Abstract
Escherichia coli O86:K61 has long been associated with outbreaks of infantile diarrhea in humans and with diarrheal disease in many animal species. Studies in the late 1990s identified E. coli O86:K61 as the cause of mortality in a variety of wild birds, and in this study, 34 E. coli O86:K61 isolates were examined. All of the isolates were nonmotile, but most elaborated at least two morphologically distinct surface appendages that were confirmed to be type 1 and curli fimbriae. Thirty-three isolates were positive for the eaeA gene encoding a gamma type of intimin. No phenotypic or genotypic evidence was obtained for elaboration of Shiga-like toxins, but most isolates possessed the gene coding for the cytolethal distending toxin. Five isolates were selected for adherence assays performed with tissue explants and HEp-2 cells, and four of these strains produced attaching and effacing lesions on HEp-2 cells and invaded the cells, as determined by transmission electron microscopy. Two of the five isolates were inoculated orally into 1-day-old specific-pathogen-free chicks, and both of these isolates colonized, invaded, and persisted well in this model. Neither isolate produced attaching and effacing lesions in chicks, although some pathology was evident in the alimentary tract. No deaths were recorded in inoculated chicks. These findings are discussed in light of the possibility that wild birds are potential zoonotic reservoirs of attaching and effacing E. coli.
Escherichia coli O86:K61 has long been associated with outbreaks of infantile diarrhea in humans (24). This serotype is classified historically as enteropathogenic E. coli (EPEC) along with serotypes O55, O111, and O127 among others and has been implicated as a major cause of acute and persistent infantile diarrhea in developing parts of the world (39). Small-bowel biopsies of children infected with EPEC revealed discrete colonies of bacteria attached to the mucosa (58). Binding of EPEC to the brush border triggers a cascade of transmembrane and intracellular signals leading to cytoskeletal reorganization and formation of a specific lesion, termed an attaching and effacing (A/E) lesion, which involves intimate bacterial attachment to the host cell and associated effacement of microvilli (43). Jerse et al. (34) first described the eaeA gene of EPEC serotype O127:H6 that encodes intimin, a surface-arrayed protein essential for intimate association with enterocytes. Subsequent analysis showed that this gene is one of a cluster of over 40 genes within the locus of enterocyte effacement (LEE) that encode a type III secretion system essential for A/E lesion formation (45). EPEC possess plasmid-encoded factors, such as the bundle-forming pili (bfp) and the plasmid-encoded regulator (per), which also play roles in intimate adherence of EPEC (45). Similar A/E lesions are produced by enterohemorrhagic E. coli (EHEC) (51, 55), Hafnia alvei (2), Citrobacter rodentium (10), and rabbit diarrheagenic E. coli (15). The deduced amino acid sequences of the EaeA protein products of these and other A/E lesion-forming E. coli strains showed a high degree of conservation in the N′-terminal region and significant diversity in the C′-terminal region (1). Four classes of EaeA that in general terms are found in either EPEC (alpha, beta, and epsilon) or EHEC (gamma) have been described (1).
E. coli O86:K61 has been associated with diarrheal disease in calves (12, 56), pigs (3), and horses (32) and on one occasion has been implicated in cellulitis in broiler chickens (47). To date, this serotype has not been isolated from commercial poultry (Rob Davies, personal communication). Wild birds have been implicated as sources of various enteric pathogens of humans (27, 31), including Salmonella and E. coli belonging to serotype O86. Recent observations showed high mortality in wild birds of the family Fringillidae (siskins, green finches, chaffinches), as well as sparrows and pheasants in Scotland (48). Postmortem analysis of these birds showed that death was due to either Salmonella enterica serovar Typhimurium DT40 or systemic colibacillosis. E. coli O86:K61 was isolated readily from 43 of 46 birds examined (25). No A/E lesions were observed in any of the birds, although autolysis did preclude a detailed examination of the intestines (48).
EPEC strains often produce cytolethal distending toxin (CLDT), which causes elongation of Chinese hamster, HeLa, and HEp-2 cells and, to a lesser extent, Vero cells (54). EPEC and other E. coli strains that produce CLDT have been associated with diarrheal disease syndromes (11, 14, 54), although the precise role of the toxin in pathogenesis remains to be elucidated. Preliminary analysis of E. coli O86:K61 strains isolated from diseased wild birds (25, 48) indicated that they may be classified as CLDT positive.
The provision of supplementary food, such as peanuts, for wild birds in gardens during the winter months has been cited as a possible cause of the spread of infection within the wild bird population (48), and the high density of infected birds in urban areas may be a public health concern. It is also possible that wild birds may be potential sources of infection with E. coli O86:K61 in the poultry industry. Thus, the aim of this study was to characterize the pathogenic potential of avian E. coli O86:K61 isolates by performing appropriate genotypic and phenotypic tests with specific reference to the in vivo behavior in a chick model.
MATERIALS AND METHODS
Bacterial strains, isolation, and inocula.
E. coli O86:K61 strains were isolated from the tissues of dead birds submitted for postmortem examinations over a 3-year period in the latter half of the 1990s from Scotland. Members of the Fringillidae were found dead in gardens where supplementary food was provided, whereas pheasants were obtained from open land. The carcasses submitted for necroscopy were representative of a larger number of cases, and living birds were often reported as being unwell (25, 48). Isolates (Table 1) were maintained on Dorset egg slopes at the ambient temperature. Derivatives of isolates used in the persistence studies were plated from overnight broth cultures (200 μl) onto Luria-Bertani (LB) agar plates supplemented with nalidixic acid (15 μg/ml) to generate spontaneous resistant mutants. Resistant colonies were subcultured on the same medium, and single resistant colonies were picked and maintained on Dorset egg slopes. For antibiotic sensitivity testing of isolates we used recognized procedures used by the VLA Enteric Bacteria Reference Laboratory.
TABLE 1.
Phenotypic and genotypic characterization of E. coli O86:K61 isolates
| Isolate | Serotype | Provenance | Cytopathic effects on Vero cells | Hemagglutination of guinea pig erythrocytes | CLDT PCR | Acid tolerancea | Colony morphologyb | FAS | eae PCR | Gamma intimin PCR | Congo red binding |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EC36198 | O86:K61 | Finch | + | + | + | + | L | + | + | + | + |
| EC36298 | O86:K61 | Finch | + | + | + | + | L | + | + | + | + |
| EC36398 | O86:K61 | Finch | + | + | − | − | S | + | + | + | − |
| EC36498 | O86:K61 | Finch | + | + | + | − | S | + | + | + | − |
| EC36598 | O86:K61 | Finch | + | + | + | + | L | + | + | + | + |
| EC36698 | O86:K61 | Finch | + | + | + | − | L | + | + | + | + |
| EC36798 | O86:K61 | Finch | + | + | + | + | L | + | + | + | + |
| EC36898 | O86:K61 | Finch | + | + | + | + | L | + | + | + | + |
| EC36998 | O86:K61 | Finch | + | + | + | + | L | + | + | + | + |
| EC37098 | O86:K61 | Finch | + | + | + | + | L | + | + | + | + |
| EC37198 | O86:K61 | Finch | + | + | + | + | L | + | + | + | + |
| EC38798 | O86:K61 | Greenfinch | + | + | + | + | L | + | + | + | + |
| EC38898 | O86:K61 | Siskin | + | + | + | + | L | − | + | + | + |
| EC41198 | O86:K61 | Siskin | + | + | + | + | S | + | + | + | + |
| EC47898 | O86:K61 | Siskin | + | + | + | − | S | + | + | + | − |
| EC47998 | O86:K61 | Siskin | + | + | + | − | S | + | + | + | − |
| EC49598 | O86:K61 | Crossbill | + | + | + | + | L | + | + | + | + |
| EC49698 | O86:K61 | Crossbill | + | + | + | + | L | − | + | + | + |
| EC65098 | O86:K61 | Pheasant | − | + | − | − | L | + | + | + | + |
| EC65298 | O86:K61 | Pheasant | − | + | − | + | S | + | + | + | + |
| EC68198 | O86:K61 | Finch | + | + | + | − | S | + | + | + | − |
| EC68298 | O86:K61 | Finch | + | + | + | + | S | − | + | − | − |
| EC74299 | O86:K61 | Greenfinch | + | + | + | + | S | + | + | + | − |
| EC74399 | O86:K61 | Siskin | + | + | − | + | L | + | + | + | + |
| EC74499 | O86:K61 | Greenfinch | + | + | + | + | L | + | + | + | + |
| EC74599 | O86:K61 | Siskin | + | + | + | + | S | + | + | + | + |
| EC74699 | O86:K61 | Siskin | + | + | + | + | L | + | + | + | + |
| EC74799 | O86:K61 | Siskin | + | + | + | + | L | + | + | + | − |
| EC74899 | O86:K61 | Greenfinch | + | + | + | + | S | + | + | + | + |
| EC75099 | O86:K61 | Siskin | + | + | + | − | S | − | + | + | − |
| EC75199 | O86:K61 | Greenfinch | + | + | + | + | L | + | + | + | + |
| EC75299 | O86:K61 | Siskin | + | + | + | + | L | − | + | + | + |
| EC75499 | O86:K61 | Siskin | + | − | + | − | S | + | + | + | − |
| EC75599 | O86:K61 | Siskin | + | + | + | + | L | + | + | + | + |
A reduction of 2 log10 or more in the total viable count after 2 h of incubation at pH 2.5 was recorded as acid sensitive (−). A reduction of less than 2 log10 in the total viable count after 2 h of incubation at pH 2.5 was recorded as acid tolerant (+).
