Abstract
The aim of this work was to generate a cyanobacterial biosensor that could be used to detect herbicides and other environmental pollutants. A representative freshwater cyanobacterium, Synechocystis sp. strain PCC6803, was chromosomally marked with the luciferase gene luc (from the firefly Photinus pyralis) to create a novel bioluminescent cyanobacterial strain. Successful expression of the luc gene during growth of Synechocystis sp. strain PCC6803 cultures was characterized by measuring optical density and bioluminescence. Bioluminescence was optimized with regard to uptake of the luciferase substrate, luciferin, and the physiology of the cyanobacterium. Bioassays demonstrated that a novel luminescent cyanobacterial biosensor has been developed which responded to a range of compounds including different herbicide types and other toxins. This biosensor is expected to provide new opportunities for the rapid screening of environmental samples or for the investigation of potential environmental damage.
Among the analytical methods used for environmental monitoring, microbial and cellular biosensors play an important role (11). Microbial biosensors have numerous advantages in ecotoxicity testing. Microorganisms are generally cheaper to culture than higher organisms, and they can be produced in large batches, subjected to stringent quality control procedures, and freeze dried for storage. They respond rapidly to toxic compounds and indicate the bioavailability of compounds in a way that chemical analysis cannot (21). Recent advances in genetic engineering have opened up new prospects in the field of microbial and cellular biosensors. An example is the use of the luciferase genes lux (from the marine bacterium Vibrio fischeri) (21) and luc (from the firefly Photinus pyralis) (12). These genes have been selected because there is a correlation between light output and the metabolic activity of the cell, so bioluminescence is a very rapid indicator of cell health (13, 21, 38). The genes encoding bioluminescence have been engineered into a range of microbes. Measurement of bioluminescence in response to exposure to samples can be used to assess the presence of a wide range of pollutants (21). Light itself is easy to measure in real time and lends itself well to automation, allowing the rapid processing of multiple samples. In addition, multiple or continuous readings can be taken, allowing both acute and chronic toxicity assessments to be made (23).
A variety of whole-cell-based bioluminescent biosensors have been constructed which respond to a wide range of pollutants while simultaneously assessing bioavailability in environmental samples (12, 16, 21, 22, 23, 41). However, existing biosensors show a poor response to herbicides, a key class of environmental pollutants (12). About one-half of the herbicides presently used in agriculture act by inhibiting the light reactions in photosynthesis, mostly by targeting the photosystem II (PSII) complex (10). A series of algal and cyanobacterial PSII-based whole-cell (5, 6, 11, 14, 27, 28, 34, 37, 43) and tissue (3, 15, 17, 18, 19, 30, 31, 32) biosensors have therefore been developed for detection of a class of herbicides which inhibit photosynthetic electron transport. In these systems, herbicides are detected by testing inhibition of the Hill reaction (6, 17, 31, 32), inhibition of 2,6-dichlorophenol indophenol photoreduction (4, 5, 15), or change in chlorophyll fluorescence (8, 11, 19, 30), which can be correlated with the pollutant concentration. Alternatively, the effect of herbicides was measured directly through inhibition of maximum growth rate (1, 34). However, the practical use of herbicide biosensors has so far been limited by their instability (15, 19), their short half-life (15, 19), the requirement for complex equipment (6, 15, 34), the time-consuming nature of the assays (15, 19), and the fact that most are specific for herbicides of the triazine-phenylurea group, which inhibit photosynthetic electron transport (11, 15, 19).
Here we report the development of a cyanobacterial Synechocystis sp. strain PCC6803 biosensor, marked with the firefly luc gene, for detection of a broad range of herbicides and other pollutant classes including heavy metals and volatile organic pollutants by a simple and quick assay system. Cyanobacteria are unusual among prokaryotes in having two distinct membrane systems: the intracellular thylakoid membrane, which is the site of photosynthesis, and the more conventional eubacterial cytoplasmic membrane system around the cell. Synechocystis sp. strain PCC6803 was chosen as a potential biosensor for its extreme ecological importance and the availability of genetic tools. In this report we describe the sensitivity and rapid reaction of this novel cyanobacterial biosensor to herbicides in an acute assay format and compare its response to a well-characterized Escherichia coli lux-marked biosensor. Also, a novel chronic toxicity test system derived from this cyanobacterial biosensor is discussed.
MATERIALS AND METHODS
Strains and culture conditions. (i) Synechocystis sp. strain PCC6803.
Wild-type Synechocystis sp. strain PCC6803 was obtained from laboratory stocks. It was grown routinely in BG-11 (29) liquid and solid medium at 30°C with constant illumination (25 μmol photons m−2 s−1 from cool-white fluorescent lamps). The antibiotic kanamycin (10 mg liter−1) was included in liquid medium for culturing transformed cyanobacteria. The growth of cyanobacterial cultures was quantified by measuring the scattering of light (optical density) at 730 nm (OD730) by use of a PU 8720 UV/VIS scanning spectrophotometer.
(ii) E. coli.
Ultracompetent XL10-Gold cells supplied by Stratagene were used as the host for all cloning experiments. E. coli was grown in Luria-Bertani medium (33) and in the presence of kanamycin at 50 mg liter−1.
Plasmids.
