Abstract
Recent reports show that contrary to common perception, branched alkyl sulfate surfactants are readily biodegradable in standard biodegradability tests. We report here the isolation of bacteria capable of biodegrading 2-butyloctyl sulfate and the identification of novel enzymes that initiate the process. Enrichment culturing from activated sewage sludge yielded several strains capable of growth on 2-butyloctyl sulfate. Of these, two were selected for further study and identified as members of the genus Pseudomonas. Strain AE-A was able to utilize either sodium dodecyl sulfate (SDS) or 2-butyloctyl sulfate as a carbon and energy source for growth, but strain AE-D utilized only the latter. Depending on growth conditions, strain AE-A produced up to three alkylsulfatases, as shown by polyacrylamide gel electrophoresis zymography. Growth on either SDS or 2-butyloctyl sulfate or in nutrient broth produced an apparently constitutive, nonspecific primary alkylsulfatase, AP1, weakly active on SDS and on 2-butyloctyl sulfate. Growth on 2-butyloctyl sulfate produced a second enzyme, AP2, active on 2-butyloctyl sulfate but not on SDS, and growth on SDS produced a third enzyme, AP3, active on SDS but not on 2-butyloctyl sulfate. In contrast, strain AE-D, when grown on 2-butyloctyl sulfate (no growth on SDS), produced a single enzyme, DP1, active on 2-butyloctyl sulfate but not on SDS. DP1 was not produced in broth cultures. DP1 was induced when residual 2-butyloctyl sulfate was present in the growth medium, but the enzyme disappeared when the substrate was exhausted. Gas chromatographic analysis of products of incubating 2-butyloctyl sulfate with DP1 in gels revealed the formation of 2-butyloctanol, showing the enzyme to be a true sulfatase. In contrast, Pseudomonas sp. strain C12B, well known for its ability to degrade linear SDS, was unable to grow on 2-butyloctyl sulfate, and its alkylsulfatases responsible for initiating the degradation of SDS by releasing the parent alcohol exhibited no hydrolytic activity on 2-butyloctyl sulfate. DP1 and the analogous AP2 are thus new alkylsulfatase enzymes with novel specificity toward 2-butyloctyl sulfate.
Synthetic surfactants are components of a variety of household and industrial detergent formulations. Other industrial applications include paints, textiles and fabrics, oil-spill dispersants, concrete, paper, lubricants, and many others (14, 15, 26). In 1990, world production of synthetic surfactants was 7 megatonnes (Mt) per annum, of which 5.3 Mt came from the United States, the European Union, and Japan (12). Of this part, 1.74 Mt per annum was anionic surfactant, a significant quantity which reflects the high demand for this type of surfactant. The anionic surfactant group embraces a range of compounds distinguished on the basis of chemical structure and includes primary alkyl sulfates, such as sodium dodecyl sulfate (SDS), and linear secondary alkyl sulfates, such as dodecyl 2-sulfate. Synthetic primary alkyl sulfates are based on feedstocks derived from long-chain olefins by use of the oxo process, which yields a mixture of linear and branched primary alcohols. Sulfation of the mixed alcohols produces a mixture of linear primary alkyl sulfates (LPAS) and branched primary alkyl sulfates (BPAS), which have excellent detergent properties and are widely used in heavy-duty detergent applications. BPAS such as 2-butyloctyl sulfate [C6H13· CH(C4H9)·CH2· OSO3−] are also produced by sulfation of the products of the Guerbet reaction (16), and these find use as low-foam, effective wetting agents mainly for textile and leather production (Isofol product data; Condea Chemicals, Hamburg, Germany). BPAS from both routes are characterized by alkyl branching at C-2, although the respective lengths of the main chain and the branching alkyl group depend on the route and raw material used.
Anionic surfactants are discharged after use in large quantities to waterways, wastewater treatment works, and marine and estuarine environments (10). The biodegradability of alkyl sulfate surfactants in such environments is well established (26). All commercially important alkyl sulfate structures so far studied are readily biodegradable. Studies on the mechanisms underlying the biodegradability of alkyl sulfate surfactants so far have focused mainly on the LPAS. As a result, the metabolic pathways and enzymologic characteristics in the bacteria involved are well understood. For example, the biodegradation of LPAS in Pseudomonas sp. strain C12B is initiated by two alkylsulfatases, designated P1 and P2 on the basis of electrophoretic mobility. The liberated long-chain primary alcohols are subsequently oxidized by alcohol dehydrogenase; for a review, see reference 9. The P1 and P2 enzymes have been purified and characterized in detail. While they have the same exclusive specificity for LPAS, they are distinguishable on the basis of the mechanism of C— O—S ester bond cleavage (O—S and C—O for P1 and P2, respectively) and the regulation of synthesis (P1 is constitutive and P2 is inducible) (2, 6, 7). Alkylsulfatase activity has also been found in Pseudomonas aeruginosa (11) and Aerobacter aerogenes (24). Comamonas terrigena produces two secondary alkylsulfatases (CS1 and CS2) active on various secondary alkyl sulfates (19).Pseudomonas putida FLA produces six alkylsulfatases, three of which are primary alkylsulfatases (FP1 to FP3) and three of which are secondary alkylsulfatases (FS1 to FS3) (17).
