Abstract
Certain 2,4-diacetylphloroglucinol-producing strains of Pseudomonas fluorescens colonize roots and suppress soilborne diseases more effectively than others from which they are otherwise phenotypically almost indistinguishable. We recovered DNA fragments present in the superior colonizer P. fluorescens Q8r1-96 but not in the less rhizosphere-competent strain Q2-87. Of the open reading frames in 32 independent Q8r1-96-specific clones, 1 was similar to colicin M from Escherichia coli, 3 resembled known regulatory proteins, and 28 had no significant match with sequences of known function. Seven clones hybridized preferentially to DNA from strains with superior rhizosphere competence, and sequences in two others were highly expressed in vitro and in the rhizosphere.
Fluorescent Pseudomonas spp. that produce antifungal metabolites have been studied extensively as potential biocontrol agents of soilborne plant pathogens causing yield-limiting diseases of food, fiber, and ornamental crops (5, 29, 35, 36). Strains that synthesize the antifungal metabolite 2,4-diacetylphloroglucinol (2,4-DAPG) are of particular interest because they suppress a wide variety of diseases, including take-all of wheat caused by Gaeumannomyces graminis var. tritici (10, 21), black root rot of tobacco caused by Thielaviopsis basicola (12, 34), damping-off of sugar beet caused by Pythium ultimum (8), and Fusarium crown and root rot of tomato caused by Fusarium oxysporum f. sp. lysopersici (7). 2,4-DAPG-producing fluorescent Pseudomonas spp. also play a major role in the development of the natural biological control of take-all disease of wheat and barley known as take-all decline (TAD) (22), which develops in soils worldwide during extended monoculture of wheat or barley (4) following a severe outbreak of the disease. Root colonization is the first critical step in the biological control of take-all and other diseases because introduced or indigenous agents must establish and maintain threshold population densities if they are to be effective.
Two distinct groups of 2,4-DAPG producers have been identified, one of which also produces pyoluteorin (11, 18, 19, 25). Fine-structure genetic analyses of over 300 isolates of worldwide origin from European and U.S. collections have further distinguished 17 distinct genotypes of 2,4-DAPG producers (11, 14, 18, 19). Representatives of genotype D, as distinguished by repetitive sequence-based PCR with the BOXA1R primer (BOX-PCR) (19), predominated among isolates from roots of wheat and pea grown in soils from multiple locations throughout the United States that had experienced long-term monoculture of wheat or pea (14, 19, 23). For example, D-genotype isolates comprised over 50% of the 2,4-DAPG producers isolated from wheat grown for multiple, successive cycles in TAD soils from Quincy, Moses Lake, and Lind, Wash. (23). In both greenhouse and field studies, P. fluorescens Q8r1-96, a D-genotype strain from the Quincy TAD soil, duplicated the suppressiveness of TAD when added to conducive soils at very low doses compared to strains of other genotypes (23). Q8r1-96 and all other D-genotype isolates tested to date display an unusually high level of rhizosphere competence when applied at very low doses (14, 23; B. B. McSpadden-Gardener and D. M. Weller, unpublished data). During a cycling experiment that lasted 8 months, introduced strain Q8r1-96 maintained population densities on wheat roots that were 10- to 1,000-fold greater than those maintained by strains Q2-87 and 1 M1-96, which are examples of B- and L-genotype strains, respectively, that are also found in monoculture wheat and pea soils. The substantial difference in rhizosphere competence among these strains was unexpected because of the absence of significant phenotypic differences among them. All three belong to biotype II and are nearly identical by fatty-methyl-ester analysis, substrate utilization profiles, and classical bacteriological tests (19, 23).
Genomic subtraction is among the best methods currently available for exploring structural differences between the genomes of closely related bacteria (13, 33), including fluorescent pseudomonads (28, 37). Here we describe the application of genomic suppressive subtractive hybridization (SSH) (1) as one approach to identifying genes that contribute to the exceptional rhizosphere competence of D-genotype strains. DNA sequences present in the superior root colonizer P. fluorescens Q8r1-96 but not in the less rhizosphere-competent strain Q2-87 were cloned, their sequences determined and analyzed, and their expression in the rhizosphere and distribution among 29 other 2,4-DAPG-producing strains representative of 17 different genotypes were assessed. Several subtracted fragments distributed primarily among isolates of the D genotype or expressed in the rhizosphere were identified as candidates for further analysis.
SSH and DNA sequence analysis.
