Abstract
The entire 127,923-bp sequence of the toxin-encoding plasmid pBtoxis from Bacillus thuringiensis subsp. israelensis is presented and analyzed. In addition to the four known Cry and two known Cyt toxins, a third Cyt-type sequence was found with an additional C-terminal domain previously unseen in such proteins. Many plasmid-encoded genes could be involved in several functions other than toxin production. The most striking of these are several genes potentially affecting host sporulation and germination and a set of genes for the production and export of a peptide antibiotic.
Isolates of Bacillus thuringiensis are the biological control agents most widely used to eradicate insect pests of crops or vectors of human disease. For the latter application, Bacillus thuringiensis subsp. israelensis is the bioinsecticide of choice in programs worldwide to control mosquitoes and blackfly vectors (29). The insect pathogenicity of this bacterium depends on the presence of the pBtoxis megaplasmid (13) that encodes all six of the previously described toxins in this isolate (Cry4Aa, Cry4Ba, Cry10Aa, Cry11Aa, Cyt1Aa, and Cyt2Ba) (7, 18). In addition, the plasmid carries several insertion sequences and encodes two further proteins (P19 and P20) with roles in promoting crystal formation and enhancing cell viability, probably by acting as chaperones (12, 27, 50). The pBtoxis plasmid has been partially mapped (6, 7), but the nucleotide sequence is limited to toxin genes and their flanking regions. Since the toxicity of the B. thuringiensis subsp. israelensis crystal is greater than that of any combination of the known toxins derived from it (9), it seems that other toxins or virulence factors may play a role in the activity of wild-type crystals. One possible source of such additional factors is the approximately 80% of the pBtoxis sequence that has not previously been analyzed. In order to understand fully this highly important virulence plasmid, we have therefore determined its entire nucleotide sequence as presented here.
MATERIALS AND METHODS
Plasmid preparation.
The pBtoxis plasmid was prepared from B. thuringiensis subsp. israelensis strain 4Q2-72 (also known as 4Q5) and purified on a CsCl-ethidium bromide density gradient as previously described (6).
Sequencing and analysis.
Plasmid DNA was sonicated and size fractionated on agarose gels. Two libraries were generated in pUC18 using insert sizes of 1.4 to 2 and 2 to 4 kb. Each clone was sequenced once from each end using ABI Big-Dye terminator chemistry on ABI3700 capillary sequencing machines. The final sequence was generated from 1,467 sequencing reads, giving 6.4-fold total coverage. All repeats were bridged by clone end read pairs or end-sequenced PCR products to confirm the assembly.
The finished sequence was annotated using Artemis software (41). Potential coding sequences were identified by codon usage (34) and positional base preference methods and compared to the nonredundant protein databases using BLAST (3) and FASTA (38) software. The entire DNA sequence was also compared in all six reading frames against the nonredundant protein databases, using BLASTX to identify any possible coding sequences previously missed. Exploration of the functions of Bacillus subtilis homologues was facilitated by the Subtilist database (33). Protein motifs were identified using InterPRO (5), transmembrane domains were identified with TMHMM (23), and signal sequences were identified with SignalP version 2.0 (35).
PCR analysis of other B. thuringiensis strains.
Oligonucleotide primers (2.5D, CAGCTTCTTTCGAACATAAGAAGTC, and 2.5R, GATCTCGAAGTATTCTTATATCTGC) were designed from part of the pBt007 sequence in order to produce a 613-bp amplicon in PCRs under the following conditions: 95°C for 180 s, 48 to 54°C for 90 s, and 72°C for 120 s for 1 cycle and 94°C for 45 s, 48 to 54°C for 50 s, and 72°C for 90 s for 29 cycles. DNAs from vegetative cells from a variety of B. thuringiensis strains were added to the PCR mixtures as template DNA, and the resulting products were analyzed by agarose gel electrophoresis. Most standard B. thuringiensis strains were kindly supplied by D. R. Zeigler (Bacillus Genetic Stock Center, Columbus, Ohio).
Nucleotide sequence accession number.
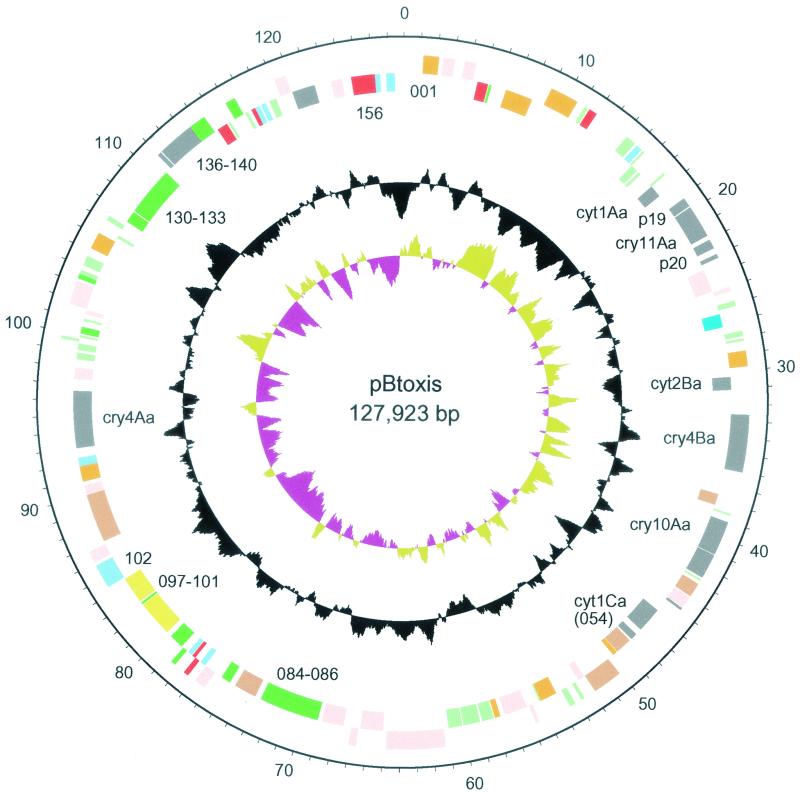
The full-length 127,923-bp pBtoxis sequence and annotation (Fig. 1) has been deposited in the EMBL database under accession number AL731825.
FIG. 1.
Circular representation of pBtoxis. The inner circle represents GC bias [(G − C)/(G + C)], with positive values in khaki and negative values in purple; the second circle represents G+C content; and the outer two circles represent predicted genes on the reverse and forward strands (selected CDSs are numbered for reference). Color coding for the genes is as follows: gray, toxin and peptide antibiotic; pink, transposon related; orange, conserved hypothetical; red, DNA metabolism; blue, regulatory; bright green, surface associated; pale green, unknown; yellow, miscellaneous metabolic genes. The outer scale is marked in kilobases.
RESULTS AND DISCUSSION
In silico restriction analyses of the complete 127,923-bp pBtoxis sequence agree with the previously published map (7), except that all of the predicted restriction fragments are slightly smaller than previously estimated, consistent with the slightly smaller overall size of the plasmid (128 kb compared to the 137 kb proposed). The placement of genes on the restriction map agreed with those detected in the sequence, with the exceptions of cyt2Ba, cry4Ba, and cry10Aa, which are in the same positions but inverted in order and orientation. pBtoxis properties are summarized in Table 1, and predicted genes are described in Table 2.
TABLE 1.
Summary of pBtoxis properties
| Property | Value |
|---|---|
| Total size | 127,923 bp |
| G + C content | 32.42% |
| Coding sequences | 125 |
| Pseudogenes | 8 |
| Coding density | 63.5% |
| Average gene length | 725 bp |
TABLE 2.
