Abstract
The mosquitocidal toxin (MTX) produced by Bacillus sphaericus strain SSII-1 is an ∼97-kDa single-chain toxin which contains a 27-kDa enzyme domain harboring ADP-ribosyltransferase activity and a 70-kDa putative binding domain. Due to cytotoxicity toward bacterial cells, the 27-kDa enzyme fragment cannot be produced in Escherichia coli expression systems. However, a nontoxic 32-kDa N-terminal truncation of MTX can be expressed in E. coli and subsequently cleaved to an active 27-kDa enzyme fragment. In vitro the 27-kDa enzyme fragment of MTX ADP-ribosylated numerous proteins in E. coli lysates, with dominant labeling of an ∼45-kDa protein. Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry combined with peptide mapping identified this protein as the E. coli elongation factor Tu (EF-Tu). ADP ribosylation of purified EF-Tu prevented the formation of the stable ternary EF-Tuaminoacyl-tRNAGTP complex, whereas the binding of GTP to EF-Tu was not altered. The inactivation of EF-Tu by MTX-mediated ADP-ribosylation and the resulting inhibition of bacterial protein synthesis are likely to play important roles in the cytotoxicity of the 27-kDa enzyme fragment of MTX toward E. coli.
The mosquitocidal toxin (MTX) from Bacillus sphaericus strain SSII-1 is a member of the family of bacterial ADP- ribosyltransferases (8, 22). These toxins transfer the ADP-ribose moiety of NAD to a protein target in eukaryotic cells, affecting essential cell functions (8). Diphtheria toxin, Pseudomonas exotoxin A, cholera toxin, pertussis toxin, C2 and iota toxins, the family of C3-like transferases, and Pseudomonas exoenzyme S are well-described members of this family of bacterial toxins. Diphtheria toxin and Pseudomonas exotoxin A block protein synthesis by ADP-ribosylation of the eukaryotic elongation factor 2 at diphthamide, a modified histidine residue (26). Cholera toxin and pertussis toxin ADP-ribosylate G proteins at specific arginine and cysteine residues, respectively, to modify signal transduction by G-protein-coupled receptors (4, 14, 25, 29). Members of the subfamily of binary ADP- ribosylating toxins, such as Clostridium botulinum C2 toxin and Clostridium perfringens iota toxin, ADP-ribosylate G actin at Arg177 (23, 24). Several bacterial exoenzymes ADP-ribosylate small GTP-binding proteins, including C. botulinum C3 exoenzyme and related C3-like transferases, which ADP-ribosylate Rho GTPases at Asn41 (2, 19, 21, 30), and Pseudomonas aeruginosa exotoxin S, which preferentially modifies Ras proteins at several arginine residues (10).
The mature MTX (without the putative signal sequence of 29 amino acid residues) is a 97-kDa protein (MTX30-870). MTX30-870 is processed into a 27-kDa N-terminal enzyme component (MTX30-264) and a 70-kDa C-terminal putative binding component (MTX265-870) by crude mosquito larval gut extracts or chymotrypsin. MTX30-870 enzyme activity is strictly dependent on proteolytic activation. Following proteolytic cleavage, the putative MTX binding component remains noncovalently bound to the enzyme component, thereby reducing enzyme activity. However, no repression is observed when the 27-kDa fragment is generated from a 32-kDa N-terminal fragment of MTX (MTX30-308), resulting in enzyme activity 150-fold higher than that of processed MTX30-870. The production of recombinant MTX30-264 in Escherichia coli is not possible due its severe toxicity toward the bacterial cell. This toxicity is dependent on the transferase activity of the enzyme, as catalytically inactive MTX30-264 constructs can be produced in E. coli (20).
In this study, we investigated the possible reasons for the toxicity of MTX toward E. coli. We identified the bacterial elongation factor Tu (EF-Tu) as one of the target proteins of MTX in E. coli cell lysates and showed the inactivation of EF-Tu by mono-ADP-ribosylation.
MATERIALS AND METHODS
Materials.
MTX30-308 and MTX30-308 with an E197Q mutation (MTX30-308E197Q) were purified and proteolytically processed as described previously (20). Cellular EF-Tu from E. coli MRE 600 and [14C]Phe-tRNAPhe were purified by published procedures (11, 18). [14C]phenylalanine was purchased from ICN (Braunschweig, Germany), and adenylate-[32P]NAD (30 Ci/mmol) was obtained from NEN (Vilvoorde, Belgium). 3′(2′)-O-(N-Methylanthraniloyl)-GTP (mant-GTP) was obtained from Molecular Probes (Leiden, The Netherlands). All other reagents were obtained from Sigma (Deisenhofen, Germany) unless otherwise indicated. Buffer A contained 10 mM MgCl2, 50 mM NH4Cl, and 25 mM Tris-HCl (pH 7.4).
Cloning and purification of EF-Tu.
