the bacterial parameters described above, the enterome samples from animals within each of the two dietary groups mimicking the Western-type diet were pooled to obtain two samples representing the control and the inulin diet (Fig. 4A). The community fractions ranging from 27 to 32%G+C (I), 44 to 49%G+C (II), and 63 to 68%G+C (III) (Fig. 4A) were subjected to 16S rDNA analysis.
FIG. 4.
16S rDNA sequence analysis from %G+C fractions characteristic of the inulin diets. (A) Pooled %G+C profiles of all mice on the control diet (dotted line) and all mice on the inulin diet (solid line). Vertical columns show the fractions of DNA selected for 16S rDNA-based analysis from each profile. Fractions corresponded to 27 to 32% G+C (I), 44 to 49% G+C (II), and 63 to 68% G+C (III). (B) The effects of inulin on the abundance of dominant phylotypes were determined for all sequence phylotypes represented at least twice. The outcome is represented as a confidence plot where each phylotype is considered to be suppressed, show no significant effect, or be favored by inulin, with the corresponding confidence interval indicated (refer to Materials and Methods). White bars, phylotypes in fraction I of panel A; gray bars, phylotypes in fraction II of panel A; black bars, phylotypes in fraction III of panel A.
PCR using universal primers was used to amplify partial 16S rDNA sequences from the three DNA fractions recovered from each enterome representing either control or inulin diet-fed mice. The resulting PCR products were cloned, and 21 randomly picked clones from each %G+C fraction were subjected to DNA sequence analysis to identify the dominant bacterial phylotypes in those fractions. Statistical analysis (see Materials and Methods for details) was performed to calculate the probability that the abundance of the bacteria representing the phylotypes observed was affected by diet (Fig. 4B).
The effect of inulin on the abundance of the bacterial phylotypes (sequences), which appeared at least twice in the study (60% of all sequences), is shown in Fig. 4B. The sequences obtained through bidirectional sequence analysis of the cloned 16S rDNA fragments were compared to a public 16S ribosomal database, RDP-II (31), to determine the closest match to aligned sequences of known species. Based on prior experience with partial 16S sequences from type strains and newly cloned sequences, when the Sab score (similarity score a versus b) of a cloned sequence was >0.95 to the type strain of any known species, the cloned sequence was assigned to that species. Whenever the Sab score of a sequence was <0.95 but >0.7 to any known sequence with at least genus level identification, the sequence was assigned to that genus. If the Sab score of a sequence was <0.7 to any known sequence, that sequence was labeled “phylotype.” Due to the relatively small number of sequences analyzed from each DNA fraction, we could not, at >95% confidence level, detect bacteria whose relative abundance was below 15% of the total bacteria in each fraction.
Most of the sequences obtained in this analysis represent novel phylotypes which, based on their low similarities to any sequence in the existing database, can be considered to represent previously unknown bacteria. This was true in all %G+C fractions analyzed. In the low %G+C fraction (Fig. 4B, fraction I), Lactobacillus gasseri was found in animals from both treatment groups, whereas a Clostridium sp. was found only in the control group. With inulin in the diet, an unknown bacterial phylotype labeled I-40 was significantly favored in the low %G+C fraction. (Fig. 4B). The genus Bacteroides was found in the mid %G+C fraction (Fig. 4B, fraction II). The major shifts observed in response to diet applied to unknown phylotypes (phylotypes II-30, II-32, and II-36). Unknowns II-30 and II-32 were not detected at all in the ceca of mice on the control diet but were abundant in the ceca of inulin-fed animals. The major shifts observed in known bacteria in the high %G+C fraction of the enterome (Fig. 4B, fraction III) were in species of Bifidobacterium and Desulfovibrio, the former of which was detected only in animals on the inulin diet and the latter only in animals on the control diet. Of the phylotypes that appeared only once in the whole study, 70% appeared to represent bacterial genera that could not be assigned into any genus based on the 16S rDNA fragment analyzed, and their proportion was similar in the two diets and all three fractions (data not shown).
