Abstract
Wastewater treatment is one of the most important commercial biotechnological processes, and yet the component bacterial populations and their associated metabolic activities are poorly understood. The novel fluorescent dye hexidium iodide allows assessment of Gram status by differential absorption through bacterial cell walls. Differentiation between gram-positive and gram-negative wastewater bacteria was achieved after flow cytometric analysis. This study shows that the relative proportions of gram-positive and gram-negative bacterial cells identified by traditional microscopy and hexidium iodide staining were not significantly different. Dual staining of cells for Gram status and activity proved effective in analyzing mixtures of cultured bacteria and wastewater populations. Levels of highly active organisms at two wastewater treatment plants, both gram positive and gram negative, ranged from 1.5% in activated sludge flocs to 16% in the activated sludge fluid. Gram-positive organisms comprised <5% of the total bacterial numbers but accounted for 19 and 55% of the highly active organisms within flocs at the two plants. Assessment of Gram status and activity within activated sludge samples over a 4-day period showed significant differences over time. This method provides a rapid, quantitative measure of Gram status linked with in situ activity within wastewater systems.
Commercial and industrial wastewater systems are of great microbiological interest, in terms of both community structure and function, yet many of the component bacteria and their metabolic activities are poorly understood (22). Cultivation-based studies have been carried out in attempts to isolate and identify the important bacteria present (3). These studies have demonstrated the diversity that exists, but because of their selective nature they cannot provide a true indication of the organisms present. Additionally, conventional plating techniques do not allow determination of in situ activity. Examination and characterization of activated sludge have proven difficult, but adaptation of conventional techniques may provide further insights into this complex microbial ecosystem (1, 26). The Gram stain is probably the most widely used stain in microbial classification and is a fundamental technique in the examination of activated sludge samples (9). Gram-positive organisms differ from gram-negative organisms in that their wall consists chiefly of peptidoglycan and lacks the complex outer membrane of lipopolysaccharides. This differentiation is based only on cell wall differences. Many Gram staining protocols for wastewater processes have been developed, but they may often be deemed unsatisfactory (9). It is well documented that the technique requires practice and should be tested with known cultures and that the decolorization step must be accurately and precisely controlled. With particular regard to the monitoring of activated sludge, Gram staining can be problematic and large, dense flocs do not decolorize effectively, thus limiting assessment (9). The Eikelboom classification of wastewater microorganisms, which is fundamental in traditional assessment of treatment systems, relies heavily on the Gram stain (9). Some gram-positive cocci have been found to cause nonfilamentous activated sludge foams that prove problematic in wastewater treatment (25). Work by Wagner et al. (29) indicates that gram-positive bacteria are important for phosphate removal from wastewater during the enhanced biological phosphate removal process. Other work has reported the use of fluorescently labeled wheat germ agglutinin, which specifically binds to the peptidoglycan layer of gram-positive bacteria (23). Hexidium iodide (HI) is a novel fluorescent nucleic acid binding dye that allows assessment of Gram status by differential absorption through bacterial cell walls (8), selectively staining gram-positive organisms in suspension (14). The use of such a fluorescent dye may provide a robust, objective, and rapid alternative to traditional Gram staining in wastewater systems.
Understanding the biochemical activity of wastewater bacterial populations may provide insight into the important organisms that are active in situ. This assessment can be made using fluorescent activity dyes (16, 18, 19, 20, 27). 5(6)-Carboxyfluorescein diacetate (CFDA) is a derivative of fluorescein diacetate. CFDA acts as a substrate for general intracellular esterases and is nonfluorescent until enzymatic cleavage produces a fluorochrome. Retention of this fluorochrome indicates an intact cell membrane, while cleavage indicates biochemical activity. CFDA is readily retained within the cell and is less susceptible to photobleaching than the parent dye, fluorescein diacetate, making it potentially useful for work of this nature (6).
This study aims to provide a rapid method for determining gram-positive and gram-negative organisms with their relative activities by combining HI and CFDA in a novel dual-staining protocol for flow cytometry (FCM). Fluorescence-based methods have been combined with FCM to analyze bacteria within wastewater systems (30). Direct staining (fluorescence) combined with FCM may help avoid cultivation bias, analyzing individual cells rapidly and accurately.
MATERIALS AND METHODS
Wastewater sampling.
Samples were taken from Countess Wear (CW) sewage treatment works, Exeter, United Kingdom. CW is a medium scale municipal plant, serving ≈150,000 population equivalents, taking domestic and industrial wastes. Up to 500 ml of primary settled influent, activated sludge, secondary settlement, and final effluent were collected in sterile vessels never more than half full. Samples were also collected from the activated sludge systems from two other sites as a comparison. One of these sites was a meat processing plant (MP) with an on-site treatment works treating fluid waste and sewage from the animal slaughter process. The second was a small town treatment works, also treating domestic and industrial wastewater. All samples were returned to the laboratory, and analysis was commenced within 60 min of collection.
Bacterial strains and culture conditions.