DNA fragments present in P. fluorescens Q8r1-96 (the tester strain) but not in P. fluorescens Q2-87 (the driver strain) were isolated by using a PCR-Select bacterial genome subtraction kit (Clontech Laboratories, Inc., Palo Alto, Calif.). Cultures of each Pseudomonas strain were grown at 28°C in Luria-Bertani (LB) broth as described previously (17). Total DNA was isolated and purified by using a cetyltrimethylammonium bromide procedure (2), digested with RsaI, and hybridized and annealed at 63°C. The pool of subtracted fragments was amplified with the Advantage 2 PCR enzyme mix (Clontech Laboratories, Inc.), cloned into pGEM-T Easy (Promega Corp., Madison, Wis.), and transformed into Escherichia coli JM109. Standard procedures (2) were used for all DNA manipulations unless noted otherwise.
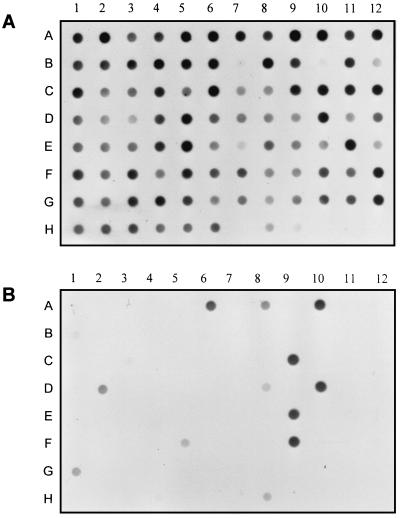
Randomly selected clones containing subtracted fragments were amplified by PCR with nested primers 1 and 2R (Clontech Laboratories, Inc.) and screened by hybridization to identify those containing tester-specific sequences. The PCR products were purified, arrayed (10 to 15 ng per spot) on duplicate BrightStar-Plus nylon membranes (Ambion, Inc., Austin, Tex.), and hybridized (2) with sheared, biotin-labeled (NEN Life Science Products, Inc., Boston, Mass.) genomic DNA from either strain Q8r1-96 or strain Q2-87. Hybrids were detected with the BrightStar detection kit (Ambion). Over 80% of the 180 screened clones contained tester-specific DNA fragments (Fig. 1) ranging in size from 0.3 to 1.5 kb that were sequenced in full by using an ABI PRISM dye terminator cycle sequencing kit (Perkin-Elmer, Norwalk, Conn.). Analysis of the data with the Genetic Computer Group and OMIGA 2.0 software packages (Accelrys, Princeton, N.J.) identified 32 DNA sequences unique to P. fluorescens Q8r1-96 (Table 1). Only 13 of these were present at least twice among the 180 clones screened, indicating that the subtracted library represents a subset of the unique sequences present in Q8r1-96.
FIG. 1.
Distribution of 90 randomly picked clones from the subtracted library in the genomes of the tester and driver strains. Plasmids were transferred to nylon filters with a dot blot manifold and hybridized with biotin-labeled genomic DNA from P. fluorescens Q8r1-96 (A) or Q2-87 (B). pGEM-T Easy with the phlD gene from strain Q2-87 (H8) and empty pGEM-T Easy cloning vector (H9) were used as positive and negative controls, respectively. Subsequent analysis revealed that clones B7 and B10 contained empty vector.
TABLE 1.
