Abstract
Citrus variegated chlorosis (CVC) is caused by Xylella fastidiosa, a phytopathogenic bacterium that can infect all Citrus sinensis cultivars. The endophytic bacterial communities of healthy, resistant, and CVC-affected citrus plants were studied by using cultivation as well as cultivation-independent techniques. The endophytic communities were assessed in surface-disinfected citrus branches by plating and denaturing gradient gel electrophoresis (DGGE). Dominant isolates were characterized by fatty-acid methyl ester analysis as Bacillus pumilus, Curtobacterium flaccumfaciens, Enterobacter cloacae, Methylobacterium spp. (including Methylobacterium extorquens, M. fujisawaense, M. mesophilicum, M. radiotolerans, and M. zatmanii), Nocardia sp., Pantoea agglomerans, and Xanthomonas campestris. We observed a relationship between CVC symptoms and the frequency of isolation of species of Methylobacterium, the genus that we most frequently isolated from symptomatic plants. In contrast, we isolated C. flaccumfaciens significantly more frequently from asymptomatic plants than from those with symptoms of CVC while P. agglomerans was frequently isolated from tangerine (Citrus reticulata) and sweet-orange (C. sinensis) plants, irrespective of whether the plants were symptomatic or asymptomatic or showed symptoms of CVC. DGGE analysis of 16S rRNA gene fragments amplified from total plant DNA resulted in several bands that matched those from the bacterial isolates, indicating that DGGE profiles can be used to detect some endophytic bacteria of citrus plants. However, some bands had no match with any isolate, suggesting the occurrence of other, nonculturable or as yet uncultured, endophytic bacteria. A specific band with a high G+C ratio was observed only in asymptomatic plants. The higher frequency of C. flaccumfaciens in asymptomatic plants suggests a role for this organism in the resistance of plants to CVC.
Brazil is the largest producer of citrus fruit in the world, holding most of the international market for concentrated orange juice. More than 80% of the country's production comes from a single state, São Paulo, where the citrus industry and associated activities generate 3.4 million jobs and up to 1.5 billion U.S. dollars each year. In recent years, several diseases have affected this economically important crop and a great deal of attention has been focused on citrus variegated chlorosis (CVC), which was first reported in Brazil in 1987 (32) and has spread over at least 90% of the orchards in São Paulo (19). It seems that CVC is solely a disease of sweet-orange (Citrus sinensis L.) cultivars and is caused by Xylella fastidiosa (5, 40), a xylem-limited gram-negative bacterial pathogen that is transmitted by xylem-feeding suctorial insects (sharpshooters) and is known to infect several other economically important plants, resulting in extensive economic losses (16). In citrus plants, symptoms of the disease include mottled chlorosis of leaves, stunting, canopy die-back, and production of small fruits with a very hard rind that are rejected by the juicing industry (21).
There is no effective way to control the disease, but a sequencing program sponsored by the State Research Foundation of São Paulo (FAPESP) has recently sequenced the genome of X. fastidiosa, which causes CVC, and brought new perspectives to the problem (36). In addition, a small population of asymptomatic plants (called escape plants) has been consistently observed in some affected orchards and these offer the opportunity to develop new approaches to the control of CVC. Because these escape plants have the same genotype as susceptible plants and both types occur in the same orchards (i.e., have developed under similar edaphic and climatic conditions) a possible explanation for the lack of CVC symptoms may lie in the nature of the microbial community associated with these plants.
Endophytes are microorganisms that do not visibly harm the host plant but can be isolated from surface-disinfected plant tissue or the inner parts of plants (13). Furthermore, they colonize an ecological niche similar to that of phytopathogens, which might favor them as candidates for biocontrol agents (13). Indeed, intensive work has shown that endophytic microorganisms can have the capacity to control pathogens (3, 8, 17, 23, 35, 38, 39), insects (3, 25), and nematodes (14, 15). In some cases, they can also accelerate seedling emergence and promote plant establishment under adverse conditions (6) and enhance plant growth and development (4, 20, 26). To our knowledge, only a few studies on the presence of endophytes in citrus plants have been performed. The fungus Physoderma citri was one of the first endophytic fungi to be reported in healthy C. sinensis plants (7). Furthermore, several bacterial species have been isolated from the xylem of lemon roots (Citrus jambhiri), including Achromobacter spp., Acinetobacter baumannii, A. lwoffii, Alcaligenes-Moraxella spp., Alcaligenes sp., Arthrobacter spp., Bacillus spp., Burkholderia cepacia, Citrobacter freundii, Corynebacterium spp., Curtobacterium flaccumfaciens, Enterobacter cloacae, E. aerogenes, Methylobacterium extorquens, Pantoea agglomerans, Pseudomonas aeruginosa, and Pseudomonas spp. (2, 11, 12, 22).
The overall aims of this work were (i) to identify the strains present in populations of culturable endophytic bacteria in asymptomatic and CVC-affected (symptomatic) citrus plants and (ii) to study the interaction between these endophytic bacterial communities and X. fastidiosa by using cultivation-based plating techniques and a cultivation-independent method involving PCR-generated 16S rRNA gene (rDNA) fragments and denaturing gradient gel electrophoresis (DGGE) (24). The DGGE method is a powerful technique for studying populations of culturable and nonculturable plant endophytes (10) and soil microorganisms (18). This method provides a fingerprint of the microbial community structure of the sampled habitat in which each band of the fingerprint represents a group of bacteria having 16S rDNA sequences with similar denaturation points (18). This paper describes the application of this methodology to the study of the association between CVC symptoms and endophytic bacteria.
MATERIALS AND METHODS
Plant material.
The diversity of endophytic bacteria was estimated in branches of sweet-orange (C. sinensis Osbeck cv. Natal) and tangerine (Citrus reticulata cv. Blanco) plants. Healthy (uninfected) sweet-orange branches were collected from asymptomatic groves, and healthy resistant escape plants (asymptomatic) and CVC-affected plants (symptomatic) were collected from diseased groves. For statistical purposes, these four categories of plants (uninfected, asymptomatic, symptomatic, and tangerine) were considered treatments in the present study. Plants from orchards located in five citrus-growing areas of the Brazilian states of São Paulo (Catanduva, Colina, Elisiário, and Novais) and Minas Gerais (Frutal) were sampled. For each treatment-location combination, branches from four plants (repetitions) were collected during the winter and summer of 1997 (two sampling periods). At one site, the Novais site, sampling was also carried out during 1998 to achieve a better overview of the annual fluctuation of the bacterial community.