DNA was amplified from plasmid pEECAHis (coding for EF-Tu) by PCR with the following primers: EF-Tu sense, 5′-CGCGGATCCTCTAAAGAAAAGTTTGAACGTAC-3′, and EF-Tu antisense, 5′-CCGGAATTCGCTCAGAACTTTTGCTACAAC-3′. The EF-Tu fragment was cut with BamHI and EcoRI, purified, and ligated to the digested pGEX-2TGL vector. This vector was previously designed in our laboratory (Freiburg) and is a modification of the pGEX-2T vector from Amersham Pharmacia Biotech, Freiburg, Germany. The vector contains an additional oligonucleotide in the multiple cloning site which codes for a glycine linker between the glutathione S-transferase (GST) residue and the inserted gene, enabling efficient cleavage of GST by thrombin. The construct was checked by DNA sequencing (ABI PRISM; Perkin-Elmer, Weiterstadt, Germany).
For the expression and purification of GST-EF-Tu, plasmids were transformed into E. coli TG 1. Cells were grown in Luria-Bertani medium to stationary phase at 37°C, harvested, resuspended in fresh medium containing 1 mM isopropyl-1-thio-β-d-galactopyranoside, and incubated for 2 h at 30°C. Then, cells were harvested, lysed by sonication in 20 mM Tris-HCl (pH 7.4)-10 mM NaCl-5 mM MgCl2-1% Triton X-100-1 mM phenylmethylsulfonyl fluoride-5 mM dithiothreitol (lysis buffer), and purified by affinity chromatography with glutathione-Sepharose beads (Amersham Pharmacia Biotech). Loaded beads were washed twice with lysis buffer and twice with thrombin cleavage buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2). EF-Tu was cleaved with thrombin directly from the beads in thrombin cleavage buffer. Thrombin was removed with benzamidine-Sepharose beads (Amersham Pharmacia Biotech).
ADP-ribosyltransferase assay.
ADP-ribosylation was performed as follows: 2.5 μM cellular or recombinant EF-Tu- GDP or 12 μg of total E. coli cell lysate protein was incubated with 100 μM [32P]NAD and 50 nM MTX fragment for various time periods at room temperature in the presence of 1 mM dithiothreitol-2 mM MgCl2-50 mM Tris-HCl (pH 7.4) in a total volume of 20 μl. Toxin-free control experiments were performed in the presence of the storage buffer for MTX proteins (thrombin cleavage buffer). For the time course experiment, 300 nM recombinant EF-Tu and 100 nM MTX30-264 were used to achieve complete ADP ribosylation of EF-Tu. The reaction was stopped by the addition of Laemmli buffer and heating for 5 min at 95°C, and the samples were subsequently subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) by the methods of Laemmli (13). [32P]ADP-ribosylated proteins were detected with a PhosphorImager from Molecular Dynamics. Quantification of PhosphorImager data was performed with ImageQuant software.
Two-dimensional gel electrophoresis.
A [32P]ADP ribosylation assay with E. coli cell lysate (150 μg of total protein) and 1 μM MTX30-264 was performed in a total volume of 200 μl, followed by isoelectric focusing (IEF) with Immobiline DryStrips (Amersham Pharmacia Biotech), pHs 3 to 10, for first-dimension separation. IEF was carried out with a MultiphorII apparatus as described in the Amersham Pharmacia Biotech manual. In the second dimension, proteins were separated by SDS-PAGE and stained with Coomassie blue. Radiolabeled proteins were detected by autoradiography.
Mass spectrometry.
The radioactively labeled proteins were excised from the gel and destained for 1 h at 50°C in 40% acetonitrile-60% ammonium hydrogen carbonate (50 mM, pH 7.8) to remove stain, gel buffer, SDS, and salts. The gel plugs were subsequently dried by vacuum centrifugation. Thereafter, 20 μl of ammonium hydrogen carbonate solution containing 0.2 μg of trypsin was added, and digestion was carried out for 12 h at 37°C.
A saturated matrix solution of 4-hydroxy-α-cyanocinnamic acid in a 1:1 solution of acetonitrile- aqueous 0.1% trifluoroacetic acid was prepared and mixed with 2 μl of the proteolytic peptide mixture in equal parts. For internal calibration, 5 pmol of a human adrenocorticotropic hormone (18 to 39 amino acids) (mass, 2,465.20 Da; Sigma) and 5 pmol of human angiotensin II (mass, 1,046.54 Da; Sigma) were added to the matrix solution. Using the dried-droplet method of matrix crystallization, 1 μl of the sample matrix solution was placed on the mass spectrometer target and dried at room temperature, resulting in a fine granular matrix layer.
Matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry (MS) was performed with a Bruker Biflex mass spectrometer equipped with a nitrogen laser (λ = 337 nm) to desorb and ionize the samples. Mass spectra were recorded in the reflector-positive mode in combination with delayed extraction. The computer program ProFound (Rockefeller University, New York, N.Y.) was used for protein identification. This program matches measured peptide masses to virtually trypsin-digested proteins in a database (peptide mapping).
Characterization of the ternary complex aminoacyl-tRNAEF-TuGTP.