Sequence analysis of Q8r1-96-specific loci
| SSH clone | Insert size (bp) | Similar protein (accession no.), organism, and BLASTX E valuea | Predicted function or property |
|---|---|---|---|
| 3 | 1,479 | Sll1503 (S74977), Synechocystis sp. PCC6803, 7e−10 | Hypothetical protein |
| 5 | 933 | None detected | |
| 6 | 549 | Colicin M (P05820), E. coli plasmid ColBM-Cl139, 1e−16 | Colicin M activity peptide |
| 8 | 353 | None detected | |
| 12 | 533 | None detected | |
| 13 | 1,296 | None detected | |
| 18 | 725 | Clostridium acetobutylicum unfinished genome, 7e−11 | Hypothetical protein |
| 25 | 1,447 | Pasteurella multocida unfinished genome, 5e−18 | Hypothetical protein |
| 26 | 596 | None detected | |
| 28 | 213 | PA0988 (AAG04377), P. aeruginosa, 3e−22 | Hypothetical protein |
| 32 | 787 | None detected | |
| 36 | 660 | None detected | |
| 41 | 692 | None detected | |
| 45 | 452 | P. putida KT2440 unfinished genome, 7e−14 | Hypothetical protein |
| 53 | 312 | Rv1507c (P71786), Mycobacterium tuberculosis, 4e−21 | Hypothetical protein |
| 58 | 387 | P. putida KT2440 unfinished genome, 6e−45 | Hypothetical protein |
| 61 | 418 | P. putida KT2440 unfinished genome, 2e−35; PA3965 (AAG07352), P. aeruginosa, 1e−08; BkdR (P42179), P. putida, 3e−08; putative Lrp PA5308 (AE004943), P. aeruginosa, 3e−08 | Putative transcriptional regulator |
| 62 | 791 | None detected | |
| 64 | 348 | Rv1505c (D70713), M. tuberculosis, 8e−29 | Hypothetical protein |
| 66 | 624 | None detected | |
| 74 | 401 | None detected | |
| 80 | 828 | None detected | |
| 81 | 318 | Yersinia pestis unfinished genome, 8e−08 | Hypothetical protein |
| 83 | 462 | None detected | |
| 85 | 381 | PA1396 (F83470), P. aeruginosa, 2e−08 | Probable two-component sensor |
| 93 | 284 | None detected | |
| 101 | 700 | PA2461 (B83339), P. aeruginosa, 2e−30 | Hypothetical protein |
| 103 | 517 | Yersinia pestis unfinished genome, 8e−17 | Hypothetical protein |
| 127 | 1,078 | PA0448 (AE004482), P. aeruginosa, 9e−45; Klebsiella pneumoniae unfinished genome, 5e−30 | Putative transcriptional regulator |
| 132 | 650 | None detected | |
| 133 | 672 | None detected | |
| 164 | 506 | YeeA (AAB66474), Bacillus subtilis, 3e−25 | Conserved hypothetical protein |
Only expectation values of 1e−05 and below were considered as significant matches during BLAST database searches.
Sequence data from the 32 Q8r1-96-specific fragments were analyzed for similarity to known nucleotide and protein sequences, including those in unfinished genomes by using OMIGA's BLAST tool and the PROSITE (EMBL, Heidelberg, Germany) and ISREC ProfileScan (Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland) probable domain homology search algorithms. Twelve of the 32 fragments (clones 3, 18, 25, 28, 45, 53, 58, 64, 81, 101, 103, and 164) resembled conserved bacterial hypothetical open reading frames (ORFs) of unknown function (Table 1), while 16 others (clones 5, 8, 12, 13, 26, 32, 36, 41, 62, 66, 74, 80, 83, 93, 132, and 133) had no significant match with known sequences. Clones 61, 85, and 127 resembled putative regulatory genes found in other Pseudomonas genomes, and clone 6 had significant protein sequence similarity with E. coli colicin M. It is not surprising that functions could not be predicted for most of the fragments recovered because the subtractive hybridization procedure would have selected against highly conserved sequences. In addition, nearly half of the genes from the recently sequenced Pseudomonas aeruginosa PAO1 genome either resembled genes of unknown function or lacked similarity to reported sequences (32). The recovery of three putative regulatory genes may be related to the fact that PAO1 has the highest proportion of regulatory genes among the bacterial genomes sequenced to date (32), and many of these genes encode members of two-component signal transduction systems. Fluorescent Pseudomonas spp. are ubiquitous microorganisms, and they may all require an unusually large number of regulatory systems in order to survive in diverse ecological niches, including the rhizosphere.
Sequence analysis of clone 61 predicted a truncated protein similar to the global response regulator Lrp from E. coli (3). A putative helix-turn-helix DNA-binding domain (27-AHNDIALKVNLSRNAVRLRIERLERDG-49), the asnC bacterial regulatory protein family signature, was identified in the N-terminal part of the sequence. Among proteins most similar to the predicted product of clone 61, only BkdR from Pseudomonas putida has been well-characterized and shown to activate the expression of genes for the branched-chain keto acid dehydrogenase multienzyme complex (16). Pathways regulated by Lrp-like proteins could affect the survival of pseudomonads in the rhizosphere. For example, amino acid prototrophy is necessary for tomato root tip colonization (30), and although amino acid concentrations in tomato root exudate may be inadequate to support the growth of auxotrophs (30), amino acids are scavenged in the rhizosphere and novel genes for amino acid permeases, including a putative permease for high-affinity transport of branched-chain amino acids, are induced during rhizosphere colonization (24). Collectively, these findings support the hypothesis that the clone 61 product could regulate the synthesis or utilization of certain amino or keto acids in the rhizosphere.
