Abstract
The dismutation of chlorite into chloride and O2 represents a central step in the reductive pathway of perchlorate that is common to all dissimilatory perchlorate-reducing bacteria and is mediated by a single enzyme, chlorite dismutase. The chlorite dismutase gene cld was isolated and sequenced from the perchlorate-reducing bacterium Dechloromonas agitata strain CKB. Sequence analysis identified an open reading frame of 834 bp that would encode a mature protein with an N-terminal sequence identical to that of the previously purified D. agitata chlorite dismutase enzyme. The predicted translation product of the D. agitata cld gene is a protein of 277 amino acids (aa), including a leader peptide of 26 aa. Primer extension analysis identified a single transcription start site directly downstream of an AT-rich region that could represent the −10 promoter region of the D. agitata cld gene. Northern blot analysis indicated that the cld gene was transcriptionally up-regulated when D. agitata cells were grown in perchlorate-reducing versus aerobic conditions. Slot blot hybridizations with a D. agitata cld probe demonstrated the conservation of the cld gene among perchlorate-reducing bacteria. This study represents the first description of a functional gene associated with microbial perchlorate reduction.
Recent concerns over the environmental contamination of water supplies with perchlorate have focused a significant amount of attention on the microbial metabolism of oxyanions of chlorine (29). Perchlorate contamination poses a significant health threat, as preliminary toxicological studies have demonstrated that it is a competitive inhibitor of iodine uptake by the thyroid gland, and at higher concentrations (6 mg per kg of body weight per day), perchlorate can result in fatal bone marrow disease. In 1998, perchlorate was added to the U.S. Environmental Protection Agency's drinking water candidate contaminant list, and a recommended regulatory concentration of 32 μg liter−1 was set, which, if exceeded, would require stoppage of water usage and remediation (http://cfpub.epa.gov/ncea/cfm/recordisplay.cfm?deid-23292). More recently, as a result of the publication of the first draft of the Environmental Protection Agency review on toxicological and risk characterization data associated with perchlorate contamination, the Californian Department of Health and Human Services revised and lowered its original provisional action level to 4 μg liter−1, which is at the limits of detection using current technologies (32; http://www.dhs.ca.gov/ps/ddwem/chemicals/perchl/actionlevel.htm).
Remediation efforts of perchlorate contamination have focused primarily on microbial processes because of the unique chemical stability and high solubility of perchlorate (29). These processes are based on the ability of perchlorate-reducing bacteria to utilize perchlorate as a physiological electron acceptor in the absence of oxygen and reduce it completely to innocuous chloride. Although it has been recognized for more than 70 years that oxyanions of chlorine are suitable electron acceptors for microbial metabolism (5), this reductive process was originally associated with nitrate-respiring organisms which simply used chlorate as an opportunistic substrate for nitrate reductase (12-14). Growth was not associated with this metabolic pathway, and chlorite was formed as a toxic end product (12-14, 23).
It is now known that specialized microorganisms have evolved that can grow by the anaerobic dissimilation of perchlorate (1, 6, 9, 10, 18, 19, 22, 24, 28, 31), and many dissimilatory perchlorate-reducing bacteria (DPRB) are now in pure culture. The known DPRB isolates represent a broad physiology and phylogeny, with members in the alpha, beta, gamma, and epsilon subclasses of the Proteobacteria (9, 31). The majority of the DPRB are members of the beta subclass of the Proteobacteria and represent two novel genera, the Dechloromonas species and the Dechlorosoma species (1). These organisms are closely related to each other and to the phototrophic Rhodocyclus species. Members of these two groups have been identified and isolated from nearly all environments screened, including both field samples and ex situ bioreactors treating perchlorate-contaminated wastes (9, 18).
Although relatively little is known about the biochemistry of perchlorate reduction, some recent studies have yielded important information. A single oxygen-sensitive perchlorate reductase enzyme of the DPRB strain GR-1 was recently purified and partially characterized (16). This enzyme was located in the periplasm and was a heterodimer in an α3β3 configuration with a total molecular mass of 420 kDa and contained iron, molybdenum, and selenium cofactors (16). In addition to perchlorate, the perchlorate reductase from strain GR-1 also catalyzed the reduction of chlorate, nitrate, iodate, and bromate (16). Perchlorate and chlorate were reduced to chlorite.
The dismutation of chlorite into chloride and O2 is now known to be a central step in the reductive pathway of perchlorate that is common to all DPRB (9). Chlorite dismutation by DPRB is mediated by a highly conserved single enzyme, chlorite dismutase (CD), which is an iron-containing homotetramer with a molecular mass of approximately 120 kDa (9, 27, 30). Phenotypic studies with the DPRB Dechloromonas agitata and Dechlorosoma suillum indicated that CD activity was only present when the organisms were grown anaerobically on perchlorate or chlorate and expression of the CD was negatively regulated by oxygen and nitrate (7). Furthermore, studies with a recently developed immunoprobe specific for purified CD from D. agitata strain CKB indicated that the CD was present on the outer membrane of all DPRB and was conserved among the DPRB, regardless of their phylogenetic affiliation (21).