Predicted genes in pBtoxis
| Name | Gene | Predicted product | Database similarity (EMBL no.) (% aa identity) | pXO1 homologue (% aa identity) |
|---|---|---|---|---|
| pBt001 | Unknown | No significant matches | pXO1-49 (78.48 in 158 aa) | |
| pBt003 | Insertion element IS240 protein | B. thuringiensis insertion element IS240-A protein TR:Q45766 (M23740) (99.57 in 235 aa) | ||
| pBt004 | Insertion element IS240 protein | B. thuringiensis insertion element IS240-A protein TR:Q45766 (M23740) (99.14 in 235 aa) | ||
| pBt005 | Integrase/recombinase family protein | Bacillus halodurans Bh2364 protein TR:Q9KAC5 (AP001515) (35.45 in 189 aa); Lactobacillus delbrueckii integrase-recombinase TR:Q48538 (Z50864) (28.88 in 90 aa); B. thuringiensis resolvase TnpI SW:TNRI_BACTU (P10020) (23.88 in 180 aa) | pXO1-18 (84.15 in 183 aa; N terminus) | |
| pBt006 | Putative integral membrane protein | No significant matches | pXO1-17 (68.08 in 47 aa) | |
| pBt007 | Conserved hypothetical protein | B. thuringiensis conserved hypothetical protein TR:CAC50562 (AJ296638) (99.67 in 310 aa) | pXO1-16 (96.13 in 569 aa) | |
| pBt009 | Conserved hypothetical protein | B. thuringiensis conserved hypothetical protein TR:CAC50561 (AJ296638) (99.28 in 280 aa) | pXO1-14 (89.71 in 564 aa) | |
| pBt010 | Hypothetical protein | No significant matches | ||
| pBt011 | Putative DNA-binding protein | B. subtilis plasmid pPOD2000 ORF4 gene TR:Q45507 (U55043) (41.48 in 94 aa); B. subtilis pTA1040 orf2c_40 TR:Q45444 (U32378) (23.07 in 169 aa); contains potential helix-turn-helix motif | ||
| pBt013 | Hypothetical protein | No significant matches | ||
| pBt014 | Probable transcriptional regulator | B. subtilis probable repressor protein YdcN TR:P96631 (AB001488) (34.61 in 130 aa); B. subtilis SinR protein SW:SINR_BACSU (P06533) (42.02 in 69 aa) | ||
| pBt015 | Hypothetical protein | No significant matches | ||
| pBt016 | Hypothetical protein | No significant matches | ||
| pBt017 | Hypothetical protein | No significant matches | ||
| pBt020 | Hypothetical protein | No significant matches | ||
| pBt021 | cyt1Aa | Type-1Aa cytolytic delta endotoxin | Previously sequenced as B. thuringiensis (subsp. israelensis) type 1Aa cytolytic delta endotoxin Cyt1Aa SW:CXAA_BACTI (P05069) (100 in 249 aa) | |
| pBt022 | p19 | 19-kDa accessory protein | Previously sequenced as B. thuringiensis 19-kDa accessory protein p19 TR:Q9R832 (AJ010753) (100 in 179 aa) | |
| pBt023 | cry11Aa | Pesticidal crystal protein Cry11Aa | Previously sequenced as B. thuringiensis pesticidal crystal protein Cry11Aa SW:CBAA_BACTI (P21256) (100 in 643 aa) | |
| pBt024 | p20 | 20-kDa accessory protein | Previously sequenced as B. thuringiensis 20-kDa accessory protein TR:Q45775 (M22860) (100 in 182 aa) | |
| pBt025 | Pesticidal crystal protein (fragment) | Similar to part of B. thuringiensis pesticidal crystal protein Cry28Aa SW:CSAA_BACTF (Q9X682) (70.96 in 31 aa); may form a deletion remnant with the downstream gene pBt026 | ||
| pBt026 | Pesticidal crystal protein (fragment) | Similar to B. thuringiensis pesticidal crystal protein Cry28Aa or SW:CSAA_BACTF (Q9X682) (61.53 in 26 aa); may form a deletion remnant with the upstream gene pBt025 | ||
| pBt027 | IS231W transposase | Identical to B. thuringiensis IS231W ORF2 TR:Q45713 (M83546) (100 in 223 aa) | pXO1-35 (58.1 in 217 aa), pXO1-36 (59.4 in 217 aa), pXO1-39 (58.9 in 151 aa) | |
| pBt028 | IS231W transposase | Identical to B. thuringiensis transposase IS231W ORF1 TR:Q45714 (M83546) (100 in 250 aa) | pXO1-35 (49.8 in 237 aa), pXO1-36 (45.1 in 244 aa), pXO1-39 (53.6 in 168 aa) | |
| pBt029 | Putative DNA-binding protein | Similar to fragment of Helicobacter pylori preprotein translocase SecA subunit hp0786 SW:SECA_HELPY (O25475) (48 in 31 aa); contains HMMPfam hit to PF02810; SEC-C motif (the motif is predicted to chelate zinc with the CXC and C[HC] pairs that constitute the most conserved feature of the motif; it is predicted to be a potential nucleic acid binding domain) | ||
| pBt030 | Hypothetical protein | No significant matches | ||
| pBt031 | Putative N-acetylmuramoyl-l-alanine amidase (peptidoglycan hydrolase) | Similar to Bacillus phage GA-1 peptidoglycan hydrolase gene 15 TR:Q9FZW0 (X96987) (42.07 in 202 aa) and to Bacillus licheniformis N-acetylmuramoyl-l-alanine amidase CwlM SW:CWLM_BACLI (P37134) (33.2 in 256 aa) | ||
| pBt032 | Hypothetical protein | No significant matches | ||
| pBt033 | Hypothetical protein | No significant matches | ||
| pBt034 | Conserved hypothetical protein | Similar to C terminus of Rickettsia conorii hypothetical protein RC1157 TR:Q92GG6 (AE008664) (37.7 in 77 aa) | pXO1-106 (C-terminal half; 62.68 in 67 aa) | |
| pBt035 | Conserved hypothetical protein | Similar to R. conorii hypothetical protein RC1156 TR:AAL03694 (AE008664) (37.5 in 80 aa) | pXO1-71 (36.98 in 73 aa) and pXO1-72 (C terminus; 32.32 in 99 aa) | |
| pBt036 | cyt2Ba | Type 2Ba cytolytic delta endotoxin | Previously sequenced as B. thuringiensis type 2Ba cytolytic delta endotoxin SW:CYBA_BACTI (Q45723) | |
| pBt038 | cry4Ba | Pesticidal crystal protein Cry4Ba | Previously sequenced as B. thuringiensis pesticidal crystal protein Cry4Ba SW:C4BA_BACTI (P05519) | |
| pBt043 | Probable Insertion element transposase (pseudogene) | Similar to Lactococcus lactis orf-w2 protein TR:Q48685 (M37396) (56.88 in 225 aa) and to Enterococcus faecium transposase TR:Q47818 (U49512) (56.75 in 222 aa); contains frameshift | ||
| pBt045 | Hypothetical protein | No significant matches | ||
| pBt047 | cry10Aa | Pesticidal crystal protein Cry10Aa | Previously sequenced as B. thuringiensis pesticidal crystal protein Cry10Aa SW:CAAA_BACTI (P09662) (99.85 in 675 aa) | |
| pBt048 | Putative pesticidal crystal protein | Similar to C terminus of B. thuringiensis pesticidal crystal protein Cry4Ba SW:C4BA_BACTI (P05519) (80.52 in 493 aa) and to C terminus of B. thuringiensis pesticidal crystal protein Cry4Aa SW:C4AA_BACTI (P16480) (79.91 in 493 aa); similar to full-length B. thuringiensis subsp. jegathesan hypothetical 60.1-kDa protein (downstream of Cry19Aa) TR:O32308 (Y07603) (75.81 in 488 aa) | ||
| pBt049 | Hypothetical protein | No significant matches | ||
| pBt051 | Transposase for IS231-like element (partial pseudogene) | Similar to B. thuringiensis transposase for insertion element IS231D SW:T23D_BACTF (Q05501) (44.32 in 273 aa); contains stop codon and is truncated by IS240 insertion | pXO1-36 (90.74 in 281 aa), pXO-35 (45.8 in 273 aa), pXO39 (45.7 in 197 aa) | |
| pBt052 | Insertion element IS240 protein | Similar to B. thuringiensis insertion element IS240-A protein TR:Q45766 (M23740) (99.14 in 235 aa) and to Mycobacterium fortuitum transposase TnpA TR:Q49185 (X53635) (48.05 in 231 aa) | ||
| pBt053 | Probable deletion remnant of pesticidal crystal protein | Similar to extreme C terminus of B. thuringiensis pesticidal crystal protein Cry26Aa SW:CQAA_BACTF (Q9X597) (65.85 in 41 aa) | ||