Surface sterilization of branches.
Branches were washed in running tap water and graded by size and surface appearance in order to exclude samples that showed symptoms of disease or superficial damage. Surface disinfection was done by stepwise washing in 70% ethanol for 5 min, sodium hypochlorite solution (2% available Cl−) for 5 min, and 70% ethanol for 30 s, followed by two rinses in sterile distilled water. To confirm that the disinfection process was successful, the branches were pressed onto tryptic soy agar (TSA) medium plates and aliquots of the sterile distilled water used in the final rinse were also plated onto the same medium and the plates were examined for growth after incubation at 28°C for 3 days.
Isolation of endophytic bacteria.
The bark of surface-disinfected branches was removed with a sterilized razor blade, and the branches were cut into pieces 4 to 6 mm long, which were placed on TSA plates amended with 50 μg of benomyl per ml to inhibit fungal growth. Incubation was carried out at 28°C for 1 to 12 days to allow growth of endophytic bacteria from the cut pieces and to determine the number of infected fragments. The isolation frequency (IF) was calculated as the frequency of pieces per branch exhibiting bacterial growth. Ten plates, each containing seven pieces of branch, were analyzed for each plant. For statistical purposes, each plant was considered as a repetition and the IF was determined for each plant.
In a further experiment, fragments of citrus branches were homogenized in 5 ml of sterile phosphate-buffered saline (containing NaCl at 8 g/liter, KCl at 0.2 g/liter, Na2HPO4 at 1.4 g/liter, and KH2PO4 at 0.24 g/liter) with a blender and serial dilutions were plated onto TSA. The plates were incubated at 28°C for 1 to 20 days or until growth was observed, upon which the CFU were counted and the population density was estimated.
Following incubation, bacteria recovered from each plant fragment and/or homogenized sample were selected at random, purified, and grouped on the basis of phenotypic characteristics, e.g., colony morphology, colony color, cell shape, motility, growth rate, and Gram reaction. Ten isolates representing each bacterial group of interest were selected for further identification. Species differentiation was made by the fatty-acid methyl ester (FAME) technique with whole-cell fatty acids derivatized to methyl esters analyzed by gas chromatography by the MIDI system (Microbial Identification System, Inc.). Isolates that could not be identified by FAME analysis were additionally tested by Biolog (Biolog Inc.) or with the analytical profile indexes API, AP 20E, and AP 50CHE (bioMérieux S.A.).
Extraction of total DNA from bacterial isolates.
DNA was extracted from the endophytic bacteria with the following protocol. A 1.5-ml sample of an overnight bacterial culture was centrifuged for 2 min at 12,000 × g and resuspended in 1 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), recentrifuged, decanted, and resuspended in 500 μl of TE buffer plus 0.5 g of 0.1-mm-diameter glass beads and 30 μl of 10% sodium dodecyl sulfate. The cells were homogenized for 30 s in a bead beater (Braun cell homogenizer; B. Braun, Melsungen, Germany). A 500-μl volume of Tris-buffered phenol was then added, the solution was mixed well and centrifuged for 10 min at 12,000 × g, the aqueous phase was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1), and the DNA was precipitated with isopropanol (5 min at room temperature) and collected by centrifugation (10 min at 12,000 × g). The pellet was washed with 70% ethanol, air dried, and resuspended in 50 μl of TE buffer.
Extraction of total DNA from plant samples.
Surface-disinfected branches (1 g) obtained as described above were cut into 4- to 6-mm-long pieces, placed in a sterile tube with 3 ml of 120 mM sodium phosphate buffer (pH 8.0), and shaken at 120 rpm for 2 h. Two milliliters of the suspension was transferred to a new tube and centrifuged for 5 min at 12,000 × g, and the resulting pellet was dissolved in 500 μl of TE buffer. DNA was extracted as described above for the bacterial isolates and further purified with the Wizard DNA cleanup system (Promega, Madison, Wis.). Total DNA was visualized by electrophoresis on a 0.8% (wt/vol) agarose gel (33).
PCR-DGGE analysis and sequencing.
The PCR mixture was made up of 2 μl of extracted DNA, 5 μl of 10× Stoffel buffer (10 mM Tris-HCl [pH 8.3], 10 mM KCl), 20 pmol of each primer (Table 1), 200 μmol of each deoxynucleoside triphosphate, 3.75 mM MgCl2, 1% (vol/vol) formamide, 0.25 μg of T4 gene 32 protein (Boehringer Mannheim, Ingelheim, Germany), and 5 U of Taq DNA polymerase Stoffel fragment (Perkin-Elmer Cetus, Nieuwerkerk, The Netherlands) in a 50-μl final volume. A negative control (PCR mixture without DNA) was included in all PCR experiments.
TABLE 1.
Primers used in this study
| Primera | Target DNA | Sequence (5′→3′) |
|---|---|---|
| F968⊥GC | Bacterial 16S rDNA | CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG-AACGCGAAGAACCTTACb |
| R1378 | Bacterial 16S rDNA | CGGTGTGTACAAGGCCCGGGAACG |
| FAlpha-U | α-Proteobacterial 16S rDNA | CCGCATACGCCCTACGGGGGAAAGATTTAT |
| FBeta-2 | β-Proteobacterial 16S rDNA | CGCACAAGCGGTGGATGA |
| CVC-1 | X. fastidiosa | AGATGAAAACAATCATGCAAA |
| 272-2-int | X. fastidiosa | GCCGCTTCGGAGAGCATTCCT |
F, forward primer; R, reverse primer.
5′ GC clamp (up to hyphen).
A nested PCR was used to investigate the structure of the α-proteobacterial community. Ribosomal sequences from environmental DNA were amplified with the FAlpha-U and R1378 (Table 1) primers and an initial denaturation step of 94°C for 7 min, followed by 25 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min and a final 10-min extension at 72°C. Two microliters of this PCR product was further amplified with the F968⊥GC and R1378 primers as described in the previous paragraph.