Although significant advances have been made in the last five years regarding the microbiology of perchlorate reduction, there is still nothing known about the genetic systems involved in this metabolism. The gene sequence for CD from Ideonella dechloratans was recently made available in the GenBank database (accession number AJ296077) by T. Nilsson (Karlstad University, Karlstad, Sweden); however, there is currently no information available on this gene in the literature. Here we report on the sequencing and transcriptional analysis of the CD gene (cld) of D. agitata strain CKB and investigate its use as a metabolic probe. This represents the first description of a functional gene involved in the microbial respiration of perchlorate.
MATERIALS AND METHODS
Growth conditions.
Both D. agitata and I. dechloratans (ATCC 51718) were grown at 30°C on basal medium (6) following standard anaerobic techniques (15). Oxygen was removed by boiling and cooling under an N2-CO2 (80/20, vol/vol) gas phase. Acetate (10 mM) and either chlorate or perchlorate (10 mM) were added, respectively, as electron donor and acceptor from sterile anoxic stock solutions. Aerobic growth was achieved utilizing the same basal medium with oxygen as the electron acceptor. Escherichia coli strains XL1-Blue MRF′ and SOLR (Stratagene, La Jolla, Calif.), used for library screening, were maintained aerobically on Luria-Bertani (LB) medium supplemented with 0.2% maltose and 10 mM MgSO4 at 32°C. SOLR excision reactions were plated on LB medium supplemented with ampicillin (50 μg/ml) to select for the pBluescript vector (Stratagene).
Nucleic acid isolation.
Genomic DNA (gDNA) was extracted using the PUREGENE DNA isolation kit (Gentra Systems Inc., Minneapolis, Minn.). For RNA extractions, mid-log-phase cultures were filtered and RNA was extracted using Lysing Matrix B tubes (Bio 101, Inc., Carlsbad, Calif.) and RNAwiz reagent (Ambion, Austin, Tex.). To assess the level of RNA degradation, samples were electrophoresed on a 1.0% (wt/vol) nondenaturing Tris-acetate-EDTA (TAE) agarose gel.
Southern blotting.
Five separate restriction enzyme digests were performed on D. agitata gDNA. Each digest was incubated at 37°C for 1 h and contained the following: 100 ng of DNA, 1 μl of buffer, 1 μl of enzyme (BamHI, EcoRI, HindIII, KpnI, or SacI), and water to 10 μl. Enzymes and buffers were from Promega, Madison, Wis. The digests were electrophoresed on a 0.7% TAE agarose gel. The gel was then denatured, neutralized, and blotted using standard techniques (26).
Genomic library construction.
EcoRI-digested D. agitata total gDNA was electrophoresed on a 0.7% TAE agarose gel. Resulting fragments ranging from 3.0 to 8.0 kb were excised from the gel, purified (GeneClean II; Bio 101), and ligated into Lambda ZAP II predigested EcoRI-calf intestine alkaline phosphatase-treated DNA (Stratagene). Recombinant lambda vectors were packaged using the Gigapack III Gold packaging extract with E. coli strain XL1-Blue MRF′ as the host strain for library plating and amplification. Lambda ligation, packaging, and library amplification methods were all performed according to the manufacturer’s instructions (Stratagene).
Library screening.
Recombinant lambda DNA was transferred to nylon membranes in accordance with the instructions of the manufacturer (Stratagene). PCR primers ICD-741F (5′-TATCTCCAAGGACAAGTCGC-3′) and ICD-1140R (5′-TCAATTGCCCATCGACAGCGT-3′) were utilized to construct a digoxigenin-labeled probe specific to 399 bases of the I. dechloratans CD gene (GenBank accession no. AJ296007; see CD probe design). Membranes were hybridized at 46°C using 5 μl of probe per 10 ml of Easy Hyb solution (Roche Molecular Biochemicals, Indianapolis, Ind.) and detected using the digoxigenin luminescence detection kit (Roche Molecular Biochemicals). Positive lambda clones were identified and used to isolate a phagemid containing the cloned D. agitata insert by in vivo excision (Stratagene).
DNA sequencing and analysis.
Recombinant phagemid DNA was sequenced using the ThermoSequenase cycle sequencing kit (U.S. Biochemical Corp., Cleveland, Ohio). A combination of vector-specific and insert-specific primers was used to completely sequence the D. agitata cld gene and flanking regions. Sequence entry and manipulation were performed using MacVector sequence analysis software for the Macintosh (version 7.0; Oxford Molecular) and the Se-Al Sequence Alignment Editor (version 1.0; created by A. Rambaut, University of Oxford). Protein sequence similarity was determined by BLAST 2.2 analysis (3).
Expression vector analysis.
