Abstract
Escherichia coli BL21 strains were found to excrete a large amount of outer membrane protein F (OmpF) into culture medium during high-cell-density cultivation. From this interesting phenomenon, a novel and efficient OmpF fusion system was developed for the excretion of recombinant proteins by E. coli. The ompF gene of E. coli BL21(DE3) was first knocked out by using the red operon of bacteriophage λ to construct E. coli MBEL-BL101. For the excretion of human β-endorphin as a model protein, the β-endorphin gene was fused to the C terminus of the E. coli ompF gene by using a linker containing the Factor Xa recognition site. To develop a fed-batch culture condition that allows efficient production of OmpF-β-endorphin fusion protein, three different feeding strategies, an exponential feeding strategy and two pH-stat strategies with defined and complex nutrient feeding solutions, were examined. Among these, the pH-stat feeding strategy with the complex nutrient feeding solution resulted in the highest productivity (0.33 g of protein per liter per h). Under this condition, up to 5.6 g of OmpF-β-endorphin fusion protein per liter was excreted into culture medium. The fusion protein was purified by anion-exchange chromatography and cleaved by Factor Xa to yield β-endorphin, which was finally purified by reverse-phase chromatography. From 2.7 liters of culture supernatant, 545.4 mg of β-endorphin was obtained.
Escherichia coli possesses two membranes, the cytoplasmic and outer membranes, which serve as penetration barriers for macromolecules. The cytoplasmic membrane is equipped with secretory machineries for the secretion of proteins into periplasm (2, 6, 28). All proteins secreted are synthesized as larger premature forms containing signal peptides at the N terminus. All of the natural proteins found in the outer membrane and the periplasmic space belong to this category. On the other hand, only a few proteins, such as hemolysin, are known to be excreted into the culture medium after crossing the outer membrane (2, 6, 7, 28). The excretion of recombinant proteins into culture medium has several advantages, a low level of proteolysis, simple purification, improved protein folding, and N-terminal authenticity, which are similar to those seen with the secretion of proteins into the periplasm (12, 22, 31). Furthermore, excretion of recombinant proteins into culture medium has at least two other advantages compared with secretion into the periplasm. First, there is no need to disrupt the outer cell membrane to recover the target proteins. Second, excretion allows continuous production of recombinant proteins since cells do not have to be collected or lysed for the recovery of proteins (14).
Due to these advantages, there have been several attempts to develop a strategy to promote the excretion of recombinant proteins by E. coli. Fusion partners such as the pelB leader, the ompA leader, the protein A leader, and maltose binding protein or dedicated translocators such as the hemolysin and pullulanase systems have been used with limited success for the excretion of specific proteins of interest (1, 3, 13, 16, 33). Another approach involves the coexpression of the kil gene, the tolAIII gene, the bacteriocin release protein gene, and the mitomycin-induced bacteriocin release protein gene (10, 24, 34, 36). However, these strategies showed relatively high levels of contamination of cellular proteins in culture supernatant. An alternative method was the use of leaky L-form E. coli cells that release periplasmic proteins into the culture medium due to the loss of outer membrane integrity (8). Kujau et al. (17) reported the excretion of functional miniantibodies with L-form E. coli strains. However, the fragility of the outer membrane has a number of deleterious consequences for the viability and growth of L-form cells. Cells are hypersensitive to detergents and EDTA and cannot be cultivated to the high density typically required for the efficient production of recombinant proteins.
Outer membrane protein F (OmpF; 36 kDa) is a porin protein in the outer membrane of E. coli that forms the pores through which small hydrophilic molecules diffuse passively. During our studies of the high-cell-density cultivation of various E. coli strains, we serendipitously found that a large amount of OmpF was excreted into the culture medium during the high-cell-density cultivation of E. coli BLR(DE3). As the cell density increased, the amount of OmpF protein in the culture supernatant also increased. Therefore, it was reasoned that the OmpF protein might be used as a fusion partner for the excretion of recombinant proteins in E. coli.