A nested PCR was also used to investigate the structure of the β-proteobacterial community. Ribosomal sequences from environmental DNA were amplified with the FBeta-2 and R1378 primers (Table 1) and a hot-start protocol, in which the primers and Taq DNA polymerase were added to the mixture at 80°C. This initial PCR entailed a denaturation step of 94°C for 5 min, followed by the addition of primers and Taq DNA polymerase and 25 cycles of 95°C for 1 min, 62°C for 1.5 min, and 72°C for 2 min and a final 10-min extension at 72°C. Two microliters of this PCR product was further amplified with the F968⊥GC and R1378 primers as described above.
The diversity of the endophytic bacterial communities was studied by the DGGE method. PCR products were obtained from 16S rDNAs with the F968⊥GC and R1378 primers (Table 1), which generated a 450-bp fragment. The amplification protocol included a touchdown series such that the annealing temperature was initially set at 60°C and decreased by 1°C every second cycle until 55°C. Twenty additional cycles were carried out with the last annealing temperature. Melting was carried out at 94°C for 1 min, primer annealing was performed in accordance with the above-described scheme for 1 min, and primer extension was at 72°C for 3 min. A final step was carried out at 72°C (10 min). Five microliters of the PCR product was analyzed by electrophoresis in a 1.4% (wt/vol) agarose gel with 0.5× TBE buffer (33) and stored at −20°C for DGGE analysis.
DGGE was performed as described previously (24) with the Ingeny phorU2 apparatus (Ingeny, Leiden, The Netherlands). PCR samples were loaded onto 6% (wt/vol) polyacrylamide gels in 0.5× TAE buffer (20 mM Tris-acetate, 0.5 mM EDTA, pH 7.4). The polyacrylamide gels were made with denaturing gradients ranging from 40 to 60% (where the 100% denaturant contained 7 M urea and 40% formamide). The gels were run for 16 h at 100 V and 60°C, after which the gels were soaked for 1 h in SYBR Green I nucleic acid stain (1:10,000 dilution; Molecular Probes, Leiden, The Netherlands) and immediately photographed under UV light. In some cases, the gels were silver stained (Bio-Rad Laboratories, Veenendaal, The Netherlands).
Prominent bands were excised from the gels, reamplified, and subjected to DGGE as previously described. The new PCR products were purified with the GFX PCR DNA and gel band purification kit (Amersham Pharmacia) to remove the unused deoxynucleoside triphosphates, and the purified PCR products were cloned into a pGEM-T Easy vector (Promega) in accordance with the manufacturer's instructions. Plasmids were isolated from Escherichia coli by using standard protocols and a QIAprep Miniprep Kit (QIAGEN). The inserts in the clones obtained were confirmed by DGGE as previously described. The purified plasmids with the correct insert (size and DGGE position) were then sequenced in both directions with universal M13 primers. Analyses of sequences were performed with the basic sequence alignment BLASTn program run against the BLAST database (National Center for Biotechnology Information website [http://www.ncbi.nlm.nih.gov]).
Detection of X. fastidiosa.
Detection of X. fastidiosa was done with specific primers for CVC-related X. fastidiosa (28) and the PCR mixture described above (excluding formamide and the T4 gene 32 protein) containing the CVC-1 and 272-2-int primers at 0.4 μM (Table 1) (28). Amplification was performed at 94°C for 4 min, followed by 30 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min and a final 10-min extension at 72°C. For electrophoresis, 5-μl samples of the PCR product were analyzed by electrophoresis in 1.2% agarose gel-TBE buffer and visualized with ethidium bromide under UV light.
Statistical analysis.
Analysis of data was carried out with the SAS software package (34) with a completely randomized analysis of variances for unequally replicated treatments (P < 0.05) (37), with the endophyte incidence values being transformed to the square root of the endophyte incidence plus 0.5 before analysis of variance. Duncan's test for unequally replicated means was used for further comparisons of means.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study have been submitted to the GenBank database and assigned accession numbers AY081206, AY081207, AY081208, and AY081209.
RESULTS
Determination of culturable endophytic bacteria in citrus branches.
The diversity of endophytic bacteria of citrus plants was assessed in samples of branches collected in the summer and winter of 1997 from five different citrus-growing areas of the Brazilian states of São Paulo and Minas Gerais. To avoid contamination and to isolate endophytic bacteria only from inner plant tissues, the branches were peeled after surface disinfection. The endophytic bacterial community that was isolated from citrus plants included Alcaligenes sp., Bacillus pumilus, Bacillus cereus, Burkholderia cepacia, Curtobacterium flaccumfaciens, Enterobacter cloacae, Methylobacterium spp. (including M. extorquens, M. fujisawaense, M. mesophilicum, M. radiotolerans, and M. zatmanii), Nocardia sp., Pantoea agglomerans, Streptomyces sp., and Xanthomonas campestris.
The numbers of culturable endophytic bacteria that we recovered with TSA medium were not significantly different within the four categories of plants (uninfected, asymptomatic, CVC symptomatic, and tangerine), ranging from 103 to 104 CFU g−1 (fresh branch weight basis). However, the frequency of endophytic bacteria recovered from branch fragments on TSA medium was significantly different within treatments and locations (Fig. 1). Endophytic bacteria were recovered less frequently from tangerine plants than from other plants (asymptomatic, uninfected, and symptomatic plants).
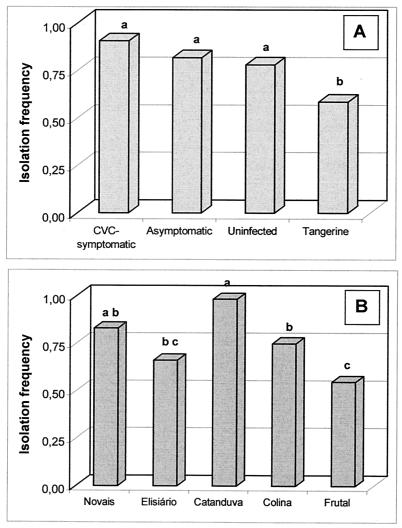
FIG. 1.
IFs of citrus endophytic bacteria from different citrus plants (A) from five citrus-growing areas (B). Means with the same letter are not significantly different by Duncan's test (P > 0.05).
The total IFs of uninfected, asymptomatic, and symptomatic sweet-orange plants were not significantly different, but the IF of species of bacteria from sweet-orange plants was significantly higher than that of tangerine plants (Fig. 1A). Statistical analysis showed differences in the total IFs of plants from different sites; the highest IF was observed in plants from the Catanduva region, and the lowest was observed in plants from the Frutal region (Fig. 1B).