Cellular EF-TuGDP (3 μM) was ADP-ribosylated by MTX30-264 (500 nM) in the presence of 100 μM NAD essentially as described above. ADP-ribosylated EF-Tu-GTP (final concentration, 1 μM) was prepared by incubation with 4 mM GTP, 5 mM phosphoenolpyruvate, and 250 μg of pyruvate kinase/ml in 40 μl of buffer A for 30 min at 37°C. Purified [14C]Phe-tRNAPhe (0.5 μM; 942 dpm/pmol) in 20 μl of buffer A was added, and the reaction mixture was placed on ice for 10 min. The reaction was carried out at 37°C, and at various time points, 10 μl of the reaction solution was transferred to trichloroacetic acid-soaked Whatman paper. After extensive washing with 5% trichloroacetic acid, the filter paper was dried, 2.5 ml of scintillation mixture was added, and nonhydrolyzed [14C]Phe-tRNAPhe was measured with a scintillation counter (Packard RT2500). Control assays were performed with wild-type EF-Tu but without ADP-ribosylation, with EF-Tu treated with catalytically defective MTX30-264E197Q (20), and in the absence of EF-Tu.
Binding of mant-GTP to ADP-ribosylated EF-Tu.
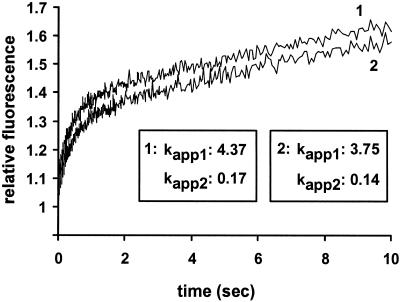
EF-TuGDP (4 μM) was incubated with 100 μM NAD and 500 nM MTX30-264. Next, ADP-ribosylated EF-Tu was incubated with 1 mM phosphoenolpyruvate and 100 μg of pyruvate kinase/ml in buffer A for 30 min at 37°C to form the ADP-ribosylated EF-TuGTP complex. The exchange of GTP with mant-GTP was accomplished by rapidly mixing 60 μl of the EF-Tu-GTP solution (1 μM) with 60 μl of a mant-GTP solution (10 μM) by using the stopped-flow technique (SX-18MV spectrometer; Applied Photophysics). The fluorescence of mant-GTP was excited via fluorescence resonance energy transfer from tryptophan 184 in EF-Tu excited at 280 nm and measured after passage through KV408 filters (Schott). Experiments were performed at 20°C, and the change in fluorescence was recorded over 10 s. Time courses (see Fig. 4) were obtained by averaging six or seven individual transients. The fluorescence at time zero was set to 1, and data were evaluated by fitting to a two-exponential function with characteristic apparent time constants (kapp) and amplitudes (A) according to the equation F = 1 + [A1 × exp(−kapp1 × t)] + [A2 × exp(−kapp2 × t)], where F is the fluorescence at time t. Calculations were performed by using SigmaPlot (Jandel Scientific).
FIG. 4.
mant-GTP binding of ADP-ribosylated EF-Tu (line 1) and native EF-Tu (line 2). The change in the fluorescence of mant-GTP upon binding of EF-Tu was monitored by using the stopped-flow technique. EF-Tu-GTP and mant-GTP solutions were preincubated with phosphoenolpyruvate and pyruvate kinase to minimize contamination with GDP. The values for kapp1 and kapp2 were calculated by fitting the data from the time courses to a two-exponential function.
Determination of the acceptor amino acid of MTX.
Platelet cytosol (150 μg) was incubated with Clostridium limosum C3 toxin (250 nM) and Clostridium botulinum C2 toxin (250 nM) in the presence of 100 μM [32P]NAD- 2 mM MgCl2- 1 mM dithiothreitol- 5 mM thymidine for 30 min at 37°C in a total volume of 200 μl. Recombinant EF-Tu (1.5 μM) was incubated with 250 nM MTX30-264 in the presence of 100 μM [32P]NAD- 2 mM MgCl2-1 mM dithiothreitol for 30 min at room temperature in a total volume of 200 μl. The reactions were stopped by adding SDS to a final concentration of 2%. The reaction mixtures were then divided into equal parts and incubated with either 0.5 M NaCl for 4 h or 0.5 M hydroxylamine (pH 7.5) for 2 or 4 h at 37°C. After precipitation of proteins with chloroform-methanol, the samples were subjected to SDS-PAGE and analyzed by phosphorimaging.
RESULTS
ADP-ribosylation of multiple proteins in E. coli cell lysates.