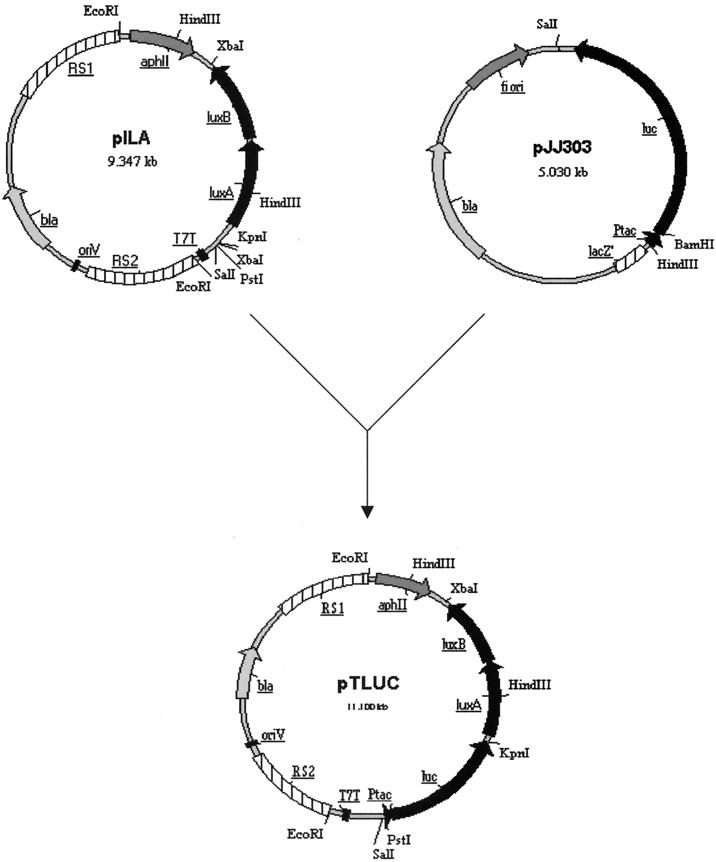
Plasmid pILA (GenBank accession no. AJ251840), a derivative of the integration vector pKW1188 (42), was used as a vector backbone to create pTLUC (Fig. 1). A PstI/KpnI fragment containing the luc gene downstream of the tac promoter was amplified by PCR from the pGEM-luc-derived plasmid pJJ303 (20). Primer tac/PstI (5′-TTACCTGCAGCTATTTAGGTGACACTATAG-3′) introduced a PstI site at the 5′ end of the tac promoter region, and a KpnI site was inserted by using primer tac-luc/KpnI (5′-TTACGGTACCGACTTTCCGCCCTTCTTG-3′) at the 3′ end of the luc gene. This PstI/KpnI PCR fragment was cloned into PstI/KpnI-digested pILA just upstream of the luxAB genes, resulting in plasmid pTLUC (Fig. 1).
FIG. 1.
Construction of plasmid pTLUC. The Synechocystis sp. strain PCC6803 integrative vector pILA (GenBank accession no. AJ251840) was used as a vector backbone for construction of pTLUC. A tac-luc PCR fragment, which introduced a PstI site at the 5′ end and a KpnI site at the 3′ end, was amplified by PCR from plasmid pJJ303 (20) and cloned into PstI/KpnI-digested pILA just upstream of the luxAB genes to create plasmid pTLUC. RS1 and RS2, a 3.0-kbp BamHI fragment of Synechocystis sp. strain PCC6803 genomic DNA spanning the region from bp 31757 to bp 31793 (GenBank accession no. D63999).
Transformation of Synechocystis sp. strain PCC6803.
To prepare a culture of Synechocystis sp. strain PCC6803 for transformation, cells were grown to an OD730 of approximately 0.6 (an OD730 of 0.25 corresponds to 108 cells ml−1) and harvested by centrifugation at 4,500 × g for 10 min at room temperature. The cell pellet was resuspended in fresh BG-11 medium at a density of 109 cells ml−1 and used immediately for transformation. Plasmid DNA (1 μg of pTLUC in 10 μl of 10 mM Tris-HCl [pH 8.5]) was added to 1 ml of cells, gently mixed, and incubated for 1 h with illumination at 30°C without agitation. Cells were spread on 50-ml BG-11 agar plates (0.1 ml per plate) and incubated under illumination for 2 days at 30°C. At this stage, the appropriate selective agent (kanamycin) was added by lifting the agar slab with an ethanol-flamed spatula and dispensing 50 μl of a solution containing kanamycin (10 mg ml−1) underneath (7, 25). After 14 days, transformant colonies were isolated and purified by restreaking three times in the presence of antibiotics as above.
Preparation of herbicides and other pollutants.
Stock solutions of the herbicides diuron, atrazine, propazine, simazine, paraquat, glyphosate, MCPA (4-chloro-2-methylphenoxyacetic acid), and mecoprop were prepared in sterile deionized water in acid-washed glass Duran bottles. Periods as long as 3 days with constant stirring were required to ensure complete solution of the herbicides, except for paraquat and glyphosate, which are very soluble. Fresh standard test solutions for these eight herbicides were prepared by diluting the stock solution with sterile deionized water in acid-washed glass Duran bottles. The test standards were used immediately after preparation.
The herbicides used in this study, with their modes of action, are listed in Table 1. The concentrations of herbicide stock solutions and the highest concentrations tested in this study are listed in Table 2.
TABLE 1.
Herbicides used in this study
| Herbicide | Type | Mode of action |
|---|---|---|
| Diuron | Phenylurea | PSII inhibitor |
| Atrazine | Triazine | PSII inhibitor |
| Propazine | Triazine | PSII inhibitor |
| Simazine | Triazine | PSII inhibitor |
| Paraquat | Bipyridyl | PSI inhibitor |
| Glyphosate | Phosphonomethyl glycine | Inhibits amino acid biosynthesis |
| MCPA | Phenoxy | Hormone mimic |
| Mecoprop | Phenoxy | Hormone mimic |
TABLE 2.