Micrococcus luteus (NCIMB 10474) was grown and maintained on nutrient agar at 37°C. Pseudomonas putida (NCIMB 9494) and Arthrobacter sp. strain NCIMB 10407 were maintained on nutrient agar at 25°C. Nutrient broth was passed through low-protein-binding 0.22-μm-pore-size filters (Millipore) to remove particulates prior to use. Heat-killed samples were prepared by heating 1 ml of overnight culture at 80°C for 15 min in a water bath.
Total heterotrophic plate counts for each environmental sample were determined using spread plates on R2A (21) after incubation at 25°C for 5 days. The Gram status of culturable bacteria were estimated using R2A supplemented with 2-phenylethanol (Sigma) at 3.06 mg/ml for the selection of gram-positive bacteria (11). Gram-negative bacterial numbers were estimated using R2A modified with crystal violet at a final concentration of 1 μg/ml or with cefsulodin (10 μg/ml) (5). Each selective plate count was expressed as a percentage of the total plate count.
Gram staining of cells for microscopy.
A smear of sample was allowed to air dry (9) before traditional Gram staining was performed with the three-step Gram stain procedure kit (Becton Dickinson Microbiology Systems, Cockeysville, Md.), in strict accordance with the manufacturers instructions, with decolorization for 20 s.
Cell labeling for direct Gram status and activity analysis.
Stock solutions of HI (Molecular Probes, Inc., Eugene, Oreg.) were prepared in dimethyl sulfoxide at 5 mg/ml. This was further diluted in 10 mM Tris-HCl (pH 7.4) to give a 100-μg/ml working stock solution. Bacterial samples were stained by incubation with 1:10 (vol/vol) working stock at room temperature for 15 min.
Stock CFDA (Sigma) was prepared at 1 mM in acetone. Samples were incubated at 37°C for 30 min after the addition of 10 μl of stock CFDA solution per ml. For dual staining, samples were incubated with CFDA before the addition of HI.
Activated sludge samples were allowed to stand for 5 min before careful removal of 5 ml of supernatant. This supernatant is referred to as activated sludge fluid. Floc cells could be analyzed along with activated sludge fluid cells after floc disruption by one of two methods. The first method involved disruption by vortexing for 3 min and allowing settlement before the supernatant was removed. Cells released by this process are referred to as activated sludge floc, vortexed. Alternatively, the sample bottle was inverted twice to gently mix the sample after settling. Approximately 15 ml of this sample was transferred into a sterile plastic 30-ml bottle before sonication for 30 s at 5-μm amplitude (peak to peak), using a Soniprep 150 (Santo, MSE) with a probe diameter of 4 mm. After sonication the sample was allowed to settle for a further 5-min, and the supernatant was removed. Samples analyzed after this process are referred to as activated sludge floc sonicated. Under these conditions, maximum floc disruption without cell lysis was achieved (data not shown).
FCM analysis.
All laboratory solutions were filtered by passage through a 0.22 μm-pore-size filter (Gelman Sciences, Ann Arbor, Mich.) at least three times before use. All cytometric analyses and cell sorting were performed with a FACS Star Plus flow cytometer-cell sorter (Becton Dickinson, Oxford, United Kingdom) set to trigger on forward scatter (threshold, 50). Focusing was performed using 0.5-μm-diameter Fluoresbrite YG fluorescent latex beads (Polysciences, Inc., Warrington, Pa.) with an argon laser with a 25-mW (488-nm) output. The laser output was 50 mW during cell analyses. Sheath fluid (cell carrying fluid) for FCM analysis was passed six times through a 0.1-μm-diameter filter (Gelman Sciences) with distilled water and was used to rinse FCM tanks prior to use. For cell sorting, phosphate-buffered saline was used as sheath fluid. The nozzle diameter was 70 μm during analysis and cell sorting. Samples were sorted into sterile polystyrene tubes (Falcon; Becton Dickinson).
CFDA fluorescence at 525 nm was analyzed through fluorescence detector 1 set at a photomultiplier tube voltage of 650V with a logarithmic gain. HI fluorescence at >565 nm was detected through fluorescence detector 2 set at a photomultiplier tube setting of 500 V with a logarithmic gain. For dual staining, fluorescence from each dye was separated using appropriate compensation settings within the machine.
Data handling.
Parallel samples were taken at each sampling time at each point in the treatment process. From each sample, two subsamples were analyzed with HI. This sampling regimen was maintained during dual staining. Data represent average readings from triplicate FCM measurements of at least 5,000 events from each subsample. FCM data files were read into WinMDI version 2.6 (Joseph Trotter, Scripps Research Institute, La Jolla, Calif.), which was used to enumerate subpopulations. Analysis of variance and comparison of means (using the least significant difference) were performed with Minitab (release 12.1) as described by Fry (7). Data are expressed as percent fluorescent cells from total bacterial events.
RESULTS
Assessment of Gram status in laboratory cultures by HI labeling.
