Abstract
Flow cytometry was used to monitor changes in the DNA content of the polychlorinated biphenyl (PCB)-degrading bacterium Comamonas testosteroni TK102 during growth in the presence or absence of PCBs. In culture medium without PCBs, the majority of stationary-phase cells contained a single chromosome. In the presence of PCBs, the percentage of cells containing two chromosomes increased from 12% to approximately 50%. In contrast, addition of PCBs did not change the DNA contents of three species that are unable to degrade PCBs. In addition, highly chlorinated PCBs that are not degraded by TK102 did not result in a change in the DNA content. These results suggest that PCBs did not affect the DNA content of the cells directly; rather, the intermediate metabolites resulting from the degradation of PCBs caused the increase in DNA content. To study the effect of intermediate metabolites on the DNA content of the cells, four bph genes, bphA1, bphB, bphC, and bphD, were disrupted by gene replacement. The resulting mutant strains accumulated intermediate metabolites when they were grown in the presence of PCBs or biphenyl (BP). When the bphB gene was disrupted, the percentage of cells containing two chromosomes increased in cultures grown with PCBs or BP. When grown with BP, cultures of this mutant accumulated two intermediate metabolites, 2-hydroxybiphenyl (2-OHBP) and 3-OHBP. Addition of 2- or 3-OHBP to a wild-type TK102 and non-PCB-degrading species culture also resulted in an increase in the percentage of cells containing two chromosomes. Electron microscopy revealed that cell-cell separation was inhibited in this culture. This is the first report that hydroxy-BPs can inhibit bacterial cell separation while allowing continued DNA replication.
Polychlorinated biphenyls (PCBs) are ubiquitous and recalcitrant environmental pollutants. Since Ahmed and Focht (1) described two species of Achromobacter capable of degrading mono- and dichlorobiphenyls, many other studies on the aerobic microbial degradation of PCBs have been reported (3, 12, 15). Comamonas testosteroni (formerly Pseudomonas alcaligenes) TK102, a PCB degrader, was isolated from soil contaminated with PCBs (32). TK102 can degrade almost all mono-, di-, and trichlorobiphenyls, as well as some tetrachlorobiphenyls, but cannot degrade PCBs that have five or more chlorine atoms. Therefore, TK102 can degrade a commercial PCB mixture, Kaneclor 300, which mainly consists of trichlorobiphenyls. Efficient degradation of Kaneclor 300 by TK102 has been achieved in batch reactors during the initial 24 h, after which the PCB degradation efficiency of the cells decreased (33).
There is evidence that the heterogeneity of growing bacterial cells in batch cultures can be high. Microbial heterogeneity may arise from three principal sources: phenotypic changes during the cell cycle, changes in the microenvironments of individual cells, and genotypic changes resulting from mutations (9, 43). Thus, growth-associated parameters, such as DNA or protein content, are nonhomogeneous in a population. Because the overall PCB-degrading activity depends on the contribution of individual cells, these population dynamics have profound implications on the efficiency of PCB degradation. Flow cytometry (FCM) can be used for measuring such dynamics.
FCM is a tool with great potential for performing both qualitative and quantitative analyses based on simultaneous measurements of structural and functional parameters of individual cells (9). These measurements are based on light scatter, which reflects cell size and structure, and fluorescence, which can reflect DNA content, enzyme activity, respiration, membrane potential, or membrane integrity depending on the fluorescent dye being used (18, 20, 21). In combination with DNA fluorescent dyes, FCM has been widely used to measure DNA content and analyze the cell cycles of eukaryotes (8, 13, 39), prokaryotes (6, 10, 37), and archaea (4). Escherichia coli has been the most extensively used organism in studies of the bacterial cell cycle. In slowly growing E. coli cells, the cell cycle is similar to that of eukaryotes (36, 37). The DNA content distribution exhibits a peak at one chromosome, which represents the B-period cells (G1 phase in eukaryotes), a ridge between one and two chromosomes, which represents the replicating C-period cells (S phase in eukaryotes), and a peak at two chromosomes, which represents the D-period cells (G2 phase in eukaryotes). However, the DNA distributions of rapidly growing cells are more complex than those of slowly growing cells because of overlapping replication cycles (37). To determine the number of chromosomes in each cell at a given time, drug treatment has been used (37). Rifampin blocks initiation of chromosome replication while allowing ongoing rounds of replication to continue to termination, and cephalexin prevents cell division, which results in replication runout and accumulation of cells with integer numbers of chromosomes (37, 42). After drug treatment, cells in a rapidly growing population contain several chromosomes (e.g., four or eight fully replicated chromosomes). In contrast, drug-treated cells in a slowly growing population contain one or two chromosomes. Thus, the DNA content distribution is very sensitive to the growth conditions of the culture. To our knowledge, there have been no reports of assessments of the effects of PCBs on the cell growth of PCB-degrading bacteria or growth-associated parameters, such as DNA content.