Toxic effects of eight herbicides on light outputs from luc-marked Synechocystis sp. strain PCC6803
| Herbicide | Stock concn (mg/liter) | Highest concn used (mg/liter) | EC20a (mg/liter) at:
|
EC50 (mg/liter) at:
|
||||
|---|---|---|---|---|---|---|---|---|
| 30 min | 6 h | 1 day | 2 days | 30 min | 1 day | |||
| Diuron | 42 | 42 | 7.47 ± 0.61 | NC | NC | NC | NDT | 29.70 ± 0.62 |
| Paraquat | 3,750 | 370 | 0.95 ± 0.01 | NC | NC | NC | 2.85 ± 0.03 | NC |
| MCPA | 825 | 412.5 | 23.60 ± 0.02 | NC | NC | NC | 54.67 ± 0.16 | NC |
| Mecoprop | 620 | 310 | 12.57 ± 0.95 | NC | NC | NC | 47.55 ± 1.42 | NC |
| Glyphosate | 2,500 | 20 | NDT | 3.62 ± 0.79 | NC | NC | NDT | 3.10 ± 0.17 |
| Atrazine | 30 | 30 | NDT | 7.39 ± 0.95 | NC | NC | NDT | NDT |
| Propazine | 4.5 | 4.5 | 3.51 ± 0.05 | NC | NC | NC | NDT | NDT |
| Simazine | 4.5 | 4.5 | NDT | NDT | NDT | 1.06 ± 0.04 | NDT | NDT |
NDT, not detected; NC, not calculated.
Stock solutions of copper, zinc and 3,5-dichlorophenol (3,5-DCP) were prepared as previously described (21, 41). Fresh standard test solutions were prepared by diluting the stock solution with sterile deionized water.
Measurement of growth and luminescence characteristics.
Correlation of luc-marked Synechocystis sp. strain PCC6803 cell growth and light output was studied by inoculating 1 ml of a late-exponential-phase culture (OD730 > 6.0) into 100 ml of BG-11 with 10 mg of kanamycin liter−1 in a 250-ml Erlenmeyer flask. Triplicate cultures were incubated on an orbital shaker at 150 rpm at 30°C with constant illumination, and 1-ml samples were taken at 24-h intervals for measurement of OD730 and luminescence. For luminescence analysis, 200 μl of culture was removed and 1 ml of McIlvaine's citrate phosphate buffer, pH 6.5 (10), containing 0.1 mM luciferin (Molecular Probes, Eugene, Oreg.) was added to the culture in each 1-ml luminometer cuvette. Following a 10-min exposure, bioluminescence was quantified in a BioOrbit 1251 luminometer by using a Multiuse software package (version 2.01; BioOrbit, Turku, Finland). Luminescence was monitored over a 10-s period, and a mean was taken which was calculated by the software. Luminescence was expressed in relative light units (RLU), which equated to millivolts per 10 s per milliliter (26).
Effect of external pH on light output.
To determine the effects of external pH on light output, a series of McIlvaine's citrate phosphate buffer (9) was prepared with a pH range from 2 to 8. Bioassays for determination of pH effect were based on the bioluminescence measurements described above, except that the pH of the citrate phosphate buffer was either 2, 3, 4, 5, 6, 7, or 8 rather than 6.5. Luminescence readings were taken at 10-min intervals for 130 min.
Bioassay procedures.
A culture of luc-marked Synechocystis sp. strain PCC6803 was grown in 100 ml of BG-11 with 10 mg of kanamycin liter−1 in a 250-ml Erlenmeyer flask on an orbital shaker at 150 rpm at 30°C with constant illumination until peak luminescence was obtained (OD730, around 3.0). The cell suspension was then removed from the Erlenmeyer flask and centrifuged at 4,500 × g for 6 min. After two washes in sterile deionized water, the pellet was resuspended in 15 ml of sterile deionized water. A two-step bioassay procedure (12) was employed. A cell suspension (50 μl) was pipetted into each 1-ml cuvette (Clinicon; catalogue no. 2174 701) which contained 450 μl of the test solution. Following a variety of exposure times (for example, 10 or 30 min, 1, 2, 4, or 6 h, or 1, 2, 3, or 4 days for herbicides and 15 min for heavy metals and volatile organic pollutants), 500 μl of McIlvaine's citrate phosphate buffer, pH 6.5 (9), containing 0.1 mM luciferin was added at room temperature. Bioluminescence was then monitored over a 10-s period at room temperature in a BioOrbit 1252 luminometer by using a Multiuse software package (version 2.01; BioOrbit), and a mean was taken which was calculated by the software and expressed as RLU. Each assay was carried out in triplicate. The units of luminescence were then amended to RLU as before to allow comparison with the bioluminescence levels of lux-marked bacterial biosensor assays reported previously (39).
Statistical treatment of data.
Bioluminescence, expressed as a percentage of that for a no-treatment control, was measured with each pollutant and plotted against the pollutant concentration for each of the time points, allowing effective concentrations (EC) to be established for each pollutant. The EC20 was taken as the effective concentration of pollutant needed to reduce bioluminescence by 20% from that of the control, while the EC50 corresponded to a 50% reduction in the bioluminescence signal. The EC20 and EC50 were calculated according to the work of Paton et al. (21).
RESULTS
Transformation of Synechocystis sp. strain PCC6803.