L, lacy; S, smooth.
Other bacteria used in this study were E. coli K-12 strain DH5α (Gibco BRL), S. enterica serotype Enteritidis strain S1400 (4), S. enterica serotype Typhimurium strain 3530 (21), E. coli O78:K80 strain EC34195 (avian pathogenic E. coli) (35), E. coli O111 (EPEC) (21), E. coli O157 strain NCTC12900 (21), and E. coli O86:H34 (1).
Inocula were prepared as follows. Stock cultures were streaked on LB agar, and discrete, isolated, single colonies were inoculated into LB broth to grow overnight with gentle agitation at 37°C aerobically or into heart infusion broth (HIB) to grow for 48 h statically at 37°C aerobically. For animal experiments, LB broth cultures were used. For tissue culture adherence and invasion assays, HIB cultures were adjusted to an optical density at 540 nm of 1.2, and 0.5 ml of each culture was diluted into 9.5 ml of Eagle's minimal essential media (EMEM) (Sigma) supplemented with 1% nonessential amino acids and 1% l-glutamine (Sigma). For tissue explant assays, HIB cultures were adjusted to an optical density at 550 nm of 0.6 in phosphate-buffered saline (PBS). Enumeration of bacteria for in vitro and in vivo experiments was accomplished by 10-fold serial dilution and plating onto LB agar and MacConkey agar, respectively.
Detection of surface antigens and their encoding genes.
The procedures used for mannose-sensitive and -resistant hemagglutination analysis, motility testing, transmission electron microscopy, description of colony morphology on media with and without Congo red, enzyme-linked immunosorbent assay (ELISA) detection of fimbriae, and Western blotting of bacterial antigens have been described previously (5-7, 19, 20).
PCR.
DNA sequences were amplified by PCR (53), and the primer sequences used are listed in Table 2. The reaction mixtures contained Thermo DNA polymerase reaction buffer (5 μl) (Promega), each deoxynucleoside triphosphate (Pharmacia) at a concentration of 200 μM, 1.5 mM MgCl2, 2.5 U of thermostable Taq polymerase (Promega), 10 pmol of each primer (Oswel), and 1 ng of total genomic DNA and were adjusted to a final volume of 50 μl with sterile distilled water. The reaction mixtures were overlaid with mineral oil (Sigma), and cycling was carried out with a thermocycler (Biometra). The cycling program used for the slt-1, slt-2, and eaeA multiplex PCR was 94°C for 2 min, followed by 25 cycles of 94°C for 1 min, 62°C for 1.5 min, and 72°C for 2 min and then 72°C for 5 min. The cycling program used for cldtAB was 94°C for 1 min, followed by 25 cycles of 94°C for 1 min, 65°C for 30 s, and 72°C for 30 s and then 72°C for 5 min. The cycling program used for hlyE was 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s and then 72°C for 5 min. The cycling program used for ial and ipaH was 94°C for 1 min, followed by 35 cycles of 94°C for 1 min, 50°C for 1.5 min, and 72°C for 1 min and then 72°C for 5 min. The cycling program used for espA/sepA was 94°C for 2 min, followed by 30 cycles of 94°C for 2 min, 54°C for 1.5 min, and 72°C for 2 min and then 72°C for 10 min.
TABLE 2.
Primers used for PCR of virulence genes and probes
| Primer | Sequence (5′-3′) | Position (bp) | Accession no. | Reference |
|---|---|---|---|---|
| fimA-F | AATGCGGATGCGACCTTCAAG | 3501-3521 | Z37500 | 41 |
| fimA-R | CATCACGCGTTGCCATATAAC | 5248-5227 | Z37500 | 41 |
| csgA-F | TATTGATCGCACACCTGACAG | 2498-2518 | X90754 | 30 |
| csgA-R | CCAAGGGTTGTGTTATCCATA | 4665-4645 | X90754 | 30 |
| fliC-F | CATGGCACAAGTCATTAATACC | 101-122 | Z36877 | 62 |
| fliC-R | GATAAGCGCAGCGCATCA | 1861-1846 | Z36877 | 62 |
| cldtAB-F | CCAACAACACTGAGTTTCCTG | 364-384 | NAa | 15 |
| cldtAB-R | CAGTCAACGTTGCAGAAGCTG | 1107-1127 | NA | 15 |
| espA-F | CCGGCTGTCAGAATGCTT | 12426-12443 | AF071034 | 49 |
| espA-R | TGGCAACATGCCAAAGGG | 14493-14476 | AF071034 | 49 |
| ial-F | CTGGTAGGTATGGTGAGG | NA | 26 | |
| ial-R | CCAGGCCAACAATTATTT | NA | 26 | |
| ipaH-F | GTTCCTTGACCGCCTTTCCGATACCGTC | 373-400 | M76443 | 59 |
| ipaH-R | GCCGGTCAGCCACCCTCTGAGAGTAC | 992-967 | M76443 | 59 |
| bfp-1 | GATTGAATCTGCAATGGTGC | 108-128 | Z12295 | 22 |
| bfp-2 | GGATTACTGTCCTCACATAT | 704-684 | Z12295 | 22 |
| slt1-F | CGCTGTTGTACCTGGAAAGG | 3313-3332 | L04539 | 46 |
| slt1-R | CGCTCTGCAATAGGTACTCC | 3567-3547 | L04539 | 46 |
| slt2-F | GCTTCTGCTGTGACAGTGAC | 385-404 | E03962 | |
| slt2-R | TCCATGACAACGGACAGCAG | 569-549 | E03962 | |
| hlyE-F | TCGGCATCCACATTAGTTG | NA | 50 | |
| hlyE-R | CCATCATCCAGCACTTG | NA | 50 | |
| ERIC1 | ATGTAAGCTCCTGGGGATTCAC | NA | 16 | |
| ERIC2 | AAGTAAGTGACTGGGGTGAGCG | NA | 16 | |
| eaeAO157-F | GCTTAGTGCTGGTTTAGGATTG | 1709-1730 | M58154 | 61 |
| eaeAO157-R | ATTCAACGACCAAATCGTCG | 1916-1896 | M58154 | 61 |
| Intimin γ F | CGTTGAAGTCGAGTACGCCA | 1867-1887 | AF081185 | 42 |
| Intimin γ R | TTCTACACAAACCGCATAGA | 2803-2782 | AF081185 | 42 |
NA, not applicable.
For rapid identification of colonies recovered from in vivo chick experiments, sterile toothpicks containing individual colonies were inoculated directly into a PCR mixture. The PCR was performed as described above except that the first cycle was an extended hot start consisting of 94°C for 2.5 min.
Enterobacterial repetitive intergenic consensus (ERIC) PCR (16) was used to assess clonality of the O86:K61 isolates used in this study. The primers are listed in Table 2, and the conditions used for PCR were standard conditions (16).
Cloning and nucleotide sequencing of eaeA PCR amplicons.
PCR products were purified from ethidium bromide (Sigma)-stained agarose gels (Sephaglas; Pharmacia) and cloned into a pCR2.1 cloning system (Invitrogen) by following the manufacturer's instructions. Plasmid DNA were extracted from positive clones, and the sequence of each insert was determined with a BigDye terminator cycle sequencing kit (Perkin-Elmer). Individual reaction mixtures (20 μl) in Eppendorf tubes contained plasmid DNA (400 ng), universal and custom primers (3.2 pmol), and ready reaction mixture (8 μl). The reaction mixtures were overlaid with mineral oil, and extension was performed with a thermocycler (Perkin-Elmer); the products were prepared for analysis with a 377 automated DNA sequencer as recommended by the manufacturer (Applied Biosystems). Trace data were analyzed and assembled by using DNAStar software (DNAStar Inc.). The test strains EC37098 (accession number AF339751) and EC74699 (accession number AY114154) were compared to database entries by using the National Center for Biotechnology Information Blast search.