In this study, we examined changes in cell growth or the DNA content of C. testosteroni TK102 cells in the presence or absence of PCBs. The effects of PCBs and their metabolites on cell-cell separation are also discussed in this paper.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. C. testosteroni strain TK102, which was isolated from PCB-contaminated soil (32), was grown and maintained at 30°C on phosphate-buffered minimal salt medium (19) with biphenyl (BP) as the sole carbon source. Four mutants of TK102 with disruptions in bph genes (bphA1, bphB, bphC, and bphD) (see below) were grown on threefold-diluted Luria-Bertani (1/3LB) medium (33) with 300 μg of kanamycin per ml at 30°C. Pseudomonas putida PpY101 and Pseudomonas aeruginosa PAO1 were grown on 1/3LB medium at 30°C. E. coli MV1184 was grown on 1/3LB medium at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference(s) |
|---|---|---|
| Strains | ||
| C. testosteroni TK102 | 32 | |
| E. coli MV1184 | ara Δ(lac-pro) str thi θ80 lacIZΔM15 Δ(srl-recA)::Tn10 (F′ traD proAB lacIqZΔM15) | 41 |
| P. aeruginosa PAO1 | Wild type | 35 |
| P. putida PpY101 | met nal | 14 |
| Plasmids | ||
| pUC18 | Cloning vector, Apr | 23, 41 |
| pBluescript II KS(+) | Cloning vector, Apr | 34 |
| pUC4K | Apr Kmr, source of Kmr cartridge | 40, 41 |
| pTKSm10 | pUC18 with a 10-kb SmaI fragment of TK102 carrying bph genes | This study |
| pTKScSI | pUC18 carrying a 2.5-kb SacI-SalI fragment (bphA1) of pTKSm10 | This study |
| pTKKpCI | pBluescript II KS(+) carrying a 2.7-kb KpnI-ClaI fragment (bphB) of pTKSm10 | This study |
| pTKSlSc | pBluescript II KS(+) carrying a 2.1-kb SalI-SacI fragment (bphC) of pTKSm10 | This study |
| pTKXSm | pBluescript II KS(+) carrying a 1.9-kb XhoI-SmaI fragment (bphD) of pTKSm10 | This study |
Apr, ampicillin resistance; Kmr, kanamycin resistance.
Mutant construction.
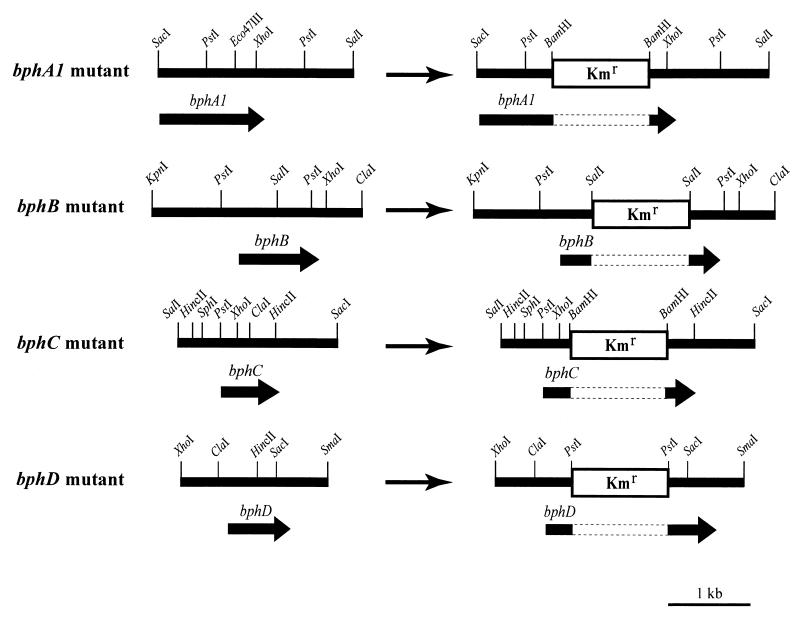
Standard molecular biology techniques, such as ligation and Southern hybridization, were carried out as described by Sambrook et al. (28). Restriction enzymes were used according to the manufacturer's instructions. Construction of the bph mutants was carried out by gene replacement (Fig. 1). A 10-kb SmaI fragment containing the bph genes from TK102 was identified by Southern hybridization by using a 1-kb XhoI-HindIII fragment containing the bphCD genes from Pseudomonas sp. strain KKS102 as the probe (19). A clone bank of SmaI-digested DNA fragments approximately 8 to 12 kb long was constructed in pUC18. This clone bank was screened by colony hybridization by using the same probe in order to isolate a clone containing the bph genes from TK102. The resulting clone was designated pTKSm10. Sequence analysis revealed that the bphA1, bphA2, bphA3, bphB, bphC, and bphD genes of TK102 were homologous to the bph genes of KKS102 (87 to 94% identity) (data not shown). To construct the bphA1 mutant, the 2.5-kb SacI-SalI fragment containing the bphA1 gene from pTKSm10 was inserted into the cloning site of pUC18 to generate pTKScSl. This recombinant was digested at the unique Eco47III site, and a BamHI linker was inserted. pUC4K was digested with BamHI, and the isolated kanamycin cartridge was inserted into the BamHI site created in the coding region of the bphA1 gene, thus disrupting it. To construct the bphB mutant, the 2.7-kb KpnI-ClaI fragment containing the bphB gene from pTKSm10 was inserted into the cloning site of pBluescript II KS(+) (Stratagene, La Jolla, Calif.) to generate pTKKpCl. pUC4K was digested with SalI, and the isolated kanamycin cartridge was inserted into the SalI site in the coding region of the bphB gene to disrupt it. To construct the bphC mutant, the 2.1-kb SalI-SacI fragment from pTKSm10 was inserted into the cloning site of pBluescript II KS(+) to generate pTKSlSc. This recombinant was digested at the unique ClaI site, which interrupts the coding region of the bphC gene, and a BamHI linker was inserted after blunt ends were generated with the Klenow fragment. A BamHI-digested kanamycin cartridge was inserted into the BamHI site created in the coding region of the bphC gene to disrupt it. To construct the bphD mutant, the 1.9-kb XhoI-SmaI fragment from pTKSm10 was inserted into pBluescript II KS(+), generating pTKXSm. This recombinant was digested at the unique HincII site, which interrupts the coding region of the bphD gene, and a PstI linker was inserted. A PstI-digested kanamycin cartridge was inserted into the PstI site created in the coding region of the bphD gene, thus disrupting it. Each mutant was analyzed by Southern hybridization by using the disrupted gene as a probe. Each of the mutants contained only one hybridizing fragment, indicating that a double-crossover event for the target gene occurred. In all cases, the size of the hybridizing fragment was found to have increased by about 1.2-kb, indicating that the kanamycin cartridge was inserted into the target gene. The plasmids were linearized with appropriate restriction enzymes and introduced into TK102 cells by electroporation. TK102 cells grown in 1/3LB medium to an optical density at 600 nm of 0.5 were washed twice with sterile ice-cold water and resuspended in 1 mM HEPES buffer (pH 7.0) containing 10% glycerol. The cells were then mixed with 200 μg of the linearized DNA fragment and electroporated with a gene pulser (Bio-Rad, Hercules, Calif.) under the following conditions: voltage, 2.5 kV; capacitance, 25 μF; and a 200-Ω pulse controller. Kanamycin-resistant colonies were selected, and gene disruption was confirmed by Southern blot analysis. The resulting mutant strains were used for further study.