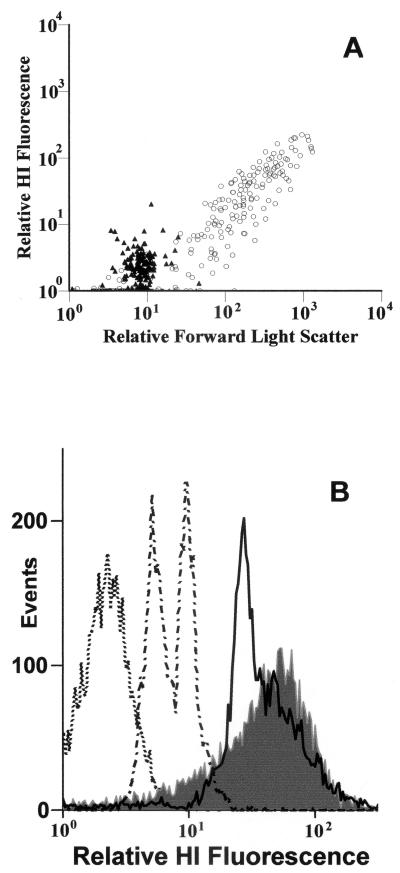
Differentiation of gram-positive and gram-negative bacteria after HI labeling and FCM analysis was achieved initially with laboratory isolates (Fig. 1A). Various levels of HI fluorescence were observed, depending on bacterial culture type and age. HI fluorescence from gram-negative cells was sufficient to distinguish them from background events (Fig. 1A).
FIG. 1.
FCM output showing Gram differentiation in laboratory cultures stained with HI. (A) M. luteus (gram positive) (○) and P. putida (gram negative) (▴) by forward scatter (x axis) and cell-associated relative fluorescence of HI (y axis). (B) Logarithmic-phase gram-negative culture (Pseudomonas spp.) (dotted line), stationary-phase gram-variable culture (Arthrobacter spp.) (dotted and dashed line), logarithmic-phase gram-variable culture (Arthrobacter spp.) (solid line), and logarithmic-phase gram-positive culture (Micrococcus spp.) (filled grey).
Relative fluorescence intensities from P. putida after HI staining showed median channel numbers of >8 (where channel number is an indicator of fluorescence intensity in arbitrary units), whether the cells were in exponential phase, were in stationary phase, or were heat killed. M. luteus cultures were also monitored. Heat-killed controls displayed median channel numbers of <10. Stationary-phase cells were 1.7 times more fluorescent by HI than heat-killed controls. Exponential-phase cells displayed fluorescence 4 times that of heat killed controls. Differentiation between gram-positive and gram-negative species was achieved throughout the growth phase. The gram-variable Arthrobacter sp. showed a fluorescence signal similar to that of gram-positive cells when analyzed during exponential growth (Fig. 1B). As the age of the gram-variable culture increased, the resulting HI fluorescence decreased to that of a gram-negative signal (Fig. 1B). This change in HI fluorescence correlated with the change in Gram staining visualized by microscopy. Heat killing of gram-negative bacterial cultures showed a doubling in relative HI fluorescence from that of live samples, but these increases did not exceed a median channel number of 8 (Fig. 1B). Gram-positive heat-killed samples showed an 80% decrease in HI fluorescence (data not shown). HI staining of Tetrahymena pyrifomis and Chlamydomonas reinhardtii was also performed (data not shown). These organisms were shown to take up HI but could be discounted from analysis on the basis of their size and associated light scatter properties (data not shown).
HI staining of environmental samples.
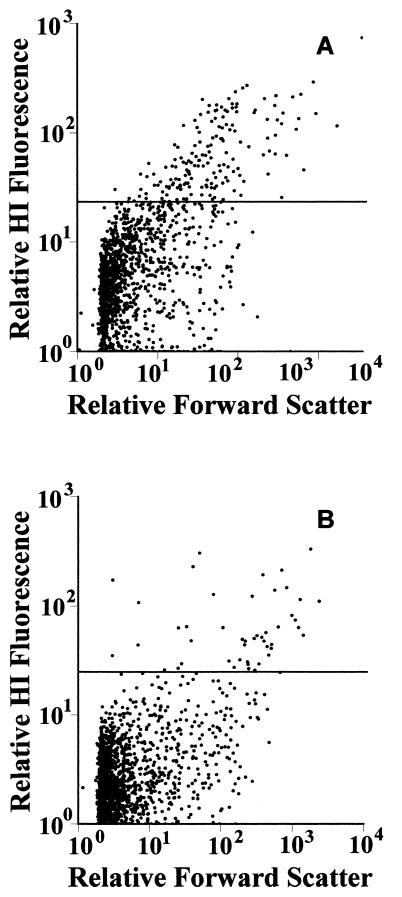
HI-stained bacterial populations were clearly distinguished throughout the wastewater treatment process (Fig. 2). Levels of gram-positive organisms varied at each stage of the treatment process and also between plants (Table 1). CW primary influent displayed a greater range and temporal fluctuation of gram-positive bacterial cell numbers than primary influent from the MP site, although levels of gram-positive organisms detected at the MP site were higher overall than those at CW. The highest levels of gram-positive organisms determined by HI staining were determined within MP primary influent, ranging from 10.5 to 13%, compared to 1.7 to 9.1% for CW primary influent.