The ORFs in clones 85 and 127 encode a putative two-component sensor kinase similar to the predicted sensor kinase PA1396 from P. aeruginosa PAO1 and a putative regulator with similarity to proteins of the LysR family of transcriptional regulators, respectively. Two-component regulators are among the most common types of regulatory systems in bacteria (27), and the P. aeruginosa genome has the greatest proportion of predicted regulatory genes of all of the bacterial genomes sequenced to date (32). Another two-component system, ColR/ColS (6), appears to influence outer membrane permeability and has an important role in root colonization by P. fluorescens WCS365. The ORF in clone 127 contains the LysR transcriptional regulator family signature, a putative helix-turn-helix DNA-binding domain (22-SFARAANELALTEGAISRQMGRLESLFLGVT-52), and resembles other proteins of the LysR family. Most LysR-like proteins are coinducer-dependent transcription activators that regulate diverse target genes or regulons or function in complex regulatory networks (27). Regulators of the LysR family are often divergently transcribed from a promoter overlapping a promoter of the target gene (27), and it is noteworthy that a divergently transcribed ORF similar to a hypothetical ORF from Klebsiella pneumoniae is situated 126 bp upstream of the putative regulatory gene in clone 127.
Subtracted clone 6 exhibited similarity to colicin M from E. coli. This pore-forming protein interferes with the biosynthesis of both peptidoglycan and O antigen by inhibiting the regeneration of the bactoprenyl-P carrier lipid, resulting in autolysis of the cell (9). Bacteriocin production is common among bacteria, but the predicted product of clone 6 is distinct from other bacteriocins produced by fluorescent pseudomonads (31) and is the first reported example of a Pseudomonas gene with similarity to colicin M. Based on the ecological role of bacteriocins (26), we speculate that the clone 6 product may function in intraspecific interactions or the competitiveness of Q8r1-96 in the rhizosphere.
Distribution among genotypes of 2,4-DAPG producers.
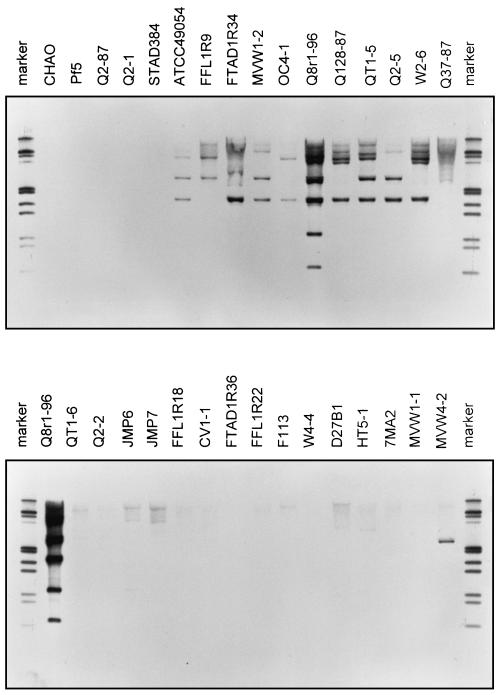
If the subtracted fragments from strain Q8r1-96 contribute to its exceptional rhizosphere competence, then they should be present among D-genotype strains that share the phenotype but not among strains of other genotypes. The distribution of the 32 tester-specific fragments was therefore determined among 31 2,4-DAPG-producing strains representing 17 distinct BOX-PCR genotypes (14, 18, 19) and including the tester, the driver, and nine other D-genotype strains. The fragments were amplified, arrayed on filters, and hybridized with biotin-labeled genomic DNA from each of the 31 strains by using the spot-blot method described above. Hybridization patterns were analyzed with MULTIV 1.2.1 software (Valerio De Patta Pillar, Department of Ecology, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil) using a simple matching coefficient which takes into account the absence as well as the presence of each of the 32 fragments in each of the 31 strains. Strains were clustered from the resulting similarity matrix (Table 2) by applying simple linkage, complete linkage, and sum of squares algorithms. All three algorithms generated similar groupings, with D-genotype isolates showing the greatest similarity to the tester strain (data not shown). Among D-genotype strains, OC4-1 and W2-6 were the least similar to Q8r1-96 (Table 2). However, W2-6 is known to exhibit superior rhizosphere competence (14), so the Q8r1-96 fragments with which it hybridized but which are not widely distributed among representatives of the less rhizosphere competent A, B, E, and L genotypes (i.e., fragments 6, 13, 18, 53, 64, and 81) are among those of particular interest. The groupings obtained by cluster analysis were consistent with the results of Southern hybridization, which showed that DNA from D-genotype strains hybridized more strongly to a mixture of subtracted fragments than did DNA from the other genotypes (Fig. 2).
