Abstract
For cultivation-independent detection of sulfate-reducing prokaryotes (SRPs) an oligonucleotide microarray consisting of 132 16S rRNA gene-targeted oligonucleotide probes (18-mers) having hierarchical and parallel (identical) specificity for the detection of all known lineages of sulfate-reducing prokaryotes (SRP-PhyloChip) was designed and subsequently evaluated with 41 suitable pure cultures of SRPs. The applicability of SRP-PhyloChip for diversity screening of SRPs in environmental and clinical samples was tested by using samples from periodontal tooth pockets and from the chemocline of a hypersaline cyanobacterial mat from Solar Lake (Sinai, Egypt). Consistent with previous studies, SRP-PhyloChip indicated the occurrence of Desulfomicrobium spp. in the tooth pockets and the presence of Desulfonema- and Desulfomonile-like SRPs (together with other SRPs) in the chemocline of the mat. The SRP-PhyloChip results were confirmed by several DNA microarray-independent techniques, including specific PCR amplification, cloning, and sequencing of SRP 16S rRNA genes and the genes encoding the dissimilatory (bi)sulfite reductase (dsrAB).
Anaerobic respiration with sulfate is a central component of the global sulfur cycle and is exhibited exclusively by prokaryotes (53). Sulfate-reducing prokaryotes (SRPs) are thus of major numerical and functional importance in many ecosystems, including marine sediments (14, 29, 30, 38, 54) and cyanobacterial microbial mats (46, 56, 70). Recently, SRPs were also identified as unculturable symbionts of gutless marine oligochetes (15) and as uncultured components of microbial aggregates catalyzing anaerobic methane oxidation (4, 10, 48, 72). In addition, some SRPs have been implicated in human disease (32, 35, 39, 43, 60, 69). More than 130 species of SRPs have been described so far, and they comprise a phylogenetically diverse assemblage of organisms consisting of members of at least four bacterial phyla and one archaeal phylum (11, 12, 66). The polyphyletic affiliation of SRPs and the fact that several SRPs are closely related to microorganisms which cannot perform anaerobic sulfate reduction for energy generation hamper cultivation-independent detection of these organisms by established 16S rRNA-based methods because many different PCR primer sets or probes would be required to target all members of this microbial guild. Consequently, previous environmental microbiology research on the composition of SRP communities performed by using specific 16S rRNA gene-targeting PCR systems or probes has focused on a few selected genera or groups (16, 24, 36, 41, 49, 50, 56, 59, 68, 71).
Nucleic acid microarrays, which have recently been introduced for bacterial identification in microbial ecology (5, 23, 37, 62, 73, 77), provide a powerful tool for parallel detection of 16S rRNA genes (23, 37, 62, 73) and thus might be particularly useful for environmental studies of phylogenetically diverse microbial groups. However, most microarrays developed so far for bacterial identification consist of a limited number of probes and are mainly used for method development and optimization. In this study, we developed and successfully used a microarray consisting of 132 16S rRNA-targeted oligonucleotide probes covering all recognized lineages of SRPs for high-resolution screening of clinical and environmental samples. For periodontal tooth pockets and a hypersaline microbial mat, microarray SRP diversity fingerprints were found to be consistent with results obtained by using well-established molecular methods for SRP community composition analysis.
MATERIALS AND METHODS
Pure cultures of SRPs.
Table 1 lists the 42 reference organisms that were obtained as lyophilized cells or active cultures from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany) and were used to evaluate our microarray (SRP-PhyloChip). Archaeoglobus veneficus SNP6T (containing plasmid XY) was deposited in the Deutsche Sammlung von Mikroorganismen und Zellkulturen by K. O. Stetter, Lehrstuhl fur Mikrobiologie, Universität Regensburg, Regensburg, Germany, as DSM 11195T.
TABLE 1.
SRP strains used in this study
| Species | Strain |
|---|---|
| Desulfovibrio cuneatus | DSM 11391T |
| Desulfovibrio aminophilus | DSM 12254T |
| Desulfovibrio gabonensis | DSM 10636T |
| Desulfovibrio alcoholivorans | DSM 5433T |
| Desulfovibrio termitidis | DSM 5308T |
| Desulfovibrio zosterae | DSM 11974T |
| Desulfovibrio halophilus | DSM 5663T |
| Desulfovibrio longus | DSM 6739T |
| “Desulfovibrio aestuarii” | DSM 1926T |
| Desulfovibrio profundus | DSM 11384T |
| Desulfomicrobium aspheronum | DSM 5918T |
| Desulfomicrobium orale | DSM 12838T |
| Desulfohalobium retbaense | DSM 5692T |
| Desulfotalea arctica | DSM 12342T |
| Desulforhopalus vacuolatus | DSM 9700T |
| Desulfobulbus propionicus | DSM 2032T |
| “Desulfobotulus sapovorans” | DSM 2055T |
| Desulfococcus multivorans | DSM 2059T |
| Desulfonema limicola | DSM 2076T |
| Desulfonema ishimotonii | DSM 9680T |
| Desulfobacterium indolicum | DSM 3383T |
| Desulfosarcina variabilis | DSM 2060T |
| Desulfofaba gelida | DSM 12344T |
| Desulfofrigus oceanense | DSM 12341T |
| “Desulfobacterium niacini” | DSM 2650T |
| Desulfobacula toluolica | DSM 7467T |
| Desulfotignum balticum | DSM 7044T |
| Desulfobacter halotolerans | DSM 11383T |
| Desulfobacter latus | DSM 3381T |
| Thermodesulforhabdus norvegica | DSM 9990T |
| Desulfomonile tiedjei | DSM 6799T |
| Desulfobacca acetoxidans | DSM 11109T |
| Desulfotomaculum aeronauticum | DSM 10349T |
| Desulfotomaculum geothermicum | DSM 3669T |
| Desulfotomaculum australicum | DSM 11792T |
| Desulfotomaculum thermobenzoicum | DSM 6193T |
| Desulfotomaculum acetoxidans | DSM 771T |
| Desulfotomaculum halophilum | DSM 11559T |
| Desulfosporosinus orientis | DSM 765T |
| Thermodesulfovibrio islandicus | DSM 12570T |
| Thermodesulfobacterium mobile (Thermodesulfobacterium thermophilum) | DSM 1276T |
| Archaeoglobus veneficus | DSM 11195T |
Solar Lake mat sample.
A core (1 by 1cm; depth, 4 cm) of a hypersaline cyanobacterial mat from Solar Lake (Sinai, Egypt) was sectioned horizontally at 200-μm intervals with a cryomicrotome (MIKROM HM500; Microm, Walldorf, Germany). The mat sections were stored at −80°C.
Peridontal tooth pocket samples.
Samples from five patients with adult periodontitis were taken by inserting a sterile medium-sized paper point into a single periodontal tooth pocket. After sampling the paper points were stored at −20°C.
DNA extraction.
Genomic DNA was isolated from reference organisms with a FastDNA kit (Bio 101, Vista, Calif.). DNA from periodontal tooth pocket material and DNA from a cryosection of Solar Lake mat from the chemocline (1,400 to 1,600 μm from the mat surface) were extracted by using a modification of the protocol of Griffiths et al. (22). In contrast to the original protocol, precipitation of nucleic acids in the aqueous phase was performed with 0.1 volume of sodium acetate (pH 5.2) and 0.6 volume of isopropanol for 2 h at room temperature.
PCR amplification of 16S rRNA and dsrAB genes.
For subsequent DNA microarray hybridization, almost complete 16S rRNA gene fragments were amplified from DNA of pure cultures of SRPs by using the 616V-630R primer pair (Table 2). 16S rRNA gene fragments of A. veneficus were amplified by using the newly designed Archaeoglobus genus-specific forward primer ARGLO36F and the universal reverse primer 1492R (Table 2). Amplification of bacterial 16S rRNA gene fragments from periodontal tooth pocket or Solar Lake mat genomic DNA was performed by using the 616V-630R and 616V-1492R primer pairs (Table 2).
TABLE 2.
16S rRNA gene-targeted primers
| Short namea | Full nameb | Annealing temp (°C) | Sequence 5′-3′ | Specificity | Reference |
|---|---|---|---|---|---|
| 616V | S-D-Bact-0008-a-S-18 | 52 | AGA GTT TGA TYM TGG CTC | Most Bacteria | 26 |
| 630R | S-D-Bact-1529-a-A-17 | 52 | CAK AAA GGA GGT GAT CC | Most Bacteria | 26 |
| 1492R | S-*-Proka-1492-a-A-19 | 52, 60c | GGY TAC CTT GTT ACG ACT T | Most Bacteria and Archaea | Modified from reference 27 |
| ARGLO36F | S-G-Arglo-0036-a-S-17 | 52 | CTA TCC GGC TGG GAC TA | Archaeoglobus spp. | This study |
| DSBAC355F | S-*-Dsb-0355-a-S-18 | 60 | CAG TGA GGA ATT TTG CGC | Most “Desulfobacterales” and “Syntrophobacterales” | 59 |
| DSM172F | S-G-Dsm-0172-a-S-19 | 56 | AAT ACC GGA TAG TCT GGC T | Desulfomicrobium spp. | This study |
| DSM1469R | S-G-Dsm-1469-a-A-18 | 56 | CAA TTA CCA GCC CTA CCG | Desulfomicrobium spp. | This study |
| DSN61F | S-*-Dsn-0061-a-S-17 | 52 | GTC GCA CGA GAA CAC CC | Desulfonema limicola, Desulfonema ishimotonii | This study |
| DSN+1201R | S-*-Dsn-1201-a-A-17 | 52 | GAC ATA AAG GCC ATG AG | Desulfonema spp. and other Bacteria | This study |
Short name used in the reference or in this study.
Name of 16S rRNA gene-targeted oligonucleotide primer based on the nomenclature of Alm et al. (1).
The annealing temperature was 52°C when the primer was used with forward primer 616V or ARGLO36F, and the annealing temperature was 60°C when the primer was used with forward primer DSBAC355F.
To confirm DNA microarray results, specific amplification of 16S rRNA gene fragments of defined SRP groups was performed with periodontal tooth pocket DNA and Solar Lake mat DNA by using previously described and newly designed primers (Table 2). In addition, an approximately 1.9-kb dsrAB fragment was amplified from periodontal tooth pockets by using primers DSR1F and DSR4R under the conditions described by Wagner et al. (76).
Positive controls containing purified DNA from suitable reference organisms were included in all of the PCR amplification experiments along with negative controls (no DNA added). For 16S rRNA gene amplification, reaction mixtures (total volume, 50 μl) containing each primer at a concentration of 25 pM were prepared by using 10× Ex Taq reaction buffer and 2.5 U of Ex Taq polymerase (Takara Biomedicals, Otsu, Shiga, Japan). Additionally, 20 mM tetramethylammonium chloride (Sigma, Deisenhofen, Germany) was added to each amplification mixture to enhance the specificity of the PCR (31). Thermal cycling was carried out by using an initial denaturation step of 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 40 s, annealing at temperatures ranging from 52 to 60°C (depending on the primer pair [Table 2]) for 40 s, and elongation at 72°C for 1 min 30 s. Cycling was completed by a final elongation step of 72°C for 10 min.
Fluorescence labeling of PCR amplificates.