Chromosomal transformation of Synechocystis sp. strain PCC6803 with the integration vector pTLUC (Fig. 1) was achieved by a natural transformation method (42). The vector backbone we used was an integrative vector (pILA; GenBank accession no. AJ251840) which contained a 3.0-kbp BamHI fragment of Synechocystis sp. strain PCC6803 genomic DNA (RS1 and RS2, representing the area from bp 31757 to bp 31793 in the Synechocystis sp. strain PCC6803 genome) (GenBank accession no. D63999). The tac-luc-luxAB-aphII genes were inserted into this fragment in pTLUC. After the transformation, the tac-luc-luxAB-aphII genes were integrated into Synechocystis sp. strain PCC6803 genomic DNA by a double-crossover event at the RS1 and RS2 sites. This result was confirmed both by PCR and by ampicillin sensitivity. For the PCR confirmation, we designed one primer using Synechocystis sp. strain PCC6803 genomic DNA information (which is not represented on plasmid pTLUC), and the other primer was designed to amplify from the luc gene. The PCR results indicated the integration of pTLUC plasmid DNA between the RSI and RS2 regions (data not shown). Similarly, the transformed strain was ampicillin sensitive and kanamycin resistant, indicating that the bla gene was not present (either through integration or as an episomal element) and that the aphII gene was present (through integration of pTLUC DNA between the RS1 and RS2 sites). The luc gene was inserted between bp 33429 and bp 33437 on the Synechocystis sp. strain PCC6803 chromosome (GenBank accession no. D63999). Sixteen different clones were analyzed for bioluminescence, and all gave very similar results, indicating a single, site-directed integration event.
Bioluminescence characteristics during batch growth.
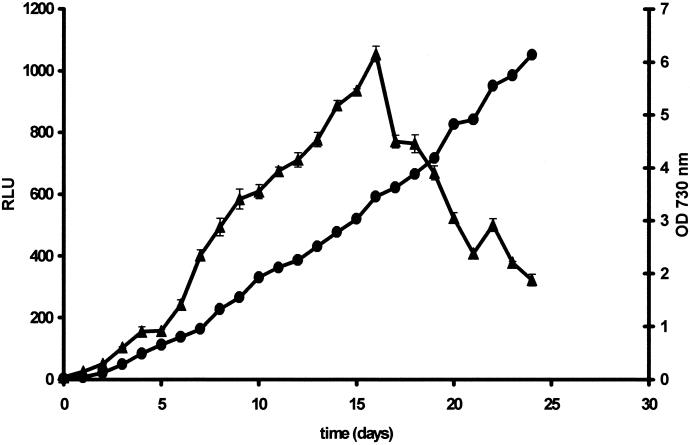
Light output during growth of transformed Synechocystis sp. strain PCC6803 in liquid batch culture is diagrammed in Fig. 2. Luminescence increased in parallel with biomass concentration until the OD730 reached 3 to 3.5 (t = 16 days) and decreased thereafter. However, even up to 24 days, when further testing was stopped, the OD730 continued to increase.
FIG. 2.
Light output during growth of luc-marked Synechocystis sp. strain PCC6803. Changes in OD730 (•) and luminescence (▴) of Synechocystis sp. strain PCC6803 during growth are means obtained from triplicate flasks ± standard errors of the means. For luminescence analysis, 200 μl of culture was removed and 1 ml of McIlvaine's citrate phosphate buffer, pH 6.5 (9), containing 0.1 mM luciferin (Molecular Probes) was added to the culture in each 1-ml luminometer cuvette. Following a 10-min exposure, bioluminescence was quantified in a BioOrbit 1251 luminometer by using a Multiuse software package (version 2.01; BioOrbit). Luminescence was monitored over a 10-s period, and a mean was taken which was calculated by the software. Luminescence was expressed in RLU, which equated to millivolts per 10 s per milliliter (26).
The growth rate of transformed Synechocystis sp. strain PCC6803 was not significantly different from that of the nontransformed parental strain (data not shown), indicating that integration and expression of the luc gene did not affect the growth rate. Similar results have been reported for Pseudomonas fluorescens (2, 35).
Effect of external pH on bioluminescence.
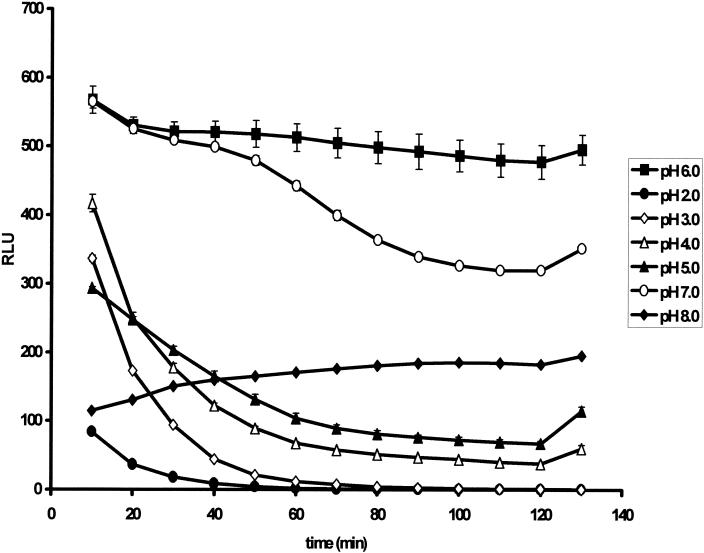
Investigations performed to examine the effect of external pH on light output demonstrated that the external pH of the assay influenced light output drastically. At pHs between 2.0 and 5.0, bioluminescence peaked at 10 min and then decreased. At pH 8.0, bioluminescence showed the reverse trend; that is, it continued to increase with the length of the assay (Fig. 3). At the optimum pH of 6.5, bioluminescence showed a marked increase and light output remained stable over 2 h.
FIG. 3.