TABLE 2.
Detection of tester-specific DNA fragments in 2,4-DAPG-producing Pseudomonas spp. by spot-blot hybridization
| SSH clone or promoter | Bacterial strainsa
|
|||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q2-87 (B) | CHA0 (A) | Pf5 (A) | STAD384 (C) | Q2-1 (B) | MVW4-2 (Q) | Q2-2 (E) | Q37-87 (E) | QT1-6 (E) | D27B1 (M) | FFL1R22 (J) | CV1-1 (H) | FTAD1R36 (I) | W4-4 (L) | 7MA12 (O) | MVW1-1 (P) | HT5-1 (N) | FFL1R18 (G) | F113 (K) | JMP6 (F) | JMP7 (F) | FTAD1R34 (D) | OC4-1 (D) | FFL1R9 (D) | MVW1-2 (D) | Q2-5 (D) | W2-6 (D) | Q128-87 (D) | ATCC49054 (D) | QT1-5 (D) | Q8r1-96 (D) | Frequency (%)b | |
| 3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 0.03 |
| 5 | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | 0.10 |
| 6 | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | − | − | − | − | − | + | + | + | + | + | 0.26 |
| 8 | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | 0.10 |
| 12 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 0.03 |
| 13 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | 0.32 |
| 18 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | + | + | + | − | + | + | + | + | + | + | + | + | 0.42 |
| 25 | − | − | − | + | − | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 0.84 |
| 26 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 0.03 |
| 28 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + | + | + | + | + | − | + | + | + | + | 0.32 |
| 32 | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | 0.94 |
| 36 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | 0.29 |
| 41 | − | − | − | − | − | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | 0.23 |
| 45 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 0.03 |
| 53 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + | + | + | 0.23 |
| 58 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 0.03 |
| 61 | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | + | + | + | + | + | − | + | + | + | + | 0.39 |
| 62 | − | + | + | + | + | − | + | + | + | + | − | − | − | − | − | − | − | − | + | + | − | + | − | − | − | − | − | + | + | + | + | 0.48 |
| 64 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + | + | + | 0.23 |
| 66 | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 0.84 |
| 74 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 0.03 |
| 80 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | + | 0.06 |
| 81 | − | − | − | − | − | − | − | − | − | − | + | + | + | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 0.55 |
| 83 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | + | 0.06 |
| 85 | − | − | − | + | − | − | − | − | − | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | 0.68 |
| 93 | − | − | − | − | + | − | − | − | − | − | + | + | + | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 0.58 |
| 101 | − | − | − | − | − | + | + | + | + | + | − | − | − | + | + | − | − | + | + | − | − | − | − | − | − | − | − | − | − | + | + | 0.35 |
| 103 | − | − | − | − | − | − | + | + | + | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | 0.26 |
| 127 | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | + | + | + | + | + | + | + | + | + | 0.42 |
| 132 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 0.03 |
| 133 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 0.03 |
| 164 | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 0.97 |
| Similarityc | 0.00 | 0.13 | 0.13 | 0.16 | 0.16 | 0.16 | 0.25 | 0.25 | 0.25 | 0.25 | 0.22 | 0.25 | 0.19 | 0.19 | 0.19 | 0.19 | 0.22 | 0.25 | 0.34 | 0.38 | 0.31 | 0.44 | 0.38 | 0.41 | 0.47 | 0.47 | 0.34 | 0.63 | 0.59 | 0.69 | 1.00 | |
Letters in parentheses correspond to BOX-PCR genotypes (18, 19). The majority of tested strains are described in references 18 and 19, except for Q2-87 (10), CHA0 (34), Pf5 (20), 7MA12 (15), Q128-87 (11), ATCC49054 (31), and Q8r1-96 (23).
Frequency values correspond to the percentage of test strains containing homologous sequences.
Similarity to Q8r1-96 was calculated using the simple matching coefficient.
FIG. 2.