Prior to labeling, PCR amplificates were purified by using a QIAquick PCR purification kit (Qiagen, Hilden, Germany). Subsequently, the amount of DNA was determined spectrophotometrically by measuring the optical density at 260 nm. Purified PCR products were labeled with Cy5 by using a DecaLabel DNA labeling kit (MBI Fermentas, Vilnius, Lithuania). Reaction mixtures (total volume, 45 μl) containing 200 ng of purified PCR product and 10 μl of decanucleotides in reaction buffer were denatured at 95°C for 10 min and immediately placed on ice. After addition of 3 μl of deoxynucleotide Mix C (containing no dCTP), 1 μl of Cy5-dCTP (Amersham Biosciences, Freiburg, Germany), and 1 μl of the Klenow fragment (Exo−; 5 U μl−1), the labeling reaction mixtures were incubated at 37°C for 45 min. For more efficient labeling, the addition of Mix C, Cy5-dCTP, and the Klenow fragment and incubation at 37°C for 45 min were repeated. Labeling was completed by addition of 4 μl of dNTP-Mix and incubation at 37°C for 60 min. To remove unincorporated deoxynucleotides and decanucleotides, the labeling mixture was purified with a QIAquick nucleotide removal kit (Qiagen) by using double-distilled water for DNA elution. Finally, the eluted DNA was vacuum dried and stored in the dark at −20°C.
Microarray manufacture and processing.
Oligonucleotides for microarray printing were obtained from MWG Biotech (Ebersberg, Germany). The sequence, specificity, and microarray position of each oligonucleotide probe are shown in Table 3. In addition, difference alignments for all probes generated with the latest ARB small-subunit rRNA database (http://www.arb-home.de) can be viewed at the probeBase website (http://www.probebase.net). The 5′ end of each oligonucleotide probe was tailed with 15 dTTP molecules (T-spacer) to increase the on-chip accessibility of spotted probes to target DNA (61, 63). In addition, the 5′-terminal nucleotide of each oligonucleotide was aminated to allow covalent coupling of the oligonucleotides to aldehyde group-coated CSS-100 glass slides (CEL Associates, Houston, Tex.). The concentration of oligonucleotide probes before printing was adjusted to 50 pmol μl−1 in 50% dimethyl sulfoxide to prevent evaporation during the printing procedure. SRP-PhyloChips were printed by using a GMS 417 contact arrayer (Affymetrix, Santa Clara, Calif.). Spotted DNA microarrays were dried overnight at room temperature to allow efficient cross-linking. Slides were washed twice at room temperature in 0.2% sodium dodecyl sulfate (SDS) and then twice with double-distilled water with vigorous agitation to remove unbound oligonucleotides and the SDS. After air drying, the slides were incubated for 5 min in a fresh sodium borohydride solution (1.0 g of NaBH4 in 300 ml of phosphate-buffered saline and 100 ml of absolute ethanol) to reduce all remaining reactive aldehyde groups on the glass. The reaction was stopped by adding ice-cold absolute ethanol. The reduced slides were washed three times (with 0.2% SDS and double-distilled water), air dried, and stored in the dark at room temperature.
TABLE 3.
16S rRNA-targeted oligonucleotide probes
| Original probe name | Short name | Full namea | Sequence (5′-3′) | Microarray position | Specificity | Reference |
|---|---|---|---|---|---|---|
| CONT | AGG AAG GAA GGA AGG AAG | A1-F1, A48-F48 | Control oligonucleotide | This study | ||
| CONT-COMP | CTT CCT TCC TTC CTT CCT | Complementary to control oligonucleotide | This study | |||
| NONSENSE | AGA GAG AGA GAG AGA GAG | F47 | Nonbinding control | This study | ||
| EUB338 | EUB338 | S-D-Bact-0338-a-A-18 | GCT GCC TCC CGT AGG AGT | D25, F2 | Most Bacteria | 2 |
| EUB338II | EUB338II | S-*-BactP-0338-a-A-18 | GCA GCC ACC CGT AGG TGT | F3 | Phylum Planctomycetes | 6 |
| EUB338III | EUB338III | S-*-BactV-0338-a-A-18 | GCT GCC ACC CGT AGG TGT | F4 | Phylum Verrucomicrobia | 6 |
| UNIV1390 | UNIV1389a | S-D-Univ-1389-a-A-18 | ACG GGC GGT GTG TAC AAG | D26, F5 | Bacteria, not “Epsilonproteobacteria” | 78b |
| UNIV1390 | UNIV1389b | S-D-Univ-1389-b-A-18 | ACG GGC GGT GTG TAC AAA | F6 | Eucarya | 78b |
| UNIV1390 | UNIV1389c | S-D-Univ-1389-c-A-18 | ACG GGC GGT GTG TGC AAG | D34, F7 | Archaea | 78b |
| ARCH915 | ARCH917 | S-D-Arch-0917-a-A-18 | GTG CTC CCC CGC CAA TTC | D35 | Archaea | 67b |
| DELTA495a | S-C-dProt-0495-a-A-18 | AGT TAG CCG GTG CTT CCT | C2, E2 | Most “Deltaproteobacteria” | This study | |
| DELTA495b | S-*-dProt-0495-b-A-18 | AGT TAG CCG GCG CTT CCT | C3, E3 | Some “Deltaproteobacteria” | This study | |
| DELTA495c | S-*-dProt-0495-c-A-18 | AAT TAG CCG GTG CTT CCT | C4, E4 | Some “Deltaproteobacteria” | This study | |
| S-*-Ntspa-712-a-A-21 | NTSPA714 | S-*-Ntspa-714-a-A-18 | CCT TCG CCA CCG GCC TTC | D30 | Phylum Nitrospira, not Thermodesulfovibrio islandicus | 7 |
| LGC354A | LGC354a | S-*-Lgc-0354-a-A-18 | TGG AAG ATT CCC TAC TGC | A2 | Probes LGC354a, LGC354b, and LGC354c target together the phylum Firmicutes but not Desulfotomaculum and Desulfosporosinus | 44 |
| LGC354B | LGC354b | S-*-Lgc-0354-b-A-18 | CGG AAG ATT CCC TAC TGC | A3 | See above | 44 |
| LGC354C | LGC354c | S-*-Lgc-0354-c-A-18 | CCG AAG ATT CCC TAC TGC | A4 | See above | 44 |
| SRB385 | SRB385 | S-*-Srb-0385-a-A-18 | CGG CGT CGC TGC GTC AGG | C5, E5 | Many but not all deltaproteobacterial SRPs, Aerothermobacter spp., Thermomonospora spp., Actinobispora spp., Actinomadura spp., Thermoanaerobacter spp., Frankia spp., Clostridium spp., Streptosporangium spp., Nitrospira spp., Geodermatophilus spp., Nocardiopsis spp., and many more | 2 |
| SRB385Db | SRB385Db | S-*-Srb-0385-b-A-18 | CGG CGT TGC TGC GTC AGG | C6, E6 | Many but not all deltaproteobacterial SRPs, Geobacter spp., Pelobacter spp., Campylobacter spp., Saccharopolyspora spp., Acetivibrio spp., Syntrophus spp., Clostridium spp., Nitrospina spp., Chlorobium spp., and many more | 52 |
| DSBAC355 | DSBAC355 | S-*-Dsbac-0355-a-A-18 | GCG CAA AAT TCC TCA CTG | C7 | Most “Desulfobacterales” and “Syntrophobacterales” | 59 |
| DSB706 | S-*-Dsb-0706-a-A-18 | ACC GGT ATT CCT CCC GAT | C8 | Desulfotalea spp., Desulfosarcina sp., Desulforhopalus sp., Desulfocapsa spp., Desulfofustis sp., Desulfobacterium sp., Desulfobulbus spp., Thermodesulforhabdus sp. | This study | |
| DSS658 | DSS658 | S-*-Dsb-0658-a-A-18 | TCC ACT TCC CTC TCC CAT | C11 | Desulfostipes sp., Desulfobacterium sp., Desulfofrigus spp., Desulfofaba sp., Desulfosarcina sp., Desulfomusa sp. | 41 |
| DSR651 | DSR651 | S-*-Dsb-0651-a-A-18 | CCC CCT CCA GTA CTC AAG | C10 | Desulforhopalus sp., Desulfobacterium sp., Desulfofustis sp., Desulfocapsa sp., Desulfobulbus spp., Spirochaeta spp. | 41 |
| probe 804 | DSB804 | S-*-Dsb-0804-a-A-18 | CAA CGT TTA CTG CGT GGA | C9 | Desulfobacter spp., Desulfobacterium spp., Desulfofrigus spp., Desulfofaba sp., Desulfosarcina sp., Desulfostipes sp., Desulfococcus sp., Desulfobotulus sp., Desulforegula sp. | 13 |
| DSB230 | S-*-Dsb-0230-a-A-18 | CTA ATG GTA CGC AAG CTC | B6 | Desulfotalea spp., Desulforhopalus sp., Desulfocapsa spp., Desulfofustis sp., Desulfobacterium sp. | This study | |
| DSTAL131 | S-G-Dstal-0131-a-A-18 | CCC AGA TAT CAG GGT AGA | B9 | Desulfotalea spp. | This study | |
| DSTAL213 | S-G-Dstal-0213-a-A-18 | CCT CCC GAT ACA ATA GCT | B8 | See above | This study | |
| DSTAL645 | S-G-Dstal-0645-a-A-18 | CCA GTA CTC AAG CTC CCC | B10 | See above | This study | |
| DSTAL732 | S-G-Dstal-0732-a-A-18 | TAT CTG GCC AGA TGG TCG | B12 | See above | This study | |
| DSTAL835 | S-G-Dstal-0835-a-A-18 | GAA GCG ATT AAC CAC TCC | B11 | See above | This study | |
| DSRHP185 | S-*-Dsrhp-0185-a-A-18 | CCA CCT TTC CTG TTT CCA | B7 | Desulforhopalus spp. | This study | |
| DSBB228 | S-G-Dsbb-0228-a-A-18 | AAT GGT ACG CAG ACC CCT | B4 | Desulfobulbus spp. | This study | |