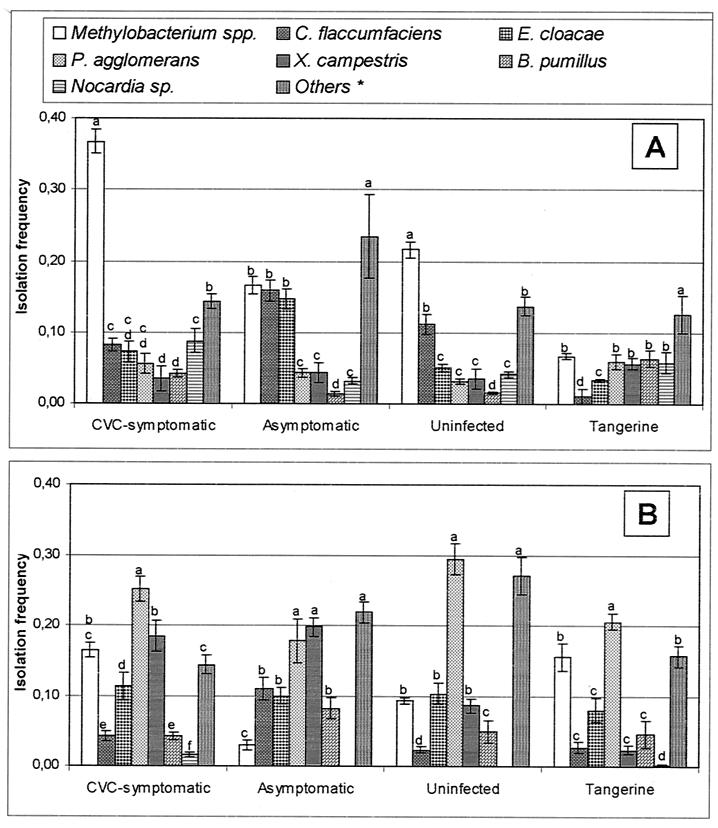
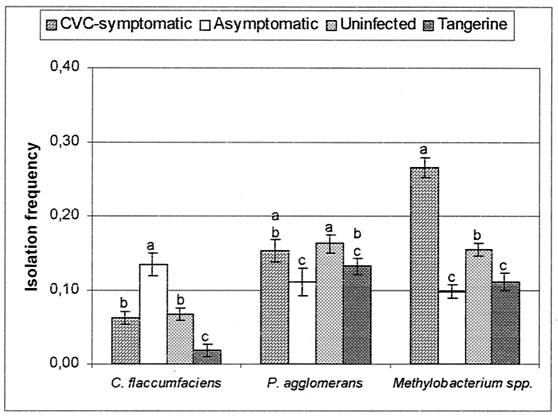
Most species occurred at low frequencies throughout the study, regardless of the plant category or sampling time. A relationship between the presence of symptoms of CVC and IF was observed for the dominant taxa; i.e., Methylobacterium spp. were the most frequent group in uninfected and symptomatic plants but were also isolated from asymptomatic plants and tangerine plants throughout the study (Fig. 2 and 3). In contrast, C. flaccumfaciens was isolated significantly (P < 0.05) more frequently from asymptomatic plants than from symptomatic plants, while P. agglomerans was frequently isolated from all plants, mainly in the second sampling period (Fig. 2 and 3).
FIG. 2.
IFs of endophytic bacteria from citrus plants in two sampling periods, March and April (A) and September and October (B) of 1997. Means within a plant category with the same letter are not significantly different by Duncan's test (P > 0.05). *, Other isolates included are Bacillus cereus, Burkholderia cepacia, Streptomyces sp., and unidentified isolates.
FIG. 3.
IFs of C. flaccumfaciens, P. agglomerans, and Methylobacterium spp. in branches of CVC-symptomatic, asymptomatic, uninfected, and tangerine plants. Means within a bacterial group with the same letter are not significantly different by Duncan's test (P > 0.05). Error bars indicate standard errors of the means (at least 40 plants were sampled).
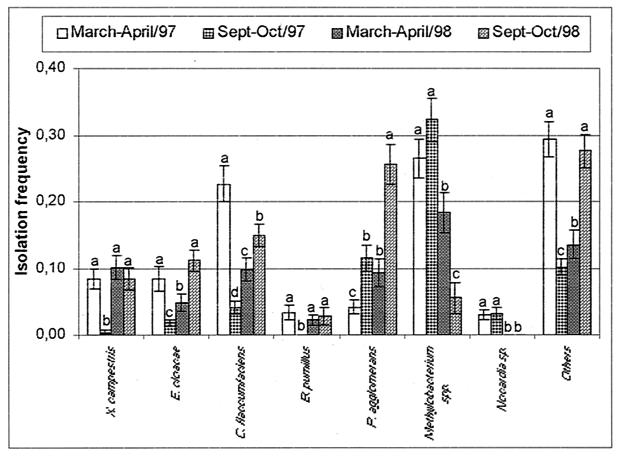
As explained above, in order to obtain a better overview of the annual fluctuation that the endophytic bacterial populations may experience, we sampled the Novais site twice. The IF of the main culturable groups during the 1997 and 1998 sampling periods is shown in Fig. 4. Significant (P < 0.05) seasonal variation can be seen, but Methylobacterium spp. were again the most frequently recovered bacteria in all samples, except those collected at the end of 1998, when P. agglomerans and other, unidentified, species were most frequently isolated (Fig. 4). At this location, C. flaccumfaciens was more frequently isolated from asymptomatic plants and Methylobacterium spp. were more frequently isolated from symptomatic plants (both significant at P < 0.05).
FIG. 4.
IFs of endophytic bacteria from peeled citrus branches sampled in March and April and in September and October of 1997 and 1998 in the Brazilian city of Novais, São Paulo. Means within a bacterial group with the same letter are not significantly different by Duncan's test (P > 0.05). Error bars indicate standard errors of the means (at least 16 plants were sampled).
Detection of X. fastidiosa.