In contrast to that of LPAS, the biochemistry of the biodegradation of BPAS is less well documented. During the period from 1950 to 1970, poor biodegradability of highly branched tetrapropylene benzene sulfonate surfactants caused significant problems of foaming in receiving waters and wastewater treatment plants (26). Elimination of the problem by switching to linear alkylbenzene sulfonates led to the tacit assumption that any branching in the hydrophobic part would result in poorly degraded and therefore environmentally unacceptable products. Recently, however, this assumption has been successfully challenged in studies which showed that alkyl sulfates with either a small number of methyl branches along the hydrophobic chain (25) or 2-alkyl branches (4) were as rapidly and completely biodegraded as their linear counterparts. However, nothing is known about the biochemical basis of the biodegradation of this group of anionic surfactants, in particular, whether they serve as substrates for existing known alkylsulfatases or whether novel enzymes are involved.
This article describes the first isolation of bacteria capable of utilizing 2-butyloctyl sulfate as a sole source of carbon and the discovery in cell extracts of new alkylsulfatases with novel substrate specificities.
MATERIALS AND METHODS
Chemicals.
SDS (AnalaR grade) was obtained from BDH, Poole, England. Nutrient broth, nutrient agar, and Noble agar were supplied by Difco Laboratories, Detroit, Mich. All other chemicals were the purest available from Fisher Chemicals, Loughborough, United Kingdom, or Sigma Chemical Co., London, United Kingdom, unless stated otherwise.
Pure 2-butyloctanol and an aqueous saturated solution-crystal suspension of 2-butyloctyl sulfate were kindly provided by Condea Chemicals. Based on the smanufacturer’s analysis, the latter contained 2-butyloctyl sulfate (28.8% [wt/vol]), sodium chloride (0.6% [wt/vol]), sodium sulfate (0.8% [wt/vol]), and unsulfated organic matter (0.8% [wt/vol]).
Purification and analysis of 2-butyloctyl sulfate.
The commercial preparation of 2-butyloctyl sulfate was thoroughly mixed and diluted with an equal volume of distilled water. The suspension was then heated to 45°C with stirring until a clear solution was obtained. Subsequent slow cooling to 5°C yielded crystals that were filtered on a Buchner filter (no. 1 paper; Whatman International, Maidstone, United Kingdom) under reduced pressure (water pump) at 5°C. The “sludge” remaining after filtration was washed with acetone and petroleum ether and dried in a rotary evaporator to yield alkyl sulfate crystals.
Infrared spectra of 2-butyloctyl sulfate (as a Nujol mull) and 2-butyloctanol (pure liquid) were determined by using a Perkin-Elmer 257 grating infrared spectrometer. The peak at 1,060 to 1,110 cm−1 in the parent alcohol, corresponding to C·OH vibrations, was absent in the alkyl sulfate and was replaced with O·S and C·O·S vibrations at 1,200 to 1,250 cm −1, 1,065 cm−1, and 810 cm−1. The water-recrystallized alkyl sulfate, after drying, showed a broad peak centered at 3,560 cm−1, indicating water of crystallization. Carbon and hydrogen analysis (performed at Warwick Analytical Service, Department of Chemistry, Warwick University) indicated a carbon content of 46.82% and a hydrogen content of 8.49% (compared to theoretical contents of 47.06 and 8.82%, respectively, based on the structure C12H25OSO3−Na+· H2O). Gravimetric analysis of duplicate samples for sulfate as BaSO4 indicated the sulfate contents of the ester to be 35.7 and 34.0% (theoretical content, 31.4%). 13C and1H nuclear magnetic resonance (NMR) spectra were consistent with literature values for particular resonance signals (20) and the spectra predicted for the 2-butyloctyl sulfate structure with ChemDraw C NMR software (CambridgeSoft Corporation, Cambridge, Mass.). For the 13C spectra, the peak at 71.7 ppm, the peak at 37.3 ppm, two peaks at 13.67 to 13.72 ppm, and eight peaks at 30.48 to 22.67 ppm were assigned to the ester CH2·O·S, the branch point C-H, two CH3 units, and eight CH2 units, respectively. For the1H spectra, the peaks at 3.7, 1.5, 1.1, and 0.7 ppm were assigned to the ester-linked CH2·O·S hydrogen atoms, the branch point C·H hydrogen atom, 16 hydrogen atoms from the eight CH2 methylene groups in the carbon chain, and 6 hydrogen atoms from the two terminal CH3 groups.