The primer set CD-79F (5′-ATGGATGCGAAGCCGCCAAT-3′) and CD-831R (5′-GCGTCCCATGGACAACGTAT-3′) was used in a standard PCR to amplify a 752-bp region of the cld open reading frame (ORF) excluding the signal peptide and stop codon. This amplified product was cloned into E. coli using the pBAD/TOPO Thiofusion expression kit (Invitrogen, Carlsbad, Calif.). Clones containing the cld insert in the correct orientation and reading frame were determined via sequencing using vector primers. To test for cld expression, single colonies were picked and grown overnight at 37°C in 5 ml of LB broth supplemented with ampicillin (50 μg/ml). The next day, 0.1 ml of the overnight cultures was inoculated into 10 ml of LB broth supplemented with ampicillin (50 μg/ml) and grown to an optical density at 600 nm of 0.5. To induce transcription from the vector's promoter, l-arabinose was added to the cultures at a final concentration of 0.2% and the cultures were incubated at 37°C for 4 h. The cells were washed and resuspended in 100 mM phosphate buffer, pH 7.3, to an optical density at 600 nm of 1.5. From the suspensions, 2-μl aliquots of whole cells from both the cld expression culture and a negative control culture were spotted onto an Immobilon-P (Millipore, Bedford, Mass.) membrane along with 5 μg of diluted pure CD enzyme as a positive control. The dot blots were probed with the CD-specific immunoprobe and detected as previously described (21).
Primer extension.
Primer extension reactions with [γ-33P]ATP were performed using the Primer Extension System-AMV reverse transcriptase kit (Promega). Primer CD-PX1 (5′-ACAGTAGTGCCATGAACGTT-3′), which is specific to the 5′ end of the CD mRNA, was developed and used in a primer extension reaction with 15 μg of total RNA from D. agitata cells grown under perchlorate-reducing conditions. Primer extension reactions were electrophoresed alongside sequencing reactions using the same primer on a standard 8 M urea-6.0% polyacrylamide gel for 3 h at 39 W.
CD probe design.
A probe corresponding to the 3′ end of the D. agitata cld gene was labeled via PCR using the primers CD-441F (5′-AAAAGATAAATCGCCAAATC-3′) and CD-834R (5′-TTAGCGTCCCATGGACAACG-3′). Digoxigenin-11-dUTP (Roche Molecular Biochemicals) was incorporated by PCR as previously described (17). All PCR reagents were from Promega. Reactions were cycled at the following parameters: 94°C for 3 min; followed by 30 cycles consisting of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min; and ending with a 10-min extension at 72°C.
Northern blot analysis.
Five micrograms of total RNA from D. agitata cells grown under various conditions was electrophoresed on a 1.0% glyoxal-agarose gel and blotted according to the NorthernMax-Gly glyoxal-based system for Northern blots manual (Ambion). Following transfer, the membrane was UV cross-linked (120 mJ/cm2) and hybridized according to the manufacturer's instructions at 50°C with 5 μl of digoxigenin-labeled D. agitata cld probe per 10 ml of Easy Hyb solution (Roche Molecular Biochemicals). Detection was performed using the digoxigenin luminescence detection kit (Roche Molecular Biochemicals).
DNA slot blotting and screening.
Two hundred fifty nanograms of gDNA from previously isolated perchlorate- and chlorate-reducing strains and close relatives unable to reduce perchlorate were blotted onto Zeta-Probe blotting membrane using the Bio-Dot SF Microfiltration Apparatus (Bio-Rad, Hercules, Calif.). Following transfer, the blot was UV cross-linked (120 mJ/cm2) and hybridized at 46°C with the D. agitata cld probe following the same method as in the Northern blot analysis.
GenBank accession numbers.
All sequence data generated for this study were submitted to the GenBank database under accession number AY124796. The GenBank accession numbers for the 16S ribosomal DNA (rDNA) sequences of the organisms in Fig. 5B are as follows: D. agitata, AF047462; Rhodocyclus tenuis, D16209; Dechloromonas aromatica, AY032610; Dechloromonas strain JJ, AY032611; Dechlorospirillum anomalous strain WD, AF170352; Magnetospirillum magnetotacticum, Y10110; Pseudomonas sp. strain PK, AF170358; Pseudomonas stutzeri, U26415; Dechloromarinus sp. strain NSS, AF170359; Dechlorosoma suillum, AF170348; Azoarcus strain LT-1, AY124797; I. dechloratans, X72724; E. coli, J01859.
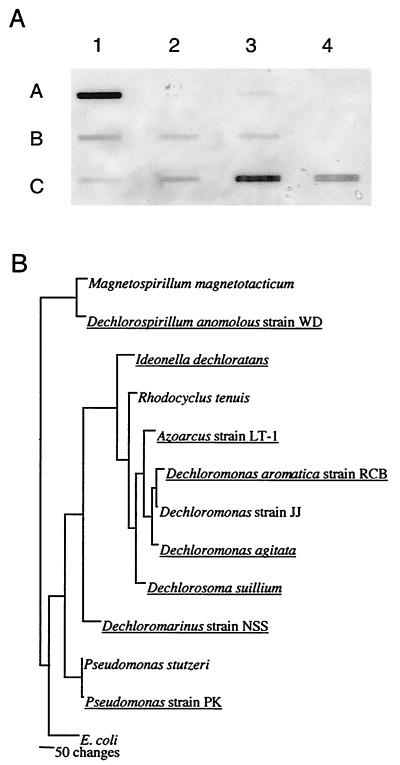
FIG. 5.