FIG. 1.
Insertion of a 1.2-kb kanamycin resistance gene (Kmr) into the bph genes of C. testosteroni TK102.
Changes in the DNA content of C. testosteroni TK102 during growth with and without PCBs.
One milliliter of a preculture of TK102 was inoculated into a 500-ml flask containing 100 ml of 1/3LB medium and incubated with 100 mg of Kaneclor 300 (equivalent to Aroclor 1242) per liter and 50 mg of Triton X-100 per liter at 30°C with shaking at 80 rpm. Cells grown without Kaneclor 300 were used as a control. Growth was measured by determining the optical density at 650 nm with a Beckman DU 650 spectrophotometer. At various times, 1 ml of the culture was collected by centrifugation at 2,500 × g for 5 min. The cell pellet was resuspended in an equal volume of fresh 1/3LB medium containing 150 μg of rifampin (Wako Pure Chemical Industries, Osaka, Japan) per ml and 20 μg of cephalexin (Sigma Chemical Co., St. Louis, Mo.) per ml. The culture was incubated for 2 h. The cells were washed once and resuspended in 100 μl of ice-cold 100 mM Tris-HCl buffer (pH 7.4). One milliliter of 77% ethanol was added to obtain a final concentration of 70%, and cells were stored at 4°C until they were used. Stationary-phase cells (after 24 h of incubation) were fixed in 70% ethanol without the drug treatment described above.
Assessment of the effects of BP, PCBs, and the intermediate metabolites on the DNA content of bacteria.
One milliliter of a preculture of P. putida PpY101, P. aeruginosa PAO1, E. coli MV1184, C. testosteroni TK102, or one of the bph mutants of TK102 described above was inoculated into a 500-ml flask containing 100 ml of 1/3LB medium and incubated with 100 mg of BP, Kaneclor 300, Kaneclor 400, Kaneclor 500, Kaneclor 600, benzoate, o-, m-, or p-chlorobenzoate, 2,3-, 2,4-, 2,5-, 2,6-, 3,4-, or 3,5-dichlorobenzoate, or 2,4,6-trichlorobenzoate per liter, 50 mg of 2-hydroxybiphenyl (2-OHBP) per liter, or 30 mg of 3-OHBP per liter. Fifty milligrams of Triton X-100 per liter was also added to each culture, and incubation was carried out at 30°C for each Comamonas and Pseudomonas culture or at 37°C for the E. coli culture with shaking at 80 rpm. One milliliter of each Comamonas or Pseudomonas culture was collected after 48 h, and 1 ml of each E. coli culture was collected after 24 h. All cultures were fixed in 70% ethanol.
FCM.
Ethanol-fixed cells were washed and resuspended in Tris buffer (pH 7.4) containing 2 μg of RNase (Nippon Gene Co., Toyama, Japan) per ml. Propidium iodide (PI) (1-mg/ml solution; Sigma) was added to a final concentration of 5 μg/ml, and the cells were incubated for 10 min at room temperature in the dark. The cell suspensions were diluted in Tris buffer to a final concentration of 106 cells per ml. FCM was carried out with a FACSCalibur instrument (Becton Dickinson, San Jose, Calif.) equipped with an argon ion laser providing 15 mW at 488 nm. The instrument was equipped with forward scatter (<15°), side scatter (>15°), and three fluorescence detectors, FL1 (530 ± 15 nm), FL2 (585 ± 21 nm), and FL3 (>605 nm). All parameters were collected as linear signals. Fluorescence emission was detected at FL2 for PI. The sheath fluid was FACSFlow (Becton Dickinson). The sample flow rate was set to low (12 μl/min), and at least 10,000 cells were acquired for analysis. Triplicate counts were obtained for each procedure. The performance of the instrument was monitored daily by using CaliBRITE beads (Becton Dickinson).
GC-MS analysis.
In order to identify metabolites of BP produced by the bphB mutant, resting cell assays were performed. The bphB mutant was grown in 1/3LB medium at 30°C for 15 h. Cells were harvested by centrifugation at 2,500 × g and resuspended in phosphate-buffered minimal salt medium (see above) to an optical density at 600 nm of 1.5. Suspensions (2 ml) were incubated with 100 mg of BP per liter for 1 h at 30°C on a gyratory shaker. Metabolites were extracted from the culture with an equal volume of ethyl acetate. The ethyl acetate layer was removed and concentrated under a stream of nitrogen. Detection of metabolites was carried out by using a gas chromatograph-mass spectrometer (GC-MS) (5890 series II GC with a 5971 series MS; Hewlett-Packard Co., Palo Alto, Calif.) equipped with an Ultra-2 capillary column (length, 50 m; diameter, 0.22 mm; thickness, 0.33 μm; Hewlett-Packard Co.) as described previously (32).
Fluorescence microscopy.
Aliquots (10 μl) of PI-stained bacterial suspensions were placed on glass slides under coverslips and observed with a Optiphot-2 inverted epifluorescence microscope (Nikon, Tokyo, Japan) fitted with a 100-W mercury arc lamp, a Nikon G-2A filter (excitation, 510 to 560 nm; emission, >590 nm), and a ×100 oil immersion objective lens. Photographs were obtained with a Nikon Coolpix 950 digital camera. The digital images were processed further by using Photoshop 5.5 (Adobe). At least 300 cells grown with and without Kaneclor 300 were counted for analysis.
Electron microscopy.