FIG. 2.
Scatter diagrams showing levels of HI fluorescent events in MP wastewaters. (A) Primary influent; (B) final effluent. The horizontal lines indicate the cutoff between gram-negative cells (below line) and gram-positive cells (above line).
TABLE 1.
Levels of gram-positive bacteria through steps in wastewater treatment plants
| Sample | Mean % gram-positive bacteria (SD) (n > 5) by:
|
||||
|---|---|---|---|---|---|
| FCM determination | Gram stain | Culture determination with:
|
|||
| Cefsulodin | Crystal violet | Phenyl ethanol | |||
| CW primary influent | 6.5 (3.1) | 6.5 (5.1) | 57.6 (27.2) | 48 (22.3) | 1.3 (0.9) |
| MP primary influent | 11.7 (1.8) | NDa | 5.8 (2.4) | 5.8 (2.4) | ND |
| CW ASb fluid | 2.9 (1.3) | 2.6 (1.1) | 31.3 (29.3) | 1.4 (2.5) | 1.2 (0.7) |
| MP AS fluid | 4.4 (1.3) | ND | 0 | 0 | 1.3 (1.0) |
| CW AS floc, vortexed | 4.1 (2.0) | 3.8 (1.2) | 7.7 (2.0) | 9.7 (4.8) | ND |
| CW AS floc, sonicated | 10.0 (0.6) | 9.7 (2.1) | 54.6 (12.8) | 91.8 (1.8) | 0.19 (0.04) |
| MP AS floc, vortexed | 9.4 (0.9) | ND | 0 | 0 | 0.3 (0.03) |
| CW settlement tank | 2.0 (0.1) | 2.3 (1.3) | 88.3 (5.4) | 70.4 (3.8) | 2.6 (0.1) |
| MP settlement tank | 4.3 (0.1) | ND | 0 | 0 | ND |
| CW final efluent | 3.4 (1.8) | 3.8 (1.2) | 48 (14.2) | 59 (20.7) | 0.24 (0.05) |
| MP final efluent | 2.1 (0.2) | ND | 0 | 0 | ND |
ND, not determined.
AS, activated sludge.
Levels of gram-positive organisms within the activated sludge fluid were lower than those displayed within primary settled influent at both sites (except for one sample from CW). Analysis showed that levels of gram-positive organisms within activated sludge fluid ranged from 2.2 to 6.8% at CW. MP showed the greatest reduction in gram-positive organisms from primary settlement to activated sludge fluid, with levels dropping to between 5.3 and 3.4%. MP gram-positive bacterial cell levels determined in the primary influent were always higher than those found in the activated sludge fluid (Table 1).
Disruption of activated sludge flocs prior to analysis produced an increased proportion of gram-positive organisms. Following disruption by the vortex method, gram-positive cell levels increased in both CW and MP. MP samples displayed higher levels, ranging from 8.7 to 10.0%. CW samples showed lower levels (2.6 to 5.6%), but these were greatly increased with sonication to levels encountered within MP activated sludge floc. Disruption by sonication did not affect the ability of cells to form colonies or affect HI fluorescence in pure cultures (data not shown).
Analysis of secondary settlement tanks for CW and MP showed minor variation within each site (Table 1). Levels of gram-positive organisms were very similar to levels found in activated sludge fluid for each site. The secondary settlement gram-positive cell levels at the MP site were 2 times greater than those at the CW site.
Gram-positive cell levels within final effluent samples varied between sites. CW showed a greater variation and a higher range than for the secondary settlement samples, with levels between 1.4 and 5.5%. The MP final effluent contained around 2% gram-positive cells. MP displayed reductions in the levels of gram-positive organisms throughout the treatment process, resulting in a fivefold reduction. CW showed a twofold reduction in the maximum levels of gram-positive bacteria found entering the treatment plant.
The proportions of gram-positive cells determined by HI staining and FCM were not significantly different from those determined by traditional microscopy (Table 1). Relating plate count estimates of Gram status to HI measurements proved to be difficult, as the selective agents tested revealed marked differences (Table 1). Two of the selective agents tested, cefsulodin and crystal violet, did not provide consistent estimations of culturable Gram status within wastewater systems (Table 1). The third gram-positive selective agent tested, 2-phenylethanol, showed results that corresponded more closely to estimations by HI staining (Table 1). A typical activated sludge sample assessed by traditional Gram staining showed gram-positive cell levels ranging from 3.8 to 11.9%, while the corresponding HI FCM data values ranged from 8.0 to 9.5%.