The critical micelle concentration for 2-butyloctyl sulfate in 50 mM Tris-HCl (pH 7.5) was determined by using a modification of the method of Carey and Small (5) which exploits spectral changes of rhodamine 6G upon its incorporation into micelles. The critical micelle concentration was determined to be 100 μg/ml (0.32 mM).
Isolation of bacteria.
Bacteria capable of utilizing 2-butyloctyl sulfate as a sole source of carbon were isolated from activated sewage sludge (from Coslech Sewage Works, Cardiff, Wales) by enrichment in basal salts (BS) medium containing (grams per liter) the following: K2HPO4, 3.5; KH2PO4, 1.5; NH4Cl, 0.5; MgCl2 · 6H2O, 0.15; NaCl, 0.5; and Na2SO4, 0.14. The medium also contained the following trace elements (1 ml of stock) (grams per liter): FeCl3 · 6H2O, 0.24; CoCl2 · 6H2O, 0.04; CuSO4 · 5H2O, 0.06; MnCl2 · 4H2O, 0.03; ZnSO4 · 7H2O, 0.31; and Na2MoO4 · 2H2O, 0.03. The medium also was supplemented with 1 g of 2-butyloctyl sulfate/liter to give a final carbon concentration of 40 mM (3.3 mM substrate). The cultures were incubated at 30°C with shaking at 150 rpm. After 6 days, 1 ml of culture was transferred to 100 ml of fresh medium. After five serial subcultures, a 1-ml sample was taken from the final enrichment flask, serially diluted, and plated on nutrient agar plates. The plates were incubated for several days at 30°C until colonies were visible. Single colonies were plated to purity and characterized by their morphology and Gram classification (23). Pure strains were tested for the ability to grow axenically on 2-butyloctyl sulfate.
16S rRNA analysis.
Cells from overnight nutrient broth cultures were harvested from 1 ml of culture by centrifugation at 17,900 × g for 3 min. The cells were washed in 1 ml of sterile distilled water, resuspended in 100 μl of sterile distilled water, and boiled for 5 min. The supernatant (0.2 μl) containing template DNA was mixed with 2.5 μl of 10× PCR buffer, (0.5 M KCl, 0.1 M Tris HCl, pH9, at 25°C, 1% [vol/vol] Triton X-100), 1.5μl of MgCl2 (25 mM), 18.25 μl of ultrapure water (MilliQ), and 0.25 μl of Taq polymerase. PCR primers 63f (5′-CAG GCC TAA CAC ATG CAA GTC-3′) and 1387r (5′-GGG CGG WGT GTA CAA GGC), designed and evaluated by Marchesi et al. (18), were used for the amplification of 16S rRNA genes. The PCR program, performed by using an MWG-Biotech Primus thermal cycler, consisted of 5 min at 96°C for 5 min; 30 cycles of 96°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and an additional extension step of 5 min at 72°C. The PCR products were purified by using a QIAquick PCR purification kit (Qiagen) in accordance with the manufacturer’s instructions and were separated on agarose gels. The DNA sequence was determined by using an ABI PRISM dRhodamine terminator cycle sequencing ready reaction kit (Perkin-Elmer Applied Biosystems, Warrington, Cheshire, United Kingdom) in accordance with the manufacturer’s instructions. Sequences were compared to known 16S rRNA gene sequences in the composite nonredundant database (includes GenBank, EMBL, DDBJ, and PDB) by using the BLAST search program (1).
Maintenance and growth of bacterial strains.
Bacterial strains were maintained on nutrient agar slopes at room temperature for routine use or stored at −20°C in PROTECT vials (beads in a cryopreservative fluid; Technical Service Consultants Ltd., Heywood, Lancs, United Kingdom) for long-term storage. To initiate batch cultures, single colonies from nutrient agar plates were inoculated into nutrient broth, and culture flasks were incubated aerobically at 30°C with shaking (150 rpm). After 18 h, cells were harvested from 1 ml of culture, washed by centrifugation, and resuspended in 1 ml of BS medium. The cell suspensions were used to inoculate (1% [vol/vol]) flasks containing BS medium and an appropriate quantity of alkyl sulfate, and the flasks were incubated as before. Growth was measured as the optical density at 540 nm by use of a Thermomax Maxline microplate reader (Molecular Devices, Wokingham, United Kingdom). In anticipation that biodegradation might produce the accumulation of insoluble intermediates (e.g., 2-butyloctanol), samples removed from flasks for growth measurements were each mixed with an equal volume of methanol to solubilize such compounds before optical densities were measured. Controls with cells grown in nutrient broth (i.e., no insoluble alcohol production) indicated that this dilution had no effect on cells other than the expected twofold reduction in cell density, and this factor has been taken into account in the data presented.