Cells grown with and without Kaneclor 300 as described above were collected on a supporting film of Butvar-98 (Monsanto, St. Louis, Mo.) and mounted on a specimen grid after 24 h of incubation. The cells were fixed with 2.5% glutaraldehyde for 1 min, washed with distilled water, and negatively stained with 2% phosphotungstic acid. Specimens were observed with a transmission electron microscope (JEM-2000EX; JEOL, Tokyo, Japan), and 300 cells grown with and without Kaneclor 300 were observed.
Chemicals.
Kaneclor 300, Kaneclor 400, Kaneclor 500, and Kaneclor 600 were obtained from GL Science (Tokyo, Japan). Kaneclor 300 consists mainly of trichlorobiphenyls (equivalent to Aroclor 1242); Kaneclor 400 consists mainly of tetrachlorobiphenyls (equivalent to Aroclor 1248); Kaneclor 500 consists mainly of pentachlorobiphenyls (equivalent to Aroclor 1254); and Kaneclor 600 consists mainly of hexachlorobiphenyls (equivalent to Aroclor 1260). 2- and 3-OHBP were obtained from Tokyo Chemical Industry Co. (Tokyo, Japan). All other chemicals were obtained from Wako Pure Chemical Industries.
Nucleotide sequence accession number.
The bph gene sequences of TK102 have been deposited in the DDBJ database under accession number AB086835.
RESULTS
Changes in the DNA content of C. testosteroni TK102 cells during growth with and without PCBs.
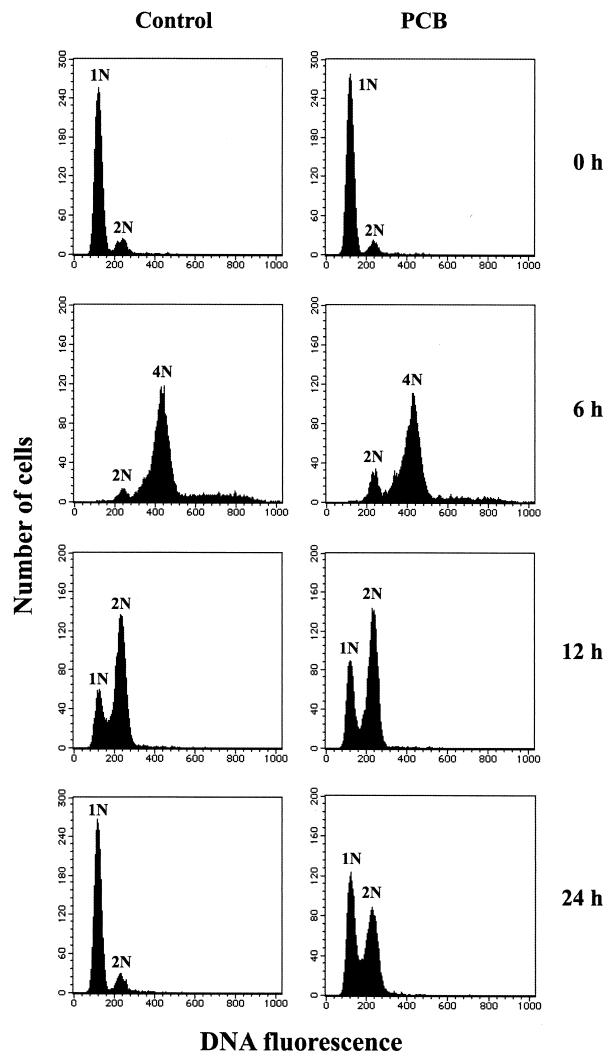
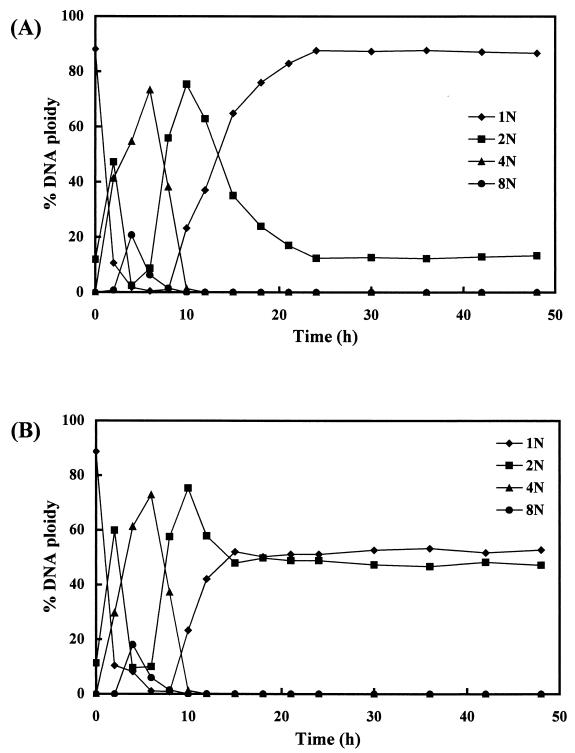
C. testosteroni TK102 cells grown in the presence or absence of Kaneclor 300 were collected at different times, and growth was measured by determining the optical density. The growth curves for TK102 grown in the presence and in the absence of Kaneclor 300 were similar (data not shown). The DNA contents of cells collected at the different times were analyzed by FCM after treatment with rifampin and cephalexin (Fig. 2 and 3). In the case of TK102 grown without Kaneclor 300, the majority of the cells contained a single chromosome when stationary-phase cells were inoculated into fresh rich medium (1/3LB medium). After 4 h of incubation (early exponential phase), most of the cells contained either four or eight chromosomes. After this, the DNA content started to decrease. After 12 h of incubation (late exponential phase), most cells contained one or two chromosomes. After 24 h of incubation, the majority of the cells contained a single chromosome again. At subsequent time points, the DNA content was almost constant, indicating that the culture had entered the stationary phase after 24 h of incubation.
FIG. 2.
FCM analysis of DNA content of C. testosteroni TK102. Cells were grown in the absence (Control) or in the presence (PCB) of Kaneclor 300 and collected after 0, 6, 12, and 24 h of incubation. After drug treatment (see Materials and Methods), cells were analyzed by FCM. The numbers before N indicate the numbers of chromosomes.