(A) Slot blot hybridization of gDNAs from DPRB and non-perchlorate-reducing close relatives using the D. agitata probe to the 3′ end of the CD gene. Organisms capable of chlorate or perchlorate reduction are underlined. Row A (left to right): lane 1, D. agitata; lane 2, Rhodocyclus tenuis; lane 3, Dechloromonas aromatica; lane 4, Dechloromonas sp. strain JJ. Row B: lane 1, Dechlorospirillum anomalous strain WD; lane 2, Magnetospirillum magnetotacticum; lane 3, Pseudomonas sp. strain PK; lane 4, Pseudomonas stutzeri. Row C: lane 1, Dechloromarinus sp. strain NSS; lane 2, Dechlorosoma suillum; lane 3, Azoarcus sp. strain LT-1; lane 4, I. dechloratans. (B) Phylogenetic tree of the organisms used in the slot blot hybridization based on 16S rDNA sequences (1, 9).
RESULTS AND DISCUSSION
I. dechloratans CD primers.
As outlined in the introduction, the gene sequence for CD (cld) from I. dechloratans was recently submitted to the GenBank database (accession number AJ296077); however, no information regarding this gene is currently available in the literature. Because it was not known which part of the putative I. dechloratans CD gene contained the most highly conserved sequence region, a series of primers was designed to specifically amplify the 5′ end, central portion, and 3′ end of the CD gene from I. dechloratans. Each primer set was used in a PCR using either I. dechloratans or D. agitata gDNA as the template. PCR amplification of I. dechloratans DNA yielded an 857-bp product using a primer set spanning the central region of the putative coding sequence for CD and yielded a 430-bp product using a primer set corresponding to the 5′ end of the cld gene. A 399-bp product corresponding to the 3′ end of the cld gene was amplified from I. dechloratans gDNA using the primer set ICD-741F and ICD-1140R. No amplification products of the desired length were obtained with any of the I. dechloratans-specific primer sets using D. agitata gDNA as the template.
Southern blot and library screening.
To determine if the I. dechloratans cld gene sequence could be used to screen a D. agitata genomic library, a Southern blot was performed using the labeled amplification products of I. dechloratans (see above) as probes. Using the probe corresponding to the 3′ end of the I. dechloratans cld gene, a positive signal was visible in each lane of digested D. agitata gDNA (data not shown). However, no hybridization signal resulted when the same Southern blot was hybridized with either the probe corresponding to the 5′ end of the I. dechloratans CD gene or the probe spanning the central portion of the gene (data not shown). This result indicates that the 3′ end of the cld gene is more conserved than the 5′ end and likely encodes the protoheme IX group binding region observed in the mature proteins from I. dechloratans (27) and the DPRB strain GR-1 (30) which is part of the active site of the CD enzyme. As such, the I. dechloratans probe corresponding to the 3′ end of the cld gene was used in all subsequent screening experiments.
Given the positive results from the Southern blot, a D. agitata gDNA lambda library was constructed and screened using the I. dechloratans cld probe. Screening of approximately 1,200 plaques resulted in a single positive clone. Following rescreening, the phagemid was produced and digested with EcoRI to yield an insert of approximately 7.5 kb.
Sequence analysis.
Previously we purified and partially characterized the CD enzyme from D. agitata (9). The purified CD was a homotetramer with a molecular mass of 120 kDa and a specific activity of 1,928 μmol of chlorite dismutated per mg of protein per min (9). These are similar to the molecular mass and specific activity observed for the CD previously purified from the perchlorate-reducing bacteria strain GR-1 (30) and I. dechloratans (27). Commercial N-terminal sequencing (Commonwealth Biotechnologies Inc., Richmond, Va.) of the CD purified from D. agitata revealed the following 20-amino acid (aa) sequence in the mature protein: DAKPPMAMPDMTKILTAPGV.
Sequencing of the D. agitata insert revealed an ORF of 834 bp with a predicted N-terminal sequence of the mature protein identical to that of the purified D. agitata CD. In addition, sequence similarity to the putative I. dechloratans cld gene sequence provided further evidence that this ORF encodes the D. agitata CD protein (Fig. 2). BLAST 2.2 analysis (3) indicated that the D. agitata CD sequence was 71% similar to I. dechloratans CD at the amino acid level. No other proteins in the GenBank database were more than 24% similar to the product encoded by the D. agitata cld gene, emphasizing the unique nature of the CD enzyme.
FIG. 2.
Amino acid sequence alignment of the mature D. agitata and putative I. dechloratans and M. magnetotacticum CD proteins. Sequence identities are shaded.
