Abstract
A multiplex PCR assay based on the 16S rRNA genes was developed for the simultaneous detection of three major fish pathogens, Aeromonas salmonicida, Flavobacterium psychrophilum, and Yersinia ruckeri. The assay proved to be specific and as sensitive as each single PCR assay, with detection limits in the range of 6, 0.6, and 27 CFU for A. salmonicida, F. psychrophilum, and Y. ruckeri, respectively. The assay was useful for the detection of the bacteria in artificially infected fish as well as in fish farm outbreaks. Results revealed that this multiplex PCR system permits a specific, sensitive, reproducible, and rapid method for the routine laboratory diagnosis of infections produced by these three bacteria.
Major pathogens involved in fish farm salmonid infections include the gram negative species Flavobacterium psychrophilum, Yersinia ruckeri, and Aeromonas salmonicida. They are the etiological agents of cold water disease, enteric red mouth disease, and furunculosis, respectively. These pathologies are common worldwide and produce considerable economic losses in the fish farming industry.
Cold water disease particularly affects juvenile fish (3), and the causal agent is F. psychrophilum, a fastidious bacterium that is difficult to grow (12). The bacterium causes saddle-like external lesions near the dorsal fin and it can also be found in the mouth, spleen, and brain tissues. The fish may darken and develop bacteremia with the microorganism present throughout the animal. Y. ruckeri is an important pathogen in intensive aquaculture of trout and salmon. Disease outbreaks are related to stress (4), and little information is available about the biology of the bacterium. Infected fish present characteristic red eyes and mouth as well as internal hemorrhages. Diagnostic methods include culturing, serology, and molecular biology techniques (5, 9, 16). A. salmonicida also produces an infection in fish that causes muscle lesions which could produce ulcers on the surface of the skin and lead to septicemia. Different diagnostic methods have been developed, including enzyme-linked immunosorbent assays (19, 20), agglutination tests (11), and PCR probes (14).
On the other hand, an efficient selective medium for reducing the growth of the background flora and facilitating the isolation and identification of each of these three bacteria species remains to be described. To combat these infections, vaccination has been shown to be an effective method for the prevention of enteric red mouth disease (17) and, less efficiently, for furunculosis (10, 13), but there is no effective control for cold water disease. In most cases, antimicrobial compounds should be used to control disease outbreaks caused by any of these microorganisms. Thus, a rapid and effective diagnostic method is essential for the application of specific treatment.
Although multiplex PCR (m-PCR) has been widely applied to the detection of multiple viruses and bacteria in clinical specimens (2, 6, 7), it has not been applied to the detection of fish pathogens (1, 15). In this investigation, an m-PCR assay for the simultaneous detection of F. psychrophilum, A. salmonicida, and Y. ruckeri was developed and compared with single PCR assays for each of the bacteria.
Bacterial strains and culture conditions.
To evaluate the m-PCR assay, the bacteria used as positive controls were (i) A. salmonicida CECT 4237, LMG 3780, and RSP70.1; (ii) F. psychrophilum NCIMB 1947T, NCIMB 1826, T1, FPC 830, 84.254, SH3-81, Tm.3.1, BC3-81, and SRCO5-90; and (iii) Y. ruckeri ATCC 29473, 146, 147, 149, and 150. The strains of other species taxonomically and/or ecologically related that were tested as negative controls were (i) Aeromonas hydrophila CECT 4588, CECT 839, and TW401, Aeromonas encheleia CECT 4341 and CECT 4342, Aeromonas jandaei CECT 4336 and CECT 4338, and Aeromonas euchrenophila CECT 4224; (ii) Flavobacterium johnsoniae UW 101 and Flavobacterium columnare LMG 13035 and LMG 10397; and (iii) Yersinia