FIG. 3.
Changes in the DNA content of C. testosteroni TK102. Cells were grown in the absence (A) or in the presence (B) of Kaneclor 300 and collected at different times during growth. After drug treatment, cells were analyzed by FCM. The data points represent the means of three independent experiments. The numbers before N indicate the numbers of chromosomes.
When Kaneclor 300 was added to the culture, there was little difference in the DNA content of the cells until the late exponential phase (12 h of incubation) compared with the DNA content of the control (Fig. 2 and 3). After 15 h of incubation, approximately 50% of the cells contained two chromosomes. After longer incubation times (up to 48 h), the DNA content did not change (Fig. 3B).
Effect of PCBs on the DNA content of bacteria that are unable to degrade PCBs.
To confirm the generality of the phenomena described above, three bacteria that are not capable of PCB degradation, P. putida PpY101, P. aeruginasa PAO1, and E. coli MV1184, were incubated with or without Kaneclor 300. There was little difference in growth curves of the organisms grown with and without Kaneclor 300 for each species (data not shown). Cells were collected in the stationary phase and analyzed by FCM. In the presence or absence of Kaneclor 300, 80 to 90% of the cells of all three strains contained one chromosome and 10 to 20% of the cells contained two chromosomes (data not shown). These results suggest that Kaneclor 300 had no effect on the DNA contents of bacteria that are unable to degrade PCBs.
Effect of highly chlorinated PCBs on the DNA content of C. testosteroni TK102.
To assess the effect of highly chlorinated PCBs (which are not degraded by TK102) on the DNA content of TK102, Kaneclor 400, Kaneclor 500, and Kaneclor 600 were added to the cell cultures, and the DNA contents of the stationary-phase cells were analyzed by FCM (Table 2). When Kaneclor 400 was added, the proportion of cells containing two chromosomes increased slightly. Addition of Kaneclor 500 or Kaneclor 600 did not result in an increase in the percentage of cells containing two chromosomes.
TABLE 2.
Effects of PCBs on the DNA content of C. testosteroni TK102
| PCBs added | % DNA ploidya
|
|
|---|---|---|
| 1N | 2N | |
| Control (none) | 87.9 | 12.1 |
| Kaneclor 300 | 51.4 | 48.6 |
| Kaneclor 400 | 83.2 | 16.8 |
| Kaneclor 500 | 90.9 | 9.1 |
| Kaneclor 600 | 91.8 | 8.2 |
C. testosteroni TK102 cells were grown with Kaneclor 300, Kaneclor 400, Kaneclor 500, or Kaneclor 600. Cells grown without PCBs were used as a control. Cells were collected after 48 h of incubation and analyzed by FCM. The values are the means of three independent experiments.
Effect of PCB metabolites on the DNA content of C. testosteroni TK102.
It is known that PCBs are aerobically metabolized to chlorobenzoates that accumulate as dead-end metabolites (16). Thus, benzoate, o-, m-, and p-chlorobenzoates, 2,3-, 2,4-, 2,5-, 2,6-, 3,4-, and 3,5-dichlorobenzoates, and 2,4,6-trichlorobenzoate were added to TK102 cultures, and changes in the DNA contents of stationary-phase cells were analyzed by FCM. These compounds had no effect on the DNA contents of stationary-phase cells (data not shown). These results suggest that PCB metabolites of the upper pathway may affect DNA content.
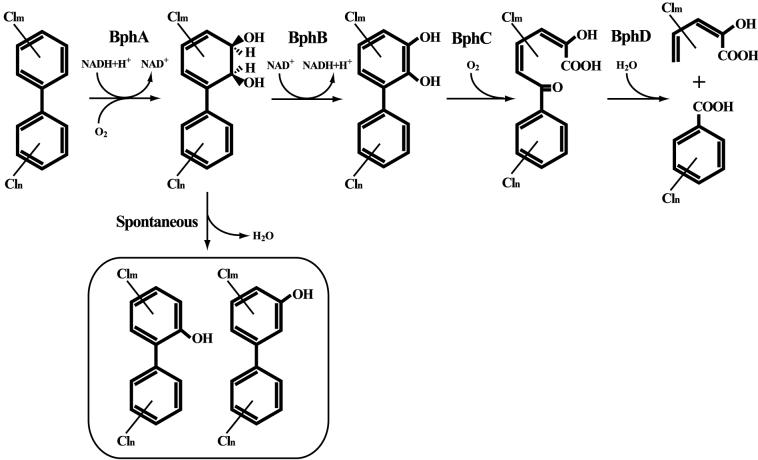
In order to generate strains that accumulated intermediate metabolites of the upper pathway (Fig. 4), four bph genes, bphA1, bphB, bphC, and bphD, were disrupted by gene replacement (see Materials and Methods). The mutants were grown with Kaneclor 300, and stationary-phase cells were analyzed by FCM (Table 3). When the bphB gene was disrupted, the percentage of cells containing two chromosomes increased from 12 to 73%. The percentages of cells in the bphC and bphD mutant cultures that contained two chromosomes were about 32 and 23%, respectively. There was no increase in the percentage of cells containing two chromosomes in the bphA1 mutant culture. When BP was added to a wild-type TK102 culture, the percentage of cells containing two chromosomes did not increase (Table 3). However, when BP was added to each bph mutant culture, the percentage of cells containing two chromosomes observed after incubation with BP was lower than the percentage observed after incubation with Kaneclor 300, except for the bphA1 mutant (Table 3).
FIG. 4.
Proposed pathway for the degradation of BP and PCBs in C. testosteroni TK102. Enzymes: BphA, biphenyl dioxygenase; BphB, cis-biphenyl-2,3-dihydrodiol dehydrogenase; BphC, 2,3-dihydroxybiphenyl 1,2-dioxygenase; BphD, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid hydrolase.
TABLE 3.