Bacterial tolerance to exposure to inorganic acid.
Acid tolerance was tested essentially as described previously (28). Overnight LB broth cultures were diluted to a concentration of 1 × 105 CFU/ml in prewarmed LB broth adjusted to pH 2.5 with inorganic acid (HCl). Preparations were incubated aerobically for 2 h at 37°C with gentle agitation. Surviving bacteria were enumerated by plating 10-fold serial dilutions onto LB agar and incubating the cultures at 37°C overnight.
Bacterial adherence and invasion assays with HEp-2 tissue cultures.
HEp-2 cells were sown at a concentration of 2 × 105 cells per well in 24-well plates (Nunc) in EMEM (Sigma) supplemented with fetal calf serum (10%, vol/vol), nonessential amino acids (1%, wt/vol), 2 mM l-glutamine, and gentamicin (50 μg/ml). Monolayers were incubated for 2 days to obtain confluence. Before use, the monolayers were washed twice with Hanks' balanced salts solution (HBSS) to remove cell debris and residual gentamicin. A bacterial inoculum was added to give a concentration of 1 × 108 CFU/ml in each well. The monolayers were then incubated at 37°C in the presence of 5% CO2 for 2 h. The inoculum was removed, and each monolayer was washed three times with HBSS (Sigma) to remove nonadherent bacteria. The monolayer was then disrupted for 10 min by using a solution containing 1% Triton X-100 (Sigma) and a 12-mm magnetic stirrer. After disruption, serial 10-fold dilutions were plated onto LB agar and incubated overnight at 37°C to determine the number of CFU per milliliter. For invasion assays, bacteria were allowed to adhere as described above for 2 h, and then the monolayers were washed three times with HBSS (Sigma) before EMEM containing gentamicin (100 μg/ml) was added. The plates were incubated at 37°C in the presence of 5% CO2 for 2 h and washed three times with HBSS (Sigma). The monolayers were then disrupted with a 1% solution of Triton X-100 (Sigma), and the numbers of CFU per milliliter were determined. All bacterial adherence and invasion assays were repeated at least twice on two separate occasions.
Giemsa staining of E. coli O86:K61 on HEp-2 cells.
Confluent HEp-2 monolayers grown in 24-well plates with 13-mm coverslips were washed twice with HBSS (Sigma), and 1 ml of fresh incomplete Eagle's medium was added to each well. Each well was inoculated with 10 μl of an overnight LB broth culture of a test isolate and incubated for 3 h at 37°C in the presence of 95% (vol/vol) air-5% (vol/vol) CO2. The inoculum was aspirated and replaced with fresh medium, and the assay mixture was incubated for an additional 3 h under the conditions described above. Cells were washed five times with HBSS and then fixed with 3% buffered formalin and washed with distilled water. The cells were stained with 10% Giemsa stain (Sigma) for 1 h and then washed three times with distilled water. The cells were differentiated with 1% acetic acid for 2 min and washed once with distilled water. The coverslips were then removed from the 24-well plates and mounted on glass slides by using xylene dibutylphthalate (DPX) (BDH). The coverslips were examined by using a high-power oil immersion light microscope (Zeiss).
FAS of E. coli O86:K61 on HEp-2 cells.
For fluorescent actin staining (FAS) of E. coli O86:K61 on HEp-2 cells, the methods used for preparation of cells and bacterial adherence were the methods described above. The medium was aspirated and replaced, and the assay mixture was incubated for an additional 3 h. After incubation the medium was aspirated from each well, and the coverslips were washed four times with PBS (pH 7.3). The cells were then fixed for 20 min with 3% (vol/vol) formalin and washed three times with PBS (pH 7.3). Cells were permeabilized by incubation with 0.1% (vol/vol) Triton X-100 (Sigma) in PBS. The Triton X-100 was removed by three washes with PBS, and the cells were covered with PBS containing fluorescein isothiocyanate-phalloidin (Sigma) at a concentration of 0.5 μg/ml. The cells were incubated at the ambient temperature in the dark and then washed three times with PBS. The coverslips were mounted on glass slides with DPX (BDH), and the slides were examined by high-power oil immersion fluorescence microscopy (Zeiss).
TEM.
For transmission electron microscopy (TEM), the methods used for preparation of cells and bacterial adherence were the methods described above. The medium was aspirated and replaced with fresh medium, and the assay mixture was incubated for an additional 3 h. All of the medium was aspirated and replaced with 3% glutaraldehyde (Sigma) in 0.1 M phosphate buffer (pH 7.4), and the cells were fixed for 10 min. The HEp-2 cells were then scraped off the coverslips with a cell scraper and resuspended in an excess of fixative in a centrifuge tube. The cells were pelleted by centrifugation at 1,620 × g for 5 min. The supernatant was aspirated and replaced with phosphate buffer, and the preparation was postfixed in 1% (wt/vol) osmium tetroxide in the same buffer, dehydrated in ethanol at concentrations up to 100%, and placed in propylene oxide before it was embedded in Araldite resin. The Araldite resin was polymerized at 60°C for 48 h. Ultrathin sections (thickness, 70 to 90 nm) on copper grids were prepared by using a diamond knife. Sections were contrasted with uranyl acetate and lead citrate prior to examination with a Philips CM10 TEM.
SEM.
For scanning electron microscopy (SEM), the methods used for preparation of cells and adherence were the methods described above for TEM. The medium was aspirated, and the specimens were fixed for 16 h in 3% (vol/vol) glutaraldehyde in 0.1 M phosphate buffer (pH 7.4). The specimens were washed in phosphate buffer and postfixed in 1% (wt/vol) osmium tetroxide in the same buffer, and then they were rinsed in six changes of phosphate buffer, dehydrated in ethanol, and placed in hexamethyldisizone for 5 min. The specimens were subjected to critical point drying with liquid carbon dioxide. Air-dried specimens were fixed to aluminum stubs with silver conductive paint, sputter coated with gold, and examined with a Stereo-scan S250 MarkIII SEM at 10 to 20 kV.
Bacterial adherence to tracheal and proximal gut explants.
One-day-old specific-pathogen-free (SPF) White Leghorn chicks were killed by cervical dislocation. Portions (ca. 2 cm) of the proximal trachea and proximal gut (duodenal loop) were removed aseptically and placed in sterile Krebs-Ringers solution. Fatty tissue and mesentery were removed, and tissues were cut to expose the ciliated surface. Individual sections were washed gently twice in prewarmed Krebs-Ringers solution and then immersed in fresh sterile Krebs-Ringers solution in a 50-ml Falcon tube (BDH) to which a bacterial inoculum (1 × 108 CFU/ml) was added to give a concentration of 1 × 107 CFU/ml. The preparations were then incubated at 37°C with orbital rotation at 225 rpm for 180 min. The sections were then rinsed three times with fresh sterile prewarmed Krebs-Ringers solution to remove all nonadherent bacteria and then homogenized (17). Serial 10-fold dilutions of each homogenate were plated in triplicate onto LB agar plates that were incubated at 37°C overnight, and bacterial counts were determined (8). All assays were repeated at least twice on at least two separate occasions.
In vivo colonization, invasion, and persistence of E. coli O86:K61 in SPF chicks.
Colonization, invasion, and persistence experiments were performed as described previously (6, 8, 17, 18, 60, 61). In separate experiments 10 1-day-old SPF White Leghorn chicks were dosed orally with 0.1 ml containing either 1 × 109 CFU (high dose) or 1 × 102 CFU (low dose) of EC37098 or EC74699. The birds were given standard rations and water ad libitum. A group of six uninoculated birds was used as a negative control. For the group of birds that received the high dose, five birds were selected at random 24 and 120 h after the dose was administered and euthanized. Liver, spleen, duodenum, jejunum, ileum, colon, and cecal tissue samples were taken aseptically for bacteriological analysis, and 10-fold serial dilutions were plated onto MacConkey agar supplemented with the appropriate antibiotic. Separate sections were placed in 10% buffered formalin for light microscopy or in glutaraldehyde (3%) for electron microscopy. Tissues were examined by light microscopy after they were stained with hematoxylin and eosin or Giemsa stain and by SEM of duplicate tissues (9). For the group of birds that received the low dose, cloacal swabs were taken from each bird at weekly intervals for 5 weeks, and they were streaked directly onto MacConkey plates supplemented with the appropriate antibiotic and enriched in LB broth for 24 h at 37°C prior to plating. The plates were incubated overnight at 37°C, and growth was scored as clear (no colonies), low (<200 colonies), medium (>200 colonies), or high (confluent).