In this paper, we report a novel OmpF fusion system for the excretion of foreign proteins. Human β-endorphin was used as a model protein. Human β-endorphin exhibits potent morphine-like activity, producing in humans a good feeling and tolerance to pain (25). This compound is hundreds or even thousands of times more potent than morphine on a molar basis. We report detailed results on the development of a new excretion system and excretory production of human β-endorphin by high-cell-density cultivation of recombinant E. coli.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1, and the oligonucleotide primers used in the PCRs are listed in Table 2. E. coli XL1-Blue was used as a host strain for the cloning and maintenance of plasmids. E. coli BL21(DE3) and E. coli MBEL-BL101 (see Results) were used for the production of human β-endorphin, which is composed of 31 amino acids (Tyr-Gly-Gly-Phe-Met-Thr-Ser-Glu-Lys-Ser-Gln-Thr-Pro-Leu-Val-Thr-Leu-Phe-Lys-Asn-Ala-Ile-Ile-Lys-Asn-Ala-Tyr-Lys-Lys-Gly-Glu).PCR was performed with a PCR thermal cycler MP TP3000 (Takara Shuzo Co., Shiga, Japan) using the High Fidelity PCR system (Boehringer Mannheim, Mannheim, Germany). Electroporation was performed with a Gene Pulser (Bio-Rad, Hercules, Calif.) at 12.5 kV/cm, 25 μF, and 200 Ω. All DNA manipulations including restriction digestion, ligation, and agarose gel electrophoresis were carried out as described previously by Sambrook et al. (29). The DNA sequences of all constructs were confirmed by an automatic DNA sequencer (ABI Prism model 377; Applied Biosystems Inc., Foster City, Calif.).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | supE44 hsdR17 recA1 endA1 gyrA96 thi relA1 lac F′ [proAB+ lacIqlacZΔM15 Tn10(Tetr)] | Stratagenea |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagenb |
| MBEL-BL101 | BL21(DE3) ΔompF::Kmr | This study |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150(Strr) relA1 flbB5301 deoC1 ptsF25 rbsR | Lab stock |
| HB101 | F−hsdS20 (rk− mk−) recA13 ara-14 proA2 lacY1 galK2 rpsL20(Strr) xyl-5 mtl-1 supE44λ | Lab stock |
| W3110 | Derived from K-12, λ−, F−, prototrophic | Lab stock |
| JM101 | F−traD36 proAB+ lacIqlacZΔM15 supE thi Δ(lac-proAB) | Lab stock |
| Plasmids | ||
| pUC18 | 2.7 kb, Apr | New England Biolabs |
| pBluescript SK(−) | 3.0 kb, Apr | Stratagene |
| pTrc99A | 4.2 kb, Apr, trc T7 promoter | Pharmacia |
| pET21c | 5.3 kb, Apr, T7 promoter | Novagen |
| pEDOmpF3 | 6.4 kb, Apr, T7 promoter, ompF gene | This study |
| pTrcOmpF4 | 5.3 kb, Apr, trc promoter, ompF gene | This study |
| pOmpF6 | 5.2 kb, Apr, ompF promoter, ompF gene | This study |
| pOmpF6βE | 5.3 kb, Apr, ompF promoter, ompF-β-endorphin fused gene | This study |
| pTrcEBG | 5.4 kb, Apr, trc promoter, red operon from bacteriophage λ | This study |
Stratagene Cloning Systems, La Jolla, Calif.
Novagen, Inc., Madison, Wis.
TABLE 2.
Oligonucleotides used in PCRsa
| Primerno. | Sequence (5′ → 3′) |
|---|---|
| 1 | CGCGCCATGGATATTAATACTGAAACTGAGATCAAGC |
| 2 | CGGGATCCTCATCGCCATTGCTCCCCAAATAC |
| 3 | GCGGATCCTTAGAACTGGTAAACGATAC |
| 4 | GCGAATTCATATGATGAAGCGCAATATTCTG |
| 5 | GCGAATTCCATGGTGAAGCGCAATATTCTGGCAG |
| 6 | CGAATTCTGGATTATACCGACGCAGTG |
| 7 | ACCGCCATACCTTCCCTCGATGAACTGGTAAACGATA |
| 8 | GGAAGGTATGGCGGTTTCATGACCAGCGAAAAAAGCCAGAC |
| 9 | CGCGTTTTTAAACAGGGTCACCAGCGGGGTCTGGCTTTTTTCGC |
| 10 | CCCTGTTTAAAAACGCGATCATCAAAAACGCGTATAAAAAAG |
| 11 | GCTCTAGACTATTATTCGCCTTTTTTATACGCGTTTTTG |
Underlining indicates the restriction enzyme sites.
Culture conditions.