Typically, higher levels of gram-positive organisms were found within primary influent samples (Fig. 2A), whereas lower levels of gram-positive bacteria were found within the final effluent at the MP site (Fig. 2B). This level of HI fluorescence provided sufficient differentiation between Gram groups to facilitate cell sorting. Sorting was undertaken with environmental samples and samples spiked with laboratory cultures of gram-positive organisms where appropriate. Removal of gram-positive organisms through FCM cell sorting was 98% effective as determined by microscopic techniques and 98.6% effective as determined by growth on selective media. HI staining affected the ability of both gram-positive and gram-negative cells to form colonies. For gram-positive cells this reduction in CFU-forming ability was approximately 100%, although washing cells prior to plating did allow some colony formation. For gram-negative cells in the presence of HI, reductions of around 70% in CFU-forming ability were noted.
Dual staining of pure cultures and environmental samples for Gram status and activity.
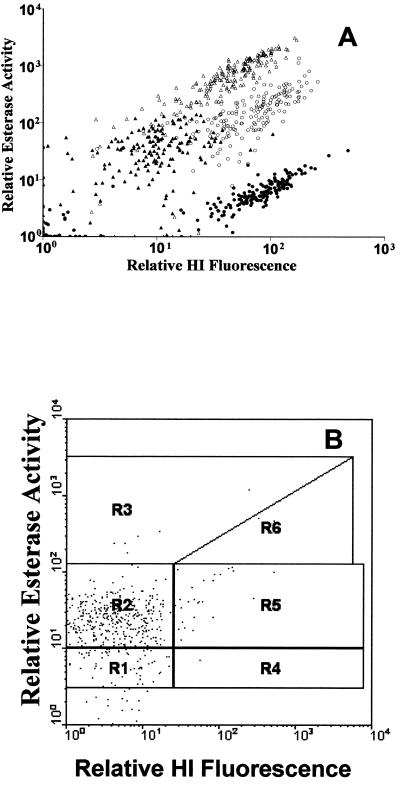
Positive differentiation between subpopulations of gram-positive and gram-negative bacteria from the laboratory was achieved following dual staining with HI and for CFDA (Fig. 3A). The levels of fluorescence displayed by each species at different stages in laboratory culture were determined. This provided the criteria for determining cellular activity (inactive, active, or highly active) as well as Gram status (Fig. 3A). Each region was then used to determine the number of bacterial events in order to analyze wastewater treatment plant population structure, defined by activity and Gram status (Fig. 3B). The mean proportions of bacterial cells in each group are displayed in Table 2. Gram-negative organisms were the most abundant group (>90%) within both the activated sludge fluid and flocs at both sites examined. Of the gram-negative cells, approximately 15% were deemed highly active in the activated sludge fluid, compared with approximately 1.6% highly active gram-negative cells within the activated sludge floc (Table 2). Gram-positive organisms were thus in a minority within these activated sludge populations, comprising <4.5% of total bacterial cells in both the fluid and flocs from both sites. However, within the gram-positive populations, 20 to 30% of the cells were classified as highly active in both activated sludge fluid and floc samples from both sites (Table 2). Overall, inactive cells ranged from 6 to 12% of the gram-negative and 2 to 6% of the gram-positive cell populations in both sites and sample types (Table 2).
FIG. 3.
Scatter diagrams of dual staining of pure cultures for Gram status and metabolic activity to determine subpopulation regions. (A) Logarithmic-phase gram-negative culture (Pseudomonas spp.) (▵), stationary-phase gram-negative culture (Pseudomonas spp.) (▴), logarithmic-phase gram-positive culture (Micrococcus spp.) (○), and stationary-phase gram-positive culture (Micrococcus spp.) (•). (B) Typical environmental sample. Regions (R) are defined for WinMDI analysis as follows: R1, gram-negative, inactive; R2, gram-negative, active; R3, gram-negative, highly active; R4, gram-positive, inactive; R5, gram-positive, active; R6, gram-positive, highly active.
TABLE 2.