| probe 660 | DSBB660 | S-G-Dsbb-0660-a-A-18 | ATT CCA CTT TCC CCT CTG | B5 | See above | 13b |
| DSB985 | DSB986 | S-*-Dsb-0986-a-A-18 | CAC AGG ATG TCA AAC CCA | C28 | Desulfobacter spp., Desulfobacula sp., Desulfobacterium sp., Desulfospira sp., Desulfotignum sp. | 41b |
| DSB1030 | S-*-Dsb-1030-a-A-18 | CTG TCT CTG TGC TCC CGA | C27 | See above | This study | |
| DSB1240 | S-*-Dsb-1240-a-A-18 | TGC CCT TTG TAC CTA CCA | C34 | Desulfobacter spp., Desulfotignum sp. | This study | |
| DSB623 | DSB623a | S-*-Dsb-0623-a-A-18 | TCA AGT GCA CTT CCG GGG | C35 | Desulfobacter curvatus, Desulfobacter halotolerans, Desulfobacter hydrogenophilus, Desulfobacter postgatei, Desulfobacter vibrioformis | 9b |
| DSB623b | S-*-Dsb-0623-b-A-18 | TCA AGT GCA CTT CCA GGG | C36 | Desulfobacter sp., strain BG8, Desulfobacter sp. strain BG23 | This study | |
| DSB623 | DSBLA623 | S-S-Dsb.la-0623-a-A-18 | TCA AGT GCT CTT CCG GGG | C37 | Desulfobacter latus | 9b |
| DSBACL143 | S-G-Dsbacl-0143-a-A-18 | TCG GGC AGT TAT CCC GGG | C29 | Desulfobacula spp. | This study | |
| DSBACL225 | S-G-Dsbacl-0225-a-A-18 | GGT CCG CAA ACT CAT CTC | C30 | See above | This study | |
| DSBACL317 | S-G-Dsbacl-0317-a-A-18 | GAC CGT GTA CCA GTT CCA | C31 | See above | This study | |
| DSBACL1268 | S-G-Dsbacl-1268-a-A-18 | AGG GAT TCG CTT ACC GTT | C32 | See above | This study | |
| DSBACL1434 | S-G-Dsbacl-1434-a-A-18 | ATA GTT AGC CCA ACG ACG | C33 | See above | This study | |
| DSF672 | DSB674 | S-*-Dsb-0674-a-A-18 | CCT CTA CAC CTG GAA TTC | C20 | Desulfofrigus spp., Desulfofaba gelida, Desulfomusa hansenii | 55b |
| DSB220 | S-*-Dsb-0220-a-A-18 | GCG GAC TCA TCT TCA AAC | C25 | Desulfobacterium niacini, Desulfobacterium vacuolatum, Desulfobacterium autotrophicum, Desulfofaba gelida | This study | |
| DSBM1239 | S-*-Dsbm-1239-a-A-18 | GCC CGT TGT ACA TAC CAT | C26 | Desulfobacterium niacini, Desulfobacterium vacuolatum, Desulfobacterium autotrophicum | This study | |
| DSFRG211 | S-G-Dsfrg-0211-a-A-18 | CCC CAA ACA AAA GCT TCC | C22 | Desulfofrigus spp. | This study | |
| DSFRG445 | S-G-Dsfrg-0445-a-A-18 | CAT GTG AGG TTT CTT CCC | C23 | See above | This study | |
| DSFRG1030 | S-G-Dsfrg-1030-a-A-18 | TGT CAT CGG ATT CCC CGA | C24 | See above | This study | |
| DCC868 | DCC868 | S-*-Dsb-0868-a-A-18 | CAG GCG GAT CAC TTA ATG | C38 | Desulfosarcina sp., Desulfonema spp., Desulfococcus sp., Desulfobacterium spp., Desulfobotulus sp., Desulfostipes sp., Desulfomusa sp. | 9 |
| DSSDBM194 | S-*-DssDbm-0194-a-A-18 | GAA GAG GCC ACC CTT GAT | C40 | Desulfosarcina variabilis, Desulfobacterium cetonicum | This study | |
| DSSDBM217 | S-*-DssDbm-0217-a-A-18 | GGC CCA TCT TCA AAC AGT | C41 | See above | This study | |
| DSSDBM998 | S-*-DssDbm-0998-a-A-18 | TTC GAT AGG ATT CCC GGG | C39 | See above | This study | |
| DSSDBM1286 | S-*-DssDbm-1286-a-A-18 | GAA CTT GGG ACG GCT TTT | C42 | See above | This study | |
| DSC193 | DSC193 | S-*-Dsb-0193-a-A-18 | AGG CCA CCC TTG ATC CAA | C43 | Desulfosarcina variabilis | 55 |
| DSBMIN218 | S-S-Dsbm.in-0218-a-A-18 | GGG CTC CTC CAT AAA CAG | C44 | Desulfobacterium indolicum | This study | |
| DCC209 | DCC209 | S-S-Dcc.mv-0209-a-A-18 | CCC AAA CGG TAG CTT CCT | B3 | Desulfococcus multivorans | 55 |
| DSNISH179 | S-S-Dsn.ish-0179-a-A-18 | GGG TCA CGG GAA TGT TAT | C45 | Desulfonema ishimotonii | This study | |
| DSNISH442 | S-S-Dsn.ish-0442-a-A-18 | CCC CAG GTT CTT CCC ACA | C46 | See above | This study | |
| DSNISH1001 | S-S-Dsn.ish-1001-a-A-18 | CGT CTC CGG AAA ATT CCC | C47 | See above | This study | |
| DNMA657 | DSN658 | S-*-Dsn-0658-a-A-18 | TCC GCT TCC CTC TCC CAT | B2 | Desulfonema limicola, Desulfonema magnum | 20b |
| DSBOSA445 | S-S-Dsbo.sa-0445-a-A-18 | ACC ACA CAA CTT CTT CCC | C21 | Desulfobotulus sapovorans | This study | |
| DSMON95 | S-*-Dsmon-0095-a-A-18 | GTG CGC CAC TTT ACT CCA | C18 | Desulfomonile spp. | This study | |
| DSMON1421 | S-*-Dsmon-1421-a-A-18 | CGA CTT CTG GTG CAG TCA | C19 | See above | This study | |
| SYBAC986 | S-*-Sybac-0986-a-A-18 | CCG GGG ATG TCA AGC CCA | C17 | Desulfovirga adipica, Desulforhabdus amnigena, Syntrophobacter spp. | This study | |
| DSACI175 | S-G-Dsaci-0175-a-A-18 | CCG AAG GGA CGT ATC CGG | C16 | Desulfacinum spp. | This study | |
| DSACI207 | S-G-Dsaci-0207-a-A-18 | CGA ACA CCA GCT TCT TCG | C15 | See above | This study | |
| TDRNO448 | S-S-Tdr.no-0448-a-A-18 | AAC CCC ATG AAG GTT CTT | C13 | Thermodesulforhabdus norvegica | This study | |
| TDRNO1030 | S-S-Tdr.no-1030-a-A-18 | TCT CCC GGC TCC CCA ATA | C12 | See above | This study | |
| TDRNO1443 | S-S-Tdr.no-1443-a-A-18 | GAC ACA ATC GCG GTT GGC | C14 | See above | This study | |
| Probe 687 | DSV686 | S-*-Dsv-0686-a-A-18 | CTA CGG ATT TCA CTC CTA | E7 | “Desulfovibrionales” and other “Deltaproteobacteria” | 13b |
| DSV1292 | DSV1292 | S-*-Dsv-1292-a-A-18 | CAA TCC GGA CTG GGA CGC | E9 | Desulfovibrio litoralis, Desulfovibrio vulgaris, Desulfovibrio longreachensis, Desulfovibrio termitidis, Desulfovibrio desulfuricans, Desulfovibrio fairfieldensis, Desulfovibrio intestinalis, Desulfovibrio inopinatus, Desulfovibrio senezii, Desulfovibrio gracilis, Desulfovibrio halophilus, Bilophila wadsworthia | 41 |
| DSV698 | DSV698 | S-*-Dsv-0698-a-A-18 | TCC TCC AGA TAT CTA CGG | E8 | Desulfovibrio caledoniensis, Desulfovibrio dechloracetivorans, Desulfovibrio profundus, Desulfovibrio aespoeensis, Desulfovibrio halophilus, Desulfovibrio gracilis, Desulfovibrio longus, Desulfovibrio salexigens, Desulfovibrio zosterae, Desulfovibrio bastinii, Desulfovibrio fairfieldensis, Desulfovibrio intestinalis, Desulfovibrio piger, Desulfovibrio desulfuricans, Desulfovibrio termitidis, Desulfovibrio longreachensis, Desulfovibrio vietnamensis, Desulfovibrio alaskensis, Bilophila wadsworthia, Lawsonia intracellularis | 41b |
| DVDAPC872 | S-*-Dv.d.a.p.c-0872-a-A-18 | TCC CCA GGC GGG ATA TTT | E33 | Desulfovibrio caledoniensis, Desulfovibrio dechloracetivorans, Desulfovibrio profundus, Desulfovibrio aespoeensis | This study | |
| DVHO130 | S-*-Dv.h.o-0130-a-A-18 | CCG ATC TGT CGG GTA GAT | E36 | Desulfovibrio halophilus, Desulfovibrio oxyclinae | This study | |
| DVHO733 | S-*-Dv.h.o-0733-a-A-18 | GAA CTT GTC CAG CAG GCC | E37 | See above | This study | |
| DVHO831 | S-*-Dv.h.o-0831-a-A-18 | GAA CCC AAC GGC CCG ACA | E35 | See above | This study | |
| DVHO1424 | S-*-Dv.h.o-1424-a-A-18 | TGC CGA CGT CGG GTA AGA | E38 | See above | This study | |
| DVAA1111 | S-*-Dv.a.a-1111-a-A-18 | GCA ACT GGC AAC AAG GGT | E30 | Desulfovibrio africanus, Desulfovibrio aminophilus | This study | |
| DVGL199 | S-*-Dv.g.l-0199-a-A-18 | CTT GCA TGC AGA GGC CAC | E26 | Desulfovibrio gracilis, Desulfovibrio longus | This study | |
| DVGL445 | S-*-Dv.g.l-0445-a-A-18 | CCT CAA GGG TTT CTT CCC | E27 | See above | This study | |
| DVGL1151 | S-*-Dv.g.l-1151-a-A-18 | AAC CCC GGC AGT CTC ACT | E28 | See above | This study | |
| DVGL1421 | S-*-Dv.g.l-1421-a-A-18 | CGA TGT CGG GTA GAA CCA | E29 | See above | This study | |
| DSD131 | DSVAE131 | S-S-Dsv.ae-0131-a-A-18 | CCC GAT CGT CTG GGC AGG | E34 | Desulfovibrio aestuarii | 41 |
| DSV820 | S-*-Dsv-0820-a-A-18 | CCC GAC ATC TAG CAT CCA | E25, E31 | Desulfovibrio salexigens, Desulfovibrio zosterae, Desulfovibrio fairfieldensis, Desulfovibrio intestinalis, Desulfovibrio piger, Desulfovibrio desulfuricans | This study | |
| DVSZ849 | S-*-Dv.s.z-0849-a-A-18 | GTT AAC TTC GAC ACC GAA | E32 | Desulfovibrio salexigens, Desulfovibrio zosterae | This study | |
| DVIG448 | S-*-Dv.i.g-0448-a-A-18 | CGC ATC CTC GGG GTT CTT | E15 | Desulfovibrio gabonensis, Desulfovibrio indonesiensis | This study | |
| DVIG468 | S-*-Dv.i.g-0468-a-A-18 | CCG TCA GCC GAA GAC ACT | E16 | See above | This study | |