enterocolitica ATCC 27729. Strains were obtained from the following sources: ATCC strains, American Type Culture Collection, Rockville, Md.; CECT strains, Coleccción Española de Cultivos Tipo, Universidad de Valencia, Valencia, Spain; LMG strains, Laboratorium voor Microbiologie Universiteit Gent, Faculteit der Wetenschappen, Ghent, Belgium; NCIMB strains, Nationals Collections of Industrial and Marine Bacteria, Aberdeen, Scotland, United Kingdom; RSP70.1 and TW401 strains, A. E. Toranzo, Departamento de Microbiologia, Universidad de Santiago de Compostela, Santiago, Spain; T1 strain, Área de Microbiologia, Universidad de Oviedo, Oviedo, Spain; FPC 830, 84.254, SH3-81, TM 3.1, BC3-81, and SRCO5-90 strains, J.-F. Bernardet, Unite de Virologie et Immunologie Moleculaires, Institut National de la Recherche Agronomique, Jouy-en-Josas, France; VW 101 strain, D. W. Hunnicutt, Department of Biological Sciences, University of Wisconsin—Milwaukee; 146, 147, 149, and 150 strains, J. L. Larsen, Danish Veterinary Laboratory, Copenhagen, Denmark. Aeromonas spp. were grown in Trypticase soy nutrient broth (TSB; Merck, Barcelona, Spain); Y. ruckeri strains were cultured in nutrient broth (Pronadisa, Madrid, Spain); and Anackel-Ordal medium supplemented with 5% horse serum (12) was used for F. psychrophilum strains. Aeromonas spp. and Yersinia spp. strains were incubated at 28°C, whereas Flavobacterium spp. strains were incubated at 12°C. F. johnsoniae and F. columnare strains were grown in 16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl (2XTY) per liter and incubated at 28°C. All of the cultures were incubated with shaking at 250 rpm in an orbital incubator.
PCR conditions.
To establish the optimal sensitivity for the m-PCR assay, four variables were examined in single experiments by using a multifactorial experimental design in which factors were tested at two levels. The variables tested were the following: two annealing temperatures (60 and 55°C), two polymerase concentrations (1 and 1.5 U), two deoxynucleoside triphosphate (dNTP) concentrations (200 and 400 μM), and two MgCl2 concentrations (2 and 3 mM). A good intensity of the amplicons for each target DNA, as well as the absence of unspecific bands, was considered in selecting the optimal m-PCR conditions. Thus, the best results were obtained with an annealing temperature of 60°C, 1.5 U of polymerase, a 200 μM concentration of each dNTP, and 2 mM MgCl2. Therefore, PCR was performed in 50-μl reaction mixtures containing 5 to 15 μl of sample as template DNA, a 200 μM concentration of each dNTP, a 1 μM concentration of each primer (Amersham Pharmacia Biotech, Barcelona, Spain), and 1.5 U of DNA polymerase and its amplification buffer (Biotools, Madrid, Spain). Primers for A. salmonicida (PAAS1, 5′ CGTTGGATATGGCTCTTCT 3′; PAAS2, 5′ CTCAAAACGGCTGCGTACCA 3′) were described by O'Brien et al. (14). The forward primer (FP1, 5′ CTTAGTTGGCATCAACAC 3′) used for detection of F. psychrophilum was described by Urdaci et al. (18), but the reverse primer (FP3, 5′ ACACTGGCAGTCTTGCTA 3′) was designed by us (nucleotides 954 to 971 in the sequence with GenBank accession number D12670). Primers used in the detection of Y. ruckeri (YER3, 5′ CGAGGAGGAAGGGTTAAGT 3′; YER4, 5′ AAGGCACCAAGGCATCTCT 3′) were modified based on primers described by Gibello et al. (5). Thermal cycling was done with a GeneAmp 9700 thermocycler (Perkin Elmer Instruments, Norwalk, Conn.) with the following conditions: an initial denaturation cycle at 94°C for 2 min, followed by 35 cycles of amplification (denaturation at 94°C for 40 s, annealing at 60°C for 40 s, and extension at 72°C for 60 s), and a final 5-min elongation period at 72°C. Reactions with or without template DNA of the three bacteria were tested in each experiment as positive and negative controls, respectively. Aliquots of 15 μl of PCR product were analyzed by electrophoresis on 1.5% agarose gels and stained with ethidium bromide.
Sensitivity and specificity of the m-PCR assay.