Hybridization of genomic DNA from 2,4-DAPG-producing Pseudomonas spp. with subtracted, biotin-labeled DNA fragments. For Southern blots, 0.5 μg of genomic DNA digested with PstI and EcoRI was separated by electrophoresis, transferred and bound to nylon membranes, and hybridized with a mixture of subtracted, biotin-labeled DNA fragments. The markers were biotin-labeled EcoRI-HindIII lambda DNA digests (Sigma-Aldrich Corp.). Variation in the intensity of the hybridization signal is due to the fact that during subtraction, some sequences are enriched in the subtracted mix because they rehybridize and amplify more efficiently.
Gene expression analysis.
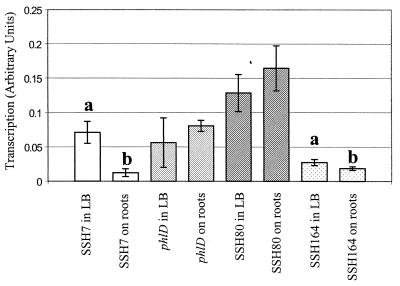
We determined whether sequences contained in the subtracted DNA fragments were expressed in cells of Q8r1-96 grown overnight in LB broth or recovered from the rhizosphere of wheat. Seeds (cv. Penawawa) were surface sterilized (21), allowed to dry, and germinated in petri dishes on moistened sterile filter paper for 1 week. Cultures of Q8r1-96 were grown overnight in King's medium B, collected by centrifugation, washed, and suspended to 10 times their original volume in M9 broth to obtain approximately 106 CFU ml−1. Seedlings were dipped into the bacterial suspension for 5 min and sown in groups of seven in sterile polypropylene jars containing 30 g of sterile quartz sand and 6.5 ml of one-fifth-strength Hoagland's solution. The roots were covered with another 30 g of moistened sand, and the containers were incubated under light at 20°C in closed acrylic boxes for 24 h. The treatment was replicated 10 times, and the experiment was repeated. After 24 h, seedlings were aseptically removed from containers and shaken to remove loosely adhering sand. Bacteria were recovered by centrifugation after the excised roots were briefly sonicated in 4 ml of sterile, cold, phosphate-buffered saline. Bacterial pellets were suspended immediately in TRIzol reagent (Invitrogen Corp., Carlsbad, Calif.) and sonicated on ice, and total RNA was extracted as recommended by the manufacturer. The same protocol was used to purify total RNA samples from 109 Q8r1-96 cells grown overnight in LB broth (referred to in this paper as in vitro growth conditions). Following the purification, all RNA samples were treated with RNase-free DNase I (Invitrogen), extracted again with TRIzol, precipitated, and suspended in RNase-free H2O. 32P-labeled cDNA probes were prepared with Superscript II reverse transcriptase (Invitrogen) by priming 5 μg of total RNA with random decamers as recommended by the manufacturer. Labeled cDNA was purified with a QIAquick nucleotide removal kit (Qiagen Inc., Valencia, Calif.) and hybridized with membrane filters on which the 32 fragments had been arrayed. Membranes were exposed to X-ray film, and the radiographs were scanned and analyzed with Lab Works analysis software (Ultra-Violet Products Ltd., Cambridge, United Kingdom). For each blot, the average volume of each dot was calculated and hybridization signals were standardized relative to a control consisting of a phzAB transcript that had been synthesized in vitro from pT7-6AB (17) with a reverse transcription-PCR competitor construction kit (Ambion) and added to each RNA sample prior to cDNA synthesis. The relative levels of transcripts were analyzed with STATISTIX 7.0 (Analytical Software, Tallahassee, Fla.) by standard analysis of variance. Mean comparisons among experiments were performed by using Fisher's protected least significant difference test at a P value of 0.05. On each membrane filter, all SSH clones were replicated twice and the experiment was repeated. The results revealed that fragments contained in tester clones 7, 80, and 164 hybridized strongly with probes prepared from total RNA isolated from cells incubated under both conditions. Clones 7 (which contains a fragment present in DNA from both Q8r1-96 and Q2-87) and 164 hybridized significantly more strongly with the probe prepared from cells grown in vitro than that from cells exposed to roots (Fig. 3). Thus, at least some of the cloned fragments contain highly expressed genes, and these genes are differentially expressed under the growth conditions examined.
FIG. 3.