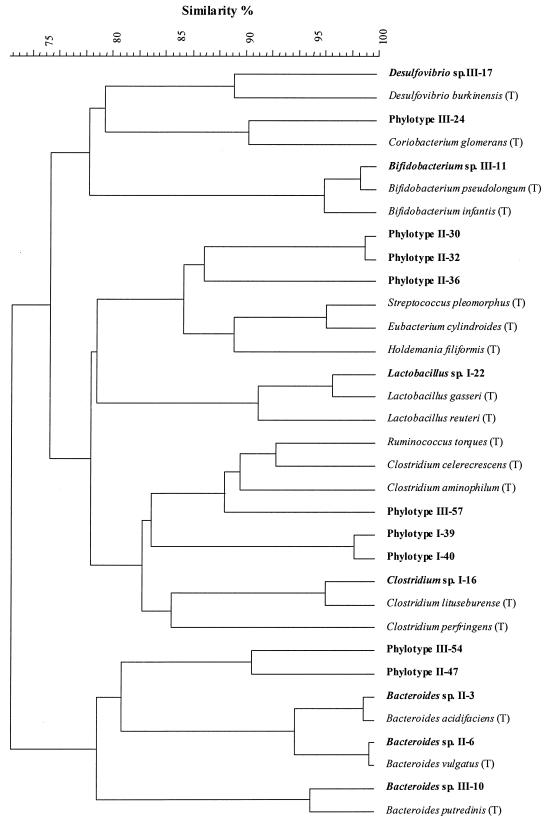
In order to obtain some sense of the phylogenetic placement of the unknown phylotypes in the context of known representatives of GI tract bacteria, we constructed phylograms containing the sequences of the phylotypes shown in Fig. 4B and the corresponding aligned sequences of type strains representative of several known GI taxa (Fig. 5). Phylograms generated by the maximum-parsimony and neighbor-joining methods produced identical branching points with very similar bootstrap values. The maximum-parsimony phylogram is presented in Fig. 5. This analysis also confirmed the identities of the phylotypes identified and assigned to the genus and species levels. In this analysis, both of the phylotypes identified to the species level, Bacteroides acidifaciens and L. gasseri, were shown to be highly related to the named species. Figure 5 also shows that all of the phylotypes identified to the genus level (Bifidobacterium, Desulfovibrio, Clostridium, and Bacteroides) were related to other members of their respective genera. Most unknown phylotypes were at least loosely associated with some known group of GI tract bacteria. For example, unknown III-24 was loosely associated with the bifidobacteria, while unknown III-57 appeared to be related to Clostridium aminophilum, Clostridium celerecrescens, and Ruminococcus torques. Unknowns II-30 and II-32, which were dominant in the inulin treatment and highly related to each other, were loosely related to unknown II-36 and the Eubacterium cylindroides-Streptococcus pleomorphus-Holdemania philiformis cluster. Unknowns III-54 and II-47 clustered with the representatives of the genus Bacteroides, while unknowns I-39 and I-40, which are clearly related to each other, were loosely associated with the genera Ruminococcus and Clostridium (Fig. 5).
FIG. 5.
Phylogenetic analysis of the most abundant bacterial phylotypes inhabiting the mouse cecum. The bacterial phylotypes shown in Fig. 4B were subjected to phylogenetic analysis together with the type strains of some known intestinal bacteria whose 16S rDNA sequences were obtained from the RDP II sequence database (31). Type strains are indicated by (T) after the species name, and phylotypes found in the present study are indicated by boldface type. Prefixes I, II, and III in strain numbers indicate the fraction of %G+C profile from which the phylotype originates. I, low %G+C; II, medium %G+C; III, high %G+C.
Principal-component analysis of the data.
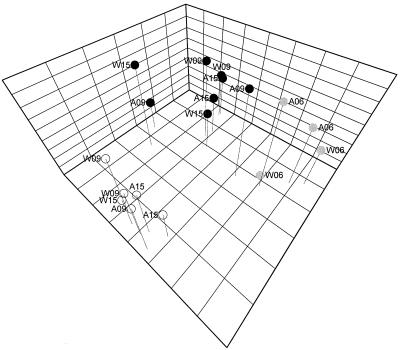
All the data presented above were subjected to principal-component analysis. The results illustrated in Fig. 6 show that the clustering of the animals was determined solely by the diet, not by age or genetic background (Fig. 6).
FIG. 6.
Principal-component analysis of the bacterial parameters. Each data point in the graph represents a pool of 8 to 12 mice, whose genetic backgrounds and ages are indicated in the labels next to each symbol. W, wild type; A, Apcmin/+; 06, 09, and 15, age of 6, 9, and 15 weeks, respectively. Clustering of the data points is based on SCFA, bacterial density, and %G+C analyses. The color of each symbol indicates the type of diet as follows: gray, starter diet; white, control diet; black, inulin diet.