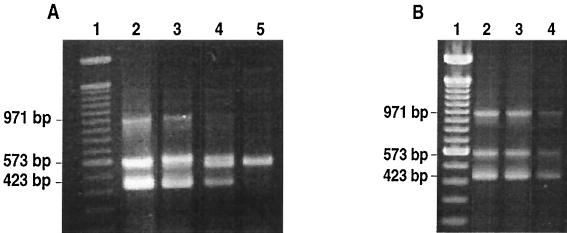
The sensitivity of the m-PCR assay was evaluated using the following strains: F. psychrophilum SH3-81, Y. ruckeri 150, and A. salmonicida CECT 4237. One milliliter of a stationary-phase culture of each strain was centrifuged for 5 min at 12,000 × g, the pellet was resuspended in 50 ml of distilled water, and cells were lysed by boiling for 10 min. Cell debris was removed by centrifugation for 30 s at 12,000 × g, and 10-fold serial dilutions of the supernatant were prepared. Aliquots of 5 μl of each dilution were removed and mixed together with the respective aliquots of the other bacteria. A mixture was used as template DNA for the m-PCR assay. To enumerate the bacteria, serial dilutions of pure cultures of each bacterium were prepared, and 0.1-ml aliquots of appropriate dilutions were spread onto TSB agar for A. salmonicida CECT 4237, nutrient agar for Y. ruckeri, and Anackel-Ordal medium supplemented with 5% horse serum with agar for F. psychrophilum. Both A. salmonicida and Y. ruckeri plates were incubated at 28°C, whereas F. psychrophilum plates were incubated at 20°C. In these experiments, detection limits of 27 CFU per m-PCR for Y. ruckeri, 6 CFU for A. salmonicida, and 0.6 CFU for F. psychrophilum were obtained when pure cultures were used (Fig. 1A).
FIG. 1.
Sensitivity of the detection of F. psychrophilum, Y. ruckeri, and A. salmonicida by m-PCR assay. (A) PCR amplification was performed on 10-fold-serially diluted cell lysates of pure cultures of the three pathogens. The size of the PCR products obtained for F. psychrophilum, Y. ruckeri, and A. salmonicida were 971, 573, and 423 bp, respectively. Lane 1, lambda DNA 100-bp molecular size marker; lane 2, 60, 2.7 × 105, and 6 × 103 CFU of F. psychrophilum, Y. ruckeri, and A. salmonicida, respectively; lane 3, 0.6, 2.7 × 103, and 60 CFU of F. psychrophilum, Y. ruckeri, and A. salmonicida, respectively; lane 4, 270 and 6 CFU of Y. ruckeri and A. salmonicida, respectively; lane 5, 27 CFU of Y. ruckeri. (B) PCR amplification on tissue samples seeded with serially diluted cultures of the three pathogens. Lane 1, lambda DNA 100-bp molecular size marker; lane 2, 104, 1.45 × 104, and 3 × 103 CFU of F. psychrophilum, Y. ruckeri, and A. salmonicida, respectively; lane 3, 103, 1.45 × 103, and 300 CFU of F. psychrophilum, Y. ruckeri, and A. salmonicida, respectively; lane 4, 100, 145, and 30 CFU of F. psychrophilum, Y. ruckeri, and A. salmonicida, respectively.
To test the sensitivity of the procedure in the presence of tissue debris, spiked samples were used. Commercial trout liver was homogenized with Tris-EDTA (TE) buffer (1 mM Tris-HCl, 0.5 mM EDTA [pH 8]) in a 1:10 ratio and seeded with serial dilutions of a mixture of pure cultures of the three bacterial species. Cell lysates were obtained by boiling and, after centrifugation at 12,000 × g for 30 s, 15 μl of each sample was used for m-PCR analysis. The limits of detection for the spiked samples were 100 CFU for F. psychrophilum, 145 CFU for Y. ruckeri, and 30 CFU for A. salmonicida (Fig. 1B). All reactions assessing limits of detection were performed in duplicate and repeated on different days to confirm the results.