Preparation of enzyme extracts.
Bacterial cells were harvested by centrifugation of the cultures at 10,000 ×g for 20 min at 4°C in a Sorvall RC-5B Superspeed centrifuge (DuPont Company, Wilmington, Del.). Cell pellets were resuspended in 10 mM Tris-HCl (pH 7.5) (1/50 the original volume). The cells were ruptured by sonication for a total duration of 3 min, consisting of intermittent sonication for 30 s on and 30 s off, by using a Lucas Dawe sonicator with a Micro-tip probe (Branson Technology) operated at level 5 and a 25% duty cycle. To minimize heat inactivation of enzymes, samples were sonicated in thin-glass vials kept on ice. Cell debris was removed by centrifugation at 12,000 × g for 40 min at 4°C in a Sorvall RC-5B Superspeed centrifuge. The cell extracts were stored at −20°C prior to being assayed for alkylsulfatase activity.
Polyacrylamide gel zymography.
To determine the multiplicity of alkylsulfatase activities in cell extracts, polyacrylamide gel electrophoresis (PAGE) was carried out by using 7% polyacrylamide rod gels prepared in 0.378 M Tris-glycine buffer (pH 8.3) and cast in glass tubes (80 mm by 20 mm [internal diameter]) (22). Per rod gel, 150 μl of sample was applied, and electrophoresis was conducted with a current of 2 mA. Stock running buffer containing 0.25 M Tris and 14.4 M glycine was diluted 1:50 (vol/vol) prior to use. Extruded gels were incubated in developing solution containing 20 mM appropriate alkyl sulfate (either 2-butyloctyl sulfate or SDS) in 0.1 M Tris-HCl (pH 7.5). Locations of active alkylsulfatases were revealed by the formation of white bands of insoluble alcohol droplets.
MBAS assay.
The concentrations of anionic surfactant (2-butyloctyl sulfate and SDS) were determined by the methylene blue active substance (MBAS) assay as described by Hayashi (13) with minor modifications. The method is based on the formation of a complex between the anionic surfactant and an excess of the cationic dye methylene blue, followed by extraction of the complex (but not excess dye) into chloroform and measurement of the absorbance of the blue chloroform layer. A stock solution of methylene blue (0.0625 g in 100 ml of distilled water) was underlaid with 10 ml of chloroform to remove any chloroform-extractable impurities and was stored in a brown bottle to minimize photochemical degradation. Aliquots of methylene blue solution (0.1 ml), 0.4 ml of 0.825 mM phosphate buffer (pH 7.2), and an appropriate volume of sample (up to 1 ml, in triplicate) were mixed in acid-washed, optically matched glass tubes (12 cm by 1 cm [internal diameter]) and vortexed intermittently five times for 3 s each time. Chloroform was added to each tube, and the contents were vigorously vortexed for 5 s. The tubes were allowed to stand at 4°C for 5 min, followed by centrifugation in an MSE Centaur 2 centrifuge at 2,000 rpm for 4 min. The tubes were allowed to warm to room temperature, and the absorbance of the chloroform layer was measured at 655 nm, against an appropriate blank, by using an LKB Novaspec spectrophotometer. Calibration curves were prepared by using standard solutions (based on weight) of pure SDS or pure 2-butyloctyl sulfate.
Gas chromatographic detection of 2-butyloctanol.
Native proteins in cell extracts of the selected isolate,Pseudomonas strain AE-D, were separated by PAGE on duplicate rod gels. The gels were incubated in a 20 mM solution of 2-butyloctyl sulfate (in 50 mM Tris-HCl [pH 7.5]) which previously had been extracted repeatedly with redistilled petroleum ether (60 to 80°C bp) in order to remove any traces of 2-butyloctanol. From each rod, the section of gel containing the single opaque white band indicative of DP1 alkylsulfatase activity was excised and washed three times with distilled water. The duplicate pieces were crushed together in 1 ml of ethanol by using a glass rod, and the mash was vortexed repeatedly for 3 min. Petroleum ether (2 ml) was added, and the mixture was intermittently vortexed for 4 min and then centrifuged (4,000× g, 5 min) to separate the layers. The top layer (containing petroleum ether and extracted material) was removed and treated with anhydrous sodium sulfate to remove residual water. For the control, the above procedure was repeated, except that no protein was included in the tracker dye sample loaded onto the gel prior to electrophoresis, incubation with substrate, and extraction of the corresponding section of the gel. Samples were analyzed for 2-butyloctanol by using a Perkin-Elmer 8410 gas chromatography system fitted with a flame ionization detector and a glass column (2 m by 2 mm) packed with 10% SP-2330. Operating conditions were as follows: oven temperature, 140°C; injection temperature, 250°C; detector temperature, 250°C; and N2 carrier gas flow rate, 20 ml/min.