For flask cultures, R/2 medium (pH 6.8) was used (26). The R/2 medium contained 2 g of (NH4)2HPO4, 6.75 g of KH2PO4, 0.85 g of citric acid, and 0.7 g of MgSO4 · 7H2O per liter and 5 ml of trace metal solution [10 g of FeSO4 · 7H2O, 2.25 g of ZnSO4 · 7H2O, 1 g of CuSO4 · 5H2O, 0.5 g of MnSO4 · 5H2O, 0.23 g of Na2B4O7 · 10H2O, 2 g of CaCl2 · 2H2O, and 0.1 g of (NH4)6MO7O24 per liter of 5 M HCl]. Glucose (10 g/liter) was used as a carbon source. Cells were cultivated in a 250-ml flask containing 50 ml of R/2 medium supplemented with 50 μg of ampicillin/ml or 25 μg of kanamycin/ml in a shaking incubator at 37°C and 200 rpm. For expression under the T7 or trc promoter, isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma Chemical Co., St. Louis, Mo.) was added to a final concentration of 1.0 mM at an optical density at 600 nm (OD600) of 0.7.
Fed-batch cultures were carried out in a 6.6-liter jar fermentor (Bioflo 3000; New Brunswick Scientific Co., Edison, N.J.) containing 2 liters of R/2 medium plus 20 g of glucose/liter. Seed culture was prepared in a 1-liter flask containing 200 ml of R/2 medium plus 20 g of glucose/liter. The pH was controlled at 6.8, except for the periods of pH rise due to glucose depletion (in the case of the pH-stat fed-batch culture), by the addition of 28% (vol/vol) ammonia water. The dissolved-oxygen concentration was controlled at 40% of air saturation by automatically increasing the agitation speed to 1,000 rpm and by changing the percentage of pure oxygen. Nutrient feeding solution was added into the fermentor by three different feeding strategies. First, a pH-stat feeding strategy with a defined nutrient feeding solution (700 g of glucose/liter plus 20 g of MgSO4 · 7H2O/liter) was used. When the pH rose to a value greater than its set point (6.8) by 0.08 due to the depletion of glucose, the appropriate volume of feeding solution was automatically added to increase the glucose concentration in the culture broth to 0.7 g/liter. Second, a pH-stat feeding strategy with a complex feeding solution (500 g of glucose/liter, 50 g of yeast extract/liter, and 20 g of MgSO4 · 7H2O/liter) was used. The feeding solution was added as described above. Last, a defined feeding solution containing 700 g of glucose/liter plus 20 g of MgSO4 · 7H2O/liter was fed exponentially into the fermentor using a computer-controlled pump to support a specific growth rate of 0.15 h−1 (19).
Fractionation of outer membrane proteins.
Culture broth (3 ml) was centrifuged at 3,500 × g for 5 min at 4°C, and the pellet was washed with 1 ml of 10 mM Na2HPO4 buffer (pH 7.2), followed by centrifugation at 3,500 × g for 5 min at 4°C. The pellet was resuspended in 0.5 ml of 10 mM Na2HPO4 buffer (pH 7.2) and sonicated thoroughly to disrupt cells. Cell debris was removed by centrifugation of sonicated samples at 12,000 × g for 2 min at room temperature. The total membrane protein fraction of the sample was isolated after centrifugation at 12,000 × g for 30 min at 4°C and resuspended in 0.5 ml of 10 mM Na2HPO4 buffer (pH 7.2) containing 0.5% (wt/vol) sarcosyl. After incubation at 37°C for 30 min, insoluble pellet was obtained by centrifugation at 12,000 × g for 30 min at 4°C. The insoluble fraction was washed with 10 mM Na2HPO4 buffer (pH 7.2) and was resuspended in 50 μl of 10 mM Tris-HCl buffer (pH 8.0).
Purification of β-endorphin.
The OmpF-β-endorphin fusion protein in the culture supernatant was purified by anion-exchange column chromatography (BioLogic HR system; Bio-Rad). The supernatant was loaded onto an anion-exchange column (Bio-Scale Q2 column; Bio-Rad) preequilibrated with 50 mM Tris-HCl (pH 7.0) and was then eluted by a linear gradient of NaCl (0 to 1.0 M in the same buffer) at 90 ml/h. The protein concentration of each fraction was monitored using a UV detector (Bio-Rad). After collection of fraction containing the fusion protein, NaCl was removed by dialysis (MWCO 3500; Spectrum Lab. Inc., Laguna Hills, Calif.) against 2 liters of 20 mM Tris-HCl buffer (pH 7.0) for 24 h with three buffer exchanges. The OmpF protein was cleaved off by incubation with 1.2 μg of Factor Xa/ml (New England Biolabs, Beverly, Mass.) at 23°C for 12 h (35). After cleavage, β-endorphin was finally purified by reverse-phase high-performance liquid chromatography (HPLC) with a 4.6- by 250-mm Microsorb-MV C18 column (Varian, Walnut Creek, Calif.) with a linear gradient elution from 100% buffer A to 50% buffer A plus 50% buffer B at 1 ml/min over 2 h (buffer A, 0.1% [vol/vol] trifluoroacetic acid; buffer B, acetonitrile containing 0.1% [vol/vol] trifluoroacetic acid). The fraction containing β-endorphin was collected and lyophilized. Electrophoretic analysis of β-endorphin on a Tricine sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel was carried out by the method of Schagger and von Jagow (30).