The predicted translation product of the D. agitata cld gene is a protein of 277 aa including a leader peptide of 26 aa (Fig. 1). The D. agitata CD amino acid sequence was manually aligned to the I. dechloratans cld gene product, and, based on this very limited comparative analysis, regions of amino acid sequence conservation could be identified within the CD protein (Fig. 2). As initially indicated by the Southern blot analysis, very low sequence similarity exists at the N terminus of the two proteins. In addition, no common promoter region could be distinguished from alignment analysis. However, regions of conservation in the central and C terminus of the mature CD protein were identified. Comparative analyses with more CD sequences are needed to refine our current view of sequence conservation and functional motifs for this protein.
FIG. 1.
Nucleotide and predicted amino acid sequence of the upstream region of the D. agitata CD gene. The asterisk indicates the transcription start site, and the putative −10 promoter region immediately upstream of the start is double-underlined. The RBS is underlined. The leader peptide is indicated; the hydrophobic region and cleavage site of the leader are shaded.
Expression vector analysis of the cloned cld gene.
To confirm that the cloned gene sequence encoded the CD enzyme, the PCR-amplified D. agitata cld gene was inserted into an E. coli expression vector which placed the gene under the control of an arabinose promoter. Whole-cell immunoprobing of the resulting E. coli transformant using a CD antibody conjugate (21) resulted in a positive signal, verifying that the transformed E. coli culture was producing the CD enzyme (data not shown). The negative control, an E. coli culture transformed with an expression vector containing a 16-kDa His-Patch thioredoxin fusion protein gene, failed to produce a signal in the immunoprobe analysis.
Transcription start site analysis of the cld gene.
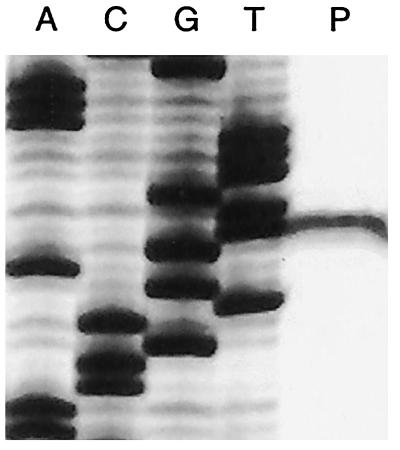
Because no information is available regarding promoter structure in the beta Proteobacterium D. agitata, primer extension reactions were performed to identify the cld gene promoter region in D. agitata. Reactions contained total RNA purified from a D. agitata culture grown under anaerobic conditions with ClO4− as the electron acceptor. Primer CD-PX1, designed to target the 5′ end of the CD mRNA, yielded a single extension product (Fig. 3). From the primer extension data, a putative −10 promoter region was identified (5′-AAATTT-3′) that is located 8 bp upstream of the transcription start site (Fig. 1). The sole transcriptional start site detected by primer extension indicates that the cld gene encodes a 277-aa product and that transcription begins 164 bp upstream of the DNA region encoding the N-terminal sequence as determined from the purified D. agitata CD protein (9).
FIG. 3.
Primer extension using RNA from a perchlorate-grown culture of D. agitata. The sequencing ladder was generated using the same primer as the primer extension reaction. The relevant sequence is 5′-GAAATTTGTTGAGTCGCCAA-3′, with the underlined nucleotide denoting the start site of transcription.
Prior to the N terminus of the mature CD protein, there is a region of DNA that, when read in-frame, could encode a 26-aa peptide (Fig. 1). This intervening amino acid sequence may correspond to a signal peptide for the CD protein that could play a role in targeting the CD to the bacterial cell membrane (6, 9, 21). In support of the membrane-bound nature of the CD enzyme, previous biochemical studies demonstrated that CD activity was associated with both the cell membrane and soluble fractions of a lysed-cell preparation of D. agitata when prepared using a French press (6, 9), which suggested that the CD was loosely bound to the membrane or was present in the periplasm. In I. dechloratans, the CD enzyme activity was located primarily in the periplasmic extract (27). Furthermore, the CD-specific immunoprobe readily bound to whole cells of both of these organisms, suggesting that the antigenic portion of the CD was present on the outer membrane (21). In contrast, van Ginkel and coworkers claimed that the CD activity of the DPRB strain GR-1 was located exclusively in the soluble fraction; however, no attempt was made to separate out the periplasmic fractions from the soluble fractions in that study (30).
The identification of a ribosome binding site (RBS) located immediately upstream of the potential leader peptide (Fig. 1) lends support to the hypothesis that the leader peptide of the D. agitata cld gene is translated and later removed to produce the mature CD protein as determined from the N-terminal sequence of the purified CD enzyme. The RBS has a sequence of 5′-AGAAAGG′-3′, which is the exact reverse complement of the conserved terminal 3′ end of the bacterial 16S rRNA.
Protein motifs and signal peptide.