Effects of PCBs and BP on the DNA content of the C. testosteroni bph mutants
| Strain | % DNA ploidy after addition ofa:
|
|||
|---|---|---|---|---|
| PCBs
|
BP
|
|||
| 1N | 2N | 1N | 2N | |
| Wild type (control) | 87.9 | 12.1 | 87.7 | 12.3 |
| Wild type | 51.4 | 48.6 | 88.1 | 11.9 |
| bphA1 mutant | 91.4 | 8.6 | 87.8 | 12.2 |
| bphB mutant | 26.6 | 73.4 | 42.7 | 57.3 |
| bphC mutant | 68.2 | 31.8 | 70.2 | 29.8 |
| bphD mutant | 76.2 | 23.3 | 81.1 | 18.9 |
Each strain was grown with Kaneclor 300 (PCBs) or BP. Cells grown without PCBs or BP were used as a control. Cells were collected after 48 h of incubation and analyzed by FCM. The values are the means of three independent experiments.
To identify the metabolites that accumulated in the culture of the bphB mutant in the presence of BP, GC-MS analysis was performed. The GC-MS analysis revealed two main products, which had retention times of 12.3 and 14.5 min (data not shown). The molecular ion peaks of both products had an m/z value of 170, indicating that they were monohydroxybiphenyls. To confirm the product identities, the retention times of authentic 2-OHBP and 3-OHBP were determined by GC-MS. The retention times of 2- and 3-OHBP were 12.3 and 14.5 min, respectively, indicating that the products that accumulated in the culture were 2- and 3-OHBP.
When 2- and 3-OHBP were added to wild-type TK102 cultures, the percentages of cells containing two chromosomes in the stationary phase increased to 48 and 50%, respectively (data not shown). To confirm the generality of this finding, 2- or 3-OHBP was added to cultures of the three non-PCB-degrading organisms (E. coli MV1184, P. putida PpY101, and P. aeruginosa PAO1). In the stationary phase, increases in the percentages of cells containing two chromosomes were observed for E. coli MV1184 and P. putida PpY101, but no increase was observed for P. aeruginosa PAO1 cells (data not shown).
Microscopic observations.
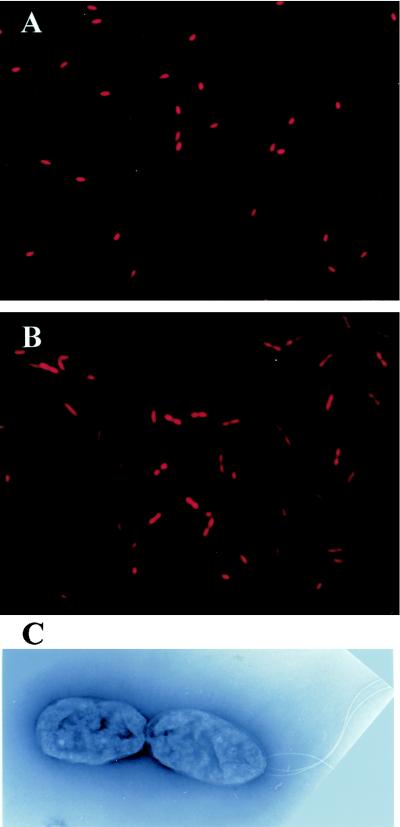
Cells grown with and without Kaneclor 300 were collected in the stationary phase, stained with PI, and observed by fluorescence microscopy. In the absence of Kaneclor 300, a majority of the cells had a single nucleoid (Fig. 5A). In the presence of Kaneclor 300, approximately 50% of the cells contained two nucleoids (Fig. 5B). Fluorescence microscopy revealed that the two nucleoids were separated completely.
FIG. 5.
Fluorescence and electron micrographs of C. testosteroni TK102. Cells were grown in the absence (A) or in the presence (B and C) of Kaneclor 300. After 24 h of incubation, cells were collected and observed by fluorescence microscopy (A and B) or transmission electron microscopy (C) (magnification, ×16,000).
To study in detail the morphology of TK102 cells, cells grown with and without Kaneclor 300 were collected in the stationary phase and observed by electron microscopy. In the absence of Kaneclor 300, a majority of the cells were rod shaped with several flagella at one pole (data not shown). In the presence of Kaneclor 300, the two daughter cells after division were attached to each other (Fig. 5C). In addition, only one of the cells had several flagella at a pole. These results indicate that the cells were blocked at a late stage in cell division (i.e., at cell separation). The proportion of cells in which cell separation was inhibited was approximately 50% (data not shown). When 2- or 3-OHBP was added to a wild-type TK102 culture, similar results were obtained (data not shown).
DISCUSSION
We used FCM to analyze changes in the DNA content of C. testosteroni TK102 cells during growth in the presence and in the absence of Kaneclor 300 (Fig. 2 and 3). Exponential-phase TK102 cells contained either four or eight chromosomes. This is probably because rapidly growing cells have several replication origins, as has been described previously for E. coli (37). In culture medium without Kaneclor 300, the majority of stationary-phase cells contained a single chromosome. In the stationary phase, initiation of chromosome replication stops, but elongation continues to termination. Thus, stationary-phase cells contain integer numbers of chromosomes (22). It is also assumed that most rounds of chromosome replication lead to cell division some time during the stationary phase. Therefore, most of the stationary-phase cells should contain a single chromosome. It has been reported that the number of chromosomes in stationary-phase E. coli cells is dependent on the growth rate of the culture during the exponential phase (2, 4). Stationary-phase E. coli cells that had been grown in rich medium had several chromosomes. In contrast, cells grown in minimal medium typically contained a single chromosome. However, Åkerlund et al. have also reported that the number of chromosomes in the stationary phase is strain dependent (2). Stationary-phase TK102 cells that had been grown in either rich or minimal medium in our study typically had one chromosome (data not shown), suggesting that the number of chromosomes was independent of the growth rate during the exponential phase. It is possible that the number of chromosomes per stationary-phase cell may be strain and species dependent.