Proportions of bacteria determined by FCM dual staining
| Sample | Mean % of total bacteria (SD)
|
|||||
|---|---|---|---|---|---|---|
| Gram negative
|
Gram positive
|
|||||
| Inactive | Active | Highly Active | Inactive | Active | Highly Active | |
| AS Fluid TW | 11.3 (0.3) | 71 (0.6) | 13.5 (1.3) | 0.2 (0.01) | 2.6 (0.15) | 1.3 (0.13) |
| AS Floc TW | 18.3 (2.5) | 76.6 (1.2) | 2.6 (0.8) | 0.05 (0.02) | 1.3 (0.1) | 0.6 (0.2) |
| AS Fluid CW | 6.1 (0.02) | 76.3 (2.0) | 16.6 (1.4) | 0.06 (0.02) | 0.7 (0.3) | 0.18 (0.18) |
| AS Floc CW | 13.2 (1.6) | 81.5 (1.6) | 0.74 (0.24) | 0.08 (0.07) | 3.5 (0.14) | 0.93 (0.14) |
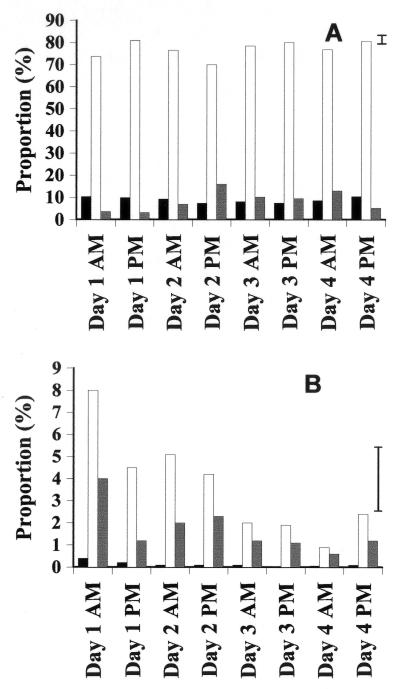
Bacterial activity and Gram status were assessed at the CW site using samples collected twice daily over a 4-day period from each of four parallel activated sludge tanks. Differences between tanks were not significant. Temporal changes in activities within the gram-positive and gram-negative populations are presented in Fig. 4. Gram-negative cells were always the most abundant group (Fig. 4). Inactive gram-negative cells remained at approximately 10% of the community and did not change significantly over the consecutive 4-day period. The proportion of gram-negative cells deemed active declined significantly on the afternoon of the second sample day, returning to their former level by the morning of day 3 (Fig. 4A). The proportion of highly active gram-negative cells showed significant fluctuations over the 4-day period (8 sample points), rising significantly on both the morning and the afternoon of day 2 before dropping on day 3 (Fig. 4A). No significant changes in the proportion of inactive gram-positive cells were recorded. The number of gram-positive cells classified as active dropped significantly on the afternoon of day 1 and again on the morning of day 3 (Fig. 4B). Levels of highly active gram-positive cells did not differ significantly until the morning of day 4, when the proportion in this category was significantly lower than that recorded at the start.
FIG. 4.
Bar charts showing the relative proportions of cells in CW activated sludge sonicated samples over a 4-day period. Error bars represent least significant differences (3%; P = 0.05). Note the y-axis scale difference. (A) Gram-negative inactive cells (black), gram-negative active cells (white), and gram-negative highly active cells (grey). (B) Gram-positive inactive cells (black), gram-positive active cells (white). and gram-positive highly active cells (grey).
DISCUSSION
The impact of microbial treatment of wastewater is of enormous commercial and environmental importance. It is crucial to obtain an understanding of the microbial populations involved if we are to improve process efficiency and reduce system failure. With the realization that traditional culture-based methods may provide a biased view of microbial community structure, it is necessary to analyze bacterial populations by using culture-independent methods (2).
This study confirms previous findings (14) that HI staining is an effective Gram status determinant that is amenable to FCM for laboratory cultures, and it extends the work to include environmental (wastewater) samples. The fluorescence characteristics of the HI dye allow its use in dual-staining protocols with traditional (fluorescein-like) green dyes such as SYTO13 (14). This study combines HI with the fluorescein-based metabolic activity stain to investigate Gram status and bacterial activity within wastewater treatment plants.
Combining HI staining with FCM allowed analysis of several thousand bacterial events in seconds, while traditional Gram staining requires growth and subsequent testing which can take days or weeks. This staining technique provides a rapid method for determining gross levels of gram-positive and gram-negative organisms within treatment plants. HI assessment was shown to classify gram-variable cells by their cell wall structure at that time. Assigning gram-variable bacteria to a specific category can be a difficult and subjective procedure (9). The use of HI avoids the difficulties associated with decolorization during traditional Gram staining. Heat killing of gram-negative cells and subsequent HI staining produced only a small increase in fluorescence. These increases did not produce fluorescence signals as intense as those from labeled gram-positive cells. Heat fixation may thus be more useful as a control than the ethanol fixation used in previous work (14), which allowed marked HI absorption into gram-negative cells. Ethanol fixation causes the disruption of bacterial lipopolysaccharides. Heat treatment may allow lipopolysaccharides to remain intact.
Floc disruption by sonication provided the means to analyze floc-bound cells through FCM. Without sonication FCM did not improve the traditional inability to assess the Gram status of floc organisms. The selectivity of such a disruption step is undetermined, but this treatment process did not affect the fluorescence of pure cultures or environmental samples.
HI staining of environmental samples.