| DSV651 | S-*-Dsv-0651-a-A-18 | CCC TCT CCA GGA CTC AAG | E39 | Desulfovibrio fructosivorans, Desulfovibrio alcoholivorans, Desulfovibrio sulfodismutans, Desulfovibrio burkinensis, Desulfovibrio inopinatus | This study | |
| DVFABS153 | S-*-Dv.f.a.b.s-0153-a-A-18 | CGG AGC ATG CTG ATC TCC | E40 | Desulfovibrio fructosivorans, Desulfovibrio alcoholivorans, Desulfovibrio sulfodismutans, Desulfovibrio burkinensis | This study | |
| DVFABS653 | S-*-Dv.f.a.b.s-0653-a-A-18 | CAC CCT CTC CAG GAC TCA | E41 | See above | This study | |
| DVFABS1351 | S-*-Dv.f.a.b.s-1351-a-A-18 | GAG CAT GCT GAT CTC CGA | E42 | See above | This study | |
| DVLVT139 | S-*-Dv.1.v.t-0139-a-A-18 | GCC GTT ATT CCC AAC TCA | E17 | Desulfovibrio termitidis, Desulfovibrio longreachensis, Desulfovibrio vulgaris | This study | |
| DVLVT175 | S-*-Dv.l.v.t-0175-a-A-18 | AAA TCG GAG CGT ATT CGG | E18 | See above | This study | |
| DVLT131 | S-*-Dv.l.t-0131-a-A-18 | TCC CAA CTC ATG GGC AGA | E22 | Desulfovibrio termitidis, Desulfovibrio longreachensis | This study | |
| DVLT986 | S-*-Dv.l.t-0986-a-A-18 | TCC CGG ATG TCA AGC CTG | E23 | See above | This study | |
| DVLT1027 | S-*-Dv.l.t-1027-a-A-18 | TCG GGA TTC TCC GAA GAG | E21 | See above | This study | |
| DSM194 | S-G-Dsm-0194-a-A-18 | GAG GCA TCC TTT ACC GAC | E11 | Desulfomicrobium spp., Desulfobacterium macestii | This study | |
| DSV214 | DSM213 | S-G-Dsm-0213-a-A-18 | CAT CCT CGG ACG AAT GCA | E10 | See above | 41b |
| DSHRE830 | S-S-Dsh.re-0830-a-A-18 | GTC CTA CGA CCC CAA CAC | E12 | Desulfohalobium retbaense | This study | |
| DSHRE995 | S-S-Dsh.re-0995-a-A-18 | ATG GAG GCT CCC GGG ATG | E13 | See above | This study | |
| DSHRE1243 | S-S-Dsh.re-1243-a-A-18 | TGC TAC CCT CTG TGC CCA | E14 | See above | This study | |
| DFM228 | DFMI227a | S-*-DfmI-0227-a-A-18 | ATG GGA CGC GGA CCC ATC | A5 | Desulfotomaculum putei, Desulfotomaculum gibsoniae, Desulfotomaculum geothermicum, Desulfotomaculum thermosapovorans, Desulfotomaculum thermoacidovorans, Desulfotomaculum thermobenzoicum, Desulfotomaculum thermoacetoxidans, Desulfotomaculum australicum, Desulfotomaculum kuznetsovii, Desulfotomaculum thermocisternum, Desulfotomaculum luciae, Sporotomaculum hydroxybenzoicum | 9b |
| DFM228 | DFMI227b | S-*-Dfml-0227-b-A-18 | ATG GGA CGC GGA TCC ATC | A6 | Desulfotomaculum aeronauticum, Desulfotomaculum nigrificans, Desulfotomaculum reducens, Desulfotomaculum ruminis, Desulfotomaculum sapomandens, Desulfotomaculum halophilum | 9b |
| S-*-Dtm(cd)-0216-a-A-19 | DFMI210 | S-*-Dfml-0210-a-A-18 | CCC ATC CAT TAG CGG GTT | A7 | Some Desulfotomaculum spp. of clusters Ic and Idc | 25b |
| S-*-Dtm(bcd)-0230-a-A-18 | DFMI229 | S-*-Dfml-0229-a-A-18 | TAA TGG GAC GCG GAC CCA | A8 | Some Desulfotomaculum spp. of clusters Ib, Ic, and Idc | 25 |
| DFMIa641 | S-*-DfmIa-0641-a-A-18 | CAC TCA AGT CCA CCA GTA | A9 | Desulfotomaculum spp., (cluster Ia)c | This study | |
| DFMIb726 | S-*-Dfmlb-0726-a-A-18 | GCC AGG GAG CCG CTT TCG | A10 | Desulfotomaculum spp., Sporotomaculum hydroxybenzoicum (cluster Ib)c | This study | |
| DFMIc841 | S-*-DfmIc-0841-a-A-18 | GGC ACT GAA GGG TCC TAT | A11 | Desulfotomaculum spp. (cluster Ic)c | This study | |
| DFMIc1012 | S-*-DfmIc-1012-a-A-18 | CGT GAA ATC CGT GTT TCC | A12 | See above | This study | |
| DFMIc1119 | S-*-DfmIc-1119-a-A-18 | ACC CGT TAG CAA CTA ACC | A13 | See above | This study | |
| DFMIc1138 | S-*-DfmIc-1138-a-A-18 | GGC TAG AGT GCT CGG CTT | A14 | See above | This study | |
| DFMId436 | S-*-DfmId-0436-a-A-18 | CTT CGT CCC CAA CAA CAG | A15 | Desulfotomaculum spp. (cluster Id)c | This study | |
| DFMId625 | S-*-DfmId-0625-a-A-18 | TTT CAA AGG CAC CCC CGC | A16 | See above | This study | |
| DFMId996 | S-*-DfmId-0996-a-A-18 | CAC AGG CTG TCA GGG GAT | A17 | See above | This study | |
| DFMId1117 | S-*-DfmId-1117-a-A-18 | CCG CTG GCA ACT AAC CGT | A18 | See above | This study | |
| DFACE199 | S-S-Df.ace-0199-a-A-18 | GCA TTG TAA AGA GGC CAC | A20 | Desulfotomaculum spp. (cluster Ie)c | This study | |
| DFACE438 | S-S-Df.ace-0438-a-A-18 | CTG TTC GTC CAA TGT CAC | A19 | See above | This study | |
| DFACE995 | S-S-Df.ace-0995-a-A-18 | CAC AGC GGT CAC GGG ATG | A21 | See above | This study | |
| D-acet1027r | DFACE1028 | S-S-Df.ace-1028-a-A-18 | CTC CGT GTG CAA GTA AAC | A22 | See above | 68 |
| DFACE1436 | S-S-Df.ace-1436-a-A-18 | TGC GAG TTA AGT CAC CGG | A23 | See above | This study | |
| DFMIf126 | S-*-DfmIf-0126-a-A-18 | CTG ATA GGC AGG TTA TCC | A24 | Desulfotomaculum spp. (cluster If)d | This study | |
| DFMII1107 | S-*-DfmII-1107-a-A-18 | CTA AAT ACA GGG GTT GCG | A29 | Desulfosporosinus spp., Desulfotomaculum auripigmentum (cluster II)c | This study | |
| DFMII1281 | S-*-DfmII-1281-a-A-18 | GAG ACC GGC TTT CTC GGA | A28 | See above | This study | |
| TDSV601 | S-*-Tdsv-0601-a-A-18 | GCT GTG GAA TTC CAC CTT | D32 | Thermodesulfovibrio spp. | This study | |
| S-*-Tdsulfo-0848-a-A-18 | TDSV849 | S-*-Tdsv-0849-a-A-18 | TTT CCC TTC GGC ACA GAG | D33 | See above | 8 |
| TDSV1326 | S-*-Tdsv-1326-a-A-18 | CGA TTC CGG GTT CAC GGA | D31 | See above | This study | |
| TDSBM1282 | S-P-Tdsbm-1282-a-A-18 | TGA GGA GGG CTT TCT GGG | D27 | Thermodesulfobacterium spp., Geothermobacterium sp. | This study | |
| TDSBM353 | S-*-Tdsbm-0353-a-A-18 | CCA AGA TTC CCC CCT GCT | D28 | Thermodesulfobacterium spp. | This study | |
| TDSBM652 | S-*-Tdsbm-0652-a-A-18 | AGC CTC TCC GGC CCT CAA | D29 | See above | This study | |
| ARGLO37 | S-G-Arglo-0037-a-A-18 | CTT AGT CCC AGC CGG ATA | D37 | Archaeoglobus spp. | This study | |
| ARGLO276 | S-G-Arglo-0276-a-A-18 | GCC CGT ACG GAT CTT CGG | D38 | See above | This study | |
| ARGLO576 | S-G-Arglo-0576-a-A-18 | CCA GCC CGG CTA CGG ACG | D39 | See above | This study | |
| ARGLO972 | S-G-Arglo-0972-a-A-18 | CCC CGG TAA GCT TCC CGG | D40 | See above | This study | |
| DSBM168e | S-*-Dsbm-0168-a-A-18 | ACT TTA TCC GGC ATT AGC | Desulfobacterium niacini, Desulfobacterium vacuolatum | This study | ||
| DVHO588e | S-*-Dv.h.o-0588-a-A-18 | ACC CCT GAC TTA CTG CGC | Desulfovibrio halophilus, Desulfovibrio oxyclinae | This study | ||
| DVIG267e | S-*-Dv.i.g-0267-a-A-18 | CAT CGT AGC CAC GGT GGG | Desulfovibrio gabonensis, Desulfovibrio indonesiensis | This study | ||
| DVLT1425e | S-*-Dv.l.t-1425-a-A-18 | TCA CCG GTA TCG GGT AAA | Desulfovibrio termitidis, Desulfovibrio longreachensis | This study | ||
| DVGL228e | S-*-Dv.g.l-0228-a-A-18 | CAG CCA AGA GGC CTA TTC | Desulfovibrio gracilis, Desulfovibrio longus | This study | ||
| ARGLO390e | S-G-Arglo-0390-a-A-18 | GCA CTC CGG CTG ACC CCG | Archaeoglobus spp. | This study | ||
| DVLVT194f | S-*-Dv.l.v.t-0194-a-A-18 | AGG CCA CCT TTC CCC CGA | Desulfovibrio termitidis, Desulfovibrio longreachensis, Desulfovibrio vulgaris | This study | ||
| DVLVT222f | S-*-Dv.l.v.t-0222-a-A-18 | ACG CGG ACT CAT CCA TGA | See above | This study | ||
| DVCL1350f | S-*-Dv.c.l-1350-a-A-18 | GGC ATG CTG ATC CAG AAT | Desulfovibrio cuneatus, Desulfovibrio litoralis | This study | ||
| DFMIf489f | S-*-DfmIf-0489-a-A-18 | CCG GGG CTT ACT CCT ATG | Desulfotomaculum spp. (cluster If)d | This study |
Name of oligonucleotide probe based on the nomenclature of Alm et al. (1).
Length of oligonucleotide probe was adapted to the microarray format (18-mer).
Cluster designation(s) of gram-positive, spore-forming SRPs according to Stackebrandt et al. (65).
Desulfotomaculum halophilum and Desulfotomaculum alkaliphilum were assigned to new cluster If.
Probe was removed from the SRP microarray because no positive signal could be detected after hybridization with fluorescently labeled 16S rRNA gene amplificate of the perfect-match reference strain.
Probe was removed from the SRP microarray because it hybridized nonspecifically to many reference organisms that have mismatches in the 16S rRNA gene target site (see supplementary web material).
Reverse hybridization on microarrays.