Analytical methods.
Protein samples were analyzed by electrophoresis on a 10% (wt/vol) SDS-PAGE gel as described previously by Laemmli (18). The protein bands on the SDS-PAGE gels were quantified by densitometry (ImagerMaster; Pharmacia Biotech, Uppsala, Sweden). The amount of soluble protein was determined by bicinchoninic acid assay with bovine serum albumin as a standard (32). The N-terminal amino acid sequence of purified β-endorphin was determined by a gas phase sequencer (model 476A; Applied Biosystems Inc.).
RESULTS
Knockout of the ompF gene in E. coli BL21(DE3).
For the development of the ompF mutant of E. coli BL21(DE3), the red operon of bacteriophage λ was used (23). The red operon containing exo, beta, and gam genes was amplified from bacteriophage λ DNA by PCR as follows. The forward primer (primer 1) was designed to contain an NcoI site upstream of the start codon of the gam gene (Table 2). The reverse primer (primer 2) was designed to contain a BamHI site downstream of the stop codon of the exo gene. The PCR product was digested with NcoI and BamHI and cloned into the same restriction sites of pTrc99A. The resulting plasmid, pTrcEBG, was transformed into E. coli BL21(DE3). Transformants were prepared as electroporation-competent cells after induction with 1 mM IPTG for the expression of the red operon. The PCR product containing a kanamycin resistance gene between the ompF promoter (PompF) and the ompF gene was mixed with electroporation-competent cells. After electroporation, transformants were selected on a Luria-Bertani medium plate with kanamycin (25 μg/ml) at 37°C. From the selected transformants, pTrcEBG was removed by the cultivation of cells in the absence of ampicillin, and then gene replacement was confirmed by PCR (data not shown). This ompF knockout mutant was named E. coli MBEL-BL101 and was used as a host strain for the excretion of the OmpF fusion protein.
Construction of ompF gene expression system.
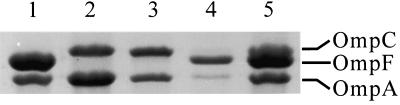
For the expression of the ompF gene, three plasmids, pEDOmpF3, pTrcOmp4, and pOmpF6, in which the ompF gene is expressed by the T7, trc, and ompF promoters, respectively, were constructed. The ompF gene was amplified from the chromosomal DNA of E. coli BL21(DE3) by PCR as follows. For expression under the T7 promoter, PCR was carried out using primers 3 and 4. The PCR product was digested with NdeI and BamHI and was then cloned into pET21c to make pEDOmpF3. For expression under the trc promoter, PCR was carried out using primers 3 and 5. The PCR product was digested with NcoI and BamHI and was then cloned into pTrc99A to yield pTrcOmpF4. For expression under the ompF promoter, primers 3 and 6 were used to amplify the DNA fragment from the region from position −960 upstream of the ompF promoter to the stop codon of the ompF structural gene. After EcoRI and BamHI digestion, the PCR product was cloned into pBluescript SK(−) to make pOmpF6. E. coli MBEL-BL101 was transformed with each of these plasmids, and the amounts of OmpF protein expressed were analyzed by SDS-PAGE of the outer membrane fraction (Fig. 1). In E. coli MBEL-BL101, OmpF protein was not produced but the level of OmpC was increased instead. Among the three expression systems, pOmpF6 showed the highest level of OmpF production. When pEDOmpF3 was used, OmpF was not produced in the outer membrane fraction. Instead, a large amount of premature OmpF was detected in the total protein fraction, which means that OmpF was produced but not secreted when a strong T7 promoter was used (data not shown). From these results, the expression system with the ompF promoter was chosen for the production of the OmpF fusion protein.
FIG. 1.
Analysis of outer membrane fraction by SDS-PAGE. Lanes: 1, E. coli BL21(DE3); 2, E. coli BL101; 3, E. coli BL101 harboring pEDOmpF3; 4, E. coli BL101 harboring pTrcOmpF4; 5, E. coli BL101 harboring pOmpF6.
Extracellular production of OmpF-β-endorphin fusion protein.