Alkylsulfatase assay.
Cell extracts (up to 0.5 ml, in duplicate) were incubated (30°C) with 0.5 ml of 2-butyloctyl sulfate at a final concentration of 50 μg/ml (0.16 mM) in 50 mM Tris-HCl (pH 7.5) and with an appropriate volume of 10 mM Tris-HCl (pH 7.5) buffer to bring the initial assay volume to 1 ml. Samples (100 μl) were removed at intervals and assayed for residual alkyl sulfate by using the MBAS method. Total enzyme activity was determined from initial rates of surfactant disappearance. One unit of enzyme activity was defined as the amount of enzyme which converted 1 μmol of substrate per minute under the assay conditions.
RESULTS
Isolation of bacteria.
Three strains (AE-A, AE-B, and AE-D), all gram-negative bacteria, were isolated for their ability to utilize 2-butyloctyl sulfate as the sole source of carbon. Isolates AE-B and AE-D were capable of growth on 2-butyloctyl sulfate, but neither could metabolize SDS. In contrast, isolate AE-A utilized either SDS or 2-butyloctyl sulfate, growing faster on the former compound. Isolates AE-A and AE-D were selected for further study.
Analysis of 16S rRNA gene sequences in isolate AE-A showed 99% sequence similarity to five sequences in the database, three of which were unidentified or uncultured strains and two of which were Pseudomonas spp. Of the 100 BLAST sequence matches, all of which showed between 96 and 99% sequence similarity, 94 were of the genus Pseudomonas. Analysis of the 16S rRNA gene in isolate AE-D showed 95% sequence similarity to five different sequences, each of which was of the genus Pseudomonas and two of which were Pseudomonas putida. As for isolate AE-A, a high proportion (94 of the 100) of the BLAST sequence matches were Pseudomonas spp. Isolates AE-A and AE-D were hereafter designated Pseudomonas sp. strain AE-A and Pseudomonas sp. strain AE-D, respectively.
Growth on surfactants.
Figures 1, 2, and 3 depict bacterial growth and surfactant disappearance in batch cultures for Pseudomonas sp. strain AE-A utilizing SDS, Pseudomonas sp. strain AE-A utilizing 2-butyloctyl sulfate, and Pseudomonas sp. strain AE-D utilizing 2-butyloctyl sulfate, respectively. For each, bacterial growth coincided with surfactant disappearance. Pseudomonas sp. strain AE-D produced a flocculated culture after 20 h, making measurements of growth unreliable for older cultures.Pseudomonas sp. strain C12B, originally isolated for its ability to grow on SDS (21), did not grow in batch cultures with 2-butyloctyl sulfate as the sole carbon source under the conditions used.
FIG. 1.
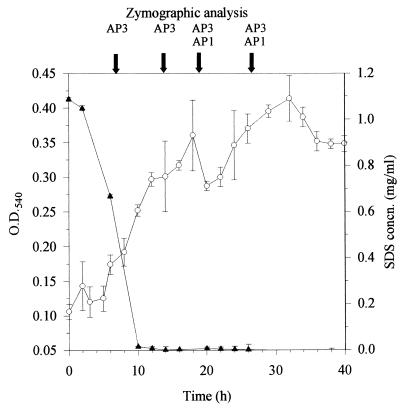
Biodegradation of SDS by Pseudomonas sp. strain AE-A. Growth (mean and standard deviation, n = 4 [○]) and residual surfactant concentration (mean and standard deviation, n = 3 [▴]) were measured with BS medium supplemented with 1 g of surfactant/liter in batch cultures. Samples were removed at the times indicated by the arrows and analyzed by gel zymography for specific alkylsulfatases. Designations above arrows, e.g., AP3, indicate alkylsulfatase bands present in the sample. O.D.540, optical density at 540 nm; concn., concentration.
FIG. 2.
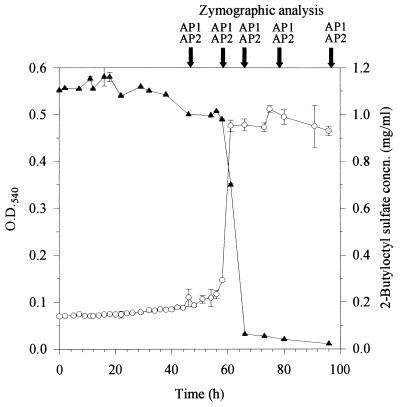
Biodegradation of 2-butyloctyl sulfate by Pseudomonas sp. strain AE-A. See the legend to Fig. 1 for details.
FIG. 3.