Effects of external pH on bioluminescence responses after 10- to 130-min exposures. At pHs between 2.0 and 5.0, bioluminescence peaked at 10 min and then decreased. At pH 8.0, bioluminescence showed the reverse trend; that is, it continued to increase with the length of the assay. At the optimum pH of 6.0 to 7.0, bioluminescence showed a marked increase and light output remained stable over 2 h.
Toxicity of herbicides for luc-marked Synechocystis sp. strain PCC6803.
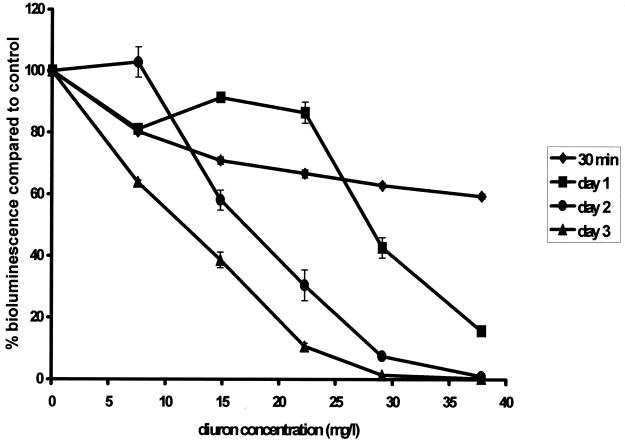
Chromosomally luc-marked Synechocystis sp. strain PCC6803 was sensitive to a range of concentrations of all eight herbicides tested. The response was dependent on both toxin concentration and the length of exposure. As an example, Fig. 4 shows the effects of different concentrations of diuron at different lengths of exposure. The results show that the decrease in bioluminescence could be correlated with the herbicide concentration and with increasing incubation time. The toxicity values (EC20 and EC50) detected with the shortest exposure times are listed in Table 2.
FIG. 4.
Luminescence responses of luc-marked Synechocystis sp. strain PCC6803 cells after 30 min (⧫), 1 day (▪), 2 days (•), and 3 days (▴) of exposure to a range of diuron concentrations. The decrease in bioluminescence could be correlated with the herbicide concentration and with increasing incubation time.
The biosensor was sensitive to the urea-based herbicide diuron (Table 2). The EC20 (7.47 ± 0.61 mg liter−1) was obtained at 30 min, while 1 day was needed to obtain an EC50 (29.70 ± 0.62 mg liter−1). The EC20 and EC50 decreased as exposure time increased; at 4 days, the EC20 was 4.08 ± 0.07 mg liter−1 and the EC50 was 10.85 ± 0.16 mg liter−1 (data not shown).
For the triazine-derived herbicides atrazine, propazine, and simazine (Table 2), EC20s (7.39 ± 0.95, 3.51 ± 0.05, and 1.06 ± 0.04 mg liter−1, respectively) were obtained at 6 h, 30 min, and 2 days, respectively. The toxicity of triazine type herbicides increased slightly with increasing exposure time, but even with an exposure time of 4 days, an EC50 could not be determined for the range of concentrations tested (data not shown).
Paraquat (Table 2) was the most toxic of all the herbicides tested. Both the EC20 (0.95 ± 0.01 mg liter−1) and the EC50 (2.85 ± 0.03 mg liter−1) could be determined at 30 min. The toxicity of paraquat increased rapidly with increasing exposure time (data not shown).
The phenoxy derivative herbicides MCPA and mecoprop (Table 2) were the least toxic of all the herbicides tested. However, the EC20s (23.60 ± 0.02 and 12.57 ± 0.95 mg liter−1, respectively) and EC50s (54.67 ± 0.16 and 47.55 ± 1.42 mg liter−1, respectively) could be easily obtained at 30 min of exposure. Increasing toxicity was also observed for this group of herbicides as exposure time increased (data not shown).
The EC20 and EC50 of the herbicide glyphosate were determined at 6 h and 1 day (Table 2), respectively. The EC20 at 6 h was 3.62 ± 0.79 mg liter−1, and the EC50 at 1 day was 3.10 ± 0.17 mg liter−1. The toxicity of glyphosate increased greatly with the exposure time; at 1 day, the EC20 was reduced to 0.41 ± 0.11 mg liter−1. The EC50 was much lower at day 2 (0.98 ± 0.02 mg liter−1) than at 1 day.
Toxicity of heavy metals and a volatile organic pollutant.
The cyanobacterial biosensor developed in this study was also very sensitive to nonherbicide toxicants, such as the heavy metals copper and zinc and a representative volatile organic, 3,5-DCP. The EC50s, 0.24 ± 0.05, 0.88 ± 0.02, and 23.39 ± 0.45 mg liter−1, respectively, were obtained at 15 min (Table 3).
TABLE 3.
EC50s of Cu, Zn, and 3,5-DCP for a range of bioluminescence-based biosensors
| Biosensor | EC50 (mg liter−1) at 15 min
|
||
|---|---|---|---|
| Cu | Zn | 3,5-DCP | |
| Synechocystis sp. strain PCC6803 (luc + luxAB) | 0.24 | 0.88 | 23.39 |
| Pseudomonas fluorescens 8866 (luxCDABE)a | 0.30 | 0.10 | 4.82 |
| Pseudomonas putida F1 (luxCDABE)a | 0.17 | 0.04 | 5.55 |
| Pseudomonas fluorescens 10586s (luxCDABE)b | 0.09 | 0.09 | NAc |
| Pseudomonas fluorescens 10586s (plasmid luxABE)b | 0.76 | 0.89 | NA |
| E. coli HB101 (luxCDABE) | 1.4d | 0.15e | 18.5d |
DISCUSSION
In this study, a novel luc-marked cyanobacterial biosensor was developed for detection of a range of classes of herbicides with different modes of action (19, 30). The biosensor described in this study has been dually marked with luc and luxAB, and both luc and luxAB could be expressed in Synechocystis sp. strain PCC6803. When we attempted to measure expression of the luxAB genes by adding the bacterial luciferase substrate aldehyde, which is required to measure luminescence, we found that aldehyde was toxic to Synechocystis sp. strain PCC6803, even at very low concentrations (data not shown). However, firefly luciferase was suitable as a reporter to detect the toxicity of pollutants. The light reaction catalyzed by the firefly luciferase enzyme relies on ATP being supplied by actively metabolizing cells. This dependence on endogenous energy supplies enables a luciferase assay system to report directly on cell health upon exposure to toxins. The optimum pH for assaying bioluminescence was found to be 6.5. This may be due to two factors: first, it may be the pH which supports the optimum physiology of the cyanobacterial biosensor, and second, it may be the pH at which the cyanobacterium is most permeable to the luciferin substrate.