DISCUSSION
The %G+C profiling technique used in this study is among the few culture-independent techniques truly capable of depicting the total bacterial community of the GI tract in a single analysis. In complex bacterial communities with many unknown members, the diagnostic value of the %G+C profiling method alone is limited. However, the three diets fed to mice produced bacterial community profiles significantly different from one another. Also, as shown here, the method can be effectively used in combination with other approaches such as 16S rDNA sequence analysis when data on specific bacterial taxa are required.
Only two previously known bacterial species were found in the enterome fractions when the criteria used demanded an Sab score >0.95 to a type strain of a species. These were L. gasseri and B. acidifaciens, which were found in the low (I) and middle (II) %G+C fractions, respectively. The reported %G+C contents of L. gasseri and B. acidifaciens are 33 to 35% and 42%, respectively (21, 32), which is consistent with the positioning of these sequences in %G+C profiles. Five other phylotypes were assigned to previously known genera Bifidobacterium, Clostridium, Bacteroides, and Desulfovibrio but did not match with any previously known species according to the criteria used (Sab > 0.95). Clostridium sp. was found in the low (I) %G+C fraction, and Desulfovibrio sp. and Bifidobacterium sp. were found in the high (III) %G+C fraction, while Bacteroides spp. were observed both in the middle (II) and high (III) %G+C fractions. The %G+C content ranges reported for these genera (12, 37, 47) are also consistent with the observations made here, but the diversity within the genera is high. Clostridium sp. and Desulfovibrio sp. were significantly suppressed by the inclusion of inulin in the diet (Fig. 4B).
In addition to the Sab approach, we also analyzed the data by using sequence alignment and calculation of the sequence similarity percentage. While the similarity percentage indicates only the homology between two fragments of DNA, the Sab approach emphasizes the divergence based on the number of individual evolutionary events (mutations) distinct for the phylotypes. Therefore, the use of the Sab algorithm appears justified when the taxonomic distance is of interest. In this study these two approaches gave largely similar results. Still, Bifidobacterium sp. III-11 and Bacteroides sp. II-6 showed high sequence similarity to the type strains of Bifidobacterium pseudolongum and Bacteroides vulgatus (97 and 99%, respectively) but did not fulfill the Sab criteria for their assignment to those species. The unknown phylotypes II-30 and II-32 were highly related to each other, as were I-39 and I-40. However, according to the Sab criteria all these represented different species.
Previously unknown bacteria appeared to dominate in the ceca of mice. Of the total number of sequences analyzed from mice on the control diet, 50% could not be assigned to any known genus according to the partial 16S rDNA sequence analyzed. For the sequences obtained from mice on the inulin diet, the proportion of potentially unknown bacterial taxa was even higher, 66% of all sequences. Perhaps the most striking individual inulin-driven effect was the outgrowth of unknown phylotypes, especially the unknowns II-30 and II-32, which likely accounted for the major peak observed at 47% in the G+C profiles. The roles of these and other previously unknown species in the health and disease of the host are unclear.
There are many studies that have monitored the effects of inulin on the bacteria of the GI tracts of humans and animals. With few exceptions (29, 30), these studies have used culture-based methods and therefore are not comparable to the results of the present study. Culture-based techniques can be very selective, effectively detecting minority populations, but likely never capture the total microbial community of anaerobic habitats such as the mouse cecum. Yet, many of the observations made here are consistent with those made before. In agreement with the present study, bifidobacteria have previously been reported to be stimulated by inulin in human and animal studies (5, 7, 10, 15, 16, 28, 30, 41, 50), clostridia have been reported to be suppressed by inulin (29), and B. acidifaciens was originally isolated from and reported to be found only in the mouse cecum (32).
The overall fermentation activity and bacterial numbers in inulin-fed mice were significantly higher than in animals on the control diet when measured as residual levels of total SCFAs and bacterial densities in the cecum. There are several previous reports on laboratory rats which showed that the level of SCFAs in the cecum became elevated and/or the pH was lowered when inulin was included in the diet (29, 44, 48). By contrast, in intervention studies with human subjects, inulin did not increase the concentration of total SCFAs when those were analyzed from fecal samples (6, 15, 30). Kleessen et al. showed that the effect of inulin on SCFAs in rats was most pronounced at the actual site of fermentation (i.e., the cecum) and became less significant when it was analyzed distal to that site or from fecal samples (29). Most likely, SCFAs become further converted to other metabolites by bacteria and are absorbed and utilized by the intestinal epithelium distal to the cecum. As a consequence, their concentrations and proportions in the feces may not reflect the residual levels in the cecum or proximal colon.