For the construction of the OmpF-β-endorphin fusion gene, five primers (primers 7, 8, 9, 10, and 11) were synthesized. Overlapping PCR was carried out using these primers and primer 4. The PCR product was digested with BglII and XbaI and was cloned into pOmpF6 to construct pOmpF6βE, in which the fused gene is expressed under the control of the ompF promoter (Fig. 2A). The Factor Xa cleavage site (Ile-Glu-Gly-Arg) was introduced between the ompF and β-endorphin genes for the removal of OmpF during purification (Fig. 2B).
FIG. 2.
Plasmid used for the production of OmpF-β-endorphin fusion protein. Shown are schematic representations of the plasmid construction (A) and the structure of fused genes (B).
For the excretory production of the OmpF-β-endorphin fusion protein, high-cell-density cultures of recombinant E. coli MBEL-BL101 harboring pOmpF6βE were carried out as described in Materials and Methods. Three different feeding strategies (pH-stat with two different feeding solutions and exponential feeding) were examined. Figure 3 shows the time profiles for cell density (OD600), dry cell weight (grams per liter), and concentrations of total proteins (grams per liter) and the OmpF-β-endorphin fusion protein (grams per liter) in the culture supernatant obtained from three fed-batch cultures. In all cultures, the OmpF-β-endorphin fusion protein was successfully excreted into culture supernatant as cells grew. Among the three cultures, the pH-stat feeding strategy with a complex feeding solution resulted in the highest productivity (0.33 g of fusion protein per liter per h). After an 8-h lag period, cells grew rapidly to reach 59 g (dry cell weight) per liter in 17 h. The final concentration of fusion protein in the culture supernatant was 5.6 g/liter, which is equivalent to 490 mg of β-endorphin/liter. Although the purity of the fusion protein decreased to 62% of total proteins in the culture supernatant towards the end of fermentation, a much higher purity of 75% could be obtained during the exponential growth phase. When the pH-stat feeding strategy with a defined feeding solution was used, a lower productivity (0.26 g of fusion protein per liter per h), a lower final concentration of fusion protein (4.5 g/liter), and a lower purity (47%) were obtained at the end of fermentation. When the exponential feeding strategy was used, the specific growth rate could be controlled successfully at a constant value of 0.15 h−1 during the exponential growth phase. In this culture, however, the lowest production of fusion protein (0.11 g of fusion protein per liter per h) was obtained.
FIG. 3.
The time profiles of cell density, dry cell weight, and the concentrations of total proteins and the OmpF-β-endorphin fusion protein in the culture supernatant during high-cell-density cultivations with the pH-stat feeding strategy with a defined feeding solution (A), the pH-stat feeding strategy with a complex feeding solution (B), and the exponential feeding strategy with a defined feeding solution (C).
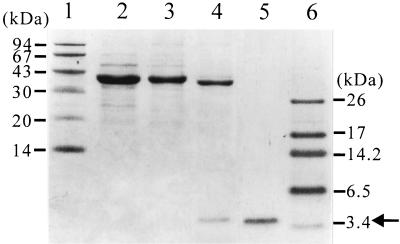
The OmpF-β-endorphin fusion protein from 50 ml of culture supernatant (Fig. 3B) was purified by anion-exchange chromatography. The OmpF protein was cleaved off by Factor Xa treatment. Finally, β-endorphin was purified by reverse-phase HPLC, producing homogeneously pure β-endorphin as shown in Fig. 4. The N-terminal amino acid sequence was found to be Tyr-Gly-Gly-Phe-Met-Thr-Ser-Glu-Lys, which was consistent with the known N-terminal amino acid sequence of β-endorphin. A total of 545.4 mg of pure β-endorphin was obtained from 2.7 liters of culture supernatant (final volume), with a purity greater than 99% and a recovery yield of 45.7% (Table 3).
FIG. 4.
SDS-PAGE analysis of the samples after each purification step. Lanes: 1 and 6, molecular mass standards; 2, the total protein in the culture supernatant; 3, after anion-exchange chromatography; 4, after Factor Xa cleavage; 5, after reverse-phase HPLC. The arrow indicates β-endorphin.
TABLE 3.
Summary of β-endorphin purification
| Purification step | Vol (ml) | Total protein (mg)a | Fusion protein (mg)b | β-Endorphin (mg) | Recovery yield (%) | Purity (%) |
|---|---|---|---|---|---|---|
| Culture supernatant | 50 | 420 | 254 | 22.1c | 100 | 60.5d |
| Anion-exchange column (Q2) | 63 | 244 | 186 | 16.1c | 72.9 | 76.4d |
| Reverse phase HPLC | 16 | 10.1 | NDe | 10.1 | 45.7 | >99f |
Protein concentration was determined by bicinchoninic acid assay.