HI staining allowed enumeration of gram-positive organisms in wastewater treatment plants at each step of the process. The data show a general decline in gram-positive numbers from primary influent to final effluent. Differences were also detected between treatment plants. The relatively high numbers of gram-positive-staining cells from the meat processing plant treatment works may be expected, as the influent will be composed of waste from the slaughter process. CW influent was variable in the proportion of gram-positive bacteria, reflecting the changes in proportions of incoming surface water drainage and domestic and industrial wastewaters. The proportions of gram-positive and gram-negative cells noted in this study are similar to those obtained from fluorescent in situ hybridization (FISH) of similar municipal wastewater samples in other studies (4, 13, 28, 29, 30). Probing these communities with oligonucleotide probes directed against the different subclasses of the proteobacteria suggests that high proportions of FISH-positive cells (generally 75 to 80% of the total cell count) are gram-negative bacteria (28). Other estimates range from 40 to 86% gram-negative cells (13, 24, 30). Studies using probes targeted to the high-GC-content gram-positive group (29) estimate the proportion of these cells to be 1 to 7% of the FISH-positive count in municipal treatment plants under normal operating conditions, increasing to 17 to 24% when the plants are operating with enhanced biological phosphate removal. Screening a 16S ribosomal DNA clone library from activated sludge suggested that around 3% of clones were from gram-positive organisms (4).
Plate count estimates of the proportions of gram-positive and-negative cells showed large variations when using cefsulodin or crystal violet as a selective agent. Data from this study would suggest using R2A amended with 2-phenylethanol to select for gram-positive colonies. Although they were considerably lower, the data consistently provided estimates closer to those obtained by HI staining than other plate methods. 2-Phenylethanol is widely used to estimate gram-positive cell numbers, but it is unlikely to prove to be completely effective in complex environmental samples. It should also be noted that HI staining removed the ability of some cells to form colonies.
Dual staining of environmental samples for Gram status and activity.
Proportions of active and inactive gram-positive and gram-negative mixtures were identified and confirmed within laboratory samples. Significant changes in the wastewater bacterial community structure and activity were recorded. Observing these changes was possible only due to the speed of the FCM analysis. This study does not assume activity on the basis of high nucleic acid content (12). The use of an independent activity assessment such as a fluorogenic esterase dye, combined with FCM cell sorting, provides a means to link metabolic activity (6) with the option of further analysis. This study defined levels of activity as inactive, active, or highly active. Inactive cells related to CFDA fluorescence obtained from heat-killed cells, while active cells equated to stationary-phase culture labeling. The high activity bracket was equivalent to CFDA activity from exponential-phase cultures. However, this activity measurement can assess only cells that respond to CFDA labeling (10). Although this should not be equated with an absolute measure of cell viability, it is an indicator of enzyme activity and thus carbon oxidation in wastewater treatment plants.
Active gram-negative cells did show some HI staining due to fluorescence carryover from fluorescence detector 1 into detector 2. This carryover was independent of HI staining. Alteration of the compensation settings within the flow cytometer allowed reduction of carryover, but in order to remove it completely, compensation compromised detection of HI-labeled cells. This arose due to the brightness of the esterase signal. Clearer discrimination may possible with alternative dyes (16, 19) or with narrower band-pass filters, but this may affect esterase signal detection.
Other studies have also demonstrated that the complex structure of activated sludge systems is difficult to assess with traditional techniques (2, 3). The range of emerging molecular techniques used to investigate systems in situ may not completely replace traditional microbiological techniques but will help to identify, monitor, and isolate important organisms in wastewater systems (24, 30). Data from such studies have also suggested that a high proportion (>80%) of the gram-negative cells found within activated sludge are active (28). HI staining may be of use in the quantification of gram-positive bacteria in natural environments, especially when the identification signal from FISH is limited.
Linking activity of key player organisms to favorable conditions within wastewater treatment may help in determining treatment efficiency. Identifying specific cells while simultaneously assessing activity may allow correlation with degradative functions. Moreover, altering environmental conditions within the treatment process may allow increased activity of specific beneficial organisms. We have also combined activity measurements with single-cell sorting (17) to remove specific cells of interest for further analysis, including denaturing gradient gel electrophoresis and 16S rRNA identification (unpublished data). This approach along with other direct methods (13, 15), such as FISH with multiple (group-specific) rRNA-targeted oligonucleotide probes (1), may determine which bacteria are responsible for an efficient activated sludge reactor. This approach may help forge the much-needed link between identity and specific activity (2), finally removing the “black box” approach to bacterial analysis of wastewater.
Acknowledgments
We thank all wastewater treatment plant staff for access and assistance in sampling.
S.F. was supported by the Biotechnological and Biological Sciences Research Council (Swindon, United Kingdom).
References
- 1.Amann, R., J. Snaidr, M. Wagner, W. Ludwig, and K. H. Schleifer. 1996. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 178:3496-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., H. Lemmer, and M. Wagner. 1998. Monitoring the community structure of wastewater treatment plants: a comparison of old and new techniques. FEMS Microbiol. Ecol. 25:205-215. [Google Scholar]
- 3.Benedict, R. G., and D. A. Carlson. 1971. Aerobic heterotrophic bacteria in activated sludge. Water Res. 5:1023-1030. [Google Scholar]
- 4.Bond, P. L., R. Erhart, M. Wagner, J. Keller, and L. L. Blackall. 1999. Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems. Appl. Environ. Microbiol. 65:4077-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman, P. A., C. A. Siddons, P. M. Zadik, and L. Jewes. 1990. An improved selective medium for the isolation of Escherichia coli O 157. J. Med. Microbiol. 32:107-110. [DOI] [PubMed] [Google Scholar]
- 6.Diaper, J. P., and C. Edwards. 1994. Use of fluorogenic esters to detect viable bacteria by flow cytometry. J. Appl. Bacteriol. 77:221-228. [Google Scholar]
- 7.Fry, J. C. 1993. Biological data analysis: a practical approach, Oxford IRL Press, Oxford, United Kingdom.