Inulin in the diet significantly increased the residual concentration of lactic acid in the cecum. This is consistent with the finding that potentially lactic acid-producing bacteria (Bifidobacterium) increased, whereas Desulfovibrio sp., a potentially lactic acid-utilizing species (20), was suppressed when diet was supplemented with inulin. Even though lactic acid is an end product of the catabolism of some members of the cecal bacterial community, in a tightly coupled bacterial food web it could be expected to be a short-lived transient intermediate. Indeed, if a strictly anaerobic, sulfidogenic, or methanogenic microbial community accumulates lactic acid, the situation is not energetically ideal for the bacterial community. A classic example of a diet-related bacterial disorder related to lactic acid accumulation is acidosis in the rumen of dairy cows. Acidosis is caused by change to a diet containing a rapidly degradable substrate for lactic acid bacteria, leading to a rapid lactic acid accumulation and, as a consequence, inhibition and washout of most members of the natural microbial community of the rumen (8, 33). Still, some lactic acid production is often considered beneficial in the GI tract because it inhibits the growth of several potential intestinal pathogens such as Salmonella, Clostridium, and Escherichia (17, 22, 23, 35, 40).
Volatile fatty acids have been shown to stimulate the proliferation of intestinal epithelial cells (46). Butyric acid in particular was found to be effective at physiological concentrations relevant for the lower GI tract of homeothermic animals (46). Furthermore, the stimulating effect of butyric acid has been found with normal rather than aberrantly growing epithelial cells (14, 18, 49). Based on such observations, butyric acid is considered beneficial for GI health. Representatives of several bacterial genera in the GI tract produce butyric acid, but it is worth noting that species of Bifidobacterium and Lactobacillus are not among these (21, 47). In the present study we found that mice on the experimental Western-type diets had significantly lower residual concentrations of butyric acid in the cecum than those of animals on the fiber-containing starter diet and that inulin amendment had little effect on these levels. In earlier animal and human studies, the effects of inulin on the level of butyric acid have been inconsistent (6, 15, 29, 30, 44).
Inulin in the diet of conventional laboratory mice was found to catalyze several significant chemical and bacteriological shifts. The results of principal-component analysis clearly showed that diet was the only significant factor affecting the structure and metabolic profile of the cecal bacterial community. Indeed, neither genetic background nor age affected the clustering of the data points in Fig. 6. Regarding all the parameters measured, the wild-type mouse and ApcMin/+ mouse were identical. This finding strongly suggests that when bacterial shifts associated with inulin amendment are studied, animals with ApcMin/+ genetic background serve as a good general model for mice. Also, it is noteworthy that the values for the bacteriological parameters in 9- and 15-week-old mice were practically identical, suggesting that conditions for cecal bacteria did not change after the first 3 weeks on the experimental diets (Fig. 6).
In a companion study with the same individual animals as studied here, it was found that inulin significantly increased the numbers and sizes of tumors in the distal small intestines of ApcMin/+ mice (A.-M. Pajari, J. Rajakangas, E. Päivärinta, V.-M. Kosma, J. Rafter, and M. Mutanen, Abstr. AACR Spec. Conf. Cancer Res., abstr. A-16, 2002). This was consistent with a study reported previously at a lower, 2.5% inclusion level of inulin (34). Although not a primary objective of the present study, the microbial parameters measured here did not indicate any direct positive or negative effect of microflora in the observed aberrant growth of intestinal adenomas. Additional studies especially designed to assess the role of individual microbes in tumor formation should be performed.