When assay conditions (such as pH) were optimized, the cyanobacterial biosensor produced reproducible responses to a wide variety of herbicides (Table 2). Only one other study (39) has reported the effects of all the herbicides we tested except glyphosate on the bioluminescence response of lux-marked bacterial biosensors. The cyanobacterial biosensor was much more sensitive to the range of herbicides tested than the lux-marked E. coli HB101 biosensor (39). The cyanobacterial biosensor sensed the toxicity of herbicides at the EC50 level for four (all except the triazine class) of the five groups tested and at the EC20 level for all five of the herbicide groups tested. In contrast, the E. coli HB101 biosensor detected toxicity at the EC50 level for only two herbicide classes (paraquat and triazine) of four tested (glyphosate not tested) and at the EC20 level for three types (all except the urea type) of four tested (39). Furthermore, the cyanobacterial biosensor was more sensitive to the toxicity of herbicides than the E. coli HB101 biosensor; the EC50s and EC20s detected with the cyanobacterial biosensor were much lower than those with the E. coli HB101 biosensor.
The cyanobacterial biosensor reacts with the urea type herbicides diuron and paraquat more rapidly and more sensitively than an E. coli HB101 biosensor. The reason for this could be related to the modes of action of diuron and paraquat, which exercise their toxicity by targeting PSII (24) and photosystem I (PSI) (36), respectively. Thus, it is not surprising that diuron and paraquat were more toxic to the cyanobacterial biosensor, which has both photosystem, than to the E. coli biosensor, which has no photosystem. With respect to environmental relevance, the cyanobacterial biosensor is sensitive to herbicides at the parts-per-million level, which is appropriate for detecting residues in groundwater or soil, and in addition, the biosensor provides information on the bioavailability of the herbicide in environmental samples.
An assay period of 30 min was suitable for detection of most of the herbicides studied. The phenoxy acid herbicides MCPA and mecoprop interfere with systemic hormone signaling pathways. They affect cellular division, activating phosphate metabolism and modifying nucleic acid metabolism (40). From the results of this study, the cyanobacterial biosensor is more sensitive to MCPA and mecoprop than the E. coli biosensor (39) and a green algal biosensor (EC50, 70 mg liter−1; t = 4 days) (1). In comparison with other herbicides, the phenoxy acid herbicides were less toxic to the cyanobacterial biosensor, but an EC20 and EC50 could still be easily determined because of their high solubility.
Compared with the green alga Selenastrum capricornutum, the cyanobacterial biosensor is more sensitive to glyphosate in terms of reaction time and sensitivity. An EC50 of 1.05 mg liter−1 at 4 days was obtained with green algae (1), while an EC50 of 0.45 mg liter−1 was obtained at 1 day with the cyanobacterial biosensor developed in this study.
Herbicides of the triazine group, which includes atrazine, propazine and simazine, exert their toxicity by targeting PSII (24). The cyanobacterial biosensor could detect triazines at the EC20 level but could not be used to determine EC50s for the triazines. One reason for this could be the low solubility of this herbicide group. If it had been possible to prepare test solutions of atrazine, propazine, or simazine at higher concentrations, it is highly likely that EC50s would have been obtained (39). However, the biosensor does detect the bioavailability of these toxins, and solubility has a major impact on bioavailability. Instead of using EC50s to describe the toxicity of the triazines, an EC10 might be a realistic indicator of the environmental health risk. Additionally, development of eukaryotic algal biosensors, which incorporate the luc gene driven by a specific inducible promoter, might provide a way to increase the sensitivity of the biosensor to chosen analytes.
The luc-marked cyanobacterial biosensor developed in this study was also very sensitive to nonherbicide toxicants, such as the heavy metals copper and zinc and a representative volatile organic, 3,5-DCP. Its sensitivity was competitive with those of a range of bioluminescence-based biosensors (Table 3).
In comparison with other bacterial biosensors, another advantage of the cyanobacterial biosensor is its suitability for monitoring the chronic toxicity of pollutants. The short doubling times of other bacterial and yeast biosensors make it difficult to detect chronic toxicity effects, because complications of cell death and potential cell division can confuse interpretation of results. The much longer doubling time of cyanobacteria makes it possible to predict the long-term effects of toxicants.
Conclusion.
A novel luminescent cyanobacterial biosensor which responds to a range of compounds, including different herbicide types and nonherbicide toxicants, has been developed. The different herbicide types show different kinetics of bioluminescence inhibition. In comparison with other methods for detecting the toxicity of herbicides such as photosystem-based whole-cell and tissue biosensors (6, 11, 15, 19, 34), the whole-cell luminescent cyanobacterial biosensor proved to be more simple, rapid, accurate, and economical. The main application of this biosensor lies in rapid screening of samples or investigation of potential environmental damage. The biosensor could also be used to indicate the type of herbicide and possibly the potential mode of action.