Vacuum-dried Cy5-labeled PCR products (400 ng) and 0.5 pmol of the Cy5-labeled control oligonucleotide CONT-COMP (Table 3) were resuspended in 20 μl of hybridization buffer (5× SSC, 1% blocking reagent [Roche, Mannheim, Germany], 0.1% n-lauryl sarcosine, 0.02% SDS, 5% formamide [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]), denatured for 10 min at 95°C, and immediately placed on ice. Then the solution was pipetted onto an SRP-PhyloChip, covered with a coverslip, and inserted into a tight custom-made hybridization chamber (http://cmgm.stanford.edu/pbrown/mguide/HybChamber.pdf) containing 50 μl of hybridization buffer for subsequent equilibration. Hybridization was performed overnight at 42°C in a water bath. After hybridization, the slides were washed immediately under stringent conditions for 5 min at 55°C in 50 ml of washing buffer (containing 3 M tetramethylammonium chloride, 50 mM Tris-HCl, 2 mM EDTA, and 0.1% SDS). To record probe-target melting curves, the temperature of the washing step was varied from 42 to 80°C. After the stringent washing, the slides were washed twice with ice-cold double-distilled water, air dried, and stored in the dark at room temperature.
Scanning of microarrays.
Fluorescence images of the SRP PhyloChips were recorded by scanning the slides with a GMS 418 array scanner (Affymetrix). The fluorescence signals were quantified by using the ImaGene 4.0 software (BioDiscovery, Inc., Los Angeles, Calif.). A grid of individual circles defining the location of each spot on the array was superimposed on the image to designate each fluorescent spot to be quantified. The mean signal intensity was determined for each spot. In addition, the mean signal intensity of the local background area surrounding the spots was determined.
Selective enrichment of nucleic acids by a capture probe approach.
Five microliters of aldehyde group-coated glass beads (diameter, 1 μm; Xenopore, Hawthorne, N.J.) was incubated overnight with 5 μl of the appropriate capture probe (100 pmol μl−1; tailed with 15 dTTP molecules; aminated with 5′-terminal nucleotide) at room temperature. Subsequently, the beads were washed once with 400 μl of 0.2% SDS and pelleted by centrifugation (1 min at 14,000 rpm; Hettich Zentrifuge type 1000, Tuttlingen, Germany), and the supernatant was decanted. After this step, the beads were washed twice with 400 μl of double-distilled water, dried, and stored at room temperature prior to hybridization. A vacuum-dried bacterial 16S rRNA gene PCR product (obtained from DNA from the Solar Lake mat with the 616V-1492R primer pair) was resuspended in 200 μl of hybridization buffer (see above), denatured for 10 min at 95°C, and immediately cooled on ice. The hybridization solution and capture probe beads were mixed in a screw-cap tube and incubated overnight at 42°C on a shaker. Subsequently, the beads were washed twice with 1.5 ml of washing buffer (see above) at 55°C for 2.5 min. After the stringent washes, the beads were washed with 1.5 ml of ice-cold double-distilled water and then with ice-cold 70% ethanol. Beads with captured nucleic acids were vacuum dried and resuspended in 50 μl of EB buffer (part of the QIAquick PCR purification kit; Qiagen) for storage at −20°C. Reamplification of bacterial 16S rRNA gene fragments from the captured nucleic acids was performed by using 5 μl of the resuspended beads for PCR performed by using the 616V-1492R primer pair and the protocols described above.
Cloning, sequencing, and phylogeny inference.
Prior to cloning, the PCR amplification products were purified by low-melting-point agarose (1.5%) gel electrophoresis (NuSieve 3:1; FMC Bioproducts, Biozym Diagnostics GmbH, Oldendorf, Germany) and stained in a SYBR Green I solution (10 μl of 1,0000× SYBR Green I stain in 100 μl of TAE buffer [40 mM Tris, 10 mM sodium acetate, 1 mM EDTA; pH 8.0]; Biozym Diagnostics GmbH) for 45 min. Bands of the expected size were excised from the agarose gel with a glass capillary and melted with 50 μl of double-distilled water for 10 min at 80°C. Four microliters of each solution was ligated as recommended by the manufacturer into the cloning vector pCR2.1 supplied with a TOPO TA cloning kit (Invitrogen Corp., San Diego, Calif.). Nucleotide sequences were determined by the dideoxynucleotide method (57) as described by Purkhold et al. (51). The new 16S rRNA sequences were added to an alignment of about 16,000 small-subunit rRNA sequences by using the alignment tool of the ARB program package (O. Strunk and W. Ludwig, http://www.arb-home.de). Alignments were refined by visual inspection. Phylogenetic analyses were performed by using distance matrix, maximum-parsimony, and maximum-likelihood methods and the appropriate tools of the ARB program package and the fastDNAml program (34). The compositions of the data sets varied with respect to the reference sequences and the alignment positions included. Variability in the individual alignment positions was determined by using the appropriate tool of the ARB package and was used as a criterion to remove or include variable positions for phylogenetic analyses. Phylogenetic consensus trees were drawn by following the recommendations of Ludwig et al. (40). The new dsrAB sequences were translated into amino acids and added to an alignment of 62 DsrAB sequences of SRPs (18, 28). Phylogenetic analyses were performed by using the procedures described by Klein et al. (28).
Nucleotide sequence accession numbers.
The sequences determined in this study are available in the GenBank database under accession numbers AY083010 to AY083027 (16S rRNA gene clones) and AY083028 to AY083029 (dsrAB gene clones). The dsrAB gene sequence of Desulfomicrobium orale DSM 12838T has been deposited under accession number AY083030.
RESULTS
SRP phylogeny.
As the basis for development of the SRP-PhyloChip, a thorough reevaluation of the phylogeny of SRPs was performed. All 16S rRNA sequences of SRPs which are available in public databases (as of October 2001) were collected, aligned, and analyzed phylogenetically by using maximum-parsimony, maximum-likelihood, and neighbor-joining methods. Figures 1 and 2 illustrate the phylogeny of the delta-proteobacterial SRPs. Figure 3 shows the phylogeny of SRPs affiliated with the Firmicutes, Nitrospira, Thermodesulfobacteria, and Euryarchaeota phyla (phylum names according to the taxonomic outline in the second edition of Bergey's Manual of Systematic Bacteriology, 2nd ed. [21]).
FIG. 1.
Phylogenetic affiliations of SRPs belonging to the orders “Desulfobacterales” and “Syntrophobacterales” of the class “Deltaproteobacteria.” The 16S rRNA consensus tree was constructed from comparative sequence analysis data by using maximum-parsimony, maximum-likelihood, and neighbor-joining methods and applying filters excluding all alignment positions which are not conserved in at least 50% of all bacterial and deltaproteobacterial 16S rRNA sequences. A collection of organisms representing all major lineages of the Archaea and Bacteria was used as an outgroup. Multifurcations connect branches for which a relative order could not be determined unambiguously. Non-SRPs are underlined. Parsimony bootstrap values (1,000 resamplings) for branches are indicated by solid circles (>90%) or an open circle (75 to 90%). Branches without circles had bootstrap values of less than 75%. The bar indicates 10% estimated sequence divergence (distance inferred by neighbor joining by using a 50% bacterial conservation filter). The colored boxes show the specificities (perfect-match target organisms) of the SRP-PhyloChip probes (indicated by short names). The numbers of probes with identical specificities for the target organisms are indicated in parentheses. Probes SRB385Db, DSS658, DSR651, and DSB804 are not shown to enhance clarity.
FIG. 2.
Phylogenetic affiliations of SRPs belonging to the order “Desulfovibrionales” of the class “Deltaproteobacteria.” The 16S rRNA consensus tree was constructed as described in the legend to Fig. 1. Non-SRPs are underlined. The colored boxes show the specificities (perfect-match target organisms) of the SRP-PhyloChip probes (indicated by short names). The numbers of probes with identical specificities for the target organisms are indicated in parentheses. Probes SRB385, DSV1292, and DSV698 are not shown to enhance clarity.
FIG. 3.
(A) Phylogenetic affiliations of SRPs belonging to the family Peptococcaceae of the phylum Firmicutes (low-G+C-content gram-positive bacteria). (B) Phylogenetic affiliations of SRPs belonging to the genus Thermodesulfovibrio of the phylum Nitrospira. (C) Phylogenetic affiliations of SRPs belonging to the phylum Thermodesulfobacteria. (D) Phylogenetic affiliations of SRPs of the genus Archaeoglobus belonging to the phylum Euryarchaeota. In all panels non-SRPs are underlined. The 16S rRNA consensus trees were constructed as described in the legend to Fig. 1. The colored boxes show the specificities (perfect-match target organisms) of the SRP-PhyloChip probes (indicated by short names). The numbers of probes with identical specificities for the target organisms are indicated in parentheses. In panel A probes DFMI210 and DFMI229 are not shown to enhance clarity.
Probe design.
Initially, the specificities of previously described probes and primers for SRPs (2, 8, 9, 13, 20, 25, 41, 52, 55, 59, 68) were reevaluated with the current 16S rRNA data set containing more than 16,000 entries. Based on this analysis, 26 probes were considered to be suitable for inclusion on the SRP-PhyloChip (Table 3). These probes were, if necessary, adjusted to a length of 18 nucleotides (not including the T-spacer). Twenty-four of these probes exclusively target SRPs. Probes SRB385 (2) and SRB385Db (52) were included on the chip because they have been widely used in previous SRP research (3, 16, 36, 49, 58, 71), although both of these probes target a considerable number of phylogenetically diverse non-SRPs. In addition, we significantly extended the SRP probe set by designing 102 probes targeting monophyletic groups of SRPs (Fig. 1 to 3 and Table 3). These probes were designed to have a minimum G+C content of 50%, a length of 18 nucleotides (not including the T-spacer), and as many centrally located mismatches with the target sites on 16S rRNA genes of nontarget organisms as possible. Several of these probes target the same SRPs, complementing several unique regions of the 16S rRNA gene, while others exhibit hierarchical specificity. For example, the genus Desulfotalea is specifically detected by five probes and is also targeted by three probes with broader specificities (Fig. 1 and Table 3). Altogether, all 134 recognized SRPs for which 16S rRNA sequences have been published are covered by the probe set which we developed. The probes were spotted onto glass slides by using a pattern roughly reflecting the phylogeny of the SRPs (Table 3). In addition, universal, bacterial, and archaeal probes, as well as a nonsense probe (NONSENSE, with a sequence having at least four mismatches with every known 16S rRNA sequence), were immobilized on the microarray for hybridization control purposes (Table 3). Furthermore, another nonsense probe (CONT) (Table 3) was spotted at the beginning and end of each probe row of the microarray. During hybridization, a fluorescently labeled oligonucleotide fully complementary to this probe was added for control of hybridization efficiency and for straightforward localization of the probe spot rows in the microarray readout.
Evaluation of the SRP-PhyloChip with pure cultures.