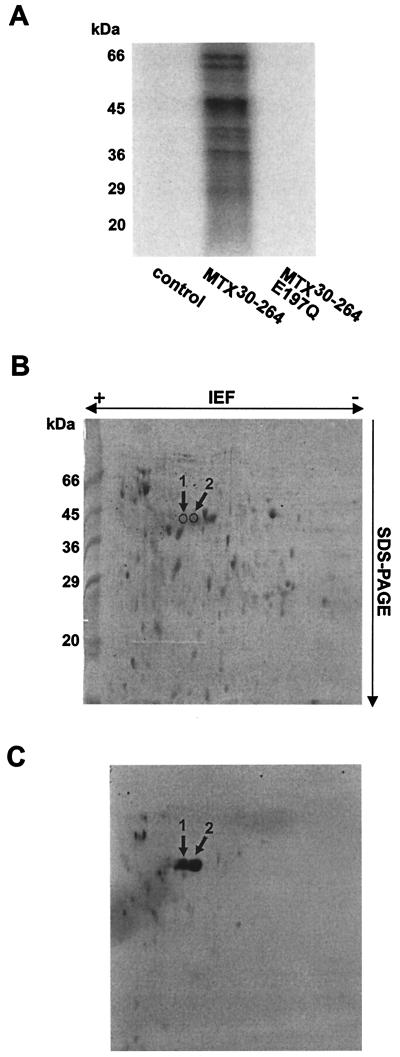
To understand why ADP-ribosylation by MTX30-264 is toxic for E. coli (20), possible protein targets of MTX30-264 were identified in E. coli cell lysates. ADP-ribosylation of total E. coli cell lysates with MTX30-264 in the presence of [32P]NAD labeled various proteins, as detected by SDS-PAGE and subsequent phosphorimaging (Fig. 1A). The labeling was dependent on MTX30-264 enzyme activity. Labeling was not detected in the absence of MTX30-264 or in the presence of catalytically inactive MTX30-264E197Q. Two-dimensional gel electrophoresis (Fig. 1B) of the 32P-labeled proteins showed that while several proteins were ADP-ribosylated by MTX30-264, two substrates of ∼45 kDa were predominantly radiolabeled (Fig. 1C).
FIG. 1.
[32P]ADP-ribosylation of E. coli proteins by MTX30-264. (A) Total E. coli cell lysates (12 μg) were incubated with thrombin cleavage buffer (control), MTX30-264 (50 nM), or MTX30-264E197Q (50 nM) in the presence of 100 μM [32P]NAD. Proteins were separated by SDS-PAGE, and labeled proteins were detected by phosphorimaging (shown). (B and C) E. coli cell lysates (150 μg) were incubated with MTX30-264 (1 μM) and [32P]NAD (100 μM) and subjected to two-dimensional gel electrophoresis. Proteins were detected with Coomassie blue (B) and then subjected to autoradiography (C). Two ∼45-kDa proteins (arrows) were excised from the gel and analyzed by MS, resulting in the identification of EF-Tu (Table 1) as a protein target for MTX.
Identification of EF-Tu as a target protein of MTX.
The ADP-ribosylated ∼45-kDa proteins from E. coli lysates were excised from the SDS gel and digested with trypsin. The resulting peptides were analyzed by MALDI-TOF MS. By use of the computer program ProFound, measured peptides of both proteins were assigned to E. coli EF-Tu (SWISS-PROT accession number P02990) with a sequence coverage of 25% (Table 1). It was not possible to elucidate the difference between the two EF-Tu protein spots by MALDI-TOF MS, as the measured peptide masses were the same. However, the differences might have arisen from altered mobility during IEF when EF-Tu was bound to different ligands.
TABLE 1.
Peptide dataa
| Peptide mass (Da) | Position in sequence | Amino acid sequence | |
|---|---|---|---|
| Measured | Calculated | ||
| 836.56 | 836.49 | 117-123 | EHILLGR |
| 1,026.54 | 1,026.59 | 270-279 | AGENVGVLLR |
| 1,128.60 | 1,128.66 | 280-288 | GIKREEIER |
| 1,217.54 | 1,217.56 | 177-187 | ALEGDAEWEAK |
| 1,375.65 | 1,375.63 | 45-56 | AFDQIDNAPEEK |
| 1,656.94 | 1,656.89 | 238-252 | VGEEVEIVGIKETQK |
| 1,802.89 | 1,802.88 | 59-74 | GITINTSHVEYDTPTR |
| 1,963.96 | 1,963.95 | 155-171 | ELLSQYDFPGDDTPIVR |
Protein spot 1 (Fig. 1) was identified as E. coli EF-Tu by MALDI-TOF MS and subsequent peptide mapping (ProFound). Monoisotopic masses are shown. Mass differences between measured and calculated peaks were below 0.1 Da. The sequence coverage of EF-Tu was 25%. Analysis of protein spot 2 yielded identical peptide masses, so that it represented EF-Tu as well.
Mono-ADP-ribosylation of purified EF-Tu.
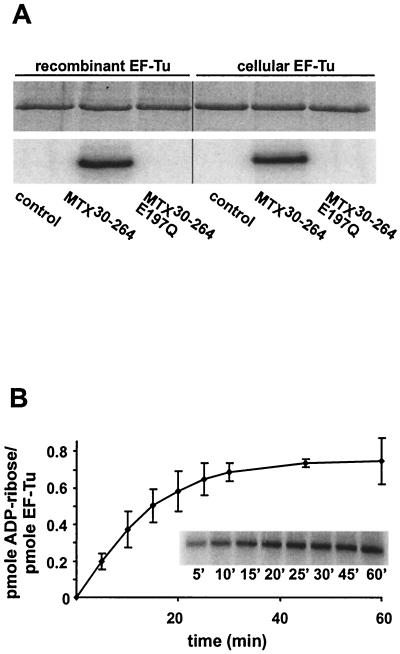
To verify that EF-Tu is a substrate for MTX30-264-catalyzed ADP-ribosylation, cellular EF-Tu-GDP and recombinant EF-Tu-GDP were tested in an ADP-ribosylation assay with MTX30-264. Both cellular EF-Tu-GDP and recombinant EF-Tu-GDP were targets for ADP-ribosylation by MTX30-264 (Fig. 2A), whereas no labeling was detected in the absence of MTX30-264 or after treatment of EF-Tu with catalytically inactive MTX30-264E197Q (Fig. 2A). A difference in the efficiency of ADP-ribosylation was not observed when EF-Tu-GTP was used instead of EF-Tu-GDP (data not shown).