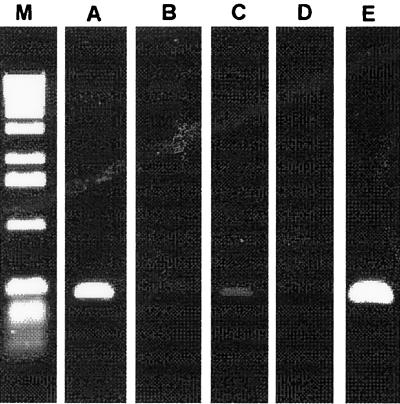
All of the plant tissue used for bacterial endophytic studies was investigated for the presence of X. fastidiosa with specific primers for CVC-related X. fastidiosa (28). This organism was detected in both symptomatic and asymptomatic plants, but the intensity of the amplified fragment was quite different in each case, with symptomatic plants showing a stronger signal than asymptomatic plants. Under the conditions employed, no amplification was observed from uninfected sweet-orange and tangerine plants (Fig. 5).
FIG. 5.
Detection of X. fastidiosa in citrus plants with specific primers for CVC-causing strains. All plants studied were evaluated for the presence of X. fastidiosa. Lanes: M, molecular size marker (Life Technologies); A, CVC-symptomatic plants; B, uninfected plants; C, asymptomatic plants; D, tangerine plants; E, positive control containing X. fastidiosa DNA.
DGGE analysis of the 968-to-1387 16S rDNA region of Methylobacterium isolates.
Forty-four strains of Methylobacterium-like bacteria were initially identified as members of the genus Methylobacterium by FAME and then genotyped by DGGE, and the strains were allocated to seven groups on the basis of the production of amplicons with different melting behavior (Fig. 6). Bacterial isolates with these bands were grouped into eight haplotypes that could not be associated with species; e.g., some M. mesophilicum, M. radiotolerans, and M. zatmanii isolates were grouped into haplotype A while some other M. mesophilicum and M. zatmanii isolates were grouped into haplotype B. Grouping of these DGGE haplotypes by plant category showed that asymptomatic plants were less diverse with respect to DGGE haplotypes than uninfected or symptomatic plants or tangerine; that is, asymptomatic plants were apparently colonized only by M. mesophilicum haplotype A (Table 2).
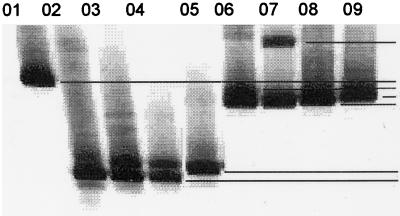
FIG. 6.
DGGE profile of Methylobacterium spp. isolated from the branches of citrus plants. Lanes: 01, M. mesophilicum haplotype H (AR3/19); 02, Methylobacterium sp. haplotype G (AR1.6/4); 03, M. fujisawaense haplotype F (PR5/4); 04, M. fujisawaense haplotype F (PR5.1/1); 05, M. extorquens haplotype E (AR1.6/11); 06, M. fujisawaense haplotype D (SR5/3); 07, M. zatmanii haplotype C (SR1.6/2); 08, M. mesophilicum haplotype B (SR1.6/6); 09, M. zatmanii haplotype B (PR3/8). The codes in parentheses indicate the strains used.
TABLE 2.
Grouping of DGGE haplotypes by plant category
| Host plant category and Methylobacterium haplotype species | Strain(s) | Isolation sitea | DGGE |
|---|---|---|---|
| CVC-symptomatic C. sinensis | |||
| M. extorquens | AR1.6/2, AR1.6/3, AR1.6/8, AR1.6/11 | Novais | E |
| M. mesophilicum | APB/1 | Bebedouro | A |
| M. mesophilicum | AR1.6/1, AR1.6/6 | Novais | A |
| M. mesophilicum | AR3/15, AR3/20 | Catanduva | A |
| M. mesophilicum | AR3/19 | Catanduva | H |
| M. mesophilicum | AR4/19 | Frutal | A |
| M. mesophilicum | AR5/1, AR5.1/4, AR5.1/5, AR5.1/6 | Colina | A |
| Methylobacterium sp. | AR1.6/4 | Novais | G |
| Asymptomatic C. sinensis | |||
| M. mesophilicum | ER1/21, ER1.6/1, ER1.6/2, ER1.6/4, ER1.6/5 | Novais | A |
| M. mesophilicum | ER5/2 | Colina | A |
| Uninfected C. sinensis | |||
| M. radiotolerans | SR1.6/3 | Novais | A |
| M. radiotolerans | SR1.4/10, SR1.6/4 | Novais | B |
| M. zatmanii | SR1.6/2 | Novais | C |
| M. zatmanii | SR1.6/9 | Novais | A |
| M. fujisawaense | SR5/3 | Colina | D |
| M. extorquens | SR1.6/1, SR1.6/15 | Novais | E |
| M. extorquens | SR5/4 | Colina | D |
| M. mesophilicum | SR1.6/6 | Novais | B |
| M. mesophilicum | SR1.6/13 | Novais | A |
| M. mesophilicum | SR3/27 | Catanduva | A |
| Tangerine (C. reticulata) | |||
| M. fujisawaense | PR5/4, PR5.1/1 | Colina | F |
| M. mesophilicum | PR1/3, PR1.4/10 | Novais | A |
| M. mesophilicum | PR2/2 | Elisiário | A |
| M. mesophilicum | PR3/5 | Catanduva | H |
| M. mesophilicum | PR3/11, PR3/15 | Catanduva | A |
| M. zatmanii | PR3/8, PR3/17 | Catanduva | B |
All of the sites are in São Paulo State except for Frutal, which is in Minas Gerais State.
DGGE analysis of the endophytic bacterial communities of citrus plants.