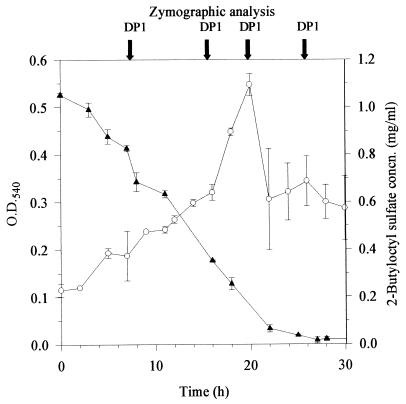
Biodegradation of 2-butyloctyl sulfate by Pseudomonas sp. strain AE-D. See the legend to Fig. 1 for details.
Alkylsulfatase multiplicity and specificity.
Previous work showed that, in Pseudomonas sp. strain C12B, P1 is produced from late exponential phase onward but P2 is produced transiently during exponential phase. To ensure that alkylsulfatases that might be produced transiently in the new strains did not pass undetected, samples were taken throughout batch culturing of strains AE-A and AE-D and extracts were examined by gel zymography. Both strains were grown in nutrient broth and on 2-butyloctyl sulfate, and strain AE-A was grown on SDS as well.
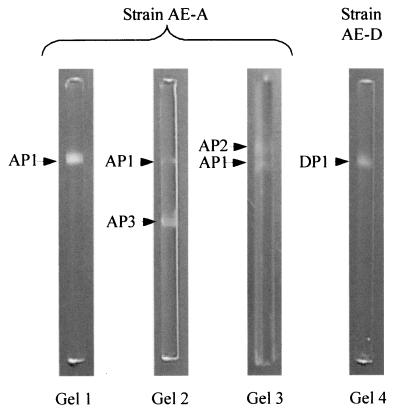
Depending on growth substrate and culture age, Pseudomonas sp. strain AE-A produced up to three primary alkylsulfatases (AP1, AP2, and AP3; Fig. 4 and Table 1). The AP1 band was revealed by incubating gels with either SDS or 2-butyloctyl sulfate. AP1 was observed throughout batch culture growth in nutrient broth as a well-defined band with an electrophoretic mobility of 0.27 relative to that of bromophenol blue. When strain AE-A was grown at the expense of SDS, the enzyme was detectable only in cells in stationary phase (Fig. 1). In contrast, when strain AE-A was grown on 2-butyloctyl sulfate, it produced the enzyme throughout batch culture growth (Fig. 2).
FIG. 4.
Examples of PAGE zymograms of primary alkylsulfatases in Pseudomonas sp. strains AE-A and AE-D. Migration of proteins is toward the bottom (anode). Gel 1, strain AE-A grown in nutrient broth and gel stained with 2-butyloctyl sulfate; gel 2, strain AE-A grown on SDS and gel stained with SDS; gels 3 and 4, strains AE-A and AE-D, respectively, both grown on and gels stained with 2-butyloctyl sulfate. Full details of the conditions under which each band was produced are shown in Table 1. “Bands” visible at the top and bottom of each gel are artifacts caused by refraction of light from the ends of the rod gels.
TABLE 1.
Alkylsulfatase specificities based on gel zymograms of cell extracts of alkyl sulfate-degrading bacteria grown with different substrates
| Pseudomonas strain | Enzyme band designationa | Occurrence of bands on gel zymograms with the following growth substrate and gel substrateb:
|
|||||
|---|---|---|---|---|---|---|---|
| 2BOS
|
SDS
|
Nutrient broth
|
|||||
| SDS | 2BOS | SDS | 2BOS | SDS | 2BOS | ||
| AE-A | AP1 (0.27) | + | + | + | + | + | + |
| AP2 (0.24) | − | + | − | − | − | − | |
| AP3 (0.55) | − | − | + | − | − | − | |
| AE-D | DP1 (0.27) | − | + | NG | NG | − | − |
| C12B | P1 (0.25) | NG | NG | + | − | + | − |
| P2 (0.38) | NG | NG | + | − | − | − | |
Numbers in parentheses are electrophoretic mobilities relative to that of the bromophenol blue tracker dye.
+, band was present on gel; −, band was absent from gel; NG, no growth. 2BOS, 2-butyloctyl sulfate. Growth substrates are listed above gel substrates.
When grown on 2-butyloctyl sulfate (but not on SDS or in nutrient broth), isolate AE-A produced a second, 2-butyloctyl sulfate-hydrolyzing enzyme (AP2) throughout batch culturing (Fig. 2). The enzyme was observed as a band with a relative mobility of 0.24. The AP2 band was detectable by incubating gels with 2-butyloctyl sulfate but not with SDS and thus appeared to be inducible by and specific for 2-butyloctyl sulfate.