| pBt054 | cyt1Ca | Possible two-domain toxin | Possible two-domain toxin; N-terminal half is similar to B. thuringiensis type 1Ab cytolytic delta endotoxin Cyt1Ab SW:CXAB_BACTV (P94594) (52.21 in 226 aa); C-terminal half is similar to several ricin-B lectin domain-containing toxins, e.g., Pieris brassicae pierisin-b TR:Q9GV36 (AB037676) (27.45 in 306 aa), B. sphaericus mosquitocidal toxin protein Mtx1 TR:Q03988 (M60446) (25.7 in 284 aa), and C. botulinum main hemagglutinin component ha-33 SW:HA33_CLOBO (P46084) (27.61 in 268 aa) | |
| pBt055 | Putative deletion pseudogene | Possible deletion pseudogene; C terminus of protein is similar to C termini of products of genes upstream of toxin genes, e.g., B. thuringiensis hypothetical 29.1-kDa protein encoded in cryB1 5′ region SW:YCR2_BACTK (P21733) (36.66 in 90 aa); N terminus is similar to N terminus of, e.g., B. thuringiensis pesticidal crystal protein Cry11Bb SW:CBBB_BACTV (Q9ZIU5) (40.47 in 84 aa) | ||
| pBt056 | Hypothetical protein (pseudogene) | Potential pseudogene; matches pBt152 with two frameshifts and an in-frame stop | ||
| pBt059 | Conserved hypothetical protein | Similar to N-termini of products of genes upstream of B. thuringiensis toxin genes, e.g., B. thuringiensis hypothetical 29.1-kDa protein encoded in cryB1 5′ region SW:YCR2_BACTK (P21733) (41.17 in 85 aa) and B. thuringiensis ORF2 TR:Q45742 (X57252) (47.76 in 67 aa) | ||
| pBt060 | Putative spore germination protein (pseudogene) | Similar to N terminus of B. cereus spore germination protein GerIA SW:GRIA_BACCE (O85467) (35.54 in 467 aa); contains two potential frameshifts | pXO1-113 (31.3 in 492 aa) | |
| pBt063 | Putative spore germination protein (pseudogene) | Similar to N terminus of B. halodurans spore germination protein bh1598 TR:Q9KCH3 (AP001512) (33 in 87 aa) and to B. cereus spore germination protein GeriB TR:O85468 (AF067645) (30.52 in 95 aa); truncated by IS240 insertion | ||
| pBt064 | IS240 protein (partial) | Similar to B. thuringiensis insertion element IS240-A protein TR:Q45766 (M23740) (99.31% in 146 aa) | ||
| pBt065 | Hypothetical methionine-rich protein | No significant matches | ||
| pBt066 | Hypothetical protein | No significant matches | ||
| pBt067 | Conserved hypothetical protein | Similar to R. conorii hypothetical protein RC1156 TR:AAL03694 (AE008664) (37.5 in 96 aa) and to R. conorii hypothetical protein RC1157 TR:AAL03695 (AE008664) (48.43 in 64 aa) | pXO1-71 (42.46 in 73 aa), pXO1-72 (32.99 in 97 aa), pXO1-106 (35 in 100 aa) | |
| pBt068 | Hypothetical protein | No significant matches | ||
| pBt070 | Insertion element transposase | Similar to E. faecium transposase TR:Q47818 (U49512) (58.66 in 225 aa); interrupted by IS231 insertion | ||
| pBt071 | Transposase for insertion sequence IS231F | Previously sequenced as B. thuringiensis transposase for insertion element IS231F SW:T23F_BACTI (Q02404) | pXO1-35 (70.5 in 478 aa), pXO1-36 (51.9 in 474 aa), pXO1-39 (68.0 in 325 aa) | |
| pBt072 | Conserved hypothetical protein | Similar to part of Streptomyces hygroscopicus subsp. varascomyceticus FkbH TR:Q9KIE6 (AF235504) (35.86 in 92 aa) | ||
| pBt073 | Hypothetical protein | No significant matches | ||
| pBt074 | Hypothetical protein | No significant matches | ||
| pBt075 | Hypothetical protein | Weakly similar to Yersinia pestis plasmid hypothetical protein Y1034 or YPMT1.21C TR:O68737 (AF053947) (22.44 in 303 aa) | ||
| pBt076 | Resolvase | Similar to B. sphaericus putative resolvase TnpR TR:Q9REE7 (Y18010) (72.13 in 183 aa) and to Staphylococcus aureus DNA invertase BinR SW:BINR_STAAU (P19241) (65.02 in 183 aa) | pXO1-115 (33 in 183 aa) | |
| pBt077 | Transposase | Similar to Shigella flexneri Tn501 transposition transposase TnpA TR:AAK18584 (AF348706) (45.74 in 986 aa) | pXO1-116 (66 in 922 aa) | |
| pBt079 | Transposase for insertion sequence IS231F | Previously sequenced as B. thuringiensis transposase for insertion element IS231F SW:T23F_BACTI (Q02404) | pXO1-35 (70.5 in 478 aa), pXO1-36 (51.9 in 474 aa), pXO1-39 (68.0 in 325 aa) | |
| pBt082 | Probable transposase (pseudogene) | Similar to Streptococcus pyogenes transposase-IS1562 TR:Q99XV1 (AE006623) (39.13 in 391 aa) | pXO1-120 (97.36 in 190 aa) | |
| pBt084 | Putative spore germination protein | Similar to B. subtilis spore germination protein A3 precursor GerAC SW:GRAC_BACSU (P07870) (23.51 in 387 aa) and to B. subtilis spore germination protein B3 precursor GerBC SW:GRBC_BACSU (P39571) (21.83 in 371 aa) | ||
| pBt085 | Putative spore germination protein | Similar to B. cereus spore germination protein GerIB TR:O85468 (AF067645) (25.56 in 352 aa) and to B. subtilis spore germination protein B2 GerBB SW:GRBB_BACSU (P39570) (22.19 in 365 aa) | ||
| pBt086 | Putative spore germination protein | Similar to B. subtilis spore germination protein GerKA SW:GRKA_BACSU (P49939) (38.21 in 458 aa) and to B. cereus spore germination protein GerIA SW:GRIA_BACCE (O85467) (35.81 in 483 aa) | pXO1-113 (29 in 439 aa) | |
| pBt087 | Putative 1-phosphatidylinositol phosphodiesterase precursor | Similar to Listeria monocytogenes 1-phosphatidylinositol phosphodiesterase precursor PlcA or LMO0201 SW:PLC_LISMO (P34024) (34.02 in 288 aa) and to B. thuringiensis 1-phosphatidylinositol phosphodiesterase precursor SW:PLC_BACTU (P08954) (27.69 in 325 aa); contains an in-frame TGA stop after aa 86 | ||
| pBt089 | Putative exported protein | No significant matches | ||
| pBt090 | Insertion element transposase | Similar to E. faecium transposase TR:Q47818 (U49512) (58.22 in 225 aa) | ||
| pBt091 | Putative transcriptional regulator; ArsR family | Similar to Vibrio cholerae transcriptional activator HlyU or VC0678 SW:HLYU_VIBCH (P52695) (32.6 in 92 aa) | pXO1-109 (PagR or TcrA) (51.8 in 83 aa) and pXO1-138 (46.06 in 89 aa) | |
| pBt092 | Small DNA-binding protein (bacterial histone-like family) | Similar to B. subtilis DNA-binding protein HU 1 (hupA) SW:DBH1_BACSU (P08821) (63.04 in 92 aa) and to Streptococcus thermophilus DNA-binding protein HU SW:DBH_STRTR (P96045) (61.36 in 88 aa) | ||
| pBt093 | HfQ protein (RNA-binding protein) | Similar to B. subtilis HfQ protein SW:HFQ_BACSU (O31796) (46.55 in 58 aa) and to E. coli Hfq protein SW:HFQ_ECOLI (P25521) (36.2 in 58 aa) | pXO1-137 (40.7 in 59 aa) | |
| pBt094 | Putative transcriptional regulator | Similar to B. subtilis transition state regulatory protein AbrB or CpsX SW:ABRB_BACSU (P08874) (62.06 in 87 aa), to B. subtilis putative transition state regulator Abh SW:ABH_BACSU (P39758) (56.32 in 87 aa), and to B. subtilis stage V sporulation protein T SpoVT SW:SP5T_BACSU (P37554) (70 in 50 aa) | pXO1-105 (45.9 in 61 aa) | |
| pBt095 | Conserved hypothetical membrane protein | Similar to B. subtilis YnzD protein TR:O31819 (Z99113) (41.86 in 43 aa) | ||
| pBt096 | Conserved hypothetical integral membrane protein | Similar to Rhizobium meliloti hypothetical transmembrane protein SMC01970 TR:CAC47083 (AL591790) (37.36 in 273 aa) and to Pseudomonas carboxydovorans CoxK protein TR:Q9KX21 (X82447) (34.71 in 265 aa) | ||