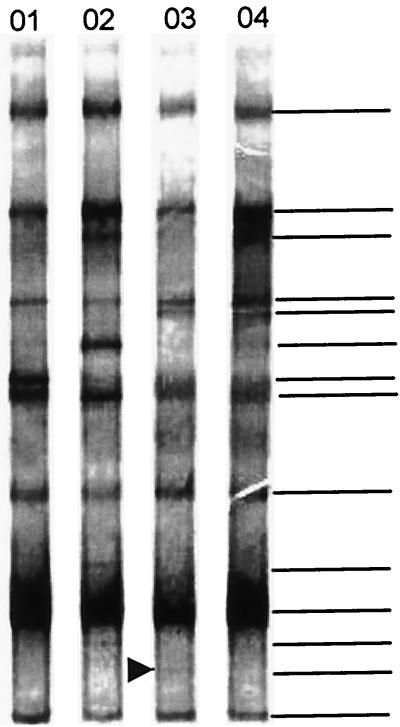
The DGGE patterns of the PCR products (amplified with universal primers for the 968-to-1387 16S rDNA fragment) from DNA extracted from peeled citrus branches revealed that the communities were composed of groups or species that were represented in all plant samples. With sequence analysis, the dominant and strongest band was found to be derived from plastid DNA, which was as expected considering that we were working with citrus samples. A high similarity among the plant categories (Fig. 7) was observed, with both universal primers and nested PCR specific for the α-proteobacterial group. However, DGGE patterns obtained by β-proteobacterium-specific PCR showed clear differences between the plant categories; asymptomatic plants had more bands than uninfected or asymptomatic plants or tangerine plants. A band with a high percentage of G+C (whose DGGE profile was not similar to those of other culturable bacteria) was observed in the DGGE profile of all asymptomatic plants but was not observed in uninfected or symptomatic plants or tangerine plants (Fig. 8). Unfortunately, attempts to clone and sequence this band were unsuccessful.
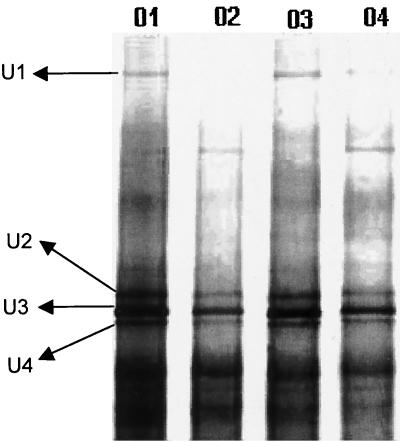
FIG. 7.
Fingerprinting of the bacterial endophytic community of citrus branches by DGGE separation and analysis of 16S rDNA fragments amplified with the bacterial primer set 968F (primer F968⊥GC without GC clamp) plus 1387R. Lanes: 01, CVC-symptomatic plants; 02, healthy plants; 03, resistant plants; 04, tangerine plants. Bands U1, U2, U3, and U4 were excised and sequenced, and the sequence was shown to have between 92 and 98% sequence similarity to Nocardia nova (98%), Methylobacterium sp. (99%), Curtobacterium sp. (92%), and Corynebacterium accolens (99%).
FIG. 8.
DGGE analysis of 16S rDNA fragments of citrus plant samples collected in the Brazilian town of Novais, São Paulo. Lanes: 01, tangerine plants; 02, CVC-symptomatic plants; 03, asymptomatic plants; 04, uninfected plants. The 16S rDNA genes were previously amplified with specific primers for β-proteobacteria and then with bacterial universal primers with a GC clamp. The band with a high percentage of G+C was observed in all of the asymptomatic plants (arrow) but not in susceptible plants. The horizontal lines indicate the bands used for analysis. At least 20 plants within each category were used.
Some DGGE bands were selected for further analysis by sequencing, and this analysis showed that the strongest bacterial band (band U3) in citrus plants is most closely related to Curtobacterium sp. and Curtobacterium luteum (Fig. 7). Other bands were sequenced and then analyzed by BLASTn, which showed similarity to Corynebacterium accolens (band U4), Nocardia nova (band U1), and Methylobacterium sp. (band U2) (Fig. 7), which was observed as the strongest band in the α-proteobacterial gel.
DISCUSSION
The plant-associated habitat is a dynamic environment in which many factors affect the structure and species composition of the microbial communities that colonize roots, stems, branches, and leaves. It has previously been shown that endophytic communities vary spatially in the plant (9) or may be dependent on the interaction with other endophytic or pathogenic bacteria (29).
Methylobacterium species, which have the capacity to fix nitrogen (1), have previously been isolated from citrus (2), Scotch pine (27), and crotalaria (1), showing the capacity of members of this bacterial genus to colonize the plant habitat. In the present work, Methylobacterium was the most frequently found genus in citrus plants and there was a positive association with the occurrence and intensity of symptoms of CVC (Fig. 2 and 3). This suggests that CVC could play a role in the establishment of endophytic bacteria in the host plant or that endophytic Methylobacterium spp. could trigger CVC by a synergistic interaction with X. fastidiosa, which showed a stronger PCR signal in symptomatic plants than in asymptomatic plants (Fig. 5). This synergistic effect of Methylobacterium sp. on the growth of X. fastidiosa had previously been noted in vitro (data not shown), suggesting that a similar interaction may occur inside the host plant. Understanding the structure and species composition of these communities is fundamental to understanding how endophytic communities are influenced by environmental factors, as well as the disease status of the host plants.
With the primers specific for CVC-causing X. fastidiosa (28), we were able to detect this pathogen in all asymptomatic and symptomatic plants. However, the intensity of the band was highest with DNA extracted from symptomatic plants, and if band intensity is proportional to the number of bacteria present in the plant, we can hypothesize that the population of X. fastidiosa bacteria remained small in asymptomatic plants. In these plants, which have been found in some diseased orchards in different regions of São Paulo State, no correlation has been found among plant genotypes, edaphic or climatic factors, and this resistance to CVC.
Some speculation can be made about the role of the endophytic bacterial community of plants in CVC, since it is interesting that C. flaccumfaciens was isolated mainly from asymptomatic plants (Fig. 2 and 3) and that the band with a high percentage of G+C was only observed in asymptomatic plants (Fig. 8), supporting the view that C. flaccumfaciens and other bacteria that produce a band with a high percentage of G+C could be associated with the resistance of citrus plants to CVC. In fact, C. flaccumfaciens has been described as a biological control agent against many pathogens and it has been reported that it acts by triggering induced systemic resistance (31) and by antibiosis (39). These endophytic bacteria could play a role in limiting the establishment of X. fastidiosa in asymptomatic plants, but the process involved is unclear and needs further study.