As determined by densitometric scanning of SDS-PAGE gels.
The amount of β-endorphin was calculated from the molecular weight composition of the fusion protein.
The purity of OmpF-β-endorphin fusion protein.
ND, not detected.
The purity of β-endorphin.
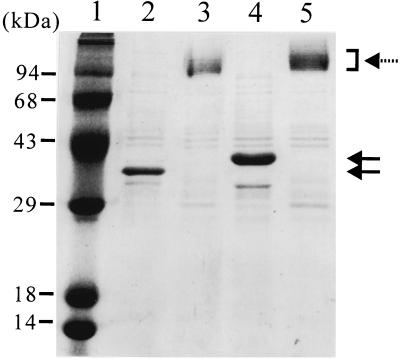
To determine the structure of the fusion protein excreted into culture medium, the culture supernatant, boiled and not boiled, was analyzed by SDS-PAGE (Fig. 5). The fusion protein appeared as a monomeric form (40 kDa) when the boiled sample was analyzed. However, when the sample was not boiled, multiple protein bands of greater than 95 kDa were observed. These bands correspond to the trimer of the fusion protein associated with lipopolysaccharide (LPS). The OmpF protein excreted into the culture medium during the fed-batch culture of E. coli BL21(DE3) also showed the same pattern on the SDS-PAGE gel. These results indicate that the OmpF-β-endorphin fusion protein also forms a trimeric porin structure on the outer membrane and is released into culture medium without disruption of its trimeric structure.
FIG. 5.
Analysis of conformation of OmpF and OmpF-β-endorphin fusion protein excreted into the culture medium. Lanes: 1, molecular mass standards; 2 and 3, culture supernatant from the high-cell-density culture of E. coli BL21(DE3); 4 and 5, culture supernatant from the high-cell-density culture of E. coli MBEL-BL101 harboring pOmpF6βE. Lanes 2 and 4 show the boiled samples, and lanes 3 and 5 show the unboiled samples. The solid arrows indicate the monomeric form of OmpF and OmpF fusion proteins, and the dashed arrow indicates the trimeric form of OmpF and OmpF fusion proteins.
DISCUSSION
Because of a number of advantages in the extracellular production of target proteins compared with cytosolic production, various methods have been developed as described earlier. Despite these efforts, excretion of foreign proteins into the culture medium during high-cell-density cultivation has rarely been demonstrated. In this study, we developed a new system for excretory production and showed that human β-endorphin as a model protein could be successfully excreted into culture medium as an OmpF fusion protein during high-cell-density cultivation. It is interesting that OmpF protein is excreted into culture medium during the high-cell-density cultivation of E. coli BL21 strains, although the reason for this is not yet understood. In general, the total amount of OmpF and OmpC in E. coli K12 is fairly constant, but the relative amounts of the two proteins are dependent on several factors, including the osmolarity of the external medium (9), concentrations of certain antibiotics (4), nutrient limitation (20), cell density (21), and growth phase (27). When flask cultures were carried out in a defined R/2 medium, the outer membrane fraction of E. coli BL21(DE3) showed a different composition from those of E. coli K12 strains such as HB101, JM101, MC4100, XL1-Blue, and W3110 (Fig. 6). In BL21(DE3), OmpF was the major outer membrane protein, with small amounts of OmpA and OmpC. On the other hand, other E. coli strains produced OmpA as a major protein. When high-cell-density cultivations of E. coli MC4100 and XL1-Blue were carried out, OmpF protein was not detected in the culture supernatant (data not shown). It seems that the expression of the ompF gene in BL21 strain is regulated by a mechanism different from that in K12 strains. When the ompF gene sequence of E. coli BL21(DE3) was compared with that of E. coli K12 from the GenBank database, two substitutions, G→T at position −18 and T→C at position −363, were found in the promoter and its upstream region. Position −363 is located within the OmpR binding site to which OmpR binds and represses ompF gene expression (11). Dairl et al. (5) also reported that a base substitution at the Pribnow box (especially A→T at position −12) rendered ompF expression independent of positive regulation. Therefore, the difference between the upstream regions of the ompF gene in the BL21 and K12 strains seems to be the reason for the large difference in OmpF levels in the two strains.
FIG. 6.
SDS-PAGE analysis of outer membrane fractions of E. coli strains BL21(DE3) (lane 1), HB101 (lane 2), JM101 (lane 3), MC4100 (lane 4), XL1-Blue (lane 5), and W3110 (lane 6).