Acknowledgments
This work was supported by BBSRC.
We thank Kunter Anja (Rostock University, Rostock, Germany) for generously providing plasmid pILA and Janet K. Jansson (Section for Natural Sciences, Sodertorns Hogskola University College, Huddinge, Sweden) for plasmid pJJ303.
REFERENCES
- 1.Abdel-Hamid, M. I. 1996. Development and application of a simple procedure for toxicity testing using immobilized algae. Water Sci. Technol. 33:129-138. [Google Scholar]
- 2.Amin-Hanjani, S., A. Meikle, L. A. Glover, J. I. Prosser, and K. Killham. 1993. Plasmid and chromosomally encoded luminescence marker systems for detection of Pseudomonas fluorescens in soil. Mol. Ecol. 2:47-54. [Google Scholar]
- 3.Arsalane, W., G. Paresys, J. C. Duval, R. Conrad, and C. Buechel. 1993. A new fluorometric device to measure the in vivo chlorophyll a fluorescence yield in microalgae and its use as a herbicide monitor. Eur. J. Phycol. 28:247-252. [Google Scholar]
- 4.Brewster, J. D., and A. R. Lightfield. 1993. Rapid biorecognition assay for herbicides in biological matrices. Anal. Chem. 65:2415-2419. [DOI] [PubMed] [Google Scholar]
- 5.Brewster, J. D., A. R. Lightfield, and P. L. Bermel. 1995. Storage and immobilization of photosystem II reaction centers used in an assay for herbicides. Anal. Chem. 67:1296-1299. [Google Scholar]
- 6.Campanella, L., F. Cubadda, M. P. Sammartino, and A. Saoncella. 2000. An algal biosensor for the monitoring of water toxicity in estuarine environments. Water Res. 35:69-76. [DOI] [PubMed] [Google Scholar]
- 7.Chauvat, F., L. De Vries, A. Van de Ende, and G. A. Van Arkel. 1986. A host-vector system for gene cloning in the cyanobacterium Synechocystis PCC 6803. Mol. Gen. Genet. 204:185-191. [Google Scholar]
- 8.Conrad, R., C. Buchel, C. Wilhelm, W. Arsalane, C. Berkaloff, and J. C. Duval. 1993. Changes in yield in in-vivo fluorescence of chlorophyll a as a tool for selective herbicide monitoring. J. Appl. Phycol. 5:505-516. [Google Scholar]
- 9.Dawson, R. M. C., D. C. Elliott, W. H. Elliott, and K. M. Jones. 1986. Data for biochemical research, 3rd ed. Clarendon Press, Oxford, United Kingdom.
- 10.Draber, W. T., K. Tietjen, J. F. Kluth, and A. Trebst. 1991. Herbicides in photosynthesis research. Angew. Chem. 3:1621-1633. [Google Scholar]
- 11.Frense, D., A. Muller, and D. Beckmann. 1998. Detection of environmental pollutants using an optical biosensor with immobilized algae cells. Sensors Actuators B 51:256-260. [Google Scholar]
- 12.Hollis, R. P., K. Killham, and L. A. Glover. 2000. Design and application of a biosensor for monitoring toxicity of compounds to eukaryotes. Appl. Environ. Microbiol. 66:1676-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isenberg, D. L. 1993. The Microtox toxicity test: a developer's commentary, p. 3-15. In M. Richardson (ed.), Ecotoxicology monitoring. VCH, Weinheim, Germany.
- 14.Jay, A. E., J. M. Ducruet, J. C. Duval, and J. P. Pelletier. 1997. A high-sensitivity chlorophyll fluorescence assay for monitoring herbicide inhibition of photosystem II in the chlorophyte Selenastrum capricornutum: comparison with effect on cell growth. Arch. Hydrobiol. 140:273-286. [Google Scholar]
- 15.Koblizek, M., J. Masojidek, J. Komenda, T. Kucera, R. Pilloton, A. K. Mattoo, and M. T. Giardi. 1998. A sensitive photosystem II based biosensor for detection of a class of herbicides. Biotechnol. Bioeng. 60:664-669. [PubMed] [Google Scholar]
- 16.Lagido, C., J. Pettitt, A. J. R. Porter, G. I. Paton, and L. A. Glover. 2001. Development and application of bioluminescent Caenorhabditis elegans as multicellular eukaryotic biosensors. FEBS Lett. 493:36-39. [DOI] [PubMed] [Google Scholar]
- 17.Loranger, C., and R. Carpentier. 1994. A fast bioassay for phytotoxicity measurements using immobilized photosynthetic membranes. Biotechnol. Bioeng. 44:178-183. [DOI] [PubMed] [Google Scholar]
- 18.Marty, J. L., D. Garcia, and R. Rouillon. 1995. Biosensors: potential in pesticide detection. Trends Anal. Chem. 14:329-333. [Google Scholar]
- 19.Merz, D., M. Geyer, D. A. Moss, and H. J. Ache. 1996. Chlorophyll fluorescence biosensor for the detection of herbicides. Fresenius J. Anal. Chem. 354:299-305. [DOI] [PubMed] [Google Scholar]
- 20.Moller, A., K. Gustafsson, and J. K. Jansson. 1994. Specific monitoring by PCR amplification and bioluminescence of firefly luciferase gene-tagged bacteria added to environmental samples. FEMS Microbiol. Ecol. 15:193-206. [Google Scholar]
- 21.Paton, G. I., C. D. Campbell, L. A. Glover, and K. Killham. 1995. Assessment of bioavailability of heavy metals using lux modified constructs of Pseudomonas fluorescens. Lett. Appl. Microbiol. 20:52-56. [Google Scholar]
- 22.Paton, G. I., E. A. S. Rattray, C. D. Campbell, M. S. Cresser, L. A. Glover, J. C. L. Meeussen, and K. Killham. 1996. Use of genetically modified microbial biosensor for soil ecotoxicity testing, p. 394-418. In C. F. Pankhurst, B. M. Doube, and V. V. S. R. Gupta (ed.), Biological indicators of soil health. CAB International Press, Oxon, United Kingdom.