- 8.Haugland, R. P. 1999. Molecular probes. Handbook of fluorescent probes and research chemicals, 7th ed. Molecular Probes, Eugene, Oreg.
- 9.Jenkins, D., M. G. Richard, and G. T. Daigger. 1993. Manual on the causes and control of activated sludge bulking and foaming, 2nd ed. Lewis Publishers, Chelsea, Mich.
- 10.Kell, D. B., A. S. Kaprelyants, D. H. Weichart, C. R. Harwood, and M. R. Barer. 1998. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Leeuwenhoek 73:169-187. [DOI] [PubMed] [Google Scholar]
- 11.Koneman, E. W. 1983. Color atlas and textbook of diagnostic microbiology, 2nd ed. J. B. Lippincott, Philadelphia, Pa.
- 12.Lebaron, P., P. Servais, H. Agogue, C. Courties, and F. Joux. 2001. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active and inactive cells in aquatic system? Appl. Environ. Microbiol. 67:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manz, W., M. Wagner, R. Amann, and K. H. Schleifer. 1994. In situ characterization of the microbial consortia active in two wastewater treatment plants. Water Res. 28:1715-1723. [Google Scholar]
- 14.Mason, D. J., S. Shanmuganathan, C. Mortimer, and V. A. Gant. 1998. A fluorescent Gram stain for flow cytometry and epifluorescence microscopy. Appl. Environ. Microbiol. 64:2681-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oerther, D. B., J. Pernthaler, A. Schramm, R. Amann, and L. Raskin. 2000. Monitoring precursor 16S rRNAs of Acinetobacter spp. in activated sludge treatment systems. Appl. Environ. Microbiol. 66:2154-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parthuisot, N., P. Catala, K. Lemarchand, J. Baudart, and P. Lebaron,. 2000. Evaluation of ChemChrome V6 for bacterial viability assessment in waters. J. Appl. Microbiol. 89:370-380. [DOI] [PubMed] [Google Scholar]
- 17.Porter, J., C. Edwards, A. W. Morgan, and R. W. Pickup. 1993. Rapid, automated separation of specific bacteria from lake water and sewage by flow cytometry and cell sorting. Appl. Environ. Microbiol. 59:3327-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter, J., C. Edwards, and R. W. Pickup. 1995. Rapid assessment of physiological status in Escherichia coli using fluorescent probes. J. Appl. Bacteriol. 79:399-408. [DOI] [PubMed] [Google Scholar]
- 19.Porter, J., D. Deere, R. W. Pickup, and C. Edwards. 1996. Fluorescent probes and flow cytometry: new insights into environmental microbiology. Cytometry 23:91-96. [DOI] [PubMed] [Google Scholar]
- 20.Porter, J., D. Deere, M. Hardman, C. Edwards, and R. W. Pickup. 1997. Go with the flow—use of flow cytometry in environmental microbiology. FEMS Microbiol. Ecol. 24:93-101. [Google Scholar]
- 21.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seviour, R. J. 1999. The normal microbial communities of activated sludge plants, p. 76-78 In R. J. Seviour and L. L. Blackall (ed.), The microbiology of activated sludge. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 23.Sizemore, R. K., J. J. Caldwell, and A. S. Kendrick. 1990. Alternate Gram staining technique using a fluorescent lectin. Appl. Environ. Microbiol. 56:2245-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K. H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sodell, J. A., and R. Seviour. 1990. Microbiology of foaming in activated sludge plants—a review. J. Appl. Bacteriol. 69:145-176. [Google Scholar]
- 26.Steen, H. B. 2000. Flow cytometry of bacteria: glimpses from the past with a view to the future. J. Microbiol. Methods 42:65-74. [DOI] [PubMed] [Google Scholar]
- 27.Turner, K., J. Porter, R. W. Pickup, and C. Edwards. 2000. Changes in viability and macromolecular content of long-term batch cultures of Salmonella typhimurium measured by flow cytometry. J. Appl. Microbiol. 89:90-99. [DOI] [PubMed] [Google Scholar]
- 28.Wagner, M., R. Amann, H. Lemmer, and K. H. Schleifer. 1993. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 59:1520-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner, M., R. Erhart, W. Manz, R. Amann, H. Lemmer, D. Wedi, and K. H. Schleifer. 1994. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl. Environ. Microbiol. 60:792-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallner, G., R. Erhart, and R. Amann. 1995. Flow cytometric analysis of activated-sludge with rRNA-targeted probes. Appl. Environ. Microbiol. 61:1859-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]