Liu and Ferenci (20) reported that the expression level of the ompF gene was higher at a specific growth rate (μ) of 0.3 h−1 than at 0.1 or 0.6 h−1. In the present study, in the pH-stat fed-batch cultures with defined and complex feeding solutions, the specific growth rates were maintained at 0.27 and 0.29 h−1, respectively, in the exponential growth phase. However, for the exponential feeding strategy, the specific growth rate was maintained at 0.15 h−1. Therefore, the different amounts of OmpF-β-endorphin fusion protein obtained in the three fed-batch cultures could be partly explained by the results reported by Liu and Ferenci (20). Even though the amount of OmpF-β-endorphin fusion protein accumulated in the culture medium increased as cell density increased in the pH-stat fed-batch cultures, the amount of OmpF-β-endorphin fusion protein in the outer membrane fraction was not increased (constantly high level) (data not shown). The other Omp proteins, including OmpA and OmpC, were also maintained at low levels during cultivation. From these results, it can be concluded that excess OmpF-β-endorphin fusion protein was released into the culture medium because of the limitation of protein accommodation in the outer membrane of E. coli.
It was interesting to observe that the OmpF-β-endorphin fusion protein still formed a trimeric porin structure on the outer membrane. The released fusion protein appeared as multiple bands on the SDS-PAGE gel, which suggests that LPS is associated with it (15). However, the release of LPS does not seem to be a problem, because β-endorphin could be recovered with high purity by simple procedures, as shown in Fig. 4.
In this study, we report a new protein excretion system based on OmpF and its use for the excretion of human β-endorphin as a fusion protein into culture medium. Excreted β-endorphin could be purified by a simple recovery method. The findings that the OmpF fusion system allowed efficient excretion of a small peptide into the culture medium, that potentially high-level production of recombinant protein is possible by high-cell-density cultivation, and that simple recovery is possible because the target protein is excreted into the culture medium should be useful for the development of strategies for the efficient extracellular production of other recombinant proteins in E. coli. We intend to examine the excretion of a larger protein by use of this system next.
Acknowledgments
This work was supported by the Basic Industrial Research Program of the Korean Ministry of Commerce, Industry and Energy. Further support from LG Chem Investment, Samchully Pharmaceutical Co., and BioLeaders Co. are also appreciated. K.J.J. was supported by the National Research Laboratory Program of the Korean Ministry of Science and Technology and is a postdoctoral fellow supported by the Brain Korea 21 project.
REFERENCES
- 1.Better, M., C. P. Chang, R. R. Robinson, and A. H. Horwitz. 1988. Escherichia coli secretion of an active chimeric antibody fragment. Science 240:1041-1043. [DOI] [PubMed] [Google Scholar]
- 2.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, M. A., and I. B. Holland. 1994. Heterologous protein secretion and the versatile Escherichia coli haemolysin translocator. Trends Biotechnol. 12:450-455. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, S. P., L. M. McMurry, and S. B. Levy. 1988. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J. Bacteriol. 170:5416-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dairl, T., K. Inokuchi, T. Mizuno, and S. Mizushima. 1985. Positive control of transcription initiation in Escherichia coli. A base substitution at the Pribnow box renders ompF expression independent of a positive regulator. J. Mol. Biol. 184:1-6. [DOI] [PubMed] [Google Scholar]
- 6.de Gier, J. W., and J. Luirink. 2001. Biogenesis of inner membrane proteins in Escherichia coli. Mol. Microbiol. 40:314-322. [DOI] [PubMed] [Google Scholar]
- 7.Gentschev, I., G. Dietrich, and W. Goebel. 2002. The E. coli alpha-hemolysin secretion system and its use in vaccine development. Trends Microbiol. 10:39-45. [DOI] [PubMed] [Google Scholar]
- 8.Gumpert, J., and C. Hoischen. 1998. Use of cell wall-less bacteria (L-forms) for efficient expression and secretion of heterologous gene products. Curr. Opin. Biotechnol. 9:506-509. [DOI] [PubMed] [Google Scholar]
- 9.Hall, M., and T. J. Silhavy. 1981. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J. Mol. Biol. 146:23-43. [DOI] [PubMed] [Google Scholar]