FIG. 2.
[32P]ADP ribosylation of cellular EF-Tu and recombinant EF-Tu. (A) Cellular EF-Tu and recombinant EF-Tu (2.5 μM) were incubated alone (control) or with MTX30-264 (50 nM) or MTX30-264E197Q (50 nM) in the presence of 100 μM [32P]NAD for 30 min at room temperature. Proteins were separated by SDS-PAGE (upper panel), and labeling was detected by phosphorimaging (lower panel). Quantification of [32P]ADP-ribose incorporation revealed that ∼50% of the total amount of EF-Tu used in this assay was modified. (B) Recombinant EF-Tu (300 nM) was incubated with 100 nM MTX30-264 in the presence of 100 μM [32P]NAD in a total volume of 20 μl for the indicated times. The incorporation of [32P]ADP-ribose was detected after SDS-PAGE by phosphorimaging, and data were quantified with ImageQuant software. The extent of ADP-ribosylation is indicated as picomoles of ADP-ribose incorporated per picomole of EF-Tu. Data are given as means and standard errors (n = 3). The inset illustrates the phosphorimaging data from a representative experiment.
EF-Tu modification by MTX30-264 was time dependent, with about 0.75 mol of ADP-ribose incorporated per mol of EF-Tu upon completion of the assay (Fig. 2B). Adding more MTX30-264 to the ADP-ribosylation mixture after 45 min of incubation did not increase the incorporation of ADP-ribose, indicating saturation of the reaction.
Functional effects of EF-Tu ADP-ribosylation.
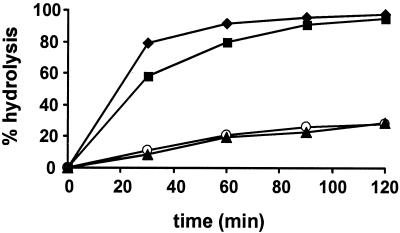
EF-Tu in its GTP-bound state forms a stable complex with aminoacyl-tRNA, protecting the 3′end of aminoacyl-tRNA from hydrolysis (28). ADP-ribosylation of EF-Tu prevented the formation of a stable ternary complex with GTP and aminoacyl-tRNA, as indicated by the rapid hydrolysis of [14C]Phe-tRNAPhe (Fig. 3). Similar time courses of [14C]Phe-tRNAPhe hydrolysis were observed in the absence of EF-Tu or in the presence of ADP-ribosylated EF-Tu, indicating that ADP-ribosylated EF-Tu is inactive in aminoacyl-tRNA binding. EF-Tu treated with catalytically inactive MTX30-264E197Q was as active as native EF-Tu in protecting [14C]Phe-tRNAPhe from hydrolysis (Fig. 3).
FIG. 3.
ADP-ribosylation of EF-Tu prevents the formation of the stable ternary EF-TuGTPaminoacyl-tRNA complex. [14C]Phe-tRNAPhe (0.5 μM) was incubated with 1 μM EF-Tu-GTP ADP-ribosylated by MTX30-264 (⧫) or 1 μM native EF-Tu-GTP (▴) in a total volume of 60 μl. As a control, the assays were performed in the absence of EF-Tu (▪) and in the presence of EF-Tu treated with catalytically inactive MTX30-264E197Q (○). At the indicated times, 10-μl aliquots of the reaction mixtures were transferred to trichloroacetic acid-soaked filter paper, and the amount of precipitated [14C]Phe-tRNAPhe was quantified by scintillation counting. Time zero was set to 0% hydrolysis.
Binding of mant-GTP to ADP-ribosylated EF-Tu is not altered.
The nucleotide binding of ADP-ribosylated EF-Tu to mant-GTP, a fluorescent GTP derivative, was studied by using fluorescence resonance energy transfer from Trp184 in EF-Tu (27). Binding of the nucleotide to native or ADP-ribosylated EF-Tu increased mant-GTP fluorescence. To verify that EF-Tu used in this assay was fully ADP-ribosylated, aliquots were tested in the hydrolysis protection assay described above. The time courses of nucleotide exchange were identical for native EF-Tu and ADP-ribosylated EF-Tu (Fig. 4). Calculation of the apparent time constants (kapp) from the time courses indicated that there was no significant difference in the velocity of nucleotide exchange between native EF-Tu and ADP-ribosylated EF-Tu (Fig. 4). Therefore, we concluded that ADP-ribosylation does not alter the GTP-binding properties of EF-Tu.
An arginine residue in EF-Tu is modified by MTX.