Collectively, inulin in comparison to the corresponding diet with no amendment stimulated overall bacterial growth and fermentation, increased the number of bifidobacteria, and increased the residual concentration of lactic acid in the cecum. The culture-independent techniques used in this study were applied for the first time in monitoring the effects of inulin on gut microflora. This approach confirmed some bacterial population shifts described previously but, perhaps more importantly, revealed that the major shifts resulting from inulin amendment concern bacterial species not previously described. Concomitantly, inulin in the diet seemed to increase the growth of intestinal adenomas. These data do not show that the observed microbial shifts are in any way connected to aberrant growth of intestinal epithelial cells. However, neither do they support the notion that suppression of clostridia and desulfovibrios or stimulation of bifidobacteria would protect against the development of adenomas. Mechanistic studies and careful analysis of host-bacteria interactions are needed to increase our understanding of the possible roles of different GI bacteria in the development of colon cancer.
Acknowledgments
We gratefully acknowledge Anne-Maria Pajari for her major role in the implementation of the animal study. We also thank Harri Mäkivuokko for constructing the phylogenetic tree and Markku Saarinen for the analysis of short-chain fatty acids. We also acknowledge Osmo Siikanen, Linda M. Schimmelpfennig, and Jaana Oksanen for excellent technical assistance and Seppo Peuranen for valuable comments.
The work was financially supported by the Finnish National Technology Agency, Tekes.
REFERENCES
- 1.Afifi, A. A., and V. Clark. 1990. Principal component analysis, p. 371-394. In A. A. Afifi and V. Clark (ed.), Computer-aided multivariate analysis. Chapman & Hall, New York, N.Y.
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apajalahti, J. H. A., L. K. Sarkilahti, B. R. Maki, J. P. Heikkinen, P. H. Nurminen, and W. E. Holben. 1998. Effective recovery of bacterial DNA and percent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl. Environ. Microbiol. 64:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apajalahti, J. H. A., A. Kettunen, M. R. Bedford, and W. E. Holben. 2001. Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens. Appl. Env. Microbiol. 67:5656-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouhnik, Y., B. Flourie, M. Riottot, N. Bisetti, M. F. Gailing, A. Guibert, F. Bornet, and J. C. Rambaud. 1996. Effects of fructo-oligosaccharides ingestion on fecal bifidobacteria and selected metabolic indexes of colon carcinogenesis in healthy humans. Nutr. Cancer 26:21-29. [DOI] [PubMed] [Google Scholar]
- 6.Brighenti, F., M. C. Casiraghi, E. Canzi, and A. Ferrari. 1999. Effect of consumption of a ready-to-eat breakfast cereal containing inulin on the intestinal milieu and blood lipids in healthy male volunteers. Eur. J. Clin. Nutr. 53:726-733. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, J. M., G. C. Fahey, Jr., and B. W. Wolf. 1997. Selected indigestible oligosaccharides affect large bowel mass, cecal and fecal short-chain fatty acids, pH and microflora in rats. J. Nutr. 127:130-136. [DOI] [PubMed] [Google Scholar]
- 8.Counotte, G. H., and R. A. Prins. 1981. Regulation of lactate metabolism in the rumen. Vet. Res. Commun. 5:101-115. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich, W. F., E. S. Lander, J. S. Smith, A. R. Moser, K. A. Gould, C. Luongo, N. Borenstein, and W. Dove. 1993. Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell 75:631-639. [DOI] [PubMed] [Google Scholar]
- 10.Djouzi, Z., and C. Andrieux. 1997. Compared effects of three oligosaccharides on metabolism of intestinal microflora in rats inoculated with a human faecal flora. Br. J. Nutr. 78:313-324. [DOI] [PubMed] [Google Scholar]
- 11.Dove, W. F., L. Clipson, K. A. Gould, C. Luongo, D. J. Marshall, A. R. Moser, M. A. Newton, and R. F. Jacoby. 1997. Intestinal neoplasia in the ApcMin mouse: independence from the microbial and natural killer (beige locus) status. Cancer Res. 57:812-814. [PubMed] [Google Scholar]
- 12.Drzyzga, O., J. Kuver, and K. H. Blotevogel. 1993. Complete oxidation of benzoate and 4-hydroxybenzoate by a new sulfate-reducing bacterium resembling Desulfoarculus. Arch. Microbiol. 159:109-113. [DOI] [PubMed] [Google Scholar]
- 13.Fritsche, T. R., M. Horn, S. Seyedirashti, R. K. Gautom, K. H. Schleifer, and M. Wagner. 1999. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl. Environ. Microbiol. 65:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.German, J. B. 1999. Butyric acid: a role in cancer prevention. BNF Nutr. Bull. 24:203-209. [Google Scholar]
- 15.Gibson, G. R., E. R. Beatty, X. Wang, and J. H. Cummings. 1995. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 108:975-982. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, G. R., and X. Wang. 1994. Bifidogenic properties of different types of fructo-oligosaccharides. Food Microbiol. 11:491-498. [Google Scholar]
- 17.Gibson, G. R., and X. Wang. 1994. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J. Appl. Bacteriol. 77:412-420. [DOI] [PubMed] [Google Scholar]
- 18.Gibson, P. R., O. Rosella, A. J. Wilson, J. M. Mariadason, K. Rickard, K. Byron, and D. H. Barkla. 1999. Colonic epithelial cell activation and the paradoxical effects of butyrate. Carcinogenesis 20:539-544. [DOI] [PubMed] [Google Scholar]
- 19.Grizard, D., and C. Barthomeuf. 1999. Non-digestible oligosaccharides used as prebiotic agents: mode of production and beneficial effects on animal and human health. Reproduction 39:563-588. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton, W. A. 1988. Energy transduction in anaerobic bacteria, p. 83-149. In C. Anthony (ed.), Bacterial energy transduction. Academic press, London, United Kingdom.