- 23.Paton, G. I., G. Palmer, M. Burton, E. A. S. Rattray, S. P. McGrath, L. A. Glover, and K. Killham. 1997. Development of an acute and chronic ecotoxicity assay using lux-marked Rhizobium leguminosarum biovar trifolii. Lett. Appl. Microbiol. 24:296-300. [DOI] [PubMed] [Google Scholar]
- 24.Pfister, K., K. E. Teinback, G. Gardner, and C. J. Arntzen. 1981. Photoaffinity labelling of a herbicide receptor protein in chloroplast membranes. Proc. Natl. Acad. Sci. USA 78:981-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porter, R. D. 1988. DNA transformation. Methods Enzymol. 167:703-712. [DOI] [PubMed] [Google Scholar]
- 26.Rattray, E. A. S., J. I. Prosser, K. Killham, and L. A. Glover. 1990. Luminescence-based non-extractive techniques for in situ detection of Escherichia coli in soil. Appl. Environ. Microbiol. 56:3368-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rawson, D. M., A. J. Willmer, and M. C. Cardosi. 1987. The development of whole cell biosensor for on line screening of herbicide pollution of surface water. Toxic. Assess. 2:325-340. [Google Scholar]
- 28.Rawson, D. M., A. J. Willmer, and A. P. F. Turner. 1989. Whole-cell biosensor for environmental monitoring. Biosensors 4:299-311. [DOI] [PubMed] [Google Scholar]
- 29.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 30.Rizzuto, M., C. Polcaro, C. Desiderio, M. Koblizek, R. Pilloton, and M. T. Giardi. 2000. Herbicide monitoring in surface water samples with a photosystem-II based biosensor, p. 346-357. In F. Mazzei and R. Pilloton (ed.), Proceedings of the second workshop on chemical sensors and biosensors, E. N. E. A., Rome, Italy.
- 31.Rouillon, R., M. Tocabens, and J. L. Marty. 1994. Stabilization of chloroplasts by entrapment in polyvinyl alcohol bearing styrylpyridinium groups. Anal. Lett. 27:2239-2248. [Google Scholar]
- 32.Rouillon, R., M. Sole, R. Carpentier, and J. L. Marty. 1995. Immobilization of thylakoids in polyvinyl alcohol for the detection of herbicides. Sensors Actuators B 26-27:477-479. [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schafer, H., H. Hettler, U. Fritsche, G. Pitzen, G. Roderer, and A. Wenzel. 1994. Biotests using unicellular algal and ciliates for predicting long-term effects of toxicants. Ecotoxicol. Environ. Safety. 27:64-81. [DOI] [PubMed] [Google Scholar]
- 35.Shaw, L. J., Y. Beaton, L. A. Glover, K. Killham, and A. A. Meharg. 1999. Development and characterization of a lux-modified 2,4-dichlorophenol-degrading Burkholderia sp. RASC. Environ. Microbiol. 1:393-399. [DOI] [PubMed] [Google Scholar]
- 36.Smith, L. L. 1988. The toxicity of paraquat. Advance Drug Reaction Account Poison Rev. 1:1-17. [PubMed] [Google Scholar]
- 37.Soukupova, J., A. Lukavska, J. Lukavsky, and L. Nedbal. 1999. Sensitivity of the algal biotest ISO 10253 to the photosystem 2 in seawater. Photosynthetica 37:209-216. [Google Scholar]
- 38.Steinberg, S. M., E. J. Poziomek, W. H. Englemann, and K. R. Rogers. 1995. A review of environmental applications of bioluminescence measurements. Chemosphere 30:2155-2197. [Google Scholar]
- 39.Strachan, G., S. Preston, H. Maciel, A. J. R. Porter, and G. I. Paton. 2001. Use of bacterial biosensors to interpret the toxicity and mixture toxicity of herbicides in freshwater. Water Res. 35:3490-3495. [DOI] [PubMed] [Google Scholar]
- 40.Thurlow, W. H., and N. S. Digby. 1981. A review of the newly recognised potential health hazards of phenoxy herbicides. Nova Scotia Med. Bull. 60:57-60. [Google Scholar]
- 41.Weitz, H. J., J. M. Ritchie, D. A. Bailey, A. M. Horsburgh, K. Killham, and L. A. Glover. 2001. Construction of a modified mini-Tn5 luxCDABE transposon for the development of bacterial biosensors for ecotoxicity testing. FEMS Microbiol. Lett. 197:159-165. [DOI] [PubMed] [Google Scholar]
- 42.Williams, J. G. K. 1988. Construction of specific mutations in photosystem II photosynthetic reaction centre by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167:766-778. [Google Scholar]
- 43.Yoneyama, K., Y. Nakajjma, N. Maejima, M. Ogasawara, M. Konnai, N. Tokutake, H. Iwamura, F. Sato, K. Ichinose, T. Asami, and S. Yoshida. 1993. Simple and rapid screening method for photosystem II inhibitory herbicides using photoautotrophically cultured plant cells with chlorophyll fluorescence monitoring. Biosci. Biotech. Biochem. 57:1389-1390. [Google Scholar]