RESULTS
Avian E. coli O86:K61 isolates were nonmotile, but not all isolates elaborated fimbriae.
Persistent colonization of the avian alimentary tract by E. coli has been shown to be dependent upon flagella and fimbriae (37). However, none of the 34 E. coli O86:K61 isolates in this study were motile by means of flagella, although each harbored the fliC gene, as demonstrated by PCR. Similarly, all the isolates harbored both fimA and csgA genes, which encode the structural monomers of type 1 and curli fimbriae, respectively. All of the isolates except one (EC75499) exhibited mannose-sensitive hemagglutination of guinea pig red blood cells, which was characteristic of organisms having type 1 fimbriae. The development of so-called lacy colonies and binding of Congo red by colonies are known phenotypes associated with the elaboration of curli fimbriae (5, 35). Twenty-three isolates generated lacy colonies that bound Congo red, whereas 10 isolates generated smooth colonies that did not. One isolate (EC74799) produced lacy colonies that did not bind Congo red (Table 1).
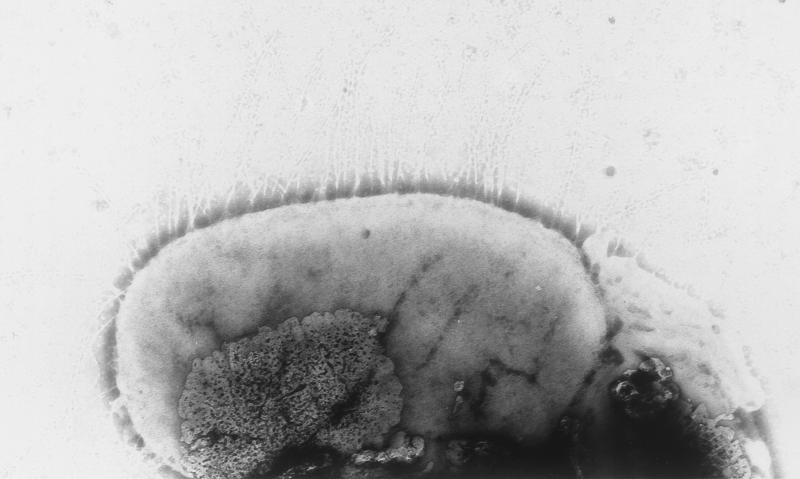
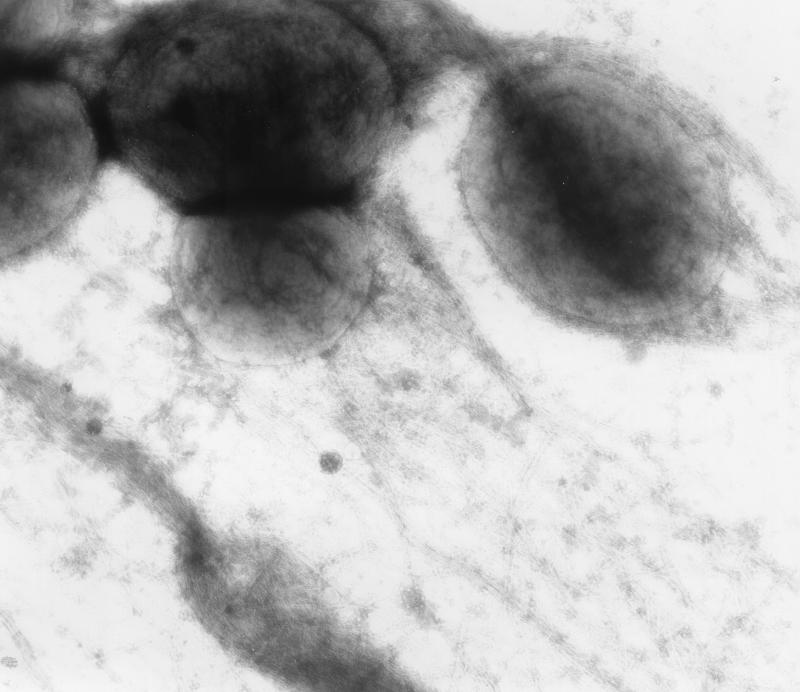
As visualized by TEM, all of the isolates except one (EC75499) elaborated type 1 fimbriae (Fig. 1) after static culture in HIB at 37°C for 48 h. The isolates that generated lacy colonies when they were cultured on colonization factor antigen agar for 72 h at 25°C were positive for curli fimbriae (Fig. 2), whereas the isolates that produced smooth colonies were not. Flagella were not observed on any isolates in this study. No other fimbrial structures were observed.
FIG. 1.
TEM of EC374899, showing peritrichous type 1 fimbriae after culture in HIB for 48 h at 37°C. Magnification, ×40,000.
FIG. 2.
TEM of EC37098, showing curli fimbriae after culture on CFA plates for 72 h at 25°C. Magnification, ×40,000.
Avian E. coli O86:K61 possessed gamma-like intimin.
It is generally accepted that to have a pathological effect upon its host, enterovirulent E. coli must adhere to the mucosal epithelium first. Preliminary data suggested that avian O86:K61 isolates possessed intimin, as determined by a generic intimin PCR. To test for the presence of an intimin gene and to determine the type of intimin gene (1) harbored by avian E. coli O86:K61, the 34 isolates were subjected to specific PCR tests (Tables 1 and 2). All of the isolates generated an amplicon with generic intimin primers, whereas all of the isolates except one, EC68298, generated an amplicon consisting of approximately 1 kb of the variable 3′ region of the eaeA gene with gamma intimin-specific primers.
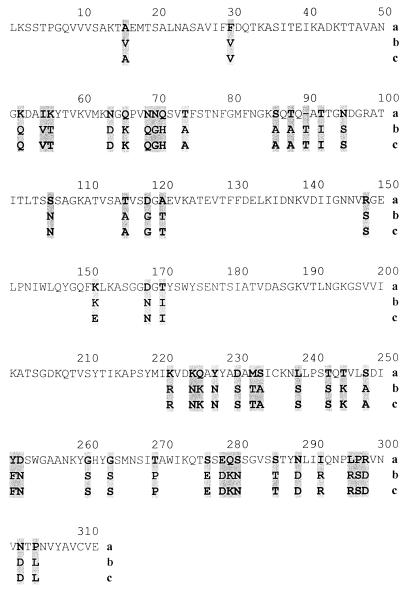
The PCR findings were unexpected because E. coli O86:K61 might be anticipated to possess the EPEC-associated alpha, beta, or epsilon intimin. Therefore, to identify the PCR product, the amplicons from two representative isolates (EC37098 and EC74699) were cloned into pCR2.1 (Invitrogen) and sequenced by using a BigDye terminator kit (Perkin-Elmer) and universal and custom-made primers. The two DNA sequences obtained exhibited 98% identity, and the deduced amino acid sequences exhibited 99.3% identity. A National Center for Biotechnology Information Blast search revealed 87% identity at the DNA level with the γ eaeA nucleotide sequence of E. coli (accession number AF081185), and the deduced amino acid sequences exhibited 83.9% identity (as determined with ClustalW) (Fig. 3) with the gamma intimin type of sequence (accession number AF081185).
FIG. 3.
Alignment of the deduced amino acid sequences of the variable regions of the intimin proteins of two O86:K61 strains (EC37098 [b] and EC74699 [c]) with the sequence of the homologous region of the gamma intimin of E. coli (GenBank accession number AF081185) (a).
Representative isolates EC75599, EC74899, EC37098, EC65298, EC49698, and EC74699 were negative for gamma intimin, as determined by using a specific ELISA and Western blotting after culture in Dulbecco's modified Eagle's medium (A. L. Cookson, personal communication and data not shown).
To test for the presence of other EPEC-associated factors, EAF plasmid-specific colony dot blot and PCR experiments were done. None of the isolates possessed such a factor, and as determined by TEM, bundle-forming pilus-like fimbriae were not observed for any isolate (Fig. 1 and 2). In addition, all isolates were negative for hlyE, ial, and ipaH.