| pBt097 | Putative class II aminotransferase | Similar to B. subtilis putative aminotransferase PatB SW:PATB_BACSU (Q08432) (43.52 in 386 aa) | ||
| pBt098 | Pyridoxal phosphate-dependent enzyme | Similar to Mycobacterium tuberculosis hypothetical protein CysM3 or Rv0848 TR:O53860 (AL022004) (38.62 in 334 aa), to Clostridium sticklandii O-acetylserine sulfhydrylase CysK TR:Q9L4R2 (AJ130879) (33.22 in 310 aa), and to Spinacia oleracea cysteine synthase chloroplast precursor CysK SW:CYSL_SPIOL (P32260) (29.96 in 317 aa) | ||
| pBt099 | Hypothetical hydrophobic protein | No significant matches | ||
| pBt100 | tRNA synthetase-related protein | Similar to part of Pseudomonas aeruginosa hypothetical protein PA2106 TR:Q9I209 (AE004638) (30.95 in 252 aa) and Homo sapiens alanyl-tRNA synthetase AarS SW:SYA_HUMAN (P49588) (29.44 in 163 aa) | ||
| pBt101 | Possible kinase | Similar to Streptomyces rishiriensis gene in coumermycin A1 biosynthetic gene cluster CouR3 TR:Q9F8T5 (AF235050) (35.29 in 272 aa), to Salmonella enterica serovar Typhimurium gene in propanediol utilization (pdu) operon PduX TR:Q9XDM4 (AF026270) (23.82 in 277 aa), and to Hyphomicrobium chloromethanicum monophosphate kinase TR:Q9APK3 (AF281259) (28.57 in 133 aa) | ||
| pBt102 | GntR family transcriptional regulator containing aminotransferase domain | Similar to B. halodurans transcriptional regulator BH0432 TR:Q9KFP6 (AP001508) (36.4 in 478 aa) and to Thermococcus profundus multiple substrate aminotransferase TR:Q9V2W5 (AB027131) (29.41 in 391 aa) | ||
| pBt103 | Insertion sequence IS240 protein | Previously sequenced as B. thuringiensis insertion element IS240-B protein TR:Q45767 (M23741) | ||
| pBt104 | Transposase (pseudogene) | Similar to Pseudomonas putida Tn4652 transposase TnpA TR:P72226 (X83686) (36.28 in 1,028 aa) and to E. faecium transposase for transposon Tn1546 SW:TNP6_ENTFC (Q06238) (25.18 in 973 aa); contains an in-frame stop codon after aa 394 | pXO1-116 (22.8 in 960 aa) | |
| pBt106 | Resolvase | Similar to E. faecium transposon Tn1546 resolvase SW:TNR6_ENTFC (Q06237) (76.72 in 189 aa) | pXO1-115 (41.6 in 185 aa) | |
| pBt107 | Conserved hypothetical protein | Weakly similar in N terminus to B. halodurans BH0264 protein TR:Q9KG49 (AP001507) (22.88 in 201 aa) | ||
| pBt108 | Putative sigma factor; ECF family | Similar to B. subtilis RNA polymerase sigma factor SigY SW:SIGY_BACSU (P94370) (23.07 in 169 aa) and to B. subtilis RNA polymerase sigma factor SigX SW:SIGX_BACSU (P35165) (23.81 in 168 aa) | ||
| pBt110 | cry4Aa | Pesticidal crystal protein Cry4Aa | Previously sequenced as B. thuringiensis pesticidal crystal protein Cry4Aa SW:C4AA_BACTI (P16480) | |
| pBt111 | Insertion sequence IS240 protein | Previously sequenced as B. thuringiensis insertion element IS240-A protein TR:Q45766 (M23740) | ||
| pBt112 | Hypothetical protein | No significant matches | ||
| pBt113 | Hypothetical protein | No significant matches | ||
| pBt114 | Hypothetical protein | No significant matches | ||
| pBt115 | Hypothetical protein | No significant matches | ||
| pBt116 | Hypothetical exported protein | No significant matches | ||
| pBt119 | Hypothetical protein | No significant matches | ||
| pBt120 | Putative DNA-binding protein | Similar to fragment of Helicobacter pylori preprotein translocase SecA subunit SW:SECA_HELPY (O25475) (48.38 in 31 aa); contains HMMPfam hit to PF02810; SEC-C motif (the motif is predicted to chelate zinc with CXC and C[HC] pairs it is predicted to be a potential nucleic acid binding domain) | ||
| pBt121 | IS231 transposase | Similar to B. thuringiensis IS231W transposase TR:Q45714 (M83546) (99.18 in 245 aa) | pXO1-35 (49.0 in 241 aa), pXO1-36 (45.7 in 243 aa), pXO1-39 (52.3 in 172 aa) | |
| pBt122 | IS231 transposase | Identical to B. thuringiensis IS231W transposase TR:Q45713 (M83546) | pXO1-35 (58.1 in 217 aa), pXO1-36 (59.4 in 217 aa), pXO1-39 (58.9 in 151 aa) | |
| pBt123 | Hypothetical membrane protein | No significant matches | ||
| pBt124 | Hypothetical protein | No significant matches | ||
| pBt125 | Hypothetical protein | No significant matches | ||
| pBt126 | Hypothetical protein | No significant matches | ||
| pBt127 | Conserved hypothetical protein | Similar to R. conorii hypothetical protein RC1156 TR:AAL03694 (AE008664) (40.96 in 83 aa) and to R. conorii hypothetical protein RC1157 TR:AAL03695 (AE008664) (32.81 in 64 aa) | pXO1-106 (80.76 in 78 aa), pXO1-71 (36.98 in 73 aa), pXO1-72 (38.77 in 98 aa) | |
| pBt128 | Hypothetical protein | No significant matches | ||
| pBt129 | Hypothetical protein | No significant matches | ||
| pBt130 | Conserved hypothetical integral membrane protein | Similar to B. subtilis YknW protein TR:O31709 (Z99111) (34.23 in 222 aa) | ||
| pBt131 | Putative ABC transporter permease protein | Similar to B. subtilis hypothetical protein encoded in moaD-fruR intergenic region YknZ SW:YKNZ_BACSU (O31712) (52.36 in 401 aa) and to Rhizobium loti predicted permease protein of ABC transporter MLRL397 TR:Q98KN3 (AP002997) (34.15 in 404 aa) | ||
| pBt132 | Putative ABC transporter ATP-binding protein | Similar to B. subtilis putative ABC transporter YvrO TR:O52857 (AJ223978) (63.22 in 223 aa) and to S. pyogenes putative ABC transporter SPY0837 TR:Q9A0C4 (AE006534) (57.33 in 225 aa) | ||
| pBt133 | Putative ABC transporter-exported solute-binding protein | Similar to B. subtilis YknX protein TR:O31710 (Z99111) (29.35 in 385 aa) and to Streptococcus cristatus ATP-binding cassette transporter-like protein TptB TR:O54498 (U96166) (24.93 in 397 aa) | ||
| pBt136 | Possible peptide antibiotic precursor? | Very weak similarity to E. faecalis peptide antibiotic AS-48 TR:Q47765 (X79542) (27.14 in 70 aa) (also called E. faecalis BacA protein TR:O52963 [D85752]). | ||
| pBt137 | Integral membrane protein (possible peptide antibiotic maturation and biosynthesis protein) | Similar to E. faecalis BacB protein TR:O52964 (D85752) (22.16 in 537 aa) (also called AS-48-B TR:O53024 [Y12234]) AS-48 maturation and biosynthesis protein) | ||
| pBt138 | Integral membrane protein (possible accessory factor in peptide antibiotic secretion) | Similar to E. faecalis AS-48C protein (putative accessory factor in AS-48 secretion) TR:O53025 (Y12234) (23.56 in 157 aa) | ||
| pBt139 | Putative ABC transporter ATP-binding protein | Similar to Thermotoga maritima ABC transporter ATP-binding protein TM0793 TR:Q9WZQ0 (AE001747) (32.64 in 193 aa) and to S. pyogenes putative ABC transporter SPY1674 TR:Q99YJ5 (AE006597) (28.77 in 212 aa) | ||
| pBt140 | Integral membrane protein | No significant matches | ||
| pBt142 | Putative DNA recombinase | Similar to E. faecalis recombinase EP0007 TR:Q9F1I8 (AE002565) (37.01 in 208 aa) and to S. thermophilus putative resolvase TR:Q9X9M6 (AJ242479) (31.7 in 205 aa) and to S. aureus potential DNA invertase Bin3 SW:BIN3_STAAU (P20384) (32.51 in 203 aa) | ||
| pBt143 | Hypothetical protein | No significant matches | ||