Both the primary and mature putative protein sequences for the D. agitata CD were submitted to the PredictProtein server (http://cubic.bioc.columbia.edu/predictprotein/submit_def.html). This server submits protein sequences to a number of modeling programs available online (25). Interestingly, the CD sequence did not match any sequence currently in the protein data bank to allow for three-dimensional modeling. As such, homology modeling was not possible for either the primary or mature CD sequence. The DAS transmembrane prediction server (11) found only one likely transmembrane region in the leader sequence (Fig. 1). This hydrophobic region spans aa 6 to 17 (GLLLTFMALLSV) and corresponds to a putative helix located in the leader sequence. The results from the SignalP server (20) further indicated that the leader sequence was indeed a signal peptide with a characteristic positively charged N terminus, a hydrophobic helix and a cleavage site motif of SQA-QQA (Fig. 1) (4).
Transcriptional regulation of CD.
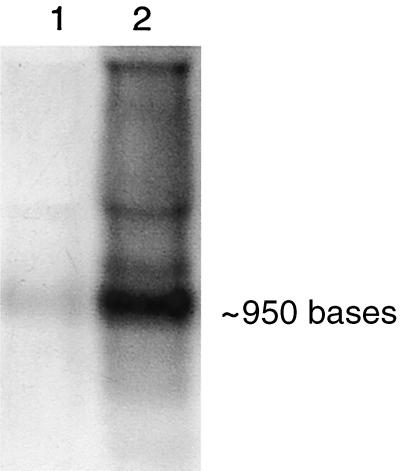
Previous physiological studies on the environmental factors that influence microbial perchlorate reduction demonstrated that perchlorate reduction was dependent on the presence of the CD enzyme which was induced during the anaerobic metabolism of perchlorate (7). In that study, CD expression was negatively regulated by oxygen and nitrate even in the presence of perchlorate (7). To characterize the transcriptional regulation of the D. agitata cld gene, a Northern blot containing RNA from D. agitata grown with O2 or ClO4− was performed using a D. agitata probe that corresponded to the 3′ end of the cld gene. Hybridization analysis detected a band of approximately 950 bases in both RNA samples (Fig. 4). A faint band was evident in the aerobic culture, but an intense signal was obtained from the RNA of the perchlorate-grown culture, indicating that the cld gene is expressed at basal levels under aerobic conditions but that transcription of this gene is greatly increased when the cells are grown under perchlorate-reducing conditions. This result supports the previous physiological studies (7) and correlates well with the results obtained using the CD-specific immunoprobe which only bound to the D. agitata cells grown under perchlorate-reducing conditions and failed to bind to cells from an aerobic culture (21). Although a faint signal was detected in the Northern blot from aerobic D. agitata cultures with the cld probe, it is possible that the amount of CD protein produced from this basal-level transcription is below the limit of detection by the immunoprobe or that there may be additional genetic regulation of CD at either the translational or posttranslational level.
FIG. 4.
Northern blot of D. agitata RNA hybridized with the D. agitata probe to the 3′ end of the CD gene. Lanes: 1, aerobic growth; 2, anaerobic growth with perchlorate as the electron acceptor.
It is also apparent from the Northern blot results that the CD gene is not transcribed as part of an operon. Only a single band representing a mRNA of approximately 950 bases was detected. Since the length of the D. agitata cld gene from the transcription start site to the stop codon is 921 bp, this suggests that the only ORF contained on this transcript is that for CD. Therefore, it appears as though the cld gene is transcribed independent of other genes that are also involved in this metabolic pathway, such as perchlorate reductase.
Utility of D. agitata CD gene probe.
Microbial dissimilatory perchlorate reduction is a phylogenetically diverse metabolism, and DPRB isolates have been placed in four of the five subclasses of the Proteobacteria (9). Furthermore, 16S rDNA sequence analysis indicates that several DPRB show less than 0.5% divergence in their 16S rDNA sequence with non-perchlorate-reducing relatives (1, 2). As such, DPRB molecular probes based on signature nucleotide sequences within the 16S rDNA sequence are of limited use. To determine the utility of the D. agitata cld gene as a metabolic probe, gDNAs of several phylogenetically distinct DPRB representing the alpha, beta, and gamma subclasses of the Proteobacteria were screened in a slot blot assay (Fig. 5A). Organisms closely related to the DPRB which do not grow by dissimilatory perchlorate reduction were also included in the analysis. All the DPRB tested positive regardless of phylogenetic affiliation using the D. agitata probe to the 3′-end of the cld gene (Fig. 5A). Strong hybridization signals were obtained from the DPRB Azoarcus sp. strain LT-1, I. dechloratans, and D. agitata. Obvious signals were also obtained from the DPRB Dechlorospirillum sp. strain WD, Dechloromarinus sp. strain NSS, and Dechlorosoma suillum, while weak, but still visible, signals were obtained from the gDNAs of the other DPRB. No signal was obtained from the non-perchlorate reducers Rhodocyclus tenuis, Dechloromonas sp. strain JJ, and Pseudomonas stutzeri (Fig. 5A).