ERIC PCR tests were performed with five representative O86:K61 isolates (EC74699, EC37098, EC75599, EC74599, and EC75299) in order to test clonality. Similar, if not identical, profiles were obtained for all isolates (data not shown).
Avian E. coli O86:K61 exhibited intimate adherence and invaded tissue culture cells.
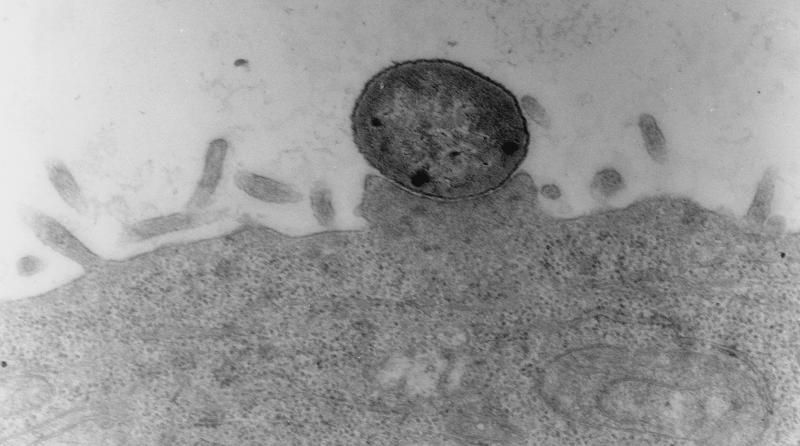
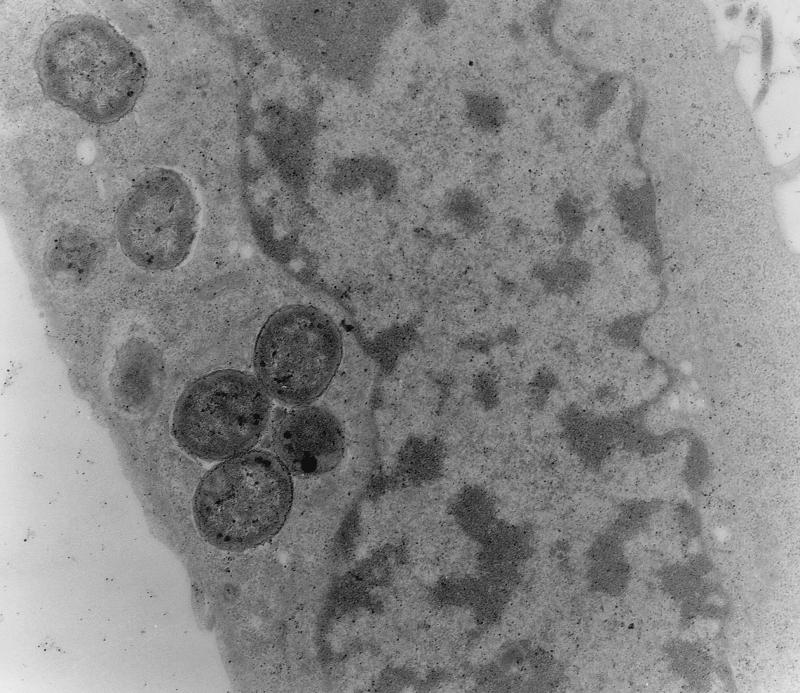
The isolates selected for further analysis included isolates that were FAS positive and isolates that were FAS negative in HEp-2 cell adherence assays and isolates that elaborated curli fimbriae and isolates that did not elaborate curli fimbriae. Five isolates (EC74699, EC37098, EC75599, EC74599, and EC75299) were analyzed to determine their interactions with HEp-2 cells and tracheal and gut explants (Tables 3 and 4). All isolates adhered to HEp-2 cells as discrete microcolonies after 6 h of incubation, as shown by Giemsa staining (data not shown), and all isolates except EC75299 induced actin rearrangements, as determined by FAS and TEM (data not shown). As determined by TEM, all isolates tested except EC75299 exhibited intimate associations with the cells, typical of A/E lesions (Fig. 4). TEM also showed that all of the isolates were internalized (Fig. 5), but surprisingly, invasion was not detected for any of the O86:K61 isolates when they were tested in a gentamicin invasion assay. In control experiments, the gentamicin assay showed that 10% of salmonellae invaded and low numbers (on the order of 0.1%) of EPEC O111, EHEC O157, and EPEC O86:H34 isolates invaded (Table 4). Adherence to gut and tracheal explants was observed, and the numbers of the five isolates adhering were broadly similar; there were no significant differences in attachment to either explant type. The control organisms used in these studies adhered and invaded in broadly similar numbers, as demonstrated previously (19, 21, 35).
TABLE 3.
Adhesion of E. coli O86:K61 isolates to tracheal and gut explant tissuea
| Isolate | Adhesion to tracheal tissue (CFU/ml)
|
Adhesion to gut tissue (CFU/ml)
|
||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| EC37098 | 1.85 × 105 | 1 × 104 | 1.55 × 105 | 8.7 × 103 |
| EC74599 | 1.8 × 105 | 4.7 × 104 | 6.2 × 105 | 1.3 × 105 |
| EC75599 | 1.6 × 105 | 5.3 × 104 | 1.5 × 106 | 2.9 × 104 |
| EC74699 | 2.4 × 105 | 1.2 × 104 | 4.3 × 105 | 1 × 105 |
| EC75299 | 2.9 × 105 | 3.1 × 105 | 6.2 × 105 | 7.6 × 104 |
Bacterial cultures for the assays were grown in HIB for 48 h statically at 37°C aerobically.
TABLE 4.
Adhesion and invasion of HEp-2 cells by E. coli O86:K61 isolatesa
| Isolate | Adhesion (CFU/ml)
|
Invasion (CFU/ml)
|
||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| E. coli K-12 strain DH5αb | 6.3 × 105 | 1.2 × 105 | 0.0 | |
| S. enterica serovar Enteritidis S1400c | 4.75 × 106 | 1.2 × 105 | 4.3 × 105 | 6.0 × 104 |
| S. enterica serovar Typhimurium 3530d | 6.3 × 106 | 1.2 × 105 | 4.75 × 105 | 1.8 × 105 |
| E. coli EC34195 (O78:K80)e | 5.3 × 106 | 1.5 × 105 | 2.8 × 102 | 4.1 × 101 |
| E. coli O111 (EPEC) strain NM B171d | 5.6 × 106 | 1.7 × 105 | 3.1 × 102 | 8.0 × 101 |
| E. coli O157 strain NCTC12900d | 5.9 × 106 | 2.3 × 105 | 5.7 × 103 | 1.6 × 103 |
| E. coli O86:H34f | 3.8 × 106 | 6.0 × 104 | 8.5 × 105 | 2.2 × 104 |
| E. coli EC37098 (O86:K61)g | 5.0 × 106 | 1.1 × 106 | 0.0 | |
| E. coli EC74599 (O86:K61)g | 7.3 × 106 | 9.3 × 105 | 0.0 | |
| E. coli EC75599 (O86:K61)g | 5.5 × 106 | 1.5 × 106 | 0.0 | |
| E. coli EC74699 (O86:K61)g | 4.25 × 106 | 9.3 × 106 | 0.0 | |
| E. coli EC75299 (O86:K61)g | 7.1 × 106 | 2.3 × 106 | 0.0 | |
FIG. 4.
TEM of EC37098 showing intimate A/E-like attachment to HEp-2 cells after 6 h of incubation. Magnification, ×40,000.
FIG. 5.
TEM of the interaction of EC37098 with HEp-2 cells showing bacterial invasion after 6 h of incubation. Magnification, ×40,000.
E. coli O86:K61 isolates were invasive in the SPF chick model.
In separate experiments 10 1-day-old SPF chicks were dosed orally with 1 × 109 CFU of each of two test isolates, EC37098 Nalr and EC74699 Nalr. At 24 and 120 h after the doses were administered, the numbers of bacteria in the ceca and in the livers and spleens were enumerated after postmortem examination (Table 5). High numbers of bacteria were recovered from livers and spleens at 24 h after oral inoculation, and there was evidence of significant clearance within 5 days. The ceca were colonized with approximately 108 CFU of E. coli O86:K61 per g of cecal lumenal contents. No background growth was observed, and swabs from uninoculated control birds gave no nalidixic acid-resistant colonies. To confirm the identity of the nalidixic acid-resistant colonies, approximately 10% of the colonies were tested by slide agglutination, and they proved to be serotype O86:K61 colonies. Additionally, PCR tests for eaeA and cldt were performed with about 1% of the colonies, all of which were positive.