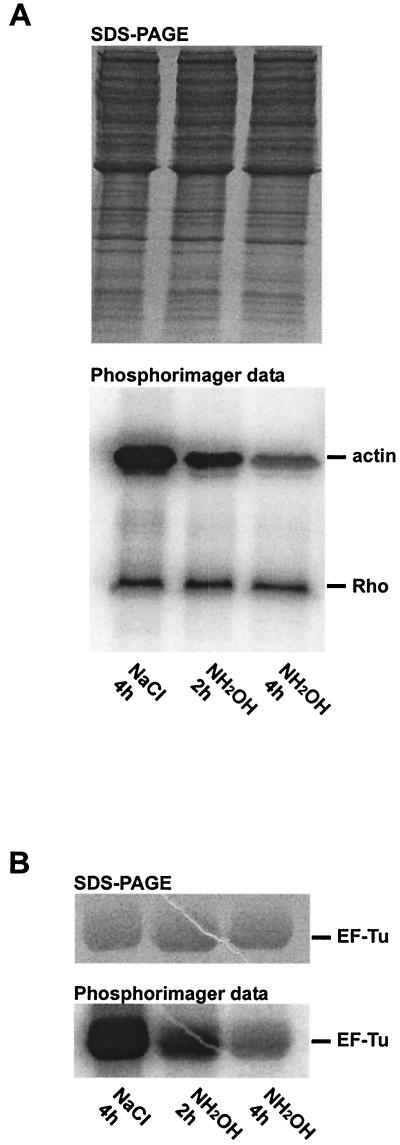
ADP- ribose-amino acid bonds can be characterized by their stability toward neutral 0.5 M hydroxylamine (1, 3). Whereas the ADP- ribose bond to an aspartate or cysteine residue is stable toward hydroxylamine, the ADP-ribose bond to an arginine residue has a half-life of ∼2 h (1, 12). The ADP-ribosylation of the small GTPase Rho (Asn41) by C3 exoenzyme (21) and that of actin (Arg177) by C2 toxin (24) were used for comparison. These toxins exclusively modify the Rho protein and actin, respectively. As shown in Fig. 5A, a time-dependent decrease in the signal of ADP-ribosylated actin was observed after 2 and 4 h of treatment with hydroxylamine, whereas the signal for ADP-ribosylated Rho remained largely unchanged. The signal intensity of ADP-ribosylated EF-Tu decreased after incubation with hydroxylamine, suggesting that an arginine residue in EF-Tu was modified (Fig. 5B).
FIG. 5.
Determination of the acceptor amino acid of MTX30-264. The stability of [32P]ADP-ribose-amino acid bonds formed by MTX30-264, C. botulinum C2 toxin (control), and Clostridium limosum C3 exoenzyme towards NH2OH was examined. (A) Platelet cytosol was incubated with C2 toxin and C3 exoenzyme in the presence of [32P]NAD. The reaction mixture was divided into equal parts andtreated with 0.5 M NaCl for 4 h or with 0.5 M NH2OH (pH 7.5) for 2 and 4 h. Proteins were separated by SDS-PAGE (upper panel), and labeled Rho and actin were detected by phosphorimaging (lower panel). (B) EF-Tu was incubated with MTX30-264 in the presence of [32P]NAD. The reaction mixture was divided into equal parts and treated with 0.5 M NaCl for 4 h or with 0.5 M NH2OH (pH 7.5) for 2 and 4 h. EF-Tu was analyzed by SDS-PAGE (upper panel) and phosphorimaging (lower panel).
DISCUSSION
In this study, we report on a target protein of MTX whose inactivation by ADP-ribosylation may play an important role in cytotoxicity toward E. coli. Separation of [32P]ADP-ribosylated E. coli proteins by two-dimensional gel electrophoresis and detection of modified proteins by autoradiography revealed two dominantly labeled proteins with apparent masses of ∼45 kDa. Both protein spots were identified as E. coli EF-Tu. Cellular EF-Tu and recombinant EF-Tu were ADP-ribosylated by MTX30-264, supporting the MS findings. ADP-ribosylation of EF-Tu by MTX30-264 led to the incorporation of about one ADP-ribose residue per molecule of EF-Tu.
EF-Tu is a bacterial translation elongation factor that delivers aminoacyl-tRNA to the A site of the ribosome during the elongation cycle of protein synthesis. ADP-ribosylation of EF-Tu prevents the formation of the stable ternary complex with GTP and aminoacyl-tRNA due to a deficiency in aminoacyl-tRNA binding. This functional inactivation of EF-Tu leads to the inhibition of protein synthesis and is likely to result in the retardation of cell growth and lethality. However, MTX30-264 ADP-ribosylated several substrates in E. coli cell lysates, an action which may have contributed to the cytotoxic effect.
All biglutamic acid ADP-ribosyltransferases so far studied attach ADP-ribose to arginine residues of their protein substrates (15). The instability of the ADP-ribose-amino acid bond in EF-Tu toward neutral hydroxylamine suggests the modification of arginine residues by MTX30-264-mediated ADP-ribosylation as well. ADP-ribose-asparagine bonds or ADP-ribose-cysteine bonds are stable toward neutral hydroxylamine, and so these amino acids can therefore be ruled out as acceptor amino acids (1, 5, 16). The exact arginine residue that is targeted by MTX30-264 in EF-Tu is not known yet, but possible candidates are currently under investigation.