Fingerprinting of endophytic bacterial communities by separation of amplified 16S rDNA fragments by DGGE provides the opportunity to compare the community structure features of multiple plant samples. In a previous study (10), the endophytic community of potato plants was evaluated by PCR-DGGE and the data obtained validated this approach to the analysis of culturable and nonculturable endophytic communities. In the present study, the genera Curtobacterium, Methylobacterium, and Nocardia were detected both by isolation and by DGGE, suggesting that the molecular approach directly reported culturable endophytic bacteria. However, Corynebacterium accolens bacteria were not detected by plating, suggesting that these endophytic bacteria are nonculturable (or not yet cultured) endophytic bacteria from citrus plants. The presence of this type of endophytes has previously been reported in potato plants (10).
With universal primers and nested PCR-DGGE for α-proteobacteria, low DGGE profile variability was observed among the plants investigated. The fact that several dominant bands were not influenced by the disease status of the plant from which they came suggests that the presence of X. fastidiosa had minor effects on the endophytic colonizers targeted by these assay systems. However, with the β-proteobacterial PCR assay combined with DGGE, a distinct band was found in asymptomatic plants and a greater diversity, as evidenced by the number of amplification products, was detected in CVC-symptomatic plants.
Previous study has shown that active penetration of cotton plants by bacterial endophytes involves the hydrolysis of cellulose, which could induce systemic resistance to pathogens (30). When present within the inner tissue of asymptomatic citrus plants, C. flaccumfaciens could produce compounds or elicit some degree of enhanced resistance in these plants that leads to enhanced resistance to X. fastidiosa. This hypothesis is supported by the results of our analysis of the diversity of Methylobacterium spp., which show that this genus was more frequently isolated from plants with symptoms of CVC (Fig. 2 and 3) and that in asymptomatic plants, only one haplotype was detected (Table 2), which could indicate that this haplotype is the only one able to colonize plants with a large population of C. flaccumfaciens. Other haplotypes, which were not able to colonize asymptomatic plants, could be sensitive to metabolites produced by the host plant (induced by endophytic bacteria) or by C. flaccumfaciens.
The work presented in this paper clearly shows the potential of a polyphasic approach based on microbial cultivation methods combined with PCR-DGGE analysis of DNA extracted from plants to help in investigating the interaction between pathogenic and endophytic bacterial communities. These results also show the power of using culture-dependent and culture-independent methods to study the interaction of endophytic bacteria and X. fastidiosa and provide preliminary evidence of possible interactions between CVC and the endophytic bacterial community of plants. In the light of this, our future work will be directed to analyzing the nature of the bacteria identified and the mechanisms involved in the resistance of citrus plants to X. fastidiosa.
Acknowledgments
This work was supported by a grant from FAPESP and by a grant from FUNDECITRUS (Fundo de Defesa Da Citricultura, Araraquara, São Paulo, Brazil).
We thank FAPESP (96/06686-4) and CAPES for the fellowship to W.L.A. We also thank Paulo T. Lacava and Siu M. Tsai (CENA, Piracicaba, São Paulo, Brazil) for performing the sequencing and providing other facilities.
REFERENCES
- 1.Abdoulaye, S. Y., E. Giraud, P. Jourand, N. Garcia, A. Willems, P. de Lajudie, Y. Prin, M. Neyra, M. Gillis, C. Bivin-Masson, and B. Dreyfus. 2001. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J. Bacteriol. 183:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araújo, W. L., H. O. Saridakis, P. A. V. Barroso, C. I. Aguilar-Vildoso, and J. L. Azevedo. 2001. Variability and interactions between endophytic bacteria and fungi isolated from leaf tissues of citrus rootstocks. Can. J. Microbiol. 47:229-236. [DOI] [PubMed] [Google Scholar]
- 3.Azevedo, J. L., W. Maccheroni, Jr., J. O. Pereira, and W. L. Araújo. 15April2000, posting date. Endophytic microorganisms: a review on insect control and recent advances on tropical plants. Bio/Technology 3:40-65. [Online.] http://www.ejb.org/content/vol3/issue1/full/3/4. [Google Scholar]
- 4.Bent, E., and C. P. Chanway. 1998. The growth-promoting effects of a bacterial endophyte on lodgepole pine are partially inhibited by the presence of other rhizobacteria. Can. J. Microbiol. 44:980-988. [Google Scholar]
- 5.Chang, C. J., M. Garnier, L. Zreik, V. Rossetti, and J. M. Bove. 1993. Culture and serological detection of the xylem-limited bacterium causing citrus variegated chlorosis and its identification as a strain of Xylella fastidiosa. Curr. Microbiol. 27:137-142. [DOI] [PubMed] [Google Scholar]
- 6.Chanway, C. P. 1997. Inoculation of tree roots with plant growth promoting soil bacteria: an emerging technology for reforestation. Forest Sci. 43:99-112. [Google Scholar]
- 7.Childs, J. F. L., L. E. Kopp, and R. E. Johnson. 1965. A species of Physoderma present in citrus and related species. Phytopathology 55:681-687. [Google Scholar]
- 8.Duijff, B. J., V. Gianinazzi-Pearsonand, and P. Lemanceau. 1997. Involvement of the outer membrane lipopolysaccharides in the endophytic colonization of tomato roots by biocontrol Pseudomonas fluorescens strain WCS417r. New Phytol. 135:325-334. [Google Scholar]
- 9.Fisher, P. J., O. Petrini, and H. M. L. Scott. 1992. The distribution of some fungal and bacterial endophytes in maize (Zea mays L.). New Phytol. 122:299-305. [DOI] [PubMed] [Google Scholar]
- 10.Garbeva, P., L. S. van Overbeek, J. W. L. van Vuurde, and J. D. van Elsas. 2001. Analysis of endophytic bacterial communities of potato by plating and denaturing gradient gel electrophoresis (DGGE) of 16S rDNA based PCR fragments. Microb. Ecol. 41:369-383. [DOI] [PubMed] [Google Scholar]