When Kaneclor 300 was added to a TK102 culture, the proportion of cells containing two chromosomes increased to about 50% in the stationary phase. We found that BP or PCBs had no direct effect on DNA content but that intermediate metabolites produced during the degradation of BP or PCBs affected the DNA content. TK102 can transform BP or PCBs aerobically by using a pathway encoded by the bph gene cluster, which has been described in many PCB-degrading bacteria (24, 26, 29, 30). In the presence of BP, the bphB mutant should theoretically accumulate cis-biphenyl-2,3-dihydrodiol. GC-MS analysis revealed, however, that 2- and 3-OHBP accumulated. It has been reported that dihydrodiols undergo a spontaneous transformation to monohydroxy compounds via dehydration (11, 27). Thus, cis-biphenyl-2,3-dihydrodiol could be transformed to 2- and 3-OHBP by a nonenzymatic reaction (Fig. 4). In addition, TK102 was unable to transform 2- and 3-OHBP (data not shown), indicating that these compounds are dead-end metabolites. Therefore, in the presence of Kaneclor 300, hydroxy-PCBs (i.e., 2-hydroxy-PCBs and 3-hydroxy-PCBs) may accumulate in the culture. This hypothesis is supported by results of a previous study which showed that hydroxy-PCBs accumulated in the culture (15).
We found that hydroxy-BPs inhibited cell separation of TK102 and some gram-negative bacteria (E. coli MV1184 and P. putida PpY101), although they did not inhibit cell separation of all gram-negative bacteria. The concentrations of hydroxy-BPs used in this study had no effect on cell growth but inhibited cell separation. When higher concentrations of hydroxy-BPs were added, the bacteria described above could not grow on the medium (data not shown). This toxic effect of hydroxy-BPs observed by us confirmed previous reports that 2- and 4-OHBP have been used as fungicides in the postharvest treatment of fruits and vegetables (17, 25). However, the mechanism of the inhibition of cell separation is not known.
It is generally accepted that intermediate metabolites of aromatic compounds can be more toxic than the original substrates (5, 38). In our study, we found that hydroxy-BPs and hydroxy-PCBs had the ability to inhibit bacterial cell separation. It has been reported that hydroxy-PCBs exhibit estrogenic or antiestrogenic activity (7) and inhibit nitric oxide synthases, which could play a role in neuroendocrine effects that cause learning and memory deficits (31). It is important, therefore, to mineralize aromatic compounds rather than transform them during biodegradation and bioremediation processes.
In conclusion, BP and PCBs did not directly affect the DNA content, but hydroxy-BPs and hydroxy-PCBs, which are metabolites of BP and PCBs, inhibited cell separation and thus increased the percentage of cells containing two chromosomes. This is the first report indicating that hydroxy-BPs and hydroxy-PCBs can inhibit bacterial cell separation while allowing continued DNA replication.
Acknowledgments
We are grateful to G. Mukerjee-Dhar (Railway Technical Research Institute, Tokyo, Japan) for providing pTKSm10, for valuable discussions, and for a critical review of the manuscript.
REFERENCES
- 1.Ahmed, M., and D. D. Focht. 1973. Degradation of polychlorinated biphenyls by two species of Achromobacter. Can. J. Microbiol. 19:47-52. [DOI] [PubMed] [Google Scholar]
- 2.Åkerlund, T., K. Nordström, and R. Bernander. 1995. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J. Bacteriol. 177:6791-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedard, D. L., R. Unterman, L. H. Bopp, M. J. Brennan, M. L. Haberl, and C. Johnson. 1986. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl. Environ. Microbiol. 51:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernander, R., and A. Poplawski. 1997. Cell cycle characteristics of thermophilic archaea. J. Bacteriol. 179:4963-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasco, R., M. Mallavarapu, R.-M. Wittich, K. N. Timmis, and D. H. Pieper. 1997. Evidence that formation of protoanemonin from metabolites of 4-chlorobiphenyl degradation negatively affects the survival of 4-chlorobiphenyl-cometabolizing microorganisms. Appl. Environ. Microbiol. 63:427-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boye, E., and A. Løbner-Olsen. 1991. Bacterial growth control studied by flow cytometry. Res. Microbiol. 142:131-135. [DOI] [PubMed] [Google Scholar]
- 7.Connor, K., K. Ramamoorthy, M. Moore, I. Chen, S. Safe, T. Zacharewski, B. Gillesby, A. Joyeux, and P. Balaguer. 1997. Hydroxylated polychlorinated biphenyls (PCBs) as estrogens and antiestrogens: structure-activity relationships. Toxicol. Appl. Pharmacol. 145:111-123. [DOI] [PubMed] [Google Scholar]
- 8.Costello, G., L. Rodgers, and D. Beach. 1986. Fission yeast enters the stationary phase G0 state from either mitotic G1 or G2. Curr. Genet. 11:119-125. [Google Scholar]
- 9.Davey, H. M., and D. B. Kell. 1996. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol. Rev. 60:641-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durodie, J., K. Coleman, and M. J. Wilkinson. 1993. Characterization of bacterial cell size and ploidy using flow cytometry and image analysis, p. 95-109. In D. Lloyd (ed.), Flow cytometry in microbiology. Springer-Verlag, London, United Kingdom.