| pBt145 | Putative spore coat-associated protein | Similar to B. subtilis spore coat-associated protein N (CotN) SW:COTN_BACSU (P54507) (35.44 in 237 aa) | ||
| pBt146 | Hypothetical protein | No significant matches | ||
| pBt147 | HfQ protein (RNA-binding protein) | Similar to B. subtilis Hfq protein SW:HFQ_BACSU (O31796) (45.45 in 55 aa) | pXO1-137 (38.98 in 59 aa) | |
| pBt148 | Putative transcriptional regulator | Similar to B. subtilis transition state regulatory protein AbrB or CpsX SW:ABRB_BACSU (P08874) (55.05 in 89 aa) and to B. subtilis putative transition state regulator Abh SW:ABH_BACSU (P39758) (55.17 in 87 aa) | ||
| pBt149 | Putative transcriptional regulator; ArsR family | Similar to Streptomyces verticillus metal-dependent regulatory protein TR:Q9FB31 (AF210249) (36.47 in 85 aa) and to Xylella fastidiosa transcriptional regulator XF0767 TR:Q9PFB1 (AE003917) (32.58 in 89 aa) | pXO1-109 PagR or TcrA (39.13 in 92 aa), pXO1-138 (43.2 in 74 aa) | |
| pBt150 | Hypothetical protein | No significant matches | ||
| pBt151 | Insertion sequence IS240 protein | Similar to B. thuringiensis insertion element IS240-A protein TR:Q45766 (M23740) (99.57 in 235 aa) | ||
| pBt152 | Hemagglutinin-related protein | Similar to C. botulinum hemagglutinin Ha-33 protein TR:Q45868 (X79103) (32.35 in 136 aa) and to C. botulinum Ha-33 protein TR:Q45871 (X79104) (25.54 in 231 aa) | ||
| pBt154 | Insertion sequence IS240 protein | Similar to B. thuringiensis insertion element IS240-A protein TR:Q45766 (M23740) (99.14 in 235 aa) | ||
| pBt156 | FtsZ/tubulin-related protein | Weakly similar to Pyrococcus kodakaraensis TubA protein TR:Q9HHD0 (AB031743) (21 in 394 aa) and to Pyrococcus horikoshii cell division protein FtsZ homologue 3 FtsZ3 SW:FTZ3_PYRHO (O59060) (23 in 304 aa) | pXO1-45 (21 in 444 aa) | |
| pBt157 | Putative DNA-binding protein | No significant matches; contains predicted helix-turn-helix motif score 1,099 (+2.93 SD) at aa 41-62 | ||
| pBt158 | Putative transcriptional regulator | Similar to Clostridium acetobutylicum transcriptional regulator; MerR family CAP0178 TR:AAK76923 (AE001438) (30 in 110 aa) |
Identification of a previously unknown toxin gene.
The pBtoxis coding sequence (CDS) pBt054 is a previously uncharacterized CDS that encodes a protein of approximately 60 kDa, which is related at its N terminus to the known Cyt toxins of B. thuringiensis. Comparison of this region of the CDS to known Cyt proteins indicates that it could represent a new subdivision of this family, Cyt1Ca according to the conventional B. thuringiensis toxin nomenclature (10; http://www.biols.susx.ac.uk/Home/Neil_Crickmore/Bt/index.html), although confirmation of this provisional name awaits further experimental evidence of its properties. The pBtoxis CDS is, however, unusual in another way. Whereas previously recognized members of the Cyt family are proteins of approximately 26 to 28 kDa, pBt054 represents a fusion between the Cyt1Ca-like region at the N terminus and an extra domain at the C terminus. The last 280 amino acids (aa) of this C-terminal domain appear to be tandem beta-trefoil modules like those found in other bacterial toxins, such as ricin, Clostridium botulinum neurotoxin, and the mosquito larvicidal Mtx1 toxin from Bacillus sphaericus (46). This superfamily of motifs is implicated as likely carbohydrate binding moieties, so one possible function for the C-terminal region of pBt054 could be recognition of carbohydrate groups on toxin receptors.
Vestigial toxin gene remnants.
In addition to the complete toxin CDSs, pBtoxis also contains short sequences encoding fragments of toxins. pBt025 and pBt026 encode two segments with homologies to the center region of a Cry28Aa-like toxin, while pBt053 appears to encode a sequence with homology to the extreme C terminus of a Cry26Aa-like protein (49). In addition, the amino acid sequence encoded by pBt055 is similar at its C terminus to proteins encoded upstream of toxin genes (e.g., a hypothetical 29.1-kDa protein in the cry2Aa 5′ region of Bacillus thuringiensis subsp. kurstaki), while its N terminus is similar to that of Cry11Bb. These apparent cry toxin gene remnants suggest that during the evolution of pBtoxis, its ancestors have been host to other toxins now lost. This suggests that toxin composition is a dynamic factor and may help to explain the great diversity in toxin composition observed in B. thuringiensis isolates. The fact that these remnants are located close to CDSs with possible roles in transposition (pBt052 is similar to IS240-A; pBt027 and pBt028 are similar to IS231W sequences) implies that transposition is the most likely mechanism for this effect, and this is consistent with previous observations that B. thuringiensis toxin genes may be flanked by transposase sequences (26). In total, over 23% of the genes on pBtoxis show similarity to transposon-related genes, indicating that a considerable amount of DNA exchange has occurred in the evolutionary history of pBtoxis. As previously reported (11, 47), the cry10Aa gene (pBt047) is similar to the 5′ end of other cry genes and encodes an ∼78-kDa protein that would appear to be truncated compared to related Cry proteins. This gene is followed by a second CDS (pBt048) with similarity to the 3′ ends of other cry genes. The intervening 67 bp contains at least two stop codons in each of the three reading frames and causes disruption of what may once have been a single CDS to produce two CDSs. However, protein derived from cry10Aa (pBt047) has been identified in B. thuringiensis subsp. israelensis inclusions (16), indicating that this CDS is not a pseudogene remnant.
Other factors.
In addition to Cry and Cyt toxins, B. thuringiensis strains, like the closely related Bacillus cereus, are known to produce other potential virulence factors, including phosphatidyl inositol-specific phospholipase C, that may contribute to the role of the spore in overall toxicity (42). The expression of the genes encoding these factors is activated by the PlcR regulator protein that binds to the palindromic sequence TATGNAN4TNCATA (2). It appears that the pBtoxis plasmid encodes a separate, extrachromosomal copy of a phosphatidyl inositol-specific phospholipase C (pBt087), although the presence of an in-frame TGA stop codon indicates that this either is a pseudogene or is expressed by translational read-through. Inspection of the upstream control region for this gene also reveals no PlcR binding site. The PlcR binding palindrome does, however, occur within pBtoxis between two divergent groups of CDSs which appear to be part of the peptide antibiotic production and export system (pBt130-134 and pBt136-140; see below). The significance of this is unclear.
Sporulation and germination.
Analysis of the plasmid revealed many other genes that may have significant effects on several aspects of the phenotype of the host organism, the most striking of which are potentially involved in sporulation and germination.