Differences in hybridization intensities for the DPRB indicate that conservation of the cld gene sequence varies among the perchlorate-reducing bacteria and that these differences do not reflect the 16S rDNA-based phylogeny of these organisms (Fig. 5B). In support of this, a similar variation in signal response based on dot blot intensity was observed in studies with the CD-specific immunoprobe for the various DPRB, suggesting that some minor differences exist in the mature CD protein in these organisms (21).
Although sequence variation of the cld gene among the DPRB likely exists, the D. agitata cld probe hybridized to all DPRB tested and failed to hybridize to any of the non-DPRB, despite their close phylogenetic relationships (Fig. 5). Predictably, the most intense signal resulted from the positive control D. agitata. Rhodocyclus tenuis, a phylogenetically close non-perchlorate-reducing relative to D. agitata (1, 6), did not produce any visible hybridization signal. Similarly, Dechloromonas sp. strain JJ, the only known Dechloromonas species that is incapable of perchlorate reduction (8) and the closest non-perchlorate-reducing relative to the DPRB Dechloromonas aromatica strain RCB, was also negative. Similar results were observed with the DPRB Pseudomonas sp. strain PK and its closest non-DPRB relative, Pseudomonas stutzeri, although these organisms share more than 99% 16S rDNA sequence similarity (9).
Interestingly, a weak hybridization signal was obtained from Magnetospirillum magnetotacticum, a bacterium closely related to the DPRB Dechlorospirillum anomalous strain WD (2, 9, 10). This was unexpected as previous studies in our laboratory demonstrated that several Magnetospirillum species, including M. magnetotacticum, did not grow by dissimilatory perchlorate reduction (10). A subsequent search of the M. magnetotacticum genome (http://genome.ornl.gov/microbial/mmag/) revealed the presence of a putative CD gene. The physiological ramifications of this discovery are as yet unknown.
CD is now known to be a central enzyme involved in microbial perchlorate reduction (9, 27, 30). The present study describes the identification and characterization of the cld gene encoding CD in D. agitata. As such, this is the first description of a functional gene associated with microbial perchlorate respiration, a ubiquitous metabolism in the environment (9). Analyses of the transcriptional regulation of the D. agitata cld gene demonstrated that transcription of the gene encoding CD significantly increased when the organism was grown on perchlorate as opposed to aerobic growth, supporting previous observations on the environmental factors that influence microbial perchlorate reduction (7).
Previous studies have indicated that the CD enzyme is unique to DPRB and is highly conserved among the phylogenetically diverse DPRB (9, 21). It has also been shown that relatives of the known DPRB that cannot grow by perchlorate respiration are incapable of the dismutation of chlorite (6, 9). In support of these observations, the CD gene probe developed in the present study hybridized to all DPRB tested, regardless of phylogenetic affiliation. In general, the gene probe was specific to DPRB, and cross hybridization with non-DPRB was not observed despite their phylogenetic similarity. However, one non-DPRB, M. magnetotacticum, did hybridize to the gene probe and examination of the M. magnetotacticum genome sequence identified the presence of a putative cld gene. As previous physiological studies with this and other Magnetospirillum species indicated that they could not grow by perchlorate or chlorate respiration (10), the functional role of the cld gene in this organism is unknown.
The CD gene probe developed in this study represents a potential tool for monitoring active perchlorate metabolism by mixed DPRB populations in environmental samples during a bioremediative process. Additional genetic analyses are needed to ascertain the specific environmental parameters and regulatory mechanisms that affect CD activity.
Acknowledgments
This work was supported by grant DACA72-00-C-0016 from the U.S. Department of Defense to J.D.C. and L.A.A.
REFERENCES
- 1.Achenbach, L. A., U. Michaelidou, R. A. Bruce, J. Fryman, and J. D. Coates. 2001. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. Evol. Microbiol. 51:527-533. [DOI] [PubMed] [Google Scholar]
- 2.Achenbach, L. A., and J. D. Coates. 2000. Disparity between bacterial phylogeny and physiology. ASM News 66:714-716. [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arkowitz, R., and M. Bassilana. 1994. Protein translocation in Escherichia coli. Biochim. Biophys. Acta 1197:311-343. [DOI] [PubMed] [Google Scholar]
- 5.Aslander, A. 1928. Experiments on the eradication of Canada thistle Cirsium arvense with chlorates and other herbicides. J. Agric. Res. 36:915-935. [Google Scholar]
- 6.Bruce, R. A., L. A. Achenbach, and J. D. Coates. 1999. Reduction of (per)-chlorate by a novel organism isolated from paper mill waste. Environ. Microbiol. 1:319-329. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri, S. K., S. M. O'Connor, R. L. Gustavson, L. A. Achenbach, and J. D. Coates. 2002. Environmental factors that control microbial perchlorate reduction. Appl. Environ. Microbiol. 68:4425-4430. [DOI] [PMC free article] [PubMed]
- 8.Coates, J. D., R. Chakraborty, J. G. Lack, S. M. O'Connor, K. A. Cole, K. S. Bender, and L. A. Achenbach. 2001. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411:1039-1043. [DOI] [PubMed] [Google Scholar]
- 9.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. Ubiquity and diversity of dissimilatory (per)-chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coates, J. D., U. Michaelidou, S. M. O'Connor, R. A. Bruce, and L. A. Achenbach. 2000. The diverse microbiology of (per)chlorate reduction, p. 257-270. In E. D. Urbansky (ed.), Perchlorate in the environment. Kluwer Academic/Plenum, New York, N.Y.