Transcriptional analysis of subtracted fragment sequences expressed by Q8r1-96 in vitro in LB broth or after exposure to plant roots. Labeled cDNA obtained from Q8r1-96 grown in vitro or exposed to plant roots were used to probe membrane filters with arrayed SSH fragments. The levels of transcripts are expressed as arbitrary units, and each value represents the ratio of the average volume of the peak of a clone to the average volume of the peak of a phzAB control. The phlD gene, which is known to be expressed both in vitro and in situ (36), was included in the experiment as an additional control. Relative levels of transcripts were analyzed with STATISTIX 7.0 (Analytical Software) by standard analysis of variance. Mean comparisons among experiments were made with Fisher's protected least significant difference test at a P value of 0.05. Only clones SSH7, SSH80, and SSH164 consistently hybridized to radiolabeled transcripts. The transcript levels of clones 7 and 164 differed significantly (P < 0.05) between RNA samples obtained from cells grown in vitro (LB broth) or exposed to roots of wheat (a, b).
Variable root colonization has been a major impediment to the use of beneficial bacteria for plant growth promotion and biological control. New approaches are needed to enhance the population densities of introduced bacteria and the longevity of those populations in the rhizosphere. P. fluorescens strain Q8r1-96 demonstrates a level of rhizosphere competence not previously reported (23). The differences between the two strains analyzed in this study may provide insight into novel genes that contribute to the superior rhizosphere competence of P. fluorescens Q8r1-96 and other genotype D strains.
Nucleotide sequence accession numbers.
The nucleotide sequences of SSH fragments 6, 28, 53, 58, 61, 64, 80, 85, 101, 127, and 164 have been deposited in GenBank under accession numbers AF461160, AF461727, AF461728, AF461729, AF461161, AF461730, AF461723, AF461724, AF461731, AF461725, and AF461726, respectively.
Acknowledgments
We thank Greg Phillips and Karen Hansen for their technical assistance in the plant experiments. We also thank Patricia Okubara for suggestions and critical review of the manuscript.
REFERENCES
- 1.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Short protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Calvo, J. M., and R. G. Matthews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook, R. J., and D. M. Weller. 1987. Enhancement of root health and plant growth by rhizobacteria. UCLA (Univ. Calif. Los Angel.) Symp. Mol. Cell. Biol. 48:125-134. [Google Scholar]
- 5.Cook, R. J., L. S. Thomashow, D. M. Weller, D. Fujimoto, M. Mazzola, G. Bangera, and D.-S. Kim. 1995. Molecular mechanisms of defense by rhizobacteria against root diseases. Proc. Natl. Acad. Sci. USA 92:4197-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekkers, L. C., C. J. P. Bloemendaal, L. A. de Weger, C. A. Wijffelman, H. P. Spaink, and B. J. J. Lugtenberg. 1998. A two-component system plays an important role in the root-colonizing ability of Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 11:45-56. [DOI] [PubMed] [Google Scholar]
- 7.Duffy, B. K., and G. Défago. 1997. Zinc improves biocontrol of Fusarium crown and root rot of tomato by Pseudomonas fluorescens and represses the production of pathogen metabolites inhibitory to bacterial antibiotic biosynthesis. Phytopathology 87:1250-1257. [DOI] [PubMed] [Google Scholar]
- 8.Fenton, A. M., P. M. Stephens, J. Crowley, M. O'Callaghan, and F. O'Gara. 1992. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl. Environ. Microbiol. 58:3873-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harkness, R. E., and T. Olschlager. 1991. The biology of colicin M. FEMS Microbiol. Rev. 88:27-41. [DOI] [PubMed] [Google Scholar]
- 10.Harrison, L. A., L. Letendre, P. Kovacevich, E. Pierson, and D. M. Weller. 1993. Purification of an antibiotic effective against Gaeumannomyces graminis var. tritici produced by biocontrol agent Pseudomonas aureofaciens. Soil Biol. Biochem. 25:215-221. [Google Scholar]
- 11.Keel, C., D. M. Weller, A. Natsch, G. Defago, R. J. Cook, and L. S. Thomashow. 1996. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 62:552-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keel, C., U. Schnider, M. Maurhofer, C. Voisard, J. Laville, U. Burger, P. Wirthner, D. Haas, and G. Defago. 1992. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant-Microbe Interact 5:4-13. [Google Scholar]