TABLE 5.
Colonization and invasion of 1-day-old SPF chicks by E. coli O86:K61 isolates
| Organ | Concn of EC37098 ata:
|
Concn of EC74699 ata:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 24 h
|
120 h
|
24 h
|
120 h
|
|||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Liver | 5.6 × 106 (3/5)b | 1.3 × 107 | 10 (5/5) | 0 | 4.7 × 105 (5/5) | 4.5 × 105 | 6.1 × 102 (5/5) | 8.4 × 102 |
| Spleen | 2.5 × 106 (4/5) | 1.0 × 105 | 10 (5/5) | 0 | 1.3 × 104 (4/5) | 2.8 × 104 | 2 (1/5) | 4 |
| Cecum | 9.9 × 108 (5/5) | 4.2 × 108 | 1.1 × 109 (5/5) | 8.0 × 108 | 6 × 108 (5/5) | 1.5 × 108 | 1 × 109 (5/5) | 4.0 × 108 |
The values are the numbers of CFU per gram for livers and the numbers of CFU per tissue for ceca and spleens.
The numbers in parentheses are number of organs positive/number of organs tested.
An experiment similar to the experiment described above was set up, and during postmortem examinations 24, 48, and 120 h postinfection tissues were taken for histopathological analysis. Mild hyperplasia of the Peyer's patches was observed. However, no classical A/E lesions were observed, although very sparse microcolonies were found attached to the mucosa in the colon. Individual bacteria were observed adhering loosely to the mucosa in the duodenum, colon, and cecum. No microcolonies were observed in negative control birds.
In a separate experiment to quantify persistence in the SPF chick model, two groups of six 1-day-old chicks were dosed orally with 1 × 102 CFU of either EC37098 Nalr or EC74699 Nalr separately. Cloacal swab samples were taken at weekly intervals for 5 weeks. Both isolates persisted for 5 weeks in relatively high numbers, and all swab-inoculated plates contained more than 200 colonies for all animals at all time points. After 5 weeks, all birds were culled, and the numbers of O86:K61 bacteria in 1-g samples of cecal contents were determined. Approximately 1 × 108 CFU of both EC37098 Nalr and EC74699 Nalr per g of lumenal contents was detected. No background growth was observed, and approximately 10% of the colonies were confirmed to be E. coli O86:K61 colonies by slide agglutination. E. coli O86:K61 was not recovered from negative control birds.
DISCUSSION
This was the first detailed study of E. coli O86:K61 of avian origin, and we demonstrated by using PCR that 33 of 34 of the isolates examined possessed a gamma-like intimin. However, although one isolate (EC68298) did not produce an amplicon with gamma intimin-specific primers, it produced an amplicon with the generic intimin primers. This requires further investigation. The 3′ region of the eaeA gene is known to be variable (1). The 3′ regions of the eaeA genes of two O86:K60 representative isolates were determined and were shown to be gamma intimin eaeA orthologues. Analysis of the deduced amino acid sequence indicated that these molecules are novel intimin polymorphs. The complete sequences of the corresponding eaeA genes were not determined because the 5′ conserved regions are relatively invariant (1). However, it would be interesting to sequence other genes within the LEE to establish differences, especially because polymorphism of these genes is considered to be associated with tissue tropism (1).
The deduced amino acid sequences of the intimins of the two O86:K61 isolates examined in detail showed single-residue differences. While the sequences were not identical, this was evidence of the close evolutionary history of these isolates. This conclusion was supported by the similarities exhibited by the ERIC PCR profiles of the O86:K61 isolates tested. However, ERIC PCR is a relatively blunt tool for clonal analysis, and clonality is worthy of additional study.
Several classes of intimins have been defined (1, 42), and these molecules are responsible along with other products of the LEE for the intimate adhesion of A/E E. coli, including EHEC and EPEC, to epithelial cells during the infective process in animals and humans (1, 23, 34). Other workers have reported that classical EPEC strains possess alpha or beta intimin, whereas E. coli O86 isolates are commonly positive for epsilon intimin (1, 42). The presence of gamma-like intimin in the isolates described in this paper confirms not only the heterogeneity of this serotype (13, 34, 57) but also the heterogeneity of the family of intimin proteins (1). It is possible that intimins, which are outer membrane proteins, may be subjected to strong selective pressure for amino acid diversity, either through adaptive shifts in tropism or through antigenic shifts to evade the immune system (42). All 34 isolates examined were negative with gamma intimin polyclonal sera in ELISA and Western blot analyses. It is possible that the conditions were not appropriate for elaboration of intimin or, more likely, that the sequence variation shown by the O86:K61 intimins resulted in an absence of epitopes in common with the recombinant C-terminal end of the EHEC O157:H7 intimin that was used to raise the polyclonal antisera. Of the 34 isolates, 29 induced a FAS reaction leading to A/E lesion formation on HEp-2 cells, whereas the remaining 5 isolates did not. Thus, the majority of the isolates possessed a fully functional LEE.
It was hypothesized that the ability to form A/E lesions in vitro is related to the formation of A/E lesions in vivo. However, there was no evidence of A/E lesions in birds experimentally infected with two isolates that were fully proficient in vitro. Additionally, no A/E lesions were observed in the original affected hosts (25, 48). Either the LEE in the avian O86:K61 isolates was not induced in vivo, at least in this model or in the original host, or avian species were not susceptible to A/E lesion formation. Alternatively, A/E lesions were sparse and not detected in this study or the original reports (25, 48). Although isolated from diseased birds, five eaeA-positive isolates did not induce FAS reactions in vitro. This indicated that these O86:K60 isolates, if they were the causes of disease in the birds, did not mediate pathogenicity through intimate attachment.
The E. coli O86:K61 isolates lacked Shiga-like toxins but possessed the CLDT (cldtAB) genes, which are characteristic of EPEC. CLDT-positive E. coli strains, including EPEC strains, have been isolated from patients with a variety of diarrheal syndromes (11, 33, 54). Unlike classic EPEC, the E. coli O86:K61 isolates in this study did not harbor the EAF plasmid or elaborate bundle-forming pilus fimbriae (22, 23). However, the evidence was probe based, and no studies were done to test for the presence of large plasmids in the O86:K61 isolates. It would be valuable to compare the isolates described here with isolates derived from humans, calves, pigs, and chickens (3, 12, 32, 56).
Five isolates, including one that was positive for A/E lesions by the fluorescent actin stain (FAS) test (EC75299), were studied to determine their abilities to adhere and invade. Both tracheal and gut explants were used as E. coli is both an enteric pathogen and a respiratory pathogen in poultry (29, 38). The O86:K61 isolates were uniformly adherent to the HEp-2 cells and tissue explants tested. However, the invasion study had an unexpected result, namely, that the gentamicin protection assay indicated that the O86:K61 isolates tested were not internalized and, by definition therefore, were noninvasive. However, electron microscopy of the cells from the adherence and invasion assays showed internalized bacteria, and furthermore, bacteria were readily isolated from internal organs of birds after oral inoculation. These data point to an invasive phenotype. With regard to the in vitro adherence and invasion assays, it is possible that the host cell membrane or cellular integrity was disrupted due to invasion by bacteria and/or production of CLDT and thereby rendered the bacteria susceptible to gentamicin killing. In the in vivo studies there was no morbidity or mortality and there was a lack of substantive pathology in the gastrointestinal mucosa. Although in vitro studies pointed to A/E lesion formation and probable cellular disruption, invasion in vivo appeared to be independent of A/E lesion formation, at least in the 1-day-old chick model. However, due to Her Majesty's Government (Animals Scientific Procedures Act, 1986) Home Office license restrictions, other birds, such as pheasants or finches, could not be used and SPF chicks were the only available model. It is possible that the E. coli O86:K61 isolates examined possessed several pathogenicity determinants that may operate differently depending upon the environment and host. There is a question concerning whether the LEE in these E. coli O86:K61 isolates was essential for pathogenicity in the birds from which they were isolated. However, original reports of the isolation of these O86:K61 isolates (25, 48) indicated that S. enterica serovar Typhimurium DT40 was also present in the birds. This definitive type was described by Morgenroth and Duguid (44) as a non-type-1-fimbriated, non-inositol-fermenting, non-rhamnose-fermenting biotype (FIRN type) of S. enterica serovar Typhimurium that is associated with wild birds and may be restricted to avian species as hosts (40, 48). Thus, it is possible that the presence of S. enterica serovar Typhimurium DT40 was predisposing and the O86:K61 bacteria were merely opportunistic pathogens. However, the array of known virulence determinants that were shown to be active in the O86:K61 isolates suggests otherwise.