- 10.Hsiung, H. M., and W. C. MacKellar. 1987. Expression of bovine growth hormone derivatives in Escherichia coli and the use of the derivatives to produce natural sequence growth hormone by cathepsin C cleavage. Methods Enzymol. 153:390-401. [DOI] [PubMed] [Google Scholar]
- 11.Huang, K. J., J. L. Schieberl, and M. M. Igo. 1994. A distant upstream site involved in the negative regulation of the Escherichia coli ompF gene. J. Bacteriol. 176:1309-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong, K. J., and S. Y. Lee. 2000. Secretory production of human leptin in Escherichia coli. Biotechnol. Bioeng. 67:398-407. [PubMed] [Google Scholar]
- 13.Kern, I., and P. Ceglowski. 1995. Secretion of streptokinase fusion proteins from Escherichia coli cells through the hemolysin transporter. Gene 163:53-57. [DOI] [PubMed] [Google Scholar]
- 14.Kim, J. K., E. L. Iannotti, and R. Bajpai. 1999. Extractive recovery of products from fermentation broth. Biotechnol. Bioprocess Eng. 4:1-11. [Google Scholar]
- 15.Klebba, P. E., S. A. Benson, S. Bala, T. Abdullah, J. Reid, S. P. Singh, and H. Nikaido. 1990. Determinants of OmpF porin antigenicity and structure. J. Biol. Chem. 265:6800-6810. [PubMed] [Google Scholar]
- 16.Ko, J. H., D. K. Park, I. C. Kim, S. H. Lee, and S. M. Byun. 1995. High level expression and secretion of streptokinase in Escherichia coli. Biotechnol. Lett. 17:1019-1024. [Google Scholar]
- 17.Kujau, M. J., C. Hoischen, D. Riesenberg, and J. Gumpert. 1998. Expression and secretion of functional miniantibodies McPC603scFvDhlx in cell-wall-less L-form strains of Proteus mirabilis and Escherichia coli: a comparison of the synthesis capacities of L-form strains with an E. coli producer strain. Appl. Microbiol. Biotechnol. 49:51-58. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S. Y. 1996. High cell density cultivation of Escherichia coli. Trends Biotechnol. 14:98-105. [DOI] [PubMed] [Google Scholar]
- 20.Liu, X., and T. Ferenci. 1998. Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli. J. Bacteriol. 180:3917-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, X., N. G. Christina, and T. Ferenci. 2000. Global adaptations resulting from high population densities in Escherichia coli cultures. J. Bacteriol. 182:4158-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makrides, S. C. 1996. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60:512-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy, K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura, S., K. Kitai, K. Soma, Y. Ichikawa, T. Kudo, R. Aono, and K. Horikoshi. 1992. Extracellular production of human immunoglobulin epsilon-chain/gamma 1-chain chimeric Fc polypeptide by Escherichia coli. Biosci. Biotechnol. Biochem. 56:349-350. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi, S., A. Inoue, T. Kita, M. Nakamura, A. C. Chang, S. N. Cohen, and S. Numa. 1979. Nucleotide sequence of cloned cDNA for bovine corticotropin-beta-lipotropin precursor. Nature 278:423-427. [DOI] [PubMed] [Google Scholar]
- 26.Park, S. J., G. Georgiou, and S. Y. Lee. 1999. Secretory production of recombinant protein by high cell density culture of protease negative mutant Escherichia coli strain. Biotechnol. Prog. 15:164-167. [DOI] [PubMed] [Google Scholar]
- 27.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. USA 93:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pugsley, A. P., and M. Schwartz. 1985. Export and secretion of proteins by bacteria. FEMS Microbiol. Rev. 32:3-38. [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 31.Shin, H. C. 2001. Protein folding, misfolding, and refolding of therapeutic proteins. Biotechnol. Bioprocess Eng. 6:237-243. [Google Scholar]
- 32.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 33.Toksoy, E., P. H. Ozdinler, Z. I. Onsan, and B. Kirdar. 1999. High level secretion of TaqI restriction endonuclease by recombinant Escherichia coli. Biotechnol. Tech. 13:803-808. [Google Scholar]
- 34.Wan, E. W., and F. Baneyx. 1998. TolAIII co-overexpression facilitates the recovery of periplasmic recombinant proteins into the growth medium of Escherichia coli. Protein Expr. Purif. 14:13-22. [DOI] [PubMed] [Google Scholar]
- 35.Wienk, H. L., and B. de Kruijff. 1999. Expression, isolation, and characterization of a chloroplast targeting peptide. Protein Expr. Purif. 17:345-350. [DOI] [PubMed] [Google Scholar]
- 36.Yu, P., A. A. Artstidou, and K. Y. San. 1991. Synergistic effects of glycine and bacteriocin release protein in the release of periplasmic protein in recombinant E. coli. Biotechnol. Lett. 13:311-316. [Google Scholar]