- 21.Hammes, W. P., and R. F. Vogel. 1995. The genus Lactobacillus, p. 19-54. In B. J. B. Wood and W. H. Holzapfel (ed.), The genera of lactic acid bacteria. Chapman & Hall, Glasgow, United Kingdom.
- 22.Hinton, A., Jr., R. J. Buhr, and K. D. Ingram. 2000. Reduction of Salmonella in the crop of broiler chickens subjected to feed withdrawal. Poult. Sci. 79:1566-1570. [DOI] [PubMed] [Google Scholar]
- 23.Hinton, A., Jr., and M. E. Hume. 1995. Synergism of lactate and succinate as metabolites utilized by Veillonella to inhibit the growth of Salmonella typhimurium and Salmonella enteritidis in vitro. Avian Dis. 39:309-316. [PubMed] [Google Scholar]
- 24.Holben, W. E., and D. Harris. 1995. DNA based monitoring of total bacterial community structure in environmental samples. Mol. Ecol. 4:627-631. [DOI] [PubMed] [Google Scholar]
- 25.Holben, W. E., K. Noto, T. Sumino, and Y. Suwa. 1998. Molecular analysis of bacterial communities in a three-compartment granular activated sludge system indicates community-level control by incompatible nitrification processes. Appl. Environ. Microbiol. 64:2528-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holben, W. E., L. K. Särkilahti, P. Williams, M. Saarinen, and J. H. A. Apajalahti. Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon. Microb. Ecol., in press. [DOI] [PubMed]
- 27.Kado, S., K. Uchida, H. Funabashi, S. Iwata, Y. Nagata, M. Ando, M. Onoue, Y. Matsuoka, M. Ohwaki, and M. Morotomi. 2001. Intestinal microflora are necessary for development of spontaneous adenocarcinoma of the large intestine in T-cell receptor beta chain and p53 double-knockout mice. Cancer Res. 61:2395-2398. [PubMed] [Google Scholar]
- 28.Kaplan, H., and R. W. Hutkins. 2000. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl. Environ. Microbiol. 66:2682-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleessen, B., L. Hartmann, and M. Blaut. 2001. Oligofructose and long-chain inulin: influence on the gut microbial ecology of rats associated with a human faecal flora. Br. J. Nutr. 86:291-300. [DOI] [PubMed] [Google Scholar]
- 30.Kruse, H. P., B. Kleessen, and M. Blaut. 1999. Effects of inulin on faecal bifidobacteria in human subjects. Br. J. Nutr. 82:375-382. [DOI] [PubMed] [Google Scholar]
- 31.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyamoto, Y., and K. Itoh. 2000. Bacteroides acidifaciens sp. nov., isolated from the cecum of mice. Int. J. Syst. Evol. Microbiol. 50:145-148. [DOI] [PubMed] [Google Scholar]
- 33.Moller, P. D., L. Diernaes, J. Sehested, J. Hyldgaard-Jensen, and E. Skadhauge. 1997. Absorption and fate of L- and D-lactic acid in ruminants. Comp. Biochem. Physiol. A Physiol. 118:387-388. [DOI] [PubMed] [Google Scholar]
- 34.Mutanen, M., A. M. Pajari, and S. I. Oikarinen. 2000. Beef induces and rye bran prevents the formation of intestinal polyps in Apc(Min) mice: relation to beta-catenin and PKC isozymes. Carcinogenesis 21:1167-1173. [DOI] [PubMed] [Google Scholar]
- 35.Nigatu, A., and B. A. Gashe. 1994. Inhibition of spoilage and food-borne pathogens by lactic acid bacteria isolated from fermenting tef (Eragrostis tef) dough. Ethiop. Med. J. 32:223-229. [PubMed] [Google Scholar]