Most ADP-ribosyltransferases are specific; i.e., they have only one specific eukaryotic target protein and exhibit no toxicity toward their host organisms. In contrast, MTX30-264 ADP- ribosylates several proteins in E. coli lysates. Also, in HeLa cell lysates (20) as well as in other eukaryotic cell lysates (unpublished observations), MTX30-264 modifies numerous proteins. Furthermore, Thanabalu et al. (22) reported that a nonactivated N-terminal MTX truncation (amino acids 30 to 493) modifies two proteins with apparent masses of 42 and 38 kDa in lysates of a Culex quinquefasciatus cell line. In contrast, the active MTX30-264 fragment used in our experiments led to the labeling of a large array of proteins in lysates of the same C. quinquefasciatus cell line, with predominant labeling of a 55-kDa protein (unpublished data).
So far, none of the labeled proteins in cell lysates other than those from E. coli could be assigned to an elongation factor. As reported previously (20), MTX30-264 leads to the rounding up of HeLa cells and the formation of actin-containing protrusions after 12 h. This effect is unlikely to be mediated by an alteration of the function of an elongation factor. Therefore, at least in HeLa cells, different protein targets of MTX seem to contribute to cytotoxicity. However, we cannot exclude the possibility that elongation factors may serve as additional substrates in eukaryotic cells as well. With respect to the sequence similarity between E. coli EF-Tu and the functionally equivalent human elongation factor 1 alpha (45% at the amino acid level), it seems possible that the latter serves as a substrate for MTX as well. Currently, investigations are ongoing to elucidate relevant substrates for MTX in eukaryotic cells.
ExoS from P. aeruginosa is another bacterial ADP-ribosyltransferase which modifies an array of proteins, with Ras as a preferred target (7). Furthermore, ExoS also targets numerous proteins in eukaryotic cells in vivo (17). However, the enzyme activity of ExoS depends on the presence of the eukaryotic 14-3-3 protein FAS (6, 9). The need for a eukaryotic activation factor may prevent possible harmful ADP-ribosylation activity in the bacterial host cell. The necessity for MTX enzyme activation by proteolytic cleavage may represent a protection mechanism for the toxin-producing B. sphaericus strain. Thus, in contrast to ADP-ribosyltransferases, which target specific proteins, the less specific toxins, such as MTX and ExoS, may use unique enzyme activation mechanisms to suppress toxicity in bacteria. Further investigations are necessary to elucidate the role of the putative binding component of MTX in the regulation of ADP-ribosyltransferase activity and target specifity. However, so far we have not found any differences in target specifity between MTX30-264 and the activated MTX holotoxin in vitro.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 388) and by the Fonds der Chemische Industrie.
We thank Kirill Gromadski for help with the stopped-flow experiments.
REFERENCES
- 1.Aktories, K., I. Just, and W. Rosenthal. 1988. Different types of ADP-ribose protein bonds formed by botulinum C2 toxin, botulinum ADP-ribosyltransferase C3 and pertussis toxin. Biochem. Biophys. Res. Commun. 156:361-367. [DOI] [PubMed] [Google Scholar]
- 2.Braun, U., B. Habermann, I. Just, K. Aktories, and J. Vandekerckhove. 1989. Purification of the 22-kDa protein substrate of botulinum ADP-ribosyltransferase C3 from porcine brain cytosol and its characterization as a GTP-binding protein highly homologous to the rho gene product. FEBS Lett. 243:70-76. [DOI] [PubMed] [Google Scholar]
- 3.Bredehorst, R., K. Wielckens, A. Gartemann, H. Lengyel, and K. Klapproth. 1978. Two different types of bonds linking single ADP-ribose residues covalently to proteins. Eur. J. Biochem. 92:129-135. [DOI] [PubMed] [Google Scholar]
- 4.Cassel, D., and T. Pfeuffer. 1978. Mechanism of cholera toxin action: covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc. Natl. Acad. Sci. USA 75:2669-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cervantes-Laurean, D., P. T. Loflin, D. E. Minter, E. L. Jacobson, and M. K. Jacobson. 1995. Protein modification by ADP-ribose via acid-labile linkages. J. Biol. Chem. 270:7929-7936. [DOI] [PubMed] [Google Scholar]
- 6.Coburn, J., A. V. Kane, L. Feig, and D. M. Gill. 1991. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J. Biol. Chem. 266:6438-6446. [PubMed] [Google Scholar]
- 7.Coburn, J., R. T. Wyatt, B. H. Iglewski, and D. M. Gill. 1989. Several GTP-binding proteins, including p21 c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J. Biol. Chem. 264:9004-9008. [PubMed] [Google Scholar]
- 8.Domenighini, M., M. Pizza, and R. Rappuoli. 1995. Bacterial ADP-ribosyltransferases, p. 59-80. In J. Moss, B. Iglewski, M. Vaughan, and A. T. Tu (ed.), Bacterial toxins and virulence factors in disease. Marcel Dekker, Inc., New York, N.Y.