A third, SDS-inducible alkylsulfatase (AP3) was observed in zymograms of extracts of Pseudomonas sp. strain AE-A throughout batch culture growth on SDS as the sole carbon source (Fig. 1). The enzyme was observed as a band with a relative mobility of approximately 0.55. It was not detected when isolate AE-A was grown at the expense of either nutrient broth or 2-butyloctyl sulfate. The AP3 band was observed when gels were incubated with SDS but not with 2-butyloctyl sulfate and thus appeared to be inducible by and specific for SDS.
Pseudomonas sp. strain AE-D grown on 2-butyloctyl sulfate produced a single alkylsulfatase which was present on gels as a band with a relative mobility of approximately 0.27 (DP1; Fig. 4) in samples taken throughout batch culture growth (Fig. 3). DP1 was not detected when isolate AE-D was grown in nutrient broth. The DP1 band appeared when the strain was incubated with 2-butyloctyl sulfate but not with SDS. This enzyme thus appeared analogous to AP2 in being inducible by and specific for 2-butyloctyl sulfate.
Extracts of Pseudomonas sp. strain C12B cells grown to late exponential phase on SDS-supplemented BS medium in batch cultures produced the anticipated (8) P1 and P2 bands when gels were incubated with SDS. When the same cell extracts were separated on gels and the gels were incubated with 2-butyloctyl sulfate instead of SDS, no alkylsulfatase bands were detectable.
Alkylsulfatase activity in cell extracts.
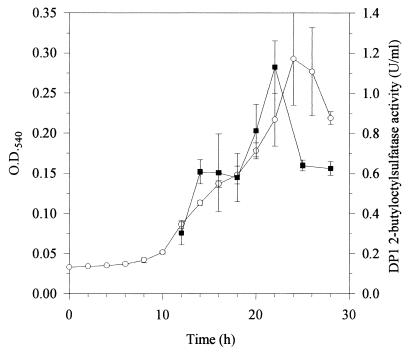
Demonstration of 2-butyloctyl sulfate sulfatase activity in cell extracts was achieved by incubating extracts of Pseudomonas sp. strain AE-D (containing the single DP1 enzyme) with 2-butyloctyl sulfate and measuring the rate of loss of the substrate. The disappearance of 2-butyloctyl sulfate was linear with time under the assay conditions until at least 50% of 2-butyloctyl sulfate had been degraded. Rates measured from these linear portions were used to assay relative enzyme concentrations. Preliminary experiments indicated that enzyme activity was proportional to the amount of extract used in the assay. Figure 5 shows the activity of DP1 in extracts at various stages of batch culture growth. Total 2-butyloctyl sulfate sulfatase activity (units per milliliter) increased with increasing culture density up to the onset of stationary phase, at which time the disappearance of 2-butyloctyl sulfate from the growth medium was almost complete (Fig. 3); thereafter, enzyme activity declined.
FIG. 5.
Production of DP1 alkylsulfatase (mean and standard deviation, n = 2 [▪]) during growth (mean and standard deviation, n = 4 [○]) of Pseudomonas sp. strain AE-D on 2-butyloctyl sulfate (initially 1 g/liter) in batch cultures. Enzyme activity was measured in duplicate as described in the text and is expressed as units per milliliter of extracts of cells which were harvested and resuspended at 50 times the culture cell density prior to disruption. O.D.540, optical density at 540 nm.
Gas chromatographic analysis of petroleum ether extracts of zymogram bands showed that material extracted from the zone of the DP1 band eluted with a retention time (7.73 min) identical to that of 2-butyloctanol. Based on peak areas and an appropriate calibration graph, the concentration of 2-butyloctanol in the extract for this band (40 ng/ml) was 60-fold higher than that in the control gel (0.07 ng/ml).
DISCUSSION
Several degraders of 2-butyloctyl sulfate were isolated at the first attempt from a single sample of activated sewage sludge from a wastewater treatment plant receiving mainly domestic sewage, indicating that such microorganisms may be widely distributed in wastewater treatment plants and discharge waters. These results are consistent with recent reports showing ready biodegradability of BPAS (4) in tests based on sewage treatment plant inocula and with the identification of the most effective degraders (isolates AE-A and AE-D) as members of the genus Pseudomonas, which is ubiquitous and well known for versatility in the biodegradation of industrial compounds. In strain AE-A, the time from inoculation to completion of 2-butyloctyl sulfate degradation (66 h; Fig. 2) was considerably longer than that for SDS (10 h; Fig. 1). However, the degradation of 2-butyloctyl sulfate involved a long lag period, and once initiated, degradation was completed within 10 h. In strain AE-D, there was no lag, and the completion of degradation of 2-butyloctyl sulfate required about 30 h. Thus, the rates of degradation of SDS and 2-butyloctyl sulfate in adapted cultures were comparable, consistent with the results of standard Organization for Economic Cooperation and Development tests (4) used to assess comparative degradation rates in wastewater treatment plants.