- 11.Cserzo, M., E. Wallin, I. Simon, G. Von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Prot. Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 12.de Groot, G. N., and A. H. Stouthamer. 1969. Regulation of reductase formation in Proteus mirabilis. I. Formation of reductases and enzymes of the formic hydrogenlyase complex in the wild type and in chlorate-resistant mutants. Arch. Microbiol. 66:220-233. [PubMed] [Google Scholar]
- 13.Hackenthal, E. 1965. Die reduktion von perchlorat durch bacterien. II. Die identität der nitratreduktase und des perchlorat reduzierenden enzyms aus B. cereus. Biochem. Pharm. 14:1313-1324. [DOI] [PubMed] [Google Scholar]
- 14.Hackenthal, E., W. Mannheim, R. Hackenthal, and R. Becher. 1964. Die reduktion von perchlorat durch bakterien. I. Untersuchungen an intaken zellen. Biochem. Pharm. 13:195-206. [DOI] [PubMed] [Google Scholar]
- 15.Hungate, R. E. 1969. A roll tube method for cultivation of strict anaerobes. Methods Microbiol. 3B:117-132. [Google Scholar]
- 16.Kengen, S. W. M., G. B. Rikken, W. R. Hagen, C. G. Van Ginkel, and A. J. M. Stams. 1999. Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J. Bacteriol. 181:6706-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanzillo, J. J. 1990. Preparation of digoxigenin-labeled probes by the polymerase chain reaction. Biotechniques 8:620-622. [PubMed] [Google Scholar]
- 18.Logan, B. E., H. Zhang, P. Mulvaney, M. G. Milner, I. M. Head, and R. F. Unz. 2001. Kinetics of perchlorate- and chlorate-respiring bacteria. Appl. Environ. Microbiol. 67:2499-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malmqvist, A., T. Welander, E. Moore, A. Ternstrom, G. Molin, and I.-M. Stenstrom. 1994. Ideonella dechloratans gen. nov., sp. nov., a new bacterium capable of growing anaerobically with chlorate as an electron acceptor. Syst. Appl. Microbiol. 17:58-64. [Google Scholar]
- 20.Nielsen, H., J. Engelbrecht, S. Brunak, and G. Von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot. Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 21.O'Connor, S. M., and J. D. Coates. 2002. Universal immunoprobe for (per)-chlorate-reducing bacteria. Appl. Environ. Microbiol. 68:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rikken, G. B., A. G. M. Kroon, and C. G. van Ginkel. 1996. Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 45:420-426. [Google Scholar]
- 23.Roldan, M. D., F. Reyes, C. Moreno-Vivian, and F. Castillo. 1994. Chlorate and nitrate reduction in the phototrophic bacteria Rhodobacter capsulatus and Rhodobacter sphaeroides. Curr. Microbiol. 29:241-245. [Google Scholar]
- 24.Romanenko, V. I., V. N. Korenkov, and S. I. Kuznetsov. 1976. Bacterial decomposition of ammonium perchlorate. Mikrobiologiia 45:204-209. [PubMed] [Google Scholar]
- 25.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Plainview, N.Y.
- 27.Stenklo, K., H. D. Thorell, H. Bergius, R. Aasa, and T. Nilsson. 2001. Chlorite dismutase from Ideonella dechloratans. J. Biol. Inorgan. Chem. 6:601-607. [DOI] [PubMed] [Google Scholar]
- 28.Stepanyuk, V. V., G. F. Smirnova, T. M. Klyushnikova, N. I. Kanyuk, L. P. Panchenko, T. M. Nogina, and V. I. Prima. 1992. New species of the Acinetobacter genus Acinetobacter thermotoleranticus sp. nov. Mikrobiologiia 61:347-356. [Google Scholar]
- 29.Urbansky, E. T. 1998. Perchlorate chemistry: implications for analysis and remediation. Bioremed. J. 2:81-95. [Google Scholar]
- 30.van Ginkel, C. G., G. B. Rikken, A. G. M. Kroon, and S. W. M. Kengen. 1996. Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch. Microbiol. 166:321-326. [DOI] [PubMed] [Google Scholar]
- 31.Wallace, W., T. Ward, A. Breen, and H. Attaway. 1996. Identification of an anaerobic bacterium which reduces perchlorate and chlorate as Wolinella succinogenes. J. Ind. Microbiol. 16:68-72. [Google Scholar]
- 32.Wirt, K., M. Laikhlman, J. Rohrer, and P. Jackson. 1998. Low-level perchlorate analysis in drinking water and groundwater by ion chromatography. Am. Environ. Lab. 10:1-5. [Google Scholar]