The apparently cotranscribed genes pBt084, pBt085, and pBt086 are similar to several operons encoding germination complex genes. pBt086 is similar to the A integral membrane component (e.g., gerAA [14]), pBt085 is similar to the B integral membrane component (e.g., gerAB [54]), and pBt084 is similar to the C lipoprotein component (e.g., gerAC [54]). These components form membrane-associated complexes that allow the spore to respond to different germination signals (32). The putative plasmid-encoded complex composed of pBt084, pBt085, and pBt086 might enhance the response of the host to known germinants or allow it to recognize a novel germination signal. The Bacillus anthracis toxin-encoding pXO1 plasmid also encodes a set of germination proteins, GerXB, -A, and -C, and these have been shown to be important for the virulence of the host (19). Of the three, only the pXO1 gerXA gene shows significant similarity to the pBtoxis gene pBt086. Intriguingly, the remnants of a second germination complex are also present in the form of two interrupted and truncated pseudogenes, pBt060 and pBt063, representing the A and B components of such a complex. This suggests that, as with the toxin genes themselves, the plasmid may have carried a different repertoire of germination genes in the past.
pBt031 shows significant similarity to many cell wall hydrolases, both phage and chromosomally encoded, and appears to contain a direct-repeat peptidoglycan binding domain at its C terminus. One homologue of this protein, CwlM, is sporulation specific in B. subtilis, raising the possibility that pBt031 might also be involved in sporulation. The product of pBt145 is a homologue of CotN, a secreted protein that has been shown to be incorporated into, and potentially be involved in the production of, the B. subtilis spore (43, 45).
pBt094 and pBt148 encode homologues of the B. subtilis transition state regulatory protein AbrB, which is known to be involved in the regulation of postexponential expression and the early events leading to sporulation (15). The B. subtilis genome also includes a second arbB-like gene, abh (24), suggesting that the putative redundancy or complementarity of these chromosomal regulators may be supplemented by additional plasmid-borne genes in B. thuringiensis. Divergently transcribed from the plasmid-borne arbB-like gene is pBt095, a homologue of the ynzD gene of B. subtilis whose product has been identified as an aspartyl phosphatase which has direct effects on sporulation efficiency (39).
Taken together, the presence of these genes indicates that pBtoxis may exert a considerable influence on the sporulation and germination processes of its B. thuringiensis host, and this possibility is under experimental analysis.
Regulation.
In addition to the putative sporulation-regulatory proteins described above, pBtoxis encodes a number of other potential transcriptional regulators. pBt108 is a predicted sigma factor that shows homology to sigma E, which is known to be associated with the transcription of cry4Aa (51, 52), cry4Ba (53), cry11Aa (12), and both cyt genes (8, 18) in B. thuringiensis subsp. israelensis. This sigma factor is involved in transcription within the early mother cell (approximately 3 h into sporulation)—the time at which crystal formation is also occurring within the mother cell. pBt091 and pBt149 are members of the ArsR family and show similarity to the pXO1 genes pXO1-109 (PagR), a regulator of transcription of the B. anthracis protective antigen (22), and pXO1-138. pBt102 contains a GntR family regulator fused to an aminotransferase domain and has homologues in B. subtilis (YdfD) and many other bacterial genomes. Other genes that are predicted to encode regulators include pBt014, a member of the PbsX/Xre family of regulators with some similarity to B. subtilis SinR, a global regulator of post-exponential-phase response genes (17, 28); pBt158, a member of the MerR family; and the genes pBt157 and pBt011, which both contain predicted helix-turn-helix domains but have no significant database similarities.
Aside from the genes for these transcriptional regulator proteins, pBtoxis contains two genes, pBt093 and pBt147, with similarity to the bacterial RNA-binding protein Hfq, a regulator of mRNA poly(A) tails (20), and a gene, pBt092, which encodes a member of the bacterial histone-like protein family, HU.
Peptide antibiotic.
One of the more surprising determinants carried by the plasmid is pBt136-140, a set of genes that appear to be involved in the production and export of a peptide antibiotic. Several of these are similar in order and orientation to those in an operon from Enterococcus faecalis responsible for the production and secretion of the ribosomally synthesized circular peptide antibiotic AS-48 (31). AS-48 is apparently produced by the circularization of a propeptide produced by the removal of a 35-aa signal sequence (30); pBt136 encodes a protein similar in length and sequence to the processed propeptide of AS-48. The next two genes, pBt137 and pBt138, encode predicted integral membrane proteins similar to AS-48B and AS-48C, which have been suggested to be involved in the maturation and secretion of the antibiotic. pBt139 encodes an ABC transporter ATP-binding protein with some similarity to AS48-D, and pBt140 is predicted to encode an integral membrane protein which is presumably part of the same system. Interestingly, there is no homologue of AS-48C1, which is the only gene shown to be indispensable for immunity to AS-48. No other potential immunity proteins could be identified.
Divergently transcribed from these genes are pBt133 to pBt130, encoding the components of an ABC transport system: an exported solute binding protein (pBt133), an ATP-binding protein (pBt132), a permease protein (pBt131), and a predicted integral membrane protein (pB130). These resemble many predicted components of ABC transporters from microbial genomes, with little evidence of their specific functions. However, the first three components do show weak similarity to BacG, BacH, and BacI, encoded by genes downstream of the bacteriocin 21 production and secretion genes from E. faecalis plasmid pPD1 (which are nearly identical to the AS-48 genes described above [48]). These bac genes are necessary for full bacteriocin 21 expression, and the pBt genes, therefore, may also be involved in the production or secretion of the putative peptide antibiotic.
Amino acid metabolism.
The genes pBt096 to pBt101 encode a series of proteins with diverse database similarities and protein motifs. All seem to be involved in some way in amino acid metabolism. The first gene in the cluster, pBt101, encodes a protein with weak similarities to diverse kinase proteins; the second, pBt102, is weakly similar to (although considerably smaller than) a number of alanyl-tRNA synthetases and contains a class II tRNA synthetase PFAM domain. Although it is highly unlikely to be a tRNA synthetase, it could potentially encode some form of amino acid transferase or ligase activity. The third gene encodes a small hydrophobic protein with no database matches, while the fourth, pBt098, is a member of the pyridoxal phosphate-dependent enzymes and has similarities to many O-acetyl serine lyases (cysteine synthases); again, this is probably not a cysteine synthase but may be involved in amino acid modification. The product of pBt097 is predicted to be an aminotransferase with similarities to many characterized and predicted aminotransferases in the database, including the Escherichia coli MalY protein, a bifunctional protein with cysteine lyase activity, and several aspartate aminotransferases. The last gene in the cluster, pBt096, encodes a predicted integral membrane protein with similarity to many predicted transporters. Divergently transcribed from these proteins is the predicted regulator pBt102, which contains an aminotransferase domain and might be involved in the regulation of these genes.
Two possibilities could be suggested for the functions of these proteins. They may enable the uptake from the environment of an amino acid or an amino acid homologue and its utilization as an energy or carbon source, or they may be responsible for the production and export of an amino acid or amino acid homologue. It is known that amino acids can act as germination signals in bacilli (44), and it is possible, therefore, that these genes are involved in producing a novel sporulation signal. Although this is speculation, it does fit well with the presence of other predicted sporulation and germination determinants on this plasmid.
Plasmid replication and partition.
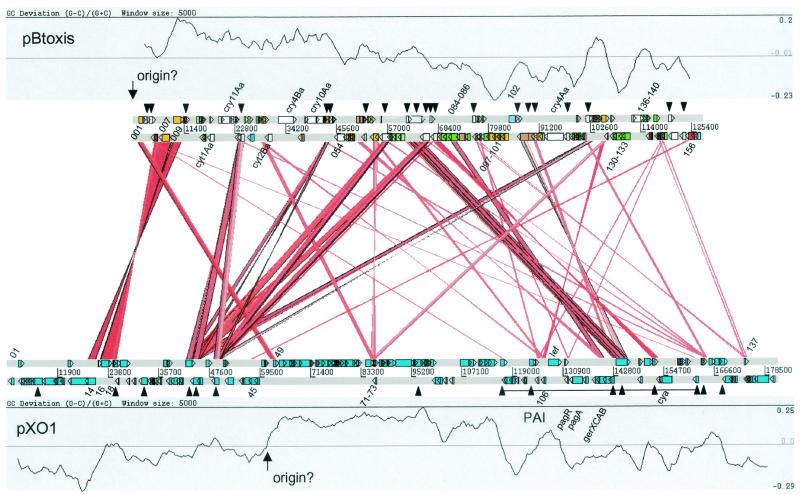
Analysis of the GC skew of the plasmid (25) indicated a potential origin of replication near base 1 of the sequence (Fig. 2). Although no replication proteins could be identified through database comparisons, the CDS to the right of this region (pBt001) showed >78% amino acid identity with pXO1-49, which is located close to a similar putative replication origin of pXO1, which we predict by GC skew analysis may be between bases 60955 and 62192 (36). pXO1-49 is shorter than pBt001, due to the predicted use of a later start codon; however, the upstream start codon equivalent to that predicted for pBt001 is present in pXO1. It is therefore possible that this protein, which has no other similarities in the database, may be involved in plasmid replication. Also close to this putative origin, on the opposite side, is pBt156, which shows weak similarities to FtsZ/tubulin-like proteins from Pyrococcus (EMBL number AB031743; 21% identity in 394 aa) and to pXO1-45 (21% identity in 444 aa), which is similarly located in pXO1. Proteins of the FtsZ family are known to be involved in cell division (21), forming a ring structure at the dividing septum, and it is therefore possible that pBt156 may play some role in plasmid partition. Previous studies have suggested that the pXO1 origin might lie between bp 86249 and 97209 (36, 40); the large majority of this region shows no similarities with pBtoxis, except in the first CDS, pXO1-72, a conserved hypothetical CDS which shows partial matches to pBt035, pBt067, and pBt127.