In the first step, the SRP-PhyloChip was hybridized with fluorescently labeled 16S rRNA gene amplificates of Desulfovibrio halophilus, Desulfomicrobium aspheronum, and Desulfohalobium retbaense under increasingly stringent conditions. For each data point, a separate microarray with nine replicate spots of each probe was hybridized, washed, and analyzed. Figure 4 shows representative melting curves of probe-target duplexes for two of the SRP-specific probes and for bacterial probe EUB338 with the labeled 16S rRNA gene amplificates of the three reference organisms. Positive hybridization signals were recorded with probe EUB338 for the three SRPs when wash temperatures between 42 and 60°C were used. However, the EUB338 hybridization signal intensities varied significantly for the three reference organisms, indicating that there were variations in the efficiency of the fluorescence labeling of the PCR amplificates (Fig. 4C). Clear discrimination between perfectly matched and mismatched duplexes was achieved for most but not all of the probes investigated (Fig. 4A and B and 5). When a wash temperature of 42°C was used, the fluorescence intensity of probe-target hybrids with mismatches was almost always lower than the fluorescence intensity of completely matched hybrids (Fig. 5A). Unexpectedly, the difference in signal intensity between completely matched and mismatched duplexes was not significantly increased by gradually increasing the wash temperature to 80°C (Fig. 5). Based on the recorded melting curves, a wash temperature of 55°C was selected for all further experiments.
FIG. 4.
Melting curves for probe SRB385 (A), probe DSV698 (B), and probe EUB338 (C) after hybridization with fluorescently labeled PCR-amplified 16S rRNA gene fragments of Desulfovibrio halophilus, Desulfomicrobium aspheronum, and Desulfohalobium retbaense. For each probe the difference alignment with these reference SRPs is shown. The observed dissociation temperature (Td) is indicated for each probe. Each data point represents the mean signal intensity value for 10 probe spots (local background was subtracted for each measurement). The error bars indicate the standard deviations. For each wash temperature and reference organism a separate microarray hybridization was performed. a.u., arbitrary units.
FIG. 5.
Hybridization intensities of probes forming perfect-match (diamonds), one-mismatch (squares), and two-mismatch (circles) duplexes after hybridization with fluorescently labeled PCR-amplified 16S rRNA gene fragments of Desulfovibrio halophilus at different stringencies. (A) Mean signal intensities (for 10 spots, with local background subtracted) for each probe and wash temperature. (B) Normalized mean signal intensity values for each probe and wash temperature. Mean intensity values were normalized for each probe separately by assuming that the highest value observed at the different wash temperatures had a value of 1.00. In panel B, probes which showed no hybridization signals at low stringencies are not shown.
In the next step, an SRP-PhyloChip with duplicate spots for each probe was evaluated by using 41 SRP reference organisms. For each SRP-specific probe, this set of reference organisms contained an SRP which has a 16S rRNA gene with a perfectly matched target site. For each reference organism, fluorescently labeled, PCR-amplified 16S rRNA gene fragments were hybridized separately with the microarray by using 55°C as the wash temperature. The array readout was quantitatively analyzed by digital image analysis to determine a signal-to-noise ratio for each probe according to the following formula:
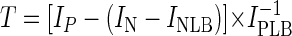 |
where T is the signal-to-noise ratio of the probe, IP is the mean pixel intensity of both specific probe spots, IN is the mean pixel intensity of both NONSENSE probe spots (note that IN − INLB must always have a lower value than IP), INLB is the mean pixel intensity of the local background area around both NONSENSE probe spots, and IPLB is the mean pixel intensity of the local background area around both specific probe spots.
Spots for which the signal-to-noise ratio was equal to or greater than 2.0 were considered positive in the pure-culture evaluation experiments and all subsequent analyses. Furthermore, the signal-to-noise ratio of each probe was divided by the signal-to-noise ratio of the bacterial EUB338 probe recorded on the same microarray in order to compare the duplex yields of the different SRP-specific probes. To do this, the following formula was used:
 |
where R is the normalized signal-to-noise ratio of the probe, IEUB is the mean pixel intensity of all EUB338 probe spots, and IEUBLB is the mean pixel intensity of the local background area around all EUB338 probe spots.
The normalized signal-to-noise ratios of the probes ranged from 0.3 for probe DFACE1028 with Desulfotomaculum acetoxidans to 16.9 for probe DSBAC355 with Desulfobacula toluolica, demonstrating that different probes exhibit very different signal intensities after hybridization with their perfectly matched target sequences.
The individual hybridization results for each of the 132 probes with each of the reference organisms can be downloaded from our website (http://www.microbial-ecology.de/srpphylochip/). Six of the probes evaluated (listed separately in Table 3) did not show a positive hybridization signal with any of the reference organisms, including the perfect-match target SRP, and thus were excluded from the microarray in subsequent experiments. In addition, four probes (listed separately in Table 3) were found to be not suitable for SRP diversity surveys due to their nonspecific binding to many nontarget organisms under stringent hybridization conditions (see supplementary web material). Under the conditions used, 75 (59%) of the probes found to be suitable for the SRP-PhyloChip hybridized exclusively to their target organisms. The other probes hybridized to rRNA gene amplificates with perfectly matched target sites, as well as to some rRNA genes with target sites having between one and six mismatches. In summary, of the 5,248 individual probe-target hybridization reactions performed (by hybridizing the 41 reference organisms with the final SRP-PhyloChip), 5,050 (96%) gave the expected results by either showing a detectable signal with the appropriate perfect-match target or showing no signal with target sequences containing mismatches.
Subsequently, the SRP-PhyloChip was hybridized in independent experiments with different amounts (1, 5, 10, 25, 50, 100, 200, and 400 ng) of PCR-amplified, labeled 16S rRNA gene fragments of Desulfovibrio halophilus. The same hybridization pattern was observed when 50 to 400 ng of labeled nucleic acids was used. When less than 50 ng of added nucleic acid was used, the signal-to-noise ratios of the hybridization signals were less than 2.0.
SRP-PhyloChip analyses of complex samples.
To evaluate the applicability of the SRP-PhyloChip for medical and environmental studies, two different samples, both containing diverse microbial communities, were analyzed. In the first experiment, tooth pocket samples from five patients suffering from adult periodontitis were investigated. While for three of the patients none of the SRP-specific probes showed a positive signal (data not shown), probe hybridization patterns indicative of the presence of members of the genus Desulfomicrobium were obtained for the other two patients (Fig. 6A). This result was confirmed independently by PCR analysis of the DNA obtained from the tooth pockets of the five patients by using primers specific for the 16S rRNA gene of members of the genus Desulfomicrobium (Table 2). Consistent with the microarray results, specific PCR amplificates were obtained for two of the five patients. Amplificates from both of these patients were cloned and sequenced. Comparative analysis of six clones demonstrated that the amplified sequences were almost identical to each other and to the corresponding 16S rRNA gene fragment of Desulfomicrobium orale (99.6 to 99.9% sequence similarity) (Fig. 6B). Furthermore, the compositions of the SRP communities in the tooth pockets of the patients were analyzed by using the genes encoding the dissimilatory (bi)sulfite reductase as a marker (28, 76). Approximately 1.9-kb dsrAB fragments could be PCR amplified from two of the five patients, and these fragments were cloned and sequenced. All 19 clones analyzed (6 clones from patient 1 and 13 clones from patient 4) had sequences almost identical to each other and to the dsrAB sequence of Desulfomicrobium orale (99.2 to 99.7% amino acid identity), which was also determined in this study.
FIG. 6.
(A) Use of the SRP-PhyloChip for surveys of SRP diversity in periodontal tooth pockets. On the microarray each probe was spotted in duplicate. For each microarray position, the probe sequence and specificity are shown in Table 3. Probe spots having a signal-to-noise ratio equal to or greater than 2.0 are indicated by boldface boxes and were considered to be positive. (B) Evaluation of the microarray results by amplification, cloning, and comparative sequence analysis of 16S rRNA gene fragments by using Desulfomicrobium-specific primers for PCR. 16S rRNA gene clones obtained from the tooth pockets are indicated by boldface type. The tree is based on a maximum-likelihood analysis performed with a 50% conservation filter for the Bacteria. Multifurcations connect branches for which a relative order could not be determined unambiguously after different treeing methods and filters were used. The bar indicates 10% estimated sequence divergence. The brackets indicate the perfect-match target organisms for the probes. The microarray position is indicated after each probe name.
In the second experiment, the SRP-PhyloChip was used to investigate the SRP community in the chemocline of a hypersaline cyanobacterial mat from Solar Lake. The SRP-PhyloChip hybridization patterns of fluorescently labeled 16S rRNA gene PCR amplificates obtained from the chemocline were more complex than those obtained from the tooth pockets (Fig. 7A). The probe hybridization patterns indicated that bacteria related to the genera Desulfonema and Desulfomonile were present. Furthermore, probe DSB220 showed signals above the threshold value which could have resulted from SRPs related to the genus Desulfofaba. However, the signal of probe DSB674, which also targets this genus, was below the threshold value. To confirm these results, 16S rRNA gene PCRs specific for most members of the “Desulfobacterales” (including the genera Desulfonema and Desulfofaba) and the “Syntrophobacterales” (primers DSBAC355F and 1492R [Table 2]), as well as for some Desulfonema species (primers DSN61F and DSN+1201R [Table 2]), were performed. Cloning and sequencing of the PCR amplificates confirmed that Desulfonema- and Desulfomonile-related organisms were present in the mat chemocline (Fig. 7B). In contrast to the microarray results, no sequences affiliated with the genus Desulfofaba were retrieved. In addition, we used glass beads coated with probe DSN658 to enrich Desulfonema-related 16S rRNA gene sequences from bacterial 16S rRNA gene amplificates from the mat chemocline. After enrichment, reamplification, and cloning, 1 of 12 cloned sequences did indeed possess the target site of probe DSN658 and was identical to Desulfonema-related sequences obtained by the specific PCR assay described above (Fig. 7B). The remaining 11 cloned sequences did not possess the probe DSN658 target site and were unrelated to recognized SRPs (data not shown).
FIG. 7.
(A) Use of the SRP-PhyloChip for surveys of SRP diversity in the chemocline of a cyanobacterial microbial mat. On the microarray each probe was spotted in duplicate. For each microarray position, the probe sequence and specificity are shown in Table 3. Probe spots having a signal-to-noise ratio equal to or greater than 2.0 are indicated by boldface boxes and were considered to be positive. The dotted boldface boxes indicate that only one of the duplicate spots had a signal-to-noise ratio equal to or greater than 2.0. (B) Evaluation of the microarray results by amplification, cloning, and comparative sequence analysis of 16S rRNA gene fragments by using primers specific for some Desulfonema species (SLM-DSN clones) and most members of the “Desulfobacterales” and “Syntrophobacterales” (SLM-DSBAC clones). Clone SLM-CP-116 was obtained from the mat chemocline by amplification, cloning, and sequencing after enrichment by using probe DSN658 as the capture probe. 16S rRNA gene clones obtained from the chemocline of the Solar Lake mat are indicated by boldface type. The tree is based on a maximum-likelihood analysis performed with a 50% conservation filter for the Bacteria. Multifurcations connect branches for which a relative order could not be determined unambiguously after different treeing methods and filters were used. The bar indicates 10% estimated sequence divergence. The brackets indicate the perfect-match target organisms of the probes. The microarray position is indicated after each probe name. The amplified and sequenced 16S rRNA gene fragment of Solar Lake mat clone SLM-DSBAC-74 (indicated by an asterisk) is outside the target site for probe DSMON95 and has one mismatch (located at position 16) within the target site for probe DSMON1421.
Software-assisted interpretation of microarray readouts.