- 11.Gardner, J. M., A. W. Feldman, and M. Zablotowicz. 1982. Identity and behavior of xylem-residing bacteria in rough lemon roots of Florida citrus trees. Appl. Environ. Microbiol. 43:1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner, J. M., J. A. Chandler, and A. W. Feldman. 1985. Growth response and vascular plugging of citrus inoculated with rhizobacteria and xylem-resident bacteria. Plant Soil 86:996-1000. [Google Scholar]
- 13.Hallmann, J., A. Quadt-Hallmann, W. F. Mahaffee, and J. W. Kloepper. 1997. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43:895-914. [Google Scholar]
- 14.Hallmann, J., A. Quadt-Hallmann, R. Rodríguez-Kábana, and J. W. Kloepper. 1998. Interactions between Meloidogyne incognita and endophytic bacteria in cotton and cucumber. Soil Biol. Biochem. 30:925-937. [Google Scholar]
- 15.Hallmann, J., R. Rodríguez-Kábana, and J. W. Kloepper. 1999. Chitin-mediated changes in bacterial communities of the soil, rhizosphere and within roots of cotton in relation to nematode control. Soil Biol. Biochem. 31:551-560. [Google Scholar]
- 16.Hopkins, D. L. 1989. Xylella fastidiosa: xylem-limited bacterial pathogen of plants. Annu. Rev. Phytopathol. 27:271-290. [Google Scholar]
- 17.Krishnamurthy, K., and S. S. Gnanamanickam. 1997. Biological control of sheath blight of rice: induction of systemic resistance in rice by plant-associated Pseudomonas spp. Curr. Sci. 72:331-334. [Google Scholar]
- 18.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambais, M. R., M. H. S. Goldman, L. E. A. Camargo, and G. H. Goldman. 2000. A genomic approach to the understanding of Xylella fastidiosa pathogenicity. Curr. Opin. Microbiol. 3:459-462. [DOI] [PubMed] [Google Scholar]
- 20.Lazarovits, G., and J. Nowak. 1997. Rhizobacteria for improvement of plant growth and establishment. Hortscience 32:188-192. [Google Scholar]
- 21.Lee, R. F., K. S. Kerrick, M. J. G. Beretta, C. M. Chagas, and V. Rossetti. 1991. Citrus variegated chlorosis: a new destructive disease of citrus in Brazil. Citrus Ind. 72:12-13, 15. [Google Scholar]
- 22.Lima, G., A. Ippolito, F. Nigro, and M. Salerno. 1994. Attempts in the biological control of citrus mal secco (Phoma tracheiphila) using endophytic bacteria. Dif. Piante 17:43-49. [Google Scholar]
- 23.M'Piga, P., R. R. Bélanger, T. C. Paulitz, and N. Benhamou. 1997. Increased resistance to Fusarium oxysporum f. sp. radicis-licopersici in tomato plants treated with the endophytic bacterium Pseudomonas fluorescens strain 63-28. Physiol. Mol. Plant Pathol. 50:301-320. [Google Scholar]
- 24.Muyzer, G., E. C. de Waal, and A. Uitterlinden. 1993. Profiling of complex microbial populations using denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrini, L. E., O. Petrini, and G. Laflamme. 1989. Recovery of endophytes of Abiens balsamea from needles and galls of Paradiplosis tumifex. Phytoprotection 70:97-103. [Google Scholar]
- 26.Pillay, V. J., and J. Nowak. 1997. Inoculum density, temperature and genotype effects on in vitro growth promotion and epiphytic and endophytic colonization of tomato (Lycopersicum esculentum L.) seedlings inoculated with a pseudomonad bacterium. Can. J. Microbiol. 43:354-361. [Google Scholar]
- 27.Pirttilä, A. M., H. Laukkane, H. Pospiech, R. Myllylä, and A. Hohtola. 2000. Detection of intracellular bacteria in buds of Scotch pine (Pinus sylvestris L.) by in situ hybridization. Appl. Environ. Microbiol. 66:3037-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pooler, M. R., and J. S. Hartung. 1995. Specific PCR detection and identification of Xylella fastidiosa strains causing citrus variegated chlorosis. Curr. Microbiol. 31:377-381. [DOI] [PubMed] [Google Scholar]
- 29.Quadt-Hallmann, A., J. Hallmann, and J. W. Kloepper. 1997. Bacterial endophytes in cotton: localization and interaction with other plant-associated bacteria. Can. J. Microbiol. 43:254-259. [Google Scholar]
- 30.Quadt-Hallmann, A., N. Benhamou, and J. W. Kloepper. 1997. Bacterial endophytes in cotton: mechanisms of entering the plant. Can. J. Microbiol. 43:577-582. [Google Scholar]
- 31.Raupach, G. S., and J. W. Kloepper. 1998. Mixtures of plant growth-promoting rhizobacteria enhance biological control of multiple cucumber pathogens. Phytopathology 88:1158-1164. [DOI] [PubMed] [Google Scholar]
- 32.Rossetti, V., M. Garnier, J. M. Bove, M. J. Beretta, A. R. R. Teixeira, J. A. Quaggio, and J. D. DeNegri. 1990. Présence de bactéries dans le xylème d'orangers atteints de chlorose variégée une nouvelle maladie des agrumes au Brésil. C. R. Acad. Sci. Paris Ser. III 310:345-349. [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.SAS Institute. 1987. SAS/STAT guide for personal computers, version 6. SAS Institute, Cary, N.C.
- 35.Sharma, V. K., and J. Nowak. 1998. Enhancement of verticillium wilt resistance in tomato transplants by in vitro co-culture of seedlings with a plant growth promoting rhizobacterium (Pseudomonas sp. strain PsJN). Can. J. Microbiol. 44:528-536. [Google Scholar]
- 36.Simpson, A. J. G., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. C. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-159. [DOI] [PubMed] [Google Scholar]
- 37.Steel, R. G. D., and J. H. Torrie. 1980. Principles and procedures of statistics: a biometrical approach. McGraw-Hill, Toronto, Ontario, Canada.
- 38.Sturz, A. V., B. R. Christie, and B. G. Matheson. 1998. Association of bacterial endophyte populations from red clover and potato crops with potential for beneficial allelopathy. Can J. Microbiol. 44:162-167. [Google Scholar]
- 39.Sturz, A. V., and B. G Matheson. 1996. Populations of endophytic bacteria which influence host-resistance to Erwinia-induced bacterial soft rot in potato tubers. Plant Soil 184:265-271. [Google Scholar]
- 40.Wells, J. M., B. C. Raju, H. Hung, W. G. Weisburg, L. Mandelco-Paul, and D. J. Brenner. 1987. Xylella fastidiosa gen. nov., sp. nov.: gram-negative, xylem-limited, fastidious plant bacteria related to Xanthomonas spp. Int. J. Syst. Bacteriol. 37:136-143. [Google Scholar]