- 13.Lan, R., and P. R. Reeves. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8:396-401. [DOI] [PubMed] [Google Scholar]
- 14.Landa, B. B., H. A. E. de Werd, B. B. McSpadden-Gardener, and D. M. Weller. 2002. Comparison of three methods for monitoring populations of different genotypes of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens in the rhizosphere. Phytopathology 92:129-137. [DOI] [PubMed] [Google Scholar]
- 15.Landa, B. B., and D. M. Weller. 2001. Crop preference by genotypes of 2,4-diacetylphloroglucinol (DAPG)-producing Pseudomonas spp. Phytopathology 91:S52. [DOI] [PubMed] [Google Scholar]
- 16.Madhusudhan, K. T., K. L. Hester, V. Friend, and J. R. Sokatch. 1997. Transcriptional activation of the bkd operon of Pseudomonas putida by BkdR. J. Bacteriol. 179:1992-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mavrodi, D. V., V. N. Ksenzenko, R. F. Bonsall, R. J. Cook, A. M. Boronin, and L. S. Thomashow. 1998. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J. Bacteriol. 180:2541-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mavrodi, O. V., B. B. McSpadden Gardener, D. V. Mavrodi, R. F. Bonsall, D. M. Weller, and L. S. Thomashow. 2001. Genetic diversity of phlD from 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. Phytopathology 91:35-43. [DOI] [PubMed] [Google Scholar]
- 19.McSpadden Gardener, B. B., K. L. Schroeder, S. E. Kalloger, J. M. Raaijmakers, L. S. Thomashow, and D. M. Weller. 2000. Genotypic and phenotypic diversity of phlD-containing Pseudomonas strains isolated from the rhizosphere of wheat. Appl. Environ. Microbiol. 66:1939-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowak-Thompson, B., S. J. Gould, J. Kraus, and J. E. Loper. 1994. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf5. Can. J. Microbiol. 40:1064-1066. [Google Scholar]
- 21.Pierson, E. A., and D. M. Weller. 1994. Use of mixtures of fluorescent pseudomonads to suppress take-all and improve the growth of wheat. Phytopathology 84:940-947. [Google Scholar]
- 22.Raaijmakers, J. M., and D. M. Weller. 1998. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol. Plant-Microbe Interact. 11:144-152. [Google Scholar]
- 23.Raaijmakers, J. M., and D. M. Weller. 2001. Exploiting genotypic diversity of 2,4-diacetylphloroglucinol-producing Pseudomonas spp.: characterization of superior root-colonizing P. fluorescens strain Q8r1-96. Appl. Environ. Microbiol. 67:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 25.Ramette, A., Y. Moenne-Loccoz, and G. Defago. 2001. Polymorphism of the polyketide synthase gene phlD in biocontrol fluorescent pseudomonads producing 2,4-diacetylphloroglucinol and comparison of PhlD with plant polyketide synthases. Mol. Plant-Microbe Interact. 14:639-652. [DOI] [PubMed] [Google Scholar]
- 26.Riley, M. A., and D. M. Gordon. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7:129-133. [DOI] [PubMed] [Google Scholar]
- 27.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt, K. D., T. Schmidt-Rose, U. Romling, and B. Tummler. 1998. Differential genome analysis of bacteria by genomic subtractive hybridization and pulsed field gel electrophoresis. Electrophoresis 19:509-514. [DOI] [PubMed] [Google Scholar]
- 29.Shanahan, P., D. J. O'Sullivan, P. Simpson, J. D. Glennon, and F. O'Gara. 1992. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons, M., H. P. Permentier, L. A. de Weger, C. A. Wijffelman, and B. J. J. Lugtenberg. 1997. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 10:102-106. [Google Scholar]
- 31.Smirnov, V. V., and E. A. Kiprianova. 1990. Bacteria of Pseudomonas genus. Naukova Dumka, Kiev, Ukraine. [PubMed]
- 32.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkmann, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 33.Straus, D., and F. M. Ausubel. 1990. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc. Natl. Acad. Sci. USA 87:1889-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stutz, E., G. Defago, and H. Kern. 1986. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76:181-185. [Google Scholar]
- 35.Thomashow, L. S., and D. M. Weller. 1996. Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites, p. 187-236. In G. Stacey and N. T. Keen (ed.), Plant-microbe interactions, vol. 1. Chapman & Hall, New York, N.Y.
- 36.Thomashow, L. S., R. F. Bonsall, and D. M. Weller. 2002. Antibiotic production by soil and rhizosphere microbes in situ, p. 638-647. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 37.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]