- 36.Ohkuma, M., and T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouattara, A. S., N. Cuzin, A. S. Traore, and J. L. Garcia. 1992. Anaerobic degradation of 1,2-propanediol by a new Desulfovibrio strain and D. alcoholovorans. Arch. Microbiol. 158:218-225. [DOI] [PubMed] [Google Scholar]
- 38.Ozaki, A., Y. Morishita, T. Oowada, S. Aoe, and T. Mizutani. 1999. Inhibitory effect of intestinal bacteria on spontaneous multiple polyps in the small intestine of gnotobiotic BALB/c mice. J. Exp. Clin. Cancer Res. 18:255-258. [PubMed] [Google Scholar]
- 39.Pajari, A. M., S. Oikarinen, S. Grasten, and M. Mutanen. 2000. Diets enriched with cereal brans or inulin modulate protein kinase C activity and isozyme expression in rat colonic mucosa. Br. J. Nutr. 84:635-643. [PubMed] [Google Scholar]
- 40.Presser, K. A., D. A. Ratkowsky, and T. Ross. 1997. Modeling the growth rate of Escherichia coli as a function of pH and lactic acid concentration. Appl. Environ. Microbiol. 63:2355-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao, A. V. 1999. Dose-response effects of inulin and oligofructose on intestinal bifidogenesis effects. J. Nutr. 129:1442S-1445S. [DOI] [PubMed] [Google Scholar]
- 42.Reddy, B. S., R. Hamid, and C. V. Rao. 1997. Effect of dietary oligofructose and inulin on colonic preneoplastic aberrant crypt foci inhibition. Carcinogenesis 18:1371-1374. [DOI] [PubMed] [Google Scholar]
- 43.Reeves, P. G., F. H. Nielsen, and G. C. Fahey, Jr. 1993. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 123:1939-1951. [DOI] [PubMed] [Google Scholar]
- 44.Roland, N., L. Nugon-Baudon, C. Andrieux, and O. Szylit. 1995. Comparative study of the fermentative characteristics of inulin and different types of fibre in rats inoculated with a human whole faecal flora. Br. J. Nutr. 74:239-249. [DOI] [PubMed] [Google Scholar]
- 45.Rowland, I. R., C. J. Rumney, J. T. Coutts, and L. C. Lievense. 1998. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis 19:281-285. [DOI] [PubMed] [Google Scholar]
- 46.Sakata, T. 1987. Stimulatory effect of short-chain fatty acids on epithelial cell proliferation in the rat intestine: a possible explanation for trophic effects of fermentable fibre, gut microbes and luminal trophic factors. Br. J. Nutr. 58:95-103. [DOI] [PubMed] [Google Scholar]
- 47.Sgorbati, B., B. Biavati, and D. Palenzona. 1995. The genus Bifidobacterium, p. 279-306. In B. J. B. Wood and W. H. Holzapfel (ed.), The genera of lactic acid bacteria. Chapman & Hall, Glasgow, United Kingdom.
- 48.Videla, S., J. Vilaseca, M. Antolin, A. Garcia-Lafuente, F. Guarner, E. Crespo, J. Casalots, A. Salas, and J. R. Malagelada. 2001. Dietary inulin improves distal colitis induced by dextran sodium sulfate in the rat. Am. J. Gastroenterol. 96:1486-1493. [DOI] [PubMed] [Google Scholar]
- 49.Wang, J., and E. A. Friedman. 1998. Short-chain fatty acids induce cell cycle inhibitors in colonocytes. Gastroenterology 114:940-946. [DOI] [PubMed] [Google Scholar]
- 50.Wang, X., and G. R. Gibson. 1993. Effects of the in vitro fermentation of oligofructose and inulin by bacteria growing in the human large intestine. J. Appl. Bacteriol. 75:373-380. [DOI] [PubMed] [Google Scholar]