FIG. 2.
Linear representation of the pBtoxis-pXO1 comparison. pBtoxis is shown above, and pXO1 is below. GC bias [(G − C)/(G + C)] plots are shown for each plasmid, and the putative origin is indicated. Protein-protein similarities (as determined by TBLASTX comparisons of the complete plasmids) are indicated in the center, with the strength of the match indicated by the intensity of the red color. The color coding for pBtoxis genes is as in Fig. 1, except that the toxin and peptide antibiotic genes are in white. The pXO1 pathogenicity island (PAI) containing the toxin genes is marked with a horizontal line, and the transposon-related genes in each plasmid are indicated with black triangles; selected pBtoxis CDSs are labeled. The representation was drawn with ACT (http://www.sanger.ac.uk/Software/ACT), which can be used to visualize the complete comparison interactively.
Similarities with other plasmids.
Possible similarities between pBtoxis and other B. thuringiensis plasmid sequences in the database were analyzed by BLAST comparisons. The only significant match was between pBtoxis pBt010 and an unannotated CDS of unknown function in pTX14-3 from B. thuringiensis subsp. israelensis (4) (44% identity in 84 aa). No other database matches for these sequences exist, so the physiological functions, if any, of the sequences cannot presently be judged. No significant matches were found between pBtoxis and pBMB9741, pBMB2062, pTX14-1, pHD2, or the miniplasmid submitted under accession number S49203, and similarity with plasmid pGI2 was limited to a transposase gene.
Overall, 29 of 125 predicted pBtoxis proteins show detectable similarity to predicted proteins from pXO1 (Table 2) (36). Excluding the transposon- or insertion sequence-related proteins, only 17 of the predicted pBtoxis proteins are similar to predicted proteins from pXO1. This corresponds to the results of a previous study looking at conservation of pXO1 genes in a variety of Bacillus species (37): between 1 and 53 pXO1 genes were found to be present in different B. thuringiensis strains by hybridization and PCR experiments.
Most isolates of B. thuringiensis, like B. thuringiensis subsp. israelensis, encode their insecticidal toxins on extrachromosomal elements. Since pBt007 was found to be conserved between pBtoxis and pXO1-16 (96% identity in 569 aa), its distribution in other B. thuringiensis strains was also investigated by PCR, as described in Materials and Methods. As expected, no amplicons were produced from the primers when the negative control B. thuringiensis subsp. israelensis strain 4Q7, a strain cured of pBtoxis, was used. PCR also produced no prod-uct from the following B. thuringiensis isolates: Bacillus thuringiensis subsp. dakota [Oats43(4R1)], Bacillus thuringiensis subsp. kyushuensis [HD541(4U1)], Bacillus thuringiensis subsp. morrisoni [HD12(4K1)], Bacillus thuringiensis subsp. tenebrionis, and Bacillus thuringiensis subsp. tohokuensis [78-FS-29-17(4V1)]. This may reflect the absence of homologous sequences in these strains, or it could be the result of an alteration in nucleotide sequence in the regions corresponding to one or both of the test primers. However, the existence of pBt007-homologous sequences was revealed by the production of ∼600-bp amplicons (results not shown) in the following B. thuringiensis isolates: Bacillus thuringiensis subsp. aegypti (from commercial Agerin powder), Bacillus thuringiensis subsp. aizawai [HD133(J3)], Bacillus thuringiensis subsp. galleriae (HD155), Bacillus thuringiensis subsp. indiana [HD521(4S2)], B. thuringiensis subsp. israelensis [IPS70(4Q3)], B. thuringiensis subsp. israelensis [HD500(4Q2)], B. thuringiensis subsp. israelensis [HD567(4Q1)], Bacillus thuringiensis subsp. jegathesan, Bacillus thuringiensis subsp. japonensis [T23001(4AT1)], Bacillus thuringiensis subsp. kenyae [HD136(4F1)], Bacillus thuringiensis subsp. kumamotoensis [HD867(4W1)], B. thuringiensis subsp. kurstaki [HD1(4D1)], B. thuringiensis subsp. kurstaki [HD73(4D4)], Bacillus thuringiensis subsp. medellin, Bacillus thuringiensis subsp. thuringiensis [HD2(4A3)], Bacillus thuringiensis subsp. tochingiensis [HD868(4Y1)], Bacillus thuringiensis subsp. tolworthy [HD125(4L1)], and Bacillus thuringiensis subsp. wuhanensis [HD525(4T1)]. This indicates that the pBt007/pXO1-16-like sequence is widespread in B. thuringiensis isolates, and we speculate that it is likely to be associated with the virulence plasmids in all of these strains. In addition, an amplicon of the same size was also produced from the house fly-toxic Bacillus cereus subsp. moritai (originally named Bacillus moritai [1]), perhaps indicating that this isolate should again be reclassified as B. thuringiensis subsp. moritai.
Acknowledgments
This project was supported by funding from the Wellcome Trust through its support of the Sanger Institute Pathogen Sequencing Unit, the Royal Society (C.B.); a grant (97-00081) from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel (A.Z.); and a postdoctoral fellowship (E.B.-D.) from the Israel Ministry of Science.
REFERENCES
- 1.Abe, K., R. M. Faust, and L. A. Bulla. 1983. Plasmid deoxyribonucleic acid in strains of Bacillus sphaericus and in Bacillus moritai. J. Invertebr. Pathol. 41:328-335. [Google Scholar]
- 2.Agaisse, H., M. Gominet, O. Økstad, A.-B. Kolstø, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Andrup, L., J. Damgaard, K. Wassermann, L. Boe, S. Madsen, and F. G. Hansen. 1994. Complete nucleotide sequence of the Bacillus thuringiensis subsp. israelensis plasmid pTX14-3 and its correlation with biological properties. Plasmid 31:72-88. [DOI] [PubMed] [Google Scholar]
- 5.Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. Lopez, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, F. Servant, C. J. Sigrist, and E. M. Zdobnov. 2000. InterPro—an integrated documentation resource for protein families, domains and functional sites. Bioinformatics 16:1145-1150. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Dov, E., M. Einav, N. Peleg, S. Boussiba, and A. Zaritsky. 1996. Restriction map of the 125-kilobase plasmid of Bacillus thuringiensis subsp. israelensis carrying the genes that encode delta-endotoxins active against mosquito larvae. Appl. Environ. Microbiol. 62:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Dov, E., G. Nissan, N. Pelleg, R. Manasherob, S. Boussiba, and A. Zaritsky. 1999. Refined, circular restriction map of the Bacillus thuringiensis subsp. israelensis plasmid carrying the mosquito larvicidal genes. Plasmid 42:186-191. [DOI] [PubMed] [Google Scholar]
- 8.Brown, K. L., and H. R. Whiteley. 1988. Isolation of a Bacillus thuringiensis RNA polymerase capable of transcribing crystal protein genes. Proc. Natl. Acad. Sci. USA 85:4166-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crickmore, N., E. J. Bone, J. A. Williams, and D. J. Ellar. 1995. Contribution of the individual components of the delta-endotoxin crystal to the mosquitocidal activity of Bacillus thuringiensis subsp. israelensis. FEMS Microbiol. Lett. 131:249-254. [Google Scholar]
- 10.Crickmore, N., D. R. Zeigler, J. Feitelson, E. Schnepf, J. Van Rie, D. Lereclus, J. Baum, and D. H. Dean. 1998. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delecluse, A., C. Bourgouin, A. Klier, and G. Rapoport. 1988. Specificity of action on mosquito larvae of Bacillus thuringiensis israelensis toxins encoded by two different genes. Mol. Gen. Genet. 214:42-47. [DOI] [PubMed] [Google Scholar]
- 12.Dervyn, E., S. Poncet, A. Klier, and G. Rapoport. 1995. Transcriptional regulation of the cryIVD gene operon from Bacillus thuringiensis subsp. israelensis. J. Bacteriol. 177:2283-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faust, R. M., K. Abe, G. A. Held, T. Iizuka, L. A. Bulla, and C. L. Meyers. 1983. Evidence for plasmid-associated crystal toxin production in Bacillus thuringiensis subsp. israelensis. Plasmid 9:98-103. [DOI] [PubMed] [Google Scholar]
- 14.Feavers, I. M., J. S. Miles, and A. Moir. 1985. The nucleotide sequence of a spore germination gene (gerA) of Bacillus subtilis 168. Gene 38:95-102. [DOI] [PubMed] [Google Scholar]