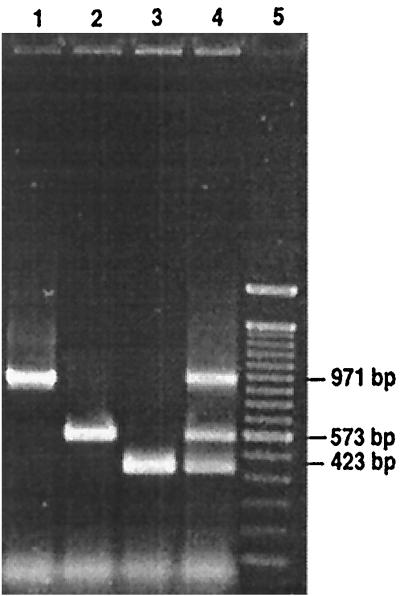
The specificity of the three primer sets was tested (separately or mixed together) using the cell lysates of the 29 strains cited above. An amplification product of the expected size was observed for the nine strains of F. psychrophilum (971 bp), five strains of Y. ruckeri (573 bp), and three strains of A. salmonicida (423 bp) (Fig. 2), but none of these three PCR products or other PCR bands occurred when the cell lysates of the other 12 representative strains were used.
FIG. 2.
Specificity of the m-PCR assay developed for the detection of F. psychrophilum, Y. ruckeri, and A. salmonicida. Agarose gel showing simultaneous m-PCR identification of F. psychrophilum (971 bp), Y. ruckeri (573 bp), and A. salmonicida (423 bp). Lane 1, F. psychrophilum alone; lane 2, Y. ruckeri alone; lane 3, A. salmonicida alone; lane 4, m-PCR with the three pathogens together; lane 5, lambda DNA 100-bp molecular size marker.
Artificially infected fish and naturally occurring outbreaks.
For artificially infected fish, three groups of 10 rainbow trout of an approximate size of 6 cm were maintained in 60-liter tanks at a temperature of 18 ± 1°C for the experiments carried out with Y. ruckeri 150 and A. salmonicida CECT 4237 and at 12 ± 1°C for fish infected with F. psychrophilum SH3-81. Each trout was intraperitoneally injected with 0.1 ml of phosphate-buffered saline (PBS; pH 7.2) containing 106 cells of the corresponding bacteria. Ten fish were inoculated with 0.1 ml of PBS for use as controls. Every day for a total of 10 days, dead fish were removed from the experimental tanks and the livers were removed aseptically and homogenized in TE buffer in a 10% (wt/vol) suspension. Samples were submitted to DNA extraction using an InstaGene matrix (Bio-Rad) following the manufacturer's instructions.
In parallel, a loopful of the fish liver was streaked onto the specific medium for each bacterium and incubated at the corresponding temperature. Fish used as controls were maintained and processed in the same way as the infected ones.
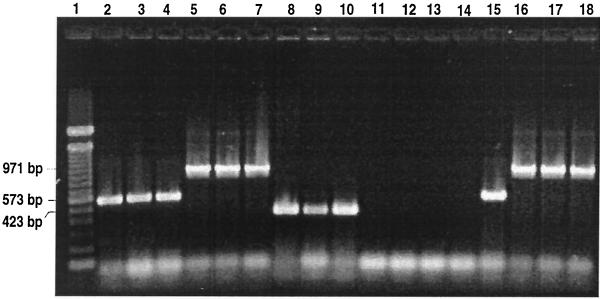
All of the fish infected with Y. ruckeri 150 and A. salmonicida CECT 4237 died within 5 days of injection, and the corresponding amplification products were seen when the DNA extracted from the tissue was submitted to m-PCR. Only 50% of the fish injected with F. psychrophilum died before the end of the experiment, and they showed an amplification product of 971 bp. This result correlates with those found with the bacteriological method. Uninfected fish used as controls did not give false positive results with either method (Fig. 3).
FIG. 3.
Detection of F. psychrophilum, Y. ruckeri, and A. salmonicida by m-PCR in artificially and naturally infected fish. Lane 1, lambda DNA 100-bp molecular size marker; lanes 2 to 4, fish artificially infected with 106 cells of Y. ruckeri 150; lanes 5 to 7, fish artificially infected with 106 cells of F. psychrophilum SH3-81; lanes 8 to 10, fish artificially infected with 106 cells of A. salmonicida CECT 4237; lanes 11 to 13, control fish inoculated with PBS; lane 14, nontemplate control; lane 15, fish isolated from a Y. ruckeri outbreak; lanes 16 to 18, fish belonging to three different F. psychrophilum outbreaks.