Interpretation of experiments performed with the SRP-PhyloChip requires translation of more or less complex probe hybridization patterns into a list of SRPs which might be present in the sample analyzed. In principle, this task can be performed manually by using Table 3 and Fig. 1 to 3 as guides, but this procedure is tedious and sometimes not straightforward when it is performed with complex hybridization patterns. Consequently, we developed a software tool termed ChipChecker, which, after the microarray readout file (output from the ImaGene software) is imported, automatically creates a list of SRPs that potential occur in a sample. To do this, the software determines for each hybridization experiment which probes were positive (signal-to-noise ratio greater than the threshold; default signal-to-noise ratio, ≥2.0) and compares this result automatically with a list which specifies for each recognized SRP all fully complementary probes. Only those SRPs for which all perfect-match probes show a positive signal are listed. The ChipChecker software can easily be adapted for interpretation of other DNA microarrays and is available together with additional information for free download (http://www.bode.cs.tum.edu/∼meierh/download_chipchecker.html).
DISCUSSION
Microarray design and hybridization strategy.
In this study an encompassing DNA microarray for analysis of SRP diversity in complex samples was developed and evaluated. A total of 132 previously described and newly designed probes for the detection of 16S rRNA genes of SRPs were immobilized on the microarray. Consistent with design formats used in previous microarray applications for identification of other bacterial groups (23, 37), a hierarchical set of oligonucleotides complementary to the 16S rRNA genes of the target microorganisms at multiple levels of specificity was developed. However, the number of 16S rRNA-targeted oligonucleotide probes used in this study is significantly higher than the numbers of probes used in previous applications of chips for bacterial identification (23, 37, 62). This difference had important implications for the strategy which we selected for optimizing the hybridization conditions to ensure maximum specificity of the probes. Initially, temperature-dependent dissociation of several probe-target duplexes with perfect matches or mismatches was measured by using labeled 16S rRNA gene amplificates of three SRP reference organisms (Fig. 4 and 5). Comparable dissociation temperatures between 58 and 62°C, at which 50% of the starting duplexes remained intact, were observed for the different duplexes. This congruence probably reflects the fact that all probes of the SRP-PhyloChip are the same length (18 nucleotides) and the fact that the wash buffer contained 3 M tetramethylammonium chloride to equalize A · T and G · C base pair stability (42). Because our setup did not allow us to determine nonequilibrium online melting curves (37), it was not feasible (due to the high number of probes used) to record melting curves for each probe with perfectly matched and suitably mismatched target nucleic acids. Based on the recorded melting curves of selected probes, a wash temperature of 55°C was chosen for all further experiments as the best compromise between signal intensity and stringency. A further increase in stringency significantly reduced the signal intensity of some probes after hybridization with the perfectly matched target molecules (Fig. 5A) and thus decreased the sensitivity of the microarray.
Evaluation of the SRP-PhyloChip with more than 40 SRP reference strains was used to determine a threshold value above which a probe hybridization signal was considered positive. In addition, for each probe the signal intensity after hybridization with a perfectly matched target was compared to the signal intensity of the EUB338 probe on the same microarray (normalized signal-to-noise ratio). Consistent with data from quantitative fluorescence in situ hybridization experiments performed with different 16S rRNA-targeted oligonucleotide probes for Escherichia coli (19), (i) some of the probes used in the first version of the SRP-PhyloChip did not hybridize to their perfect-match targets and (ii) the signal intensities measured for the other probes on the SRP-PhyloChip varied significantly, by factors of up to 56. Dramatic differences in duplex yield arising from different regions of the target were also observed in other microarray applications (45, 64) and probably reflect either accessibility differences for the different probe target sites due to secondary structures of the target DNA or different steric hindrances of the different nucleic acid hybrids formed on the microarrays after hybridization.
The evaluation of the microarray with SRP pure cultures demonstrated (i) that false-negative hybridization never occurred (within the detection limit of the microarray method) but (ii) that some of the probes still hybridized to nontarget organisms under the hybridization and washing conditions used, leading to false-positive results (see supplementary web material). As expected, the nucleotide composition of the mismatch, the mismatch position (67, 73), and possibly other variables, such as the influence of an adjacent nucleotide stacking interaction (17), were the major factors determining the duplex yields of probes with mismatched target nucleic acids. Most of the mismatched duplexes with signal intensities above the threshold value (used to differentiate between positive and negative hybridization results) had a signal intensity (and normalized signal-to-noise ratio) lower than that of the corresponding perfect-match duplex (Fig. 5). However, this difference cannot be exploited for interpretation of microarray hybridization results for environmental samples because a low hybridization signal of a probe can be caused not only by mismatched duplex formation but also by low abundance of the perfect-match target nucleic acid.
Misinterpretation of microarray hybridization patterns caused by the nonperfect specificity of some of the probes could be avoided at least partially by using the multiple-probe concept. While hybridization patterns consistent with the hierarchical or parallel specificity of the probes increase the reliability of detection, inconsistent probe hybridization patterns must be interpreted with caution. In complex samples, inconsistent hybridization patterns can be caused either by nonspecific binding of one or several probes or by previously unrecognized prokaryotes with unusual combinations of perfect-match probe target sites in their 16S rRNA gene sequences.
Microarray-based SRP diversity surveys of complex samples.
In this study, periodontal tooth pocket material and a cyanobacterial microbial mat were used to demonstrate the suitability of using the microarray developed for SRP diversity analysis of medical and environmental samples. For the tooth pocket material of two patients suffering from adult periodontitis the SRP-PhyloChip hybridization pattern indicated the presence of members of the genus Desulfomicrobium. Colonization of the tooth pockets analyzed by these SRPs, which is consistent with a previous report of isolation of Desulfomicrobium orale from periodontal tooth pockets (33), was independently confirmed by retrieval of 16S rRNA and dsrAB gene sequences of Desulfomicrobium orale, demonstrating the reliability of the microarray results.
The microarray hybridization patterns obtained by reverse hybridization of 16S rRNA gene fragments amplified from the chemocline of a Solar Lake microbial mat suggested that several phylogenetically different SRPs, including bacteria related to the genera Desulfonema, Desulfomonile, and Desulfofaba, were present. By using specific PCR assays, 16S rRNA gene sequences related to sequences of members of the genera Desulfonema and Desulfomonile were obtained from the mat material analyzed, while the presence of Desulfofaba-like organisms could not be confirmed. The failure to detect Desulfofaba-like bacteria with the PCR assay might mean that a relatively limited number of 16S rRNA gene clones was sequenced or that the microarray hybridization pattern indicative of Desulfofaba was caused by the presence of bacteria that have not been recognized yet. The detection of Desulfonema-like bacteria in the chemocline of the Solar Lake mat is consistent with findings of previous studies (46, 47, 70) and further supports the importance of these SRPs in hypersaline mat ecosystems.
In conclusion, we developed an encompassing 16S rRNA gene-targeting oligonucleotide microarray suitable for SRP diversity analyses of complex environmental and clinical samples. The microarray was used to screen samples in order to rapidly obtain indications of the presence of distinct lineages of SRPs. Subsequently, this information was used to select appropriate PCR-based techniques for confirmation of the microarray results and for retrieval of sequence information for phylogenetic analysis. In contrast to previously available tools for cultivation-independent SRP identification (13, 18, 41, 56, 75, 76), the SRP-PhyloChip allowed us to obtain a phylogenetically informative, high-resolution fingerprint of the SRP diversity in a given sample within 48 h (including all experimental work from DNA extraction to hybridization pattern interpretation). However, keeping in mind that (i) most environmental microbial communities contain a high percentage of bacteria not yet sequenced on the 16S rRNA level and (ii) not all probes on the microarray are absolutely specific under the conditions used, the SRP-PhyloChip experiments should always be supplemented with microarray-independent techniques to confirm the phylogenetic affiliations of the SRPs detected. Furthermore, it should be noted that the microarray approach described here did not allow us to obtain quantitative data on the compositions of SRP communities because of the recognized biases introduced by using PCR for 16S rRNA gene amplification (74). In addition, the duplex yield of a probe on the microarray is dependent not only on the actual abundance of its perfect-match target nucleic acid in the PCR amplificate mixture but also on a variety of other factors, including the labeling efficiency of the specific target nucleic acid, the secondary structure of the target region, and the inherent variations associated with microarray fabrication. Despite these limitations, the microarray which we developed has great potential for rapid screening of SRP diversity in complex samples. The SRP diversity microarray fingerprint technique should allow workers to identify the probes which have relevance for further characterization of a sample by PCR or quantitative hybridization experiments. This option should be particularly valuable if large numbers of samples are to be analyzed to study temporal or spatial variations in SRP diversity.
Acknowledgments
Yehuda Cohen is acknowledged for kindly providing cyanobacterial mat material from Solar Lake. The excellent technical assistance of Claudia Schulz, Helga Gaenge, Susanne Thiemann, and Sibylle Schadhauser is acknowledged. We also thank Bernhard Loy for providing the tooth pocket samples and Josef Reischenbeck for fabrication of hybridization chambers.
This research was supported by grants from bmb+f (01 LC 0021 subproject 2, in the framework of the BIOLOG program) (to M.W.), DFG (trilateral cyanobacterial mat project RU458/18-4) (to M.W.), and Bayerischen Forschungsstiftung (Development of Oligonucleotide DNA Chips, in cooperation with MWG Biotech; project 368/99) (to M.W. and K.-H.S.).
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., J. Stromley, R. Devereux, R. Key, and D. A. Stahl. 1992. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl. Environ. Microbiol. 58:614-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 5.Cho, J.-C., and J. M. Tiedje. 2002. Quantitative detection of microbial genes by using DNA microarrays. Appl. Environ. Microbiol. 68:1425-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 7.Daims, H., J. L. Nielsen, P. H. Nielsen, K. H. Schleifer, and M. Wagner. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daims, H., P. H. Nielsen, J. L. Nielsen, S. Juretschko, and M. Wagner. 2000. Novel Nitrospira-like bacteria as dominant nitrite-oxidizers in biofilms from wastewater treatment plants: diversity and in situ physiology. Water Sci. Technol. 41:85-90. [Google Scholar]
- 9.Daly, K., R. J. Sharp, and A. J. McCarthy. 2000. Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiology 146:1693-1705. [DOI] [PubMed] [Google Scholar]
- 10.DeLong, E. F. 2000. Resolving a methane mystery. Nature 407:577-579. [DOI] [PubMed] [Google Scholar]
- 11.Devereux, R., M. Delaney, F. Widdel, and D. A. Stahl. 1989. Natural relationships among sulfate-reducing eubacteria. J. Bacteriol. 171:6689-6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereux, R., S. H. He, C. L. Doyle, S. Orkland, D. A. Stahl, J. LeGall, and W. B. Whitman. 1990. Diversity and origin of Desulfovibrio species: phylogenetic definition of a family. J. Bacteriol. 172:3609-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devereux, R., M. D. Kane, J. Winfrey, and D. A. Stahl. 1992. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst. Appl. Microbiol. 15:601-609. [Google Scholar]
- 14.Devereux, R., and G. W. Mundfrom. 1994. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in a sandy marine sediment. Appl. Environ. Microbiol. 60:3437-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubilier, N., C. Mulders, T. Ferdelman, D. de Beer, A. Pernthaler, M. Klein, M. Wagner, C. Erseus, F. Thiermann, J. Krieger, O. Giere, and R. Amann. 2001. Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411:298-302. [DOI] [PubMed] [Google Scholar]
- 16.Edgcomb, V. P., J. H. McDonald, R. Devereux, and D. W. Smith. 1999. Estimation of bacterial cell numbers in humic acid-rich salt marsh sediments with probes directed to 16S ribosomal DNA. Appl. Environ. Microbiol. 65:1516-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fotin, A. V., A. L. Drobyshev, D. Y. Proudnikov, A. N. Perov, and A. D. Mirzabekov. 1998. Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res. 26:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedrich, M. W. 2002. Phylogenetic analysis reveals multiple lateral transfers of adenosine-5′-phosphosulfate reductase genes among sulfate-reducing microorganisms. J. Bacteriol. 184:278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukui, M., A. Teske, B. Aßmus, G. Muyzer, and F. Widdel. 1999. Physiology, phylogenetic relationships, and ecology of filamentous sulfate-reducing bacteria (genus Desulfonema). Arch. Microbiol. 172:193-203. [DOI] [PubMed] [Google Scholar]
- 21.Garrity, G. M., and J. G. Holt. 2001. The road map to the manual, p. 119-166. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y.