- 15.Fujita, M., and Y. Sadaie. 1998. Feedback loops involving Spo0A and AbrB in in vitro transcription of the genes involved in the initiation of sporulation in Bacillus subtilis. J. Biochem. (Tokyo) 124:98-104. [DOI] [PubMed] [Google Scholar]
- 16.Garduno, F., L. Thorne, A. M. Walfield, and T. J. Pollock. 1988. Structural relatedness between mosquitocidal endotoxins of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 54:277-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaur, N. K., J. Oppenheim, and I. Smith. 1991. The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J. Bacteriol. 173:678-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerchicoff, A., R. A. Ugalde, and C. P. Rubinstein. 1997. Identification and characterization of a previously undescribed cyt gene in Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 62:2716-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guidi-Rontani, C., Y. Pereira, S. Ruffie, J. C. Sirard, M. Weber-Levy, and M. Mock. 1999. Identification and characterization of a germination operon on the virulence plasmid pXO1 of Bacillus anthracis. Mol. Microbiol. 33:407-414. [DOI] [PubMed] [Google Scholar]
- 20.Hajnsdorf, E., and P. Regnier. 2000. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. USA 97:1501-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harry, E. J. 2001. Bacterial cell division: regulating Z-ring formation. Mol. Microbiol. 40:795-803. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmaster, A. R., and T. M. Koehler. 1999. Autogenous regulation of the Bacillus anthracis pag operon. J. Bacteriol. 181:4485-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 24.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Düsterhöft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S.-Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Hénaut, H. Hilbert, S. Holsappel, S. Hosono, M.-F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S.-M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauël, C. Médigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S.-H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B.-S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H.-F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 25.Lobry, J. R. 1996. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 13:660-665. [DOI] [PubMed] [Google Scholar]
- 26.Mahillon, J., R. Rezsöhazy, B. Hallet, and J. Delcour. 1994. IS231 and other Bacillus thuringiensis transposable elements: a review. Genetica 93:13-26. [DOI] [PubMed] [Google Scholar]
- 27.Manasherob, R., A. Zaritsky, E. Ben-Dov, D. Saxena, D. Barak, and M. Einav. 2001. Effect of accessory proteins P19 and P20 on cytolytic activity of Cyt1Aa from Bacillus thuringiensis subsp. israelensis in Escherichia coli. Curr. Microbiol. 43:355-364. [DOI] [PubMed] [Google Scholar]
- 28.Mandic-Mulec, I., L. Doukhan, and I. Smith. 1995. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J. Bacteriol. 177:4619-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margalith, Y., and E. Ben-Dov. 2000. Biological control by Bacillus thuringiensis subsp. israelensis, p. 243-301. In J. E. Rechcigl, and N. A. Rechcigl (ed.), Insect pest management: techniques for environmental protection. CRC Press, Boca Raton, Fla.
- 30.Martinez-Bueno, M., M. Maqueda, A. Galvez, B. Samyn, J. Van Beeumen, J. Coyette, and E. Valdivia. 1994. Determination of the gene sequence and the molecular structure of the enterococcal peptide antibiotic AS-48. J. Bacteriol. 176:6334-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Bueno, M., E. Valdivia, A. Galvez, J. Coyette, and M. Maqueda. 1998. Analysis of the gene cluster involved in production and immunity of the peptide antibiotic AS-48 in Enterococcus faecalis. Mol. Microbiol. 27:347-358. [DOI] [PubMed] [Google Scholar]
- 32.Moir, A., and D. A. Smith. 1990. The genetics of bacterial spore germination. Annu. Rev. Microbiol. 44:531-553. [DOI] [PubMed] [Google Scholar]
- 33.Moszer, I., L. M. Jones, S. Moreira, C. Fabry, and A. Danchin. 2002. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res. 30:62-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 36.Okinaka, R. T., K. Cloud, O. Hampton, A. R. Hoffmaster, K. K. Hill, P. Keim, T. M. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pannucci, J., R. T. Okinaka, R. Sabin, and C. R. Kuske. 2002. Bacillus anthracis pXO1 plasmid sequence conservation among closely related bacterial species. J. Bacteriol. 184:134-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perego, M. 2001. A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol. Microbiol. 42:133-143. [DOI] [PubMed] [Google Scholar]
- 40.Robertson, D., T. Bragg, R. Simpson, W. Kaspar, W. Xie, and M. Tippetts. 1990. Mapping and characterization of the Bacillus anthracis plasmids pXO1 and pXO2. Salisbury Med. Bull. 68(Suppl.):55-58. [Google Scholar]
- 41.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 42.Salamitou, S., F. Ramisse, M. Brehélin, D. Bourguet, N. Gilois, M. Gominet, E. Hernandez, and D. Lereclus. 2000. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146:2825-2832. [DOI] [PubMed] [Google Scholar]
- 43.Serrano, M., R. Zilhao, E. Ricca, A. J. Ozin, C. P. Moran, Jr., and A. O. Henriques. 1999. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J. Bacteriol. 181:3632-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Setlow, P. 1975. Energy and small-molecule metabolism during germination of Bacillus spores, p. 443-450. In P. Gerhardt, R. N. Costilow, and H. L. Sadoff (ed.), Spores VI. American Society for Microbiology, Washington, D.C.
- 45.Stover, A. G., and A. Driks. 1999. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J. Bacteriol. 181:1664-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thanabalu, T., J. Hindley, J. Jackson-Yap, and C. Berry. 1991. Cloning, sequencing and expression of a gene encoding a 100-kilodalton mosquitocidal toxin from Bacillus sphaericus SSII-1. J. Bacteriol. 173:2776-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorne, L., F. Garduno, T. Thompson, D. Decker, M. Zounes, M. Wild, A. M. Walfield, and T. J. Pollock. 1986. Structural similarity between the lepidoptera- and diptera-specific insecticidal endotoxin genes of Bacillus thuringiensis subsp. kurstaki and israelensis. J. Bacteriol. 166:801-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1996. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J. Bacteriol. 178:3585-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wojciechowska, J. A., E. Lewitin, L. P. Revina, I. A. Zalunin, and G. G. Chestukhina. 1999. Two novel delta-endotoxin gene families cry26 and cry28 from Bacillus thuringiensis ssp. finitimus. FEBS Lett. 453:46-48. [DOI] [PubMed] [Google Scholar]
- 50.Wu, D., and B. A. Federici. 1993. A 20-kilodalton protein preserves cell viability and promotes CytA crystal formation during sporulation in Bacillus thuringiensis. J. Bacteriol. 175:5276-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshisue, H., T. Fukada, K.-I. Yoshida, K. Sen, S.-I. Kurosawa, H. Sakai, and T. Komano. 1993. Transcriptional regulation of Bacillus thuringiensis subsp. israelensis mosquito larvicidal crystal protein gene cryIVA. J. Bacteriol. 175:2750-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshisue, H., K. Ihara, T. Nishimoto, H. Sakai, and T. Komano. 1995. Expression of genes for insecticidal crystal proteins in Bacillus thuringiensis: cryIVA, not cryIVB, is transcribed by RNA polymerase containing σH and that containing σE. FEMS Microbiol. Lett. 127:65-72. [DOI] [PubMed] [Google Scholar]
- 53.Yoshisue, H., T. Nishimoto, H. Sakai, and T. Komano. 1993. Identification of a promoter for the crystal protein-encoding gene cryIVB from Bacillus thuringiensis subsp. israelensis. Gene 137:247-251. [DOI] [PubMed] [Google Scholar]
- 54.Zuberi, A. R., A. Moir, and I. M. Feavers. 1987. The nucleotide sequence and gene organization of the gerA spore germination operon of Bacillus subtilis 168. Gene 51:1-11. [DOI] [PubMed] [Google Scholar]