Samples of diseased juvenile fish with an infection from different salmonid fish farms in Asturias (Spain) were analyzed for the presence of these three pathogens by m-PCR. Each sample was processed as described for artificially infected fish, and a final diagnosis of the affected fish was made by identification of the bacteria growing in the media under the conditions previously described for each species. In the m-PCR assay the amplification products corresponding to Y. ruckeri (573 bp) and F. psychrophilum (971 bp) were obtained (Fig. 3).
m-PCR assays have been developed for the detection of fish viruses (22), but only a single similar assay has been established for other pathogens of fish (1). We have developed an m-PCR assay that can detect three of the main fish pathogens: F. psychrophilum, Y. ruckeri, and A. salmonicida.
The specificity of our assay was verified by performing the m-PCR with lysates from F. psychrophilum, Y. ruckeri, A. salmonicida, and 12 different taxonomically and/or ecologically related bacteria. None of these related bacteria gave the corresponding amplification product. Therefore, the primers described here proved to be specific under the conditions assayed. These results are in agreement with those obtained by Urdaci et al. (18) for F. psychrophilum, Gibello et al. (5) for Y. ruckeri, and O'Brien et al. (14) for A. salmonicida when using single PCR.
Our m-PCR assay could detect as few as 27 CFU of Y. ruckeri, 6 CFU of A. salmonicida, and 0.6 CFU of F. psychrophilum in pure cultures and could detect 145 Y. ruckeri, 30 A. salmonicida, and 102 F. psychrophilum CFU in spiked samples. These results are similar to those previously reported in the literature, in which detection limits of 60 to 65 cells for Y. ruckeri (5) and 0.4 cells for F. psychrophilum (21) in pure cultures were found. For A. salmonicida, a limit of detection of 2.4 cells was reported when using a slot blot hybridization technique (8). In experiments with spiked samples, sensitivities of 2 × 104 CFU/g of tissue for Y. ruckeri (5) and 200 cells/g of sample for A. salmonicida (14) have been described.
These results, together with those obtained in in vivo experiments, indicate that this procedure is a highly sensitive and specific method for detecting these three bacterial species in fish farm outbreaks. Thus, a rapid and reliable diagnosis of disease could be carried out in a single PCR assay.
Acknowledgments
This research was supported by grant 1FD97-0426 to J.A.G.
We thank J.-F. Bernadet, J. L. Larsen, D. W. Hunnicut, and A. E. Toranzo for sending us different strains. Finally, we extend our thanks to the “Asociación de Piscicultores de Asturias.”
REFERENCES
- 1.Brasher, C. W., A. DePaola, D. D. Jones, and A. K. Bej. 1998. Detection of microbial pathogens in shellfish with multiplex PCR. Curr. Microbiol. 37:101-107. [DOI] [PubMed] [Google Scholar]
- 2.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, A. J. Fox, and E. B. Kaczmarski. 2001. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39:1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalsgaard, I. 1993. Virulence mechanisms in Cytophaga psychrophila and Cytophaga-like bacteria pathogenic for fish. Annu. Rev. Fish Dis. 3:127-144. [Google Scholar]
- 4.Everlyn, T. P. T. 1996. Infection and disease, p. 339-362. In G. Iwana and T. Nakanishi (ed.), The fish immune system: organism, pathogen and environment. Academic Press, San Diego, Calif.