- 22.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hines, M. E., R. S. Evans, B. R. Sharak Genthner, S. G. Willis, S. Friedman, J. N. Rooney-Varga, and R. Devereux. 1999. Molecular phylogenetic and biogeochemical studies of sulfate-reducing bacteria in the rhizosphere of Spartina alterniflora. Appl. Environ. Microbiol. 65:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hristova, K. R., M. Mau, D. Zheng, R. I. Aminov, R. I. Mackie, H. R. Gaskins, and L. Raskin. 2000. Desulfotomaculum genus- and subgenus-specific 16S rRNA hybridization probes for environmental studies. Environ. Microbiol. 2:143-159. [DOI] [PubMed] [Google Scholar]
- 26.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Roser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kane, M. D., L. K. Poulsen, and D. A. Stahl. 1993. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoblauch, C., B. B. Jorgensen, and J. Harder. 1999. Community size and metabolic rates of psychrophilic sulfate-reducing bacteria in arctic marine sediments. Appl. Environ. Microbiol. 65:4230-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knoblauch, C., K. Sahm, and B. B. Jorgensen. 1999. Psychrophilic sulfate-reducing bacteria isolated from permanently cold arctic marine sediments: description of Desulfofrigus oceanense gen. nov., sp. nov., Desulfofrigus fragile sp. nov., Desulfofaba gelida gen. nov., sp. nov., Desulfotalea psychrophila gen. nov., sp. nov. and Desulfotalea arctica sp. nov. Int. J. Syst. Bacteriol. 49:1631-1643. [DOI] [PubMed] [Google Scholar]
- 31.Kovárová, M., and P. Dráber. 2000. New specificity and yield enhancer of polymerase chain reactions. Nucleic Acids Res. 28:E70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langendijk, P. S., J. T. J. Hanssen, and J. S. van der Hoeven. 2000. Sulfate-reducing bacteria in association with human periodontitis. J. Clin. Periodontol. 27:943-950. [DOI] [PubMed] [Google Scholar]
- 33.Langendijk, P. S., E. M. Kulik, H. Sandmeier, J. Meyer, and J. S. van der Hoeven. 2001. Isolation of Desulfomicrobium orale sp. nov. and Desulfovibrio strain NY682, oral sulfate-reducing bacteria involved in human periodontal disease. Int. J. Syst. Evol. Microbiol. 51:1035-1044. [DOI] [PubMed] [Google Scholar]
- 34.Larsen, N., G. J. Olsen, B. L. Maidak, M. J. McCaughey, R. Overbeek, T. J. Macke, T. L. Marsh, and C. R. Woese. 1993. The Ribosomal Database Project. Nucleic Acids Res. 21:3021-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Scola, B., and D. Raoult. 1999. Third human isolate of a Desulfovibrio sp. identical to the provisionally named Desulfovibrio fairfieldensis. J. Clin. Microbiol. 37:3076-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, J.-H., K. J. Purdy, S. Takii, and H. Hayashi. 1999. Seasonal changes in ribosomal RNA of sulfate-reducing bacteria and sulfate reducing activity in a freshwater lake sediment. FEMS Microbiol. Ecol. 28:31-39. [Google Scholar]
- 37.Liu, W. T., A. D. Mirzabekov, and D. A. Stahl. 2001. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol. 3:619-629. [DOI] [PubMed] [Google Scholar]
- 38.Llobet-Brossa, E., R. Rossello-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loubinoux, J., F. Mory, I. A. Pereira, and A. E. Le Faou. 2000. Bacteremia caused by a strain of Desulfovibrio related to the provisionally named Desulfovibrio fairfieldensis. J. Clin. Microbiol. 38:931-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 41.Manz, W., M. Eisenbrecher, T. R. Neu, and U. Szewzyk. 1998. Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol. 25:43-61. [Google Scholar]
- 42.Maskos, U., and E. M. Southern. 1992. Parallel analysis of oligodeoxyribonucleotide (oligonucleotide) interactions. I. Analysis of factors influencing oligonucleotide duplex formation. Nucleic Acids Res. 20:1675-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDougall, R., J. Robson, D. Paterson, and W. Tee. 1997. Bacteremia caused by a recently described novel Desulfovibrio species. J. Clin. Microbiol. 35:1805-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meier, H., R. Amann, W. Ludwig, and K.-H. Schleifer. 1999. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst. Appl. Microbiol. 22:186-196. [DOI] [PubMed] [Google Scholar]
- 45.Milner, N., K. U. Mir, and E. M. Southern. 1997. Selecting effective antisense reagents on combinatorial oligonucleotide arrays. Nat. Biotechnol. 15:537-541. [DOI] [PubMed] [Google Scholar]
- 46.Minz, D., S. Fishbain, S. J. Green, G. Muyzer, Y. Cohen, B. E. Rittmann, and D. A. Stahl. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl. Environ. Microbiol. 65:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orphan, V. J., K. U. Hinrichs, W. Ussler 3rd, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. Delong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oude Elferink, S. J. W. H., W. J. C. Vorstman, A. Sopjes, and A. J. M. Stams. 1998. Characterization of the sulfate-reducing and syntrophic population in granular sludge from a full-scale anaerobic reactor treating papermill wastewater. FEMS Microbiol. Ecol. 27:185-194. [Google Scholar]
- 50.Purdy, K. J., D. B. Nedwell, T. M. Embley, and S. Takii. 1997. Use of 16S rRNA-targeted oligonucleotide probes to investigate the occurrence and selection of sulfate-reducing bacteria in response to nutrient addition to sediment slurry microcosms from a Japanese estuary. FEMS Microbiol. Ecol. 24:221-234. [Google Scholar]
- 51.Purkhold, U., A. Pommering-Röser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rabus, R., M. Fukui, H. Wilkes, and F. Widdle. 1996. Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes from crude oil. Appl. Environ. Microbiol. 62:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabus, R., T. Hansen, and F. Widdel. 2000. Dissimilatory sulfate- and sulfur-reducing prokaryotes. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer-Verlag, New York, N.Y.
- 54.Ravenschlag, K., K. Sahm, and R. Amann. 2001. Quantitative molecular analysis of the microbial community in marine arctic sediments (Svalbard). Appl. Environ. Microbiol. 67:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ravenschlag, K., K. Sahm, C. Knoblauch, B. B. Jorgensen, and R. Amann. 2000. Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine arctic sediments. Appl. Environ. Microbiol. 66:3592-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Risatti, J. B., W. C. Capman, and D. A. Stahl. 1994. Community structure of a microbial mat: the phylogenetic dimension. Proc. Natl. Acad. Sci. USA 91:10173-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santegoeds, C. M., T. G. Ferdelman, G. Muyzer, and D. de Beer. 1998. Structural and functional dynamics of sulfate-reducing populations in bacterial biofilms. Appl. Environ. Microbiol. 64:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheid, D., and S. Stubner. 2001. Structure and diversity of Gram-negative sulfate-reducing bacteria on rice roots. FEMS Microbiol. Ecol. 36:175-183. [DOI] [PubMed] [Google Scholar]
- 60.Schoenborn, L., H. Abdollahi, W. Tee, M. Dyall-Smith, and P. H. Janssen. 2001. A member of the delta subgroup of Proteobacteria from a pyogenic liver abscess is a typical sulfate reducer of the genus Desulfovibrio. J. Clin. Microbiol. 39:787-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shchepinov, M. S., S. C. Case-Green, and E. M. Southern. 1997. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 25:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Southern, E., K. Mir, and M. Shchepinov. 1999. Molecular interactions on microarrays. Nat. Genet. 21:5-9. [DOI] [PubMed] [Google Scholar]
- 64.Southern, E. M., S. C. Case-Green, J. K. Elder, M. Johnson, K. U. Mir, L. Wang, and J. C. Williams. 1994. Arrays of complementary oligonucleotides for analysing the hybridisation behaviour of nucleic acids. Nucleic Acids Res. 22:1368-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stackebrandt, E., C. Sproer, F. A. Rainey, J. Burghardt, O. Pauker, and H. Hippe. 1997. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Int. J. Syst. Bacteriol. 47:1134-1139. [DOI] [PubMed] [Google Scholar]
- 66.Stackebrandt, E., D. A. Stahl, and R. Devereux. 1995. Taxonomic relationships, p. 49-87. In L. L. Barton (ed.), Sulfate-reducing bacteria. Plenum Press, New York, N.Y.
- 67.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, England.
- 68.Stubner, S., and K. Meuser. 2000. Detection of Desulfotomaculum in an Italian rice paddy soil by 16S ribosomal nucleic acid analyses. FEMS Microbiol. Ecol. 34:73-80. [DOI] [PubMed] [Google Scholar]
- 69.Tee, W., M. Dyall-Smith, W. Woods, and D. Eisen. 1996. Probable new species of Desulfovibrio isolated from a pyogenic liver abscess. J. Clin. Microbiol. 34:1760-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teske, A., N. B. Ramsing, K. Habicht, M. Fukui, J. Kuver, B. B. Jorgensen, and Y. Cohen. 1998. Sulfate-reducing bacteria and their activities in cyanobacterial mats of Solar Lake (Sinai, Egypt). Appl. Environ. Microbiol. 64:2943-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teske, A., C. Wawer, G. Muyzer, and N. B. Ramsing. 1996. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomsen, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Urakawa, H., P. A. Noble, S. El Fantroussi, J. J. Kelly, and D. A. Stahl. 2002. Single-base-pair discrimination of terminal mismatches by using oligonucleotide microarrays and neural network analyses. Appl. Environ. Microbiol. 68:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 75.Voordouw, G., J. K. Voordouw, R. R. Karkhoff-Schweizer, P. M. Fedorak, and D. W. S. Westlake. 1991. Reverse sample genome probing, a new technique for identification of bacteria in environmental samples by DNA hybridization, and its application to the identification of sulfate-reducing bacteria in oil field samples. Appl. Environ. Microbiol. 57:3070-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng, D., E. W. Alm, D. A. Stahl, and L. Raskin. 1996. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl. Environ. Microbiol. 62:4504-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]