We have shown in previous studies that flagella and both type 1 and curli fimbriae contribute collectively to adherence of other avian pathogenic E. coli strains to the gastrointestinal epithelium (37). Additionally, these appendages contribute collectively to long-term persistence in the gastrointestinal tracts of infected birds. It was evident that the E. coli O86:K61 isolates studied in vivo here were well adapted to colonize, invade, and persist in the chick model used. Interestingly, all of the isolates were nonmotile, a feature shared with some other enteropathogens, such as E. coli O157:H- and S. enterica serovars Gallinarum and Pullorum. Not all O86:K61 strains elaborated curli fimbriae, yet they bound cells and tissue explants in vitro. It would have been interesting to test whether noncurliated isolates were as persistent as the two curliated isolates tested in vivo. By definition, nine E. coli O86:K61 isolates were acid sensitive, and seven of these isolates were noncurliated. The significance of this observation is unclear, but the two phenotypes may be linked through a common global regulatory pathway. Interestingly, RpoS mutants that are deficient in the elaboration of curli fimbriae are also acid sensitive (38, 52). However, the catalase test that is indicative of RpoS proficiency (38, 52) was performed on all the isolates, and they were all positive.
This study focused on E. coli O86:K61 isolates obtained over a 3-year period in the latter half of the 1990s from members of the Fringillidae from Scotland (25, 48). It is possible that the LEE harboring a gamma intimin variant was acquired by this serotype recently. It is also possible that this study focused on a single clonal source, but there was phenotypic variation shown in the library of isolates tested that suggested that this may not be the case. This possibility requires further investigation.
Wild birds have been implicated as sources of various enteric pathogens of humans (27, 31). While there is little evidence which suggests that wild birds play a role in incidents of E. coli O86:K61 infection in humans or animals, since the strains tested here possess several putative virulence factors associated with human and animal disease, the potential exists for this pathogen to be zoonotic. Practices that encourage large numbers of birds to congregate in urban areas may increase the zoonotic risk. Suitable precautions should be taken when fecally contaminated material is handled. In addition to posing a threat to other wild birds and people, infected populations of wild birds may also act as reservoirs for domestic livestock and companion animals.
Acknowledgments
We thank S. Done for histology, S. Farrelly for electron microscopy, J. Graham for serotyping, A. L. Cookson for advice on eaeA detection, and T. Pennycott and T. Patterson for primary isolation of E. coli isolates.
This work was supported by the Department for Environment Food and Rural Affairs, United Kingdom.
REFERENCES
- 1.Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Doucel, S. Knutton, and G. Dougan. 1998. Detection of intimins α, β, γ, and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert, M. J., S. M. Farruque, M. Ansaruzzaman, M. M. Islam, K. Haider, K. Alam, I. Kabir, and R. Robins-Browne. 1992. Sharing of virulence associated properties at the phenotypic and genotypic levels between enteropathogenic Escherichia coli and Hafnia alvei. J. Med. Microbiol. 37:310-314. [DOI] [PubMed] [Google Scholar]
- 3.Alexa, P., E. Salajka, J. Hamrik, and A. Nejezchleb. 1995. Pathogenic strains of Escherichia coli in weaned piglets in the Czech Republic. Pig News Inform. 16:81N-84N. [Google Scholar]
- 4.Allen-Vercoe, E., and M. J. Woodward. 1999. Colonisation of the chicken caecum by afimbriate and aflagellate derivatives of Salmonella enterica serotype Enteritidis. Vet. Microbiol. 69:265-275. [DOI] [PubMed] [Google Scholar]
- 5.Allen-Vercoe, E., M. P. Dibb-Fuller, C. J. Thorns, and M. J. Woodward. 1997. SEF17 fimbriae are essential for the convoluted colonial morphology of Salmonella enteritidis. FEMS Microbiol. Lett. 153:33-42. [DOI] [PubMed] [Google Scholar]
- 6.Allen-Vercoe, E., A. R. Sayers, and M. J. Woodward. 1999. Virulence of Salmonella enterica serotype Enteritidis aflagellate and afimbriate mutants in a day-old chick model. Epidemiol. Infect. 122:395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen-Vercoe, E., and M. J. Woodward. 1997. Construction of improved suicide vectors and their use in site directed mutagenesis of fimbrial genes of Salmonella enteritidis, p. 197-200. In Salmonella & salmonellosis proceedings. St. Brieuc, Ploufragan, France.
- 8.Amara, A., S. El Had, T. Jirrari, and K. Bouzoubaa. 1996. The lethality, haemagglutination and adhesion of E. coli strains (serotype O1) isolated in Morocco from chickens with colibacillosis. Avian Dis. 40:540-545. [PubMed] [Google Scholar]
- 9.Anonymous. 1957. Manual of histological and special staining techniques. Armed Forces Institute of Pathology, Washington, D.C.
- 10.Barthold, S. W., G. L. Coleman, P. N. Bhatt, G. W. Osbaldiston, and A. M. Jones. 1976. The etiology of transmissible murine colonic hyperplasia. Lab. Anim. Sci. 26:889-894. [PubMed] [Google Scholar]
- 11.Bisicchia, R., R. Ciammarughi, A. Caprioloi, V. Falbo, and F. M. Ruggeri. 1985. Toxin production and haemagglutination in strains of Escherichia coli from diarrhoea in Brescia, Italy. J. Hyg. 95:353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanco, M., J. Blanco, J. E. Blanco, and J. Ramos. 1993. Enterotoxigenic, verotoxigenic and necrotoxigenic Escherichia coli isolated from cattle in Spain. Am. J. Vet. Res. 54:1446-1451. [PubMed] [Google Scholar]
- 13.Blanco, M., J. E. Blanco, A. Mora, and J. Blanco. 1998. Distribution and characterization of faecal necrotoxigenic Escherichia coli CNF1+ and CNF2+ isolated from healthy cows and calves. Res. Microbiol. 59:183-192. [DOI] [PubMed] [Google Scholar]
- 14.Bouzari, S., and A. Varghese. 1990. Cytolethal distending toxin (CLDT) production by enteropathogenic Escherichia coli (EPEC). FEMS Microbiol. Lett. 59:193-198. [DOI] [PubMed] [Google Scholar]
- 15.Cantey, J. R., and R. K. Blake. 1977. Diarrhoea due to Escherichia coli in the rabbit, a novel mechanism. J. Infect. Dis. 135:454-462. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho de Moura, A. C., K. Irino, and M. C. Vidotto. 2001. Genetic variability of avian Escherichia coli strains evaluated by enterobacterial repetitive intergenic consensus and repetitive extragenic palindromic polymerase chain reaction. Avian Dis. 45:173-181. [PubMed] [Google Scholar]
- 17.Cooper, G. L., L. M. Venables, R. A. J. Nicholas, G. A. Cullen, and C. E. Hormaeche. 1992. Vaccination of chickens with chicken derived Salmonella enteritidis phage-type 4 aroA live oral Salmonella vaccines. Vaccine 10:247-254. [DOI] [PubMed] [Google Scholar]
- 18.Dho, M., and J. P. Lafont. 1982. Escherichia coli colonisation of the trachea in poultry: comparison of virulent and avirulent strains in gnotoxenic chickens. Avian Dis. 26:787-797. [PubMed] [Google Scholar]
- 19.Dibb-Fuller, M., E. Allen-Vercoe, C. J. Thorns, and M. J. Woodward. 1999. Fimbriae and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145:1023-1031. [DOI] [PubMed] [Google Scholar]
- 20.Dibb-Fuller, M., E. Allen-Vercoe, M. J. Woodward, and C. J. Thorns. 1997. Expression of SEF17 fimbriae by Salmonella enteritidis. Lett. Appl. Microbiol. 25:447-452. [DOI] [PubMed] [Google Scholar]
- 21.Dibb-Fuller, M. P., A. Best, D. A. Stagg, W. A. Cooley, and M. J. Woodward. 2001. Bovine epithelial primary cell cultures; an in vitro model for the study of E. coli O157:H7 and other entero-pathogens. J. Med. Microbiol. 50:1-11. [DOI] [PubMed] [Google Scholar]
- 22.Donnenberg, M. S., J. A. Giron, J. P. Nataro, and J. B. Kaper. 1992. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localised adherence. Mol. Microbiol. 6:3427-3447. [DOI] [PubMed] [Google Scholar]
- 23.Donnenberg, M. S., and J. B. Kaper. 1992. Enteropathogenic Escherichia coli. Infect. Immun. 60:3953-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewing, W. H. 1986. Edwards & Ewing identification of Enterobacteriaceae, 4th ed. Elsevier Science Publishing. New York, N.Y.