- 5.Gibello, A., M. M. Blanco, M. A. Moreno, M. T. Cutuli, A. Doménech, L. Domínguez, and J. F. Fernández-Garayzábal. 1999. Development of a PCR assay for detection of Yersinia ruckeri in tissues of inoculated and naturally infected trout. Appl. Environ. Microbiol. 65:346-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendolin, P. H., A. Markkanen, J. Ylikoski, and J. J. Wahlfors. 1997. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J. Clin. Microbiol. 35:2854-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heredia, A., V. Soriano, S. H. Weiss, R. Bravo, A. Vallejo, T. N. Denny, J. S. Epstein, and I. K. Hewlett. 1996. Development of a multiplex PCR assay for the simultaneous detection and discrimination of HIV-1, HIV-2, HTLV-I and HTLV-II. Clin. Diagn. Virol. 7:85-92. [DOI] [PubMed] [Google Scholar]
- 8.Hiney, M., M. T. Dawson, D. M. Heery, P. R. Smith, F. Gannon, and R. Powell. 1992. DNA probe for Aeromonas salmonicida. Appl. Environ. Microbiol. 58:1039-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeJeune, J. T., and F. R. Rurangirwa. 2000. Polymerase chain reaction for definitive identification of Yersinia ruckeri. J. Vet. Diagn. Investig. 12:558-561. [DOI] [PubMed] [Google Scholar]
- 10.Lutwyche, P., M. M. Exner, R. E. W. Hancock, and T. J. Trust. 1995. A conserved Aeromonas salmonicida porin provides protective immunity to rainbow trout. Infect. Immun. 63:3137-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarthy, D. H. 1975. Detection of Aeromonas salmonicida antigen in diseased fish tissue. J. Gen. Microbiol. 88:384-386. [DOI] [PubMed] [Google Scholar]
- 12.Michel, C., D. Antonio, and R. P. Hedrick. 1999. Production of viable cultures of Flavobacterium psychrophilum: approach and control. Res. Microbiol. 150:351-358. [DOI] [PubMed] [Google Scholar]
- 13.Noonan, B., P. J. Enzmann, and T. J. Trust. 1996. Recombinant infectious hematopoietic necrosis virus and viral hemorrhagic septicemia virus glycoprotein epitopes expressed in Aeromonas salmonicida induce protective immunity in rainbow trout (Oncorhynchus mykiss). Appl. Environ. Microbiol. 61:3586-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Brien, D., J. Mooney, D. Ryan, E. Powell, M. Hiney, P. R. Smith, and R. Powell. 1994. Detection of Aeromonas salmonicida, causal agent of furunculosis in salmonid fish, from the tank effluent of hatchery-reared Atlantic salmon smolts. Appl. Environ. Microbiol. 60:3874-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osorio, C. R., A. E. Toranzo, J. L. Romalde, and J. L. Barja. 2000. Multiplex PCR assay for ureC and 16S rRNA genes clearly discriminates between both subspecies of Photobacterium damselae. Dis. Aquat. Org. 40:177-183. [DOI] [PubMed] [Google Scholar]
- 16.Romalde, J. L., B. Magariños, J. L. Barja, and A. E. Toranzo. 1993. Antigenic and molecular characterization of Yersinia ruckeri: proposal for a new intraspecies classification. Syst. Appl. Microbiol. 16:411-419. [Google Scholar]
- 17.Stevenson, R. M. W. 1997. Immunization with bacterial antigens: yersiniosis. Fish Vaccinol. Dev. Biol. Standard. 90:117-124. [PubMed] [Google Scholar]
- 18.Urdaci, M. C., C. Chakroun, D. Faure, and J.-F. Bernardet. 1998. Development of a polymerase chain reaction assay for identification and detection of the fish pathogen Flavobacterium psychrophilum. Res. Microbiol. 149:519-530. [DOI] [PubMed] [Google Scholar]
- 19.Wagner, U., B. K. Gudmundsdottir, and K. Drossler. 1999. Monoclonal antibodies against AsaP1, a major exotoxin of the fish pathogen Aeromonas salmonicida subsp. chromogenes, and their application in ELISA. J. Appl. Microbiol. 87:620-629. [DOI] [PubMed] [Google Scholar]
- 20.Wagner, U., D. Hadge, B. K. Gudmundsdottir, K. Nold, and K. Drossler. 2001. Antibody response in salmonids against the 70 kDa serine protease of Aeromonas salmonicida studied by a monoclonal antibody-based ELISA. Vet. Immunol. Immunopathol. 82:121-135. [DOI] [PubMed] [Google Scholar]
- 21.Wiklund, T., L. Madsen, M. S. Bruun, and I. Dalsgaard. 2000. Detection of Flavobacterium psychrophilum from fish tissue and water samples by PCR amplification. J. Appl. Microbiol. 88:299-307. [DOI] [PubMed] [Google Scholar]
- 22.Williams, K., S. Blake, A. Sweeney, J. T. Singer, and B. L. Nicholson. 1999. Multiplex reverse transcriptase PCR assay for simultaneous detection of three fish viruses. J. Clin. Microbiol. 37:4139-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]