- 9.Fu, H., J. Coburn, and R. J. Collier. 1993. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc. Natl. Acad. Sci. USA 90:2320-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganesan, A. K., D. W. Frank, R. P. Misra, G. Schmidt, and J. T. Barbieri. 1998. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J. Biol. Chem. 273:7332-7337. [DOI] [PubMed] [Google Scholar]
- 11.Gromadski, K. B., H. J. Wieden, and M. V. Rodnina. 2002. Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry 41:162-169. [DOI] [PubMed] [Google Scholar]
- 12.Hsia, J. A., S.-C. Tsai, R. Adamik, D. A. Yost, E. L. Hewlett, and J. Moss. 1985. Amino acid-specific ADP-ribosylation. J. Biol. Chem. 260:16187-16191. [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Locht, C., and R. Antoine. 1997. Pertussis toxin, p. 33-45. In K. Aktories (ed.), Bacterial toxins: tools in cell biology and pharmacology. Chapman & Hall, Weinheim, Germany.
- 15.Masignani, V., M. Pizza, and R. Rappuoli. 2000. Common features of ADP-ribosyltransferases, p. 21-44. In G. V. R. Born, D. Ganten, H. Herken, K. Starke, and P. Taylor (ed.), Handbook of experimental pharmacology. Springer-Verlag KG, Berlin, Germany.
- 16.Mayer, T., R. Koch, W. Fanick, and H. Hilz. 1988. ADP-ribosyl proteins formed by pertussis toxin are specifically cleaved by mercury ions. Biol. Chem. Hoppe-Seyler 369:579-583. [DOI] [PubMed] [Google Scholar]
- 17.Riese, M. J., U.-M. Goehring, M. E. Ehrmantraut, J. Moss, J. T. Barbieri, K. Aktories, and G. Schmidt. 2002. Auto-ADP-ribosylation of Pseudomonas aeruginosa ExoS. J. Biol. Chem. 277:12082-12088. [DOI] [PubMed] [Google Scholar]
- 18.Rodnina, M. V., R. Fricke, and W. Wintermeyer. 1994. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry 33:12267-12275. [DOI] [PubMed] [Google Scholar]
- 19.Rubin, E. J., D. M. Gill, P. Boquet, and M. R. Popoff. 1988. Functional modification of a 21-kilodalton G protein when ADP ribosylated by exoenzyme C3 of Clostridium botulinum. Mol. Cell. Biol. 8:418-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schirmer, J., I. Just, and K. Aktories. 2002. The ADP-ribosylating mosquitocidal toxin (MTX) from Bacillus sphaericus—proteolytic activation, enzyme activity and cytotoxic effects. J. Biol. Chem. 277:11941-11948. [DOI] [PubMed] [Google Scholar]
- 21.Sekine, A., M. Fujiwara, and S. Narumiya. 1989. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. J. Biol. Chem. 264:8602-8605. [PubMed] [Google Scholar]
- 22.Thanabalu, T., C. Berry, and J. Hindley. 1993. Cytotoxicity and ADP-ribosylating activity of the mosquitocidal toxin from Bacillus sphaericus SSII-1: possible roles of the 27- and 70-kilodalton peptides. J. Bacteriol. 175:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandekerckhove, J., B. Schering, M. Bärmann, and K. Aktories. 1987. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett. 225:48-52. [DOI] [PubMed] [Google Scholar]
- 24.Vandekerckhove, J., B. Schering, M. Bärmann, and K. Aktories. 1988. Botulinum C2 toxin ADP-ribosylates cytoplasmic β/g-actin in arginine 177. J. Biol. Chem. 263:696-700. [PubMed] [Google Scholar]
- 25.Van den Akker, F., E. A. Merritt, and W. G. J. Hol. 2000. Structure and function of cholera toxin and related enterotoxins, p. 109-131. In K. Aktories and I. Just (ed.), Bacterial protein toxins. Springer-Verlag KG, Berlin, Germany.
- 26.Van Ness, B. G., J. B. Howard, and J. W. Bodley. 1980. ADP-ribosylation of elongation factor 2 by diphtheria toxin. J. Biol. Chem. 255:10717-10720. [PubMed] [Google Scholar]
- 27.Wagner, A., I. Simon, M. Sprinzl, and R. S. Goody. 1995. Interaction of guanosine nucleotides and their analogs with elongation factor Tu from Thermus thermophilus. Biochemistry 34:12535-12542. [DOI] [PubMed] [Google Scholar]
- 28.Weijland, A., K. Harmark, R. H. Cool, P. H. Anborgh, and A. Parmeggiani. 1992. Elongation factor Tu: a molecular switch in protein biosynthesis. Mol. Microbiol. 6:683-688. [DOI] [PubMed] [Google Scholar]
- 29.West, R. E., J. Moss, M. Vaughan, T. Liu, and T.-Y. Liu. 1985. Pertussis toxin-catalyzed ADP-ribosylation of transducin. J. Biol. Chem. 260:14428-14430. [PubMed] [Google Scholar]
- 30.Wilde, C., G. S. Chhatwal, G. Schmalzing, K. Aktories, and I. Just. 2001. A novel C3-like ADP-ribosyltransferase from Staphylococcus aureus modifying RhoE and Rnd3. J. Biol. Chem. 276:9537-9542. [DOI] [PubMed] [Google Scholar]