- 11.Ensley, B. D., B. J. Ratzkin, T. D. Osslund, M. J. Simon, L. P. Wackett, and D. T. Gibson. 1983. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science 222:167-169. [DOI] [PubMed] [Google Scholar]
- 12.Erickson, B. D., and F. J. Mondello. 1992. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J. Bacteriol. 174:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsburg, S. L., and P. Nurse. 1991. Cell cycle regulation in the yeasts Saccharomyces cerevisiae and Sulfolobus acidocaldarius. Annu. Rev. Cell Biol. 7:227-256. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda, M., and K. Yano. 1985. Construction of broad host range cloning vectors for gram-negative bacteria. Agric. Biol. Chem. 49:2719-2724. [Google Scholar]
- 15.Furukawa, K., N. Tomizawa, and A. Kamibayashi. 1979. Effect of chlorine substitution on the bacterial metabolism of various polychlorinated biphenyls. Appl. Environ. Microbiol. 38:301-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furukawa, K. 1994. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation 5:289-300. [DOI] [PubMed] [Google Scholar]
- 17.Gesell, M., E. Hammer, M. Specht, W. Francke, and F. Schauer. 2001. Biotransformation of biphenyl by Paecilomyces lilacinus and characterization of ring cleavage products. Appl. Environ. Microbiol. 67:1551-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jepras, R. I., J. Carter, S. C. Pearson, F. E. Paul, and M. J. Wilkinson. 1995. Development of a robust flow cytometric assay for determining numbers of viable bacteria. Appl. Environ. Microbiol. 61:2696-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimbara, K., T. Hashimoto, M. Fukuda, T. Koana, M. Takagi, M. Oishi, and K. Yano. 1989. Cloning and sequencing of two tandem genes involved in the degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J. Bacteriol. 171:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebaron, R., and F. Joux. 1994. Flow cytometric analysis of the cellular DNA content of Salmonella typhimurium and Alteromonas haloplanktis during starvation and recovery in seawater. Appl. Environ. Microbiol. 60:4345-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López-Amorós, R., J. Comas, and J. Vives-Rego. 1995. Flow cytometric assessment of Escherichia coli and Salmonella typhimurium starvation-survival in seawater using rhodamine 123, propidium iodide, and oxonol. Appl. Environ. Microbiol. 61:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mason, C. A., and T. Egli. 1993. Dynamics of microbial growth in the deceleration and stationary phase of batch culture, p. 91-93. In S. Kjelleberg (ed.), Starvation in bacteria. Plenum Press, New York, N.Y.
- 23.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20-78. [DOI] [PubMed] [Google Scholar]
- 24.Mukerjee-Dhar, G., T. Hatta, M. Shimura, and K. Kimbara. 1998. Analysis of changes in congener selectivity during PCB degradation by Burkholderia sp. strain TSN101 with increasing concentrations of PCB and characterization of the bphBCD genes and gene products. Arch. Microbiol. 169:61-70. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa, Y., and G. A. Moore. 1995. Cytotoxic effects of postharvest fungicides, ortho-phenylphenol, thiabendazole and imazall, on isolated rat hepatocytes. Life Sci. 57:1433-1440. [DOI] [PubMed] [Google Scholar]
- 26.Ohtsubo, Y., Y. Nagata, K. Kimbara, M. Takagi, and A. Ohta. 2000. Expression of the bph genes involved in biphenyl/PCB degradation in Pseudomonas sp. KKS102 induced by the biphenyl degradation intermediate, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid. Gene 256:223-228. [DOI] [PubMed] [Google Scholar]
- 27.Resnick, S. M., D. S. Torok, and D. T. Gibson. 1993. Oxidation of carbazole to 3-hydroxycarbazole by naphthalene 1,2-dioxygenase and biphenyl 2,3-dioxygenase. FEMS Microbiol. Lett. 113:297-302. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Seah, S. Y. K., G. Labbè, S. Nerdinger, M. R. Johnson, V. Snieckus, and L. D. Eltis. 2000. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem. 275:15701-15708. [DOI] [PubMed] [Google Scholar]
- 30.Seah, S. Y. K., G. Labbè, S. R. Kaschabek, F. Reifenrath, W. Reineke, and L. D. Eltis. 2001. Comparative specificities of two evolutionarily divergent hydrolases involved in microbial degradation of polychlorinated biphenyls. J. Bacteriol. 183:1511-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma, R., and P. R. Kodavanti. 2002. In vitro effects of polychlorinated biphenyls and hydroxy metabolites on nitric oxide synthases in rat brain. Toxicol. Appl. Pharmacol. 178:127-136. [DOI] [PubMed] [Google Scholar]
- 32.Shimura, M., T. Koana, M. Fukuda, and K. Kimbara. 1996. Complete degradation of polychlorinated biphenyls by a combined method of ultraviolet and biological treatments. J. Ferment. Bioeng. 81:573-576. [Google Scholar]
- 33.Shimura, M., T. Hayakawa, G. Mukerjee-Dhar, M. Fukuda, and K. Kimbara. 1998. Characterization of polychlorinated biphenyl degradation in a fermentor by Comamonas testosteroni strain TK102. Jpn. J. Water Treat. Biol. 34:57-65. [Google Scholar]
- 34.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. Huse. 1988. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shortridge, V. D., A. Lazdunski, and M. V. Vasil. 1992. Osmoprotectants and phosphate regulate expression of phospholipase C in Pseudomonas aeruginosa. Mol. Microbiol. 6:863-871. [DOI] [PubMed] [Google Scholar]
- 36.Skarstad, K., H. B. Steen, and E. Boye. 1983. Cell cycle parameters of slowly growing Escherichia coli B/r studied by flow cytometry. J. Bacteriol. 154:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skarstad, K., R. Bernander, S. Wold, H. B. Steen, and E. Boye. 1996. Cell cycle analysis of microorganisms, p. 241-255. In M. Al-Rubeai and A. N. Emery (ed.), Flow cytometry applications in cell culture. Marcel Dekker, Inc., New York, N.Y.
- 38.Snyder, R., G. Witz, and B. D. Goldstein. 1993. The toxicology of benzene. Environ. Health Perspect. 100:293-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sumner, E. R., and S. V. Avery. 2002. Phenotypic heterogeneity: differential stress resistance among individual cells of the yeast Saccharomyces cerevisiae. Microbiology 148:345-351. [DOI] [PubMed] [Google Scholar]
- 40.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 41.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 42.Withers, H. L., and R. Bernander. 1998. Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J. Bacteriol. 180:1624-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, R., A. Natarajan, and F. Srienc. 1999. A flow injection flow cytometry system for on-line monitoring of bioreactors. Biotechnol. Bioeng. 62:609-617. [DOI] [PubMed] [Google Scholar]