- 25.Foster, G., H. M. Ross, T. W. Pennycott, G. F. Hopkins, and I. M. McLaren. 1998. Isolation of Escherichia coli O86:K61 producing cyto-lethal distending toxin from wild birds of the finch family. Lett. Appl. Microbiol. 26:395-398. [DOI] [PubMed] [Google Scholar]
- 26.Frankel, G., L. Riley, J. A. Giron, et al. 1990. Detection of Shigella in faeces using DNA amplification. J. Infect. Dis. 161:1252-1256. [DOI] [PubMed] [Google Scholar]
- 27.Girwood, R. W. A., C. R. Fricker, D. Munro, C. B. Shedden, and P. Monaghan. 1985. The incidence and significance of Salmonella carriage by gulls (Larus spp.) in Scotland. J. Hyg. Camb. 95:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorden, J., and P. L. C. Small. 1993. Acid resistance in enteric bacteria. Infect. Immun. 61:364-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gross, W. B. 1991. Colibacillosis, p. 138-144. In B. W. Calnek (ed.), Diseases of poultry, 9th ed. Wolfe Publishing Ltd., Iowa State University Press, Ames, Iowa.
- 30.Hammer, M., A. Arnqvist, Z. Bian, A. Olsen, and S. Normark. 1995. Expression of two csg operons is required for the production of fibronectin and Congo red binding curli polymers in E. coli K-12. Mol. Microbiol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 31.Healing, T. D., M. H. Greenwood, and A. D. Pearson. 1992. Campylobacters and enteritis. Med. Microbiol. 3:159-167. [Google Scholar]
- 32.Holland, R. E., A. Schmidt, N. Sriranganathan, et al. 1996. Characterisation of Escherichia coli isolated from foals. Vet. Microbiol. 48:243-255. [DOI] [PubMed] [Google Scholar]
- 33.Iyoda, S., K. Tamura, K. Itoh, H. Ixumiya, N. Ueno, K. Nagata, M. Togo, J. Terajima, and H. Watanabe. 2000. Inducible stx2 phages are lysogenised in the enteroaggregative and other phenotypic Escherichia coli O86:HNM isolated from patients. FEMS Microbiol. Lett. 191:7-10. [DOI] [PubMed] [Google Scholar]
- 34.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Ragione, R. M., E. J. Collighan, and M. J. Woodward. 1999. Non-curliation of Escherichia coli O78:K80 isolates associated with IS1 insertion in csgB and reduced persistence in poultry infection. FEMS Microbiol. Lett. 175:247-253. [DOI] [PubMed] [Google Scholar]
- 36.La Ragione, R. M., W. A. Cooley, and M. J. Woodward. 2000. The role of fimbriae and flagella in the adherence of avian strains of Escherichia coli O78:K80 to tissue culture cells and tracheal and gut explants. J. Med. Microbiol. 4:327-338. [DOI] [PubMed] [Google Scholar]
- 37.La Ragione, R. M., A. R. Sayers, and M. J. Woodward. 2000. The role of fimbriae and flagella in the colonization, invasion and persistence of Escherichia coli O78:K80 in the day-old-chick model. Epidemiol. Infect. 124:351-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.La Ragione, R. M., K. E. Coles, F. Jorgensen, T. J. Humphrey, and M. J. Woodward. 2000. Virulence in the chick model and stress tolerance of Salmonella enterica servovar Orion var. 15+. J. Med. Microbiol. 290: 707-718. [DOI] [PubMed] [Google Scholar]
- 39.Levine, M. M. 1987. Escherichia coli that cause diarrhoea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohaemorrhagic and enteroadherent. J. Infect. Dis. 155:377-389. [DOI] [PubMed] [Google Scholar]
- 40.MacDonald, J. W., and D. D. Brown. 1974. Salmonella infection in wild birds in Britain. Vet. Rec. 94:321-322. [DOI] [PubMed] [Google Scholar]
- 41.Marc, D., and M. Dho-Moulin. 1996. Analysis of the fim cluster of an avian O2 strain of Escherichia coli: serogroup-specific sites within fimA and nucleotide sequence of fimI. J. Med. Microbiol. 44:444-452. [DOI] [PubMed] [Google Scholar]
- 42.McGraw, E. A., J. Li, R. K. Selander, and T. S. Whittam. 1999. Molecular evolution and mosaic structure of alpha, beta and gamma intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12-22. [DOI] [PubMed] [Google Scholar]
- 43.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgenroth, A., and J. P. Duguid. 1968. Demonstration of different mutational sites controlling rhamnose fermentation in FIRN and non-FIRN rha− strains of Salmonella typhimurium. Genet. Res. 11:151-169. [DOI] [PubMed] [Google Scholar]
- 45.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paton, A. W., J. C. Paton, P. N. Goldwater, M. W. Heuzenroeder, and P. A. Manning. 1993. Sequence of variant Shiga-like toxin type-1 operon of Escherichia coli O11:H-. Gene 129:87-92. [DOI] [PubMed] [Google Scholar]
- 47.Peighambari, S. M., R. J. Julian, J. P. Vaillancourt, and C. L. Gyles. 1995. Escherichia coli cellulitis: experimental infections in broiler chickens. Avian Dis. 39:125-134. [PubMed] [Google Scholar]
- 48.Pennycott, T. W., H. M. Ross, I. M. McLaren, A. Park, G. F. Hopkins, and G. Foster. 1998. Causes of death of wild birds of the family Fringillidae in Britain. Vet. Rec. 143:155-156. [DOI] [PubMed] [Google Scholar]
- 49.Perna, N. T., G. F. Mayhew, G. Pósfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reingold, J., N. Starr, J. Maurer, and M. D. Lee. 1999. Identification of a new Escherichia coli She haemolysin homolog in avian E. coli. Vet. Microbiol. 66:125-134. [DOI] [PubMed] [Google Scholar]
- 51.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Weels, B. R. Davis, R. J. Herbert, E. S. Olcott, L. M. Johnson, N. T. Hargett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Eng. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 52.Romling, U., Z. Bian, M. Hammer, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scarf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermo-stable DNA polymerase. Science 238:487-491. [DOI] [PubMed] [Google Scholar]
- 54.Scott, D. A., and J. B. Kaper. 1994. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect. Immun. 62:244-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherman, P., R. Soni,and M. Karmali. 1988. Attaching and effacing adherence of Vero-cytotoxin-producing Escherichia coli O157:H7 to rabbit intestinal epithelium in vivo. Infect. Immun. 56:756-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sojka, W. J. 1965. E. coli in domestic animals and poultry. Commonwealth Agricultural Bureau, Wallingford, Oxfordshire, United Kingdom.
- 57.Tozzi, A. E., A. Niccolini, A. Capriolo, I. Luzzi, G. Montini, G. Zacchello, A. Gianviti, F. Principato, and G. Rizzoni. 1994. A community outbreak of haemolytic-uraemic syndrome in children occurring in a large area of northern Italy over a period of several months. Epidemiol. Infect. 113:209-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulsen, M. H., and J. L. Rollo. 1980. Pathogenesis of Escherichia coli gastroenteritis in man: another mechanism. N. Eng. J. Med. 302:99-101. [DOI] [PubMed] [Google Scholar]
- 59.Venkatesan, M. M., J. M. Buysse, and A. B. Hartman. 1991. Sequence variation in two ipaH genes of Shigella flexneri 5 and homology to the LRG-like family of proteins. Mol. Microbiol. 5:2435-2446. [DOI] [PubMed] [Google Scholar]
- 60.Williams-Smith, H. 1965. The development of the flora of the alimentary tract in young animals. J. Pathol. Bacteriol. 80:495-513. [PubMed] [Google Scholar]
- 61.Williams-Smith, H., and J. E. Tucker. 1975. The effect of antibiotic therapy on faecal excretion of Salmonella typhimurium by experimentally infected chickens. J. Hyg. 75:275-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheleznaia, L. A., O. V. Fedorov, A. F. Brik, and N. I. Matvienko. 1994. Nucleotide sequence of the Escherichia coli B38 flagellin gene. Biokhimiya 59:1621-1637. [PubMed] [Google Scholar]