Degraders of LPAS are also known from previous studies to be widely distributed both geographically and across bacterial genera, with a strong representation of Pseudomonas spp. (26). However, the following observations now show that degraders of LPAS are not necessarily degraders of BPAS. First, Pseudomonas sp. strain C12B, well known and extensively studied for its ability to biodegrade SDS and related alkyl sulfates, failed to grow at the expense of 2-butyloctyl sulfate, and its alkylsulfatases, P1 and P2, active on a range of linear primary alkyl sulfates, were inactive on 2-butyloctyl sulfate (Table 1 shows a summary of specificities). One of the new strains, Pseudomonas sp. strain AE-A, was also able to grow on SDS and produced an SDS-inducible and SDS-specific alkylsulfatase (AP3) analogous to P2. However, this enzyme also was neither active on nor induced by growth on 2-butyloctyl sulfate. Moreover, the growth of Pseudomonas sp. strain AE-A on 2-butyloctyl sulfate induced a novel alkylsulfatase (AP2) specific for that substrate and inactive on SDS. An analogous enzyme (DP1) which was specifically induced by and active on 2-butyloctyl sulfate was produced by Pseudomonas sp. strain AE-D. Gas chromatographic analysis of material in opaque white bands on zymograms for this enzyme showed the presence of 2-butyloctanol, thus indicating that the catalytic role of the enzyme is probably as a sulfatase.
Thus, collectively these microorganisms produce two sets of alkylsulfatases with mutually exclusive specificities for either SDS or 2-butyloctyl sulfate. Against the background of exclusive specificities for LPAS (P1, P2, and AP3) and 2-butyloctyl sulfate (AP2 and DP1), alkylsulfatase AP1 was unique in showing activity toward both substrates (Table 1).
In addition, in contrast to AP2, AP3, and DP1, which were all inducible specifically by their respective substrates, AP1 was also the only enzyme from the new strains which was produced constitutively. There were also indications from its late appearance during batch culturing on SDS that its expression may be repressible. However, despite its dual specificity and constitutive expression, it appears that AP1 provides insufficient activity for Pseudomonas sp. strain AE-A to utilize either SDS or 2-butyloctyl sulfate, because cells challenged with either substrate induced the respective specific enzymes AP3 and AP2.
The combined occurrence of constitutive and inducible enzymes with overlapping specificities (AP1 and AP2 during growth on 2-butyloctyl sulfate and AP1 and AP3 on SDS) has a parallel in Pseudomonas sp. strain C12B, in which the constitutive and repressible P1 (2) and inducible P2 (7) are both present during growth on SDS. P1 and P2 have very similar chain-length substrate specificities but are clearly distinguishable by the mechanism of C—O—S ester bond cleavage, i.e., O—S in P1 and C—O in P2 (3, 6). It will be interesting to determine in closer detail the substrate specificities and mechanisms of ester bond cleavage for the new enzymes AP1, AP2, AP3, and DP1; to this end, attempts are currently under way to purify and characterize DP1.
Acknowledgments
We thank C. Nöthe, Condea Chemicals, for supplying Guerbet alcohols and sulfate esters; M. J. E. Hewlins, School of Chemistry, Cardiff University, for conducting the NMR analysis; and G. Lewis, School of Biosciences, Cardiff University, for DNA sequence analysis.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 2.Bateman, T. J. 1985. Ph.D. thesis. University of Wales, Cardiff, Wales.
- 3.Bateman, T. J., K. S. Dodgson, and G. F. White. 1986. Primary alkylsulphatase activities of the detergent-degrading bacterium Pseudomonas C12B. Purification and properties of the P1 enzyme. Biochem. J. 236:401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battersby, N. S., L. Kravetz, and J. P. Salanitro. 2000. Effect of branching on the biodegradability of alcohol-based surfactants, p.1397–1407. In 5th World Surfactant Congress. Federchimica, Milan, Italy.
- 5.Carey, M. C., and D. M. Small. 1969. Micellular properties of dihydroxy and trihydroxy bile salts: effects of counterion and temperature. J. Colloid Interface Sci. 31:383–396. [DOI] [PubMed] [Google Scholar]
- 6.Cloves, J. M., K. S. Dodgson, D. E. Games, D. J. Shaw, and G. F. White. 1977. The mechanism of action of primary alkylsulphohydrolase and arylsulphohydrolase from a detergent degrading micro-organism. Biochem. J. 167:843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloves, J. M., K. S. Dodgson, G. F. White, and J. W. Fitzgerald. 1980. Specificity of P2 primary alkylsulphohydrolase induction in the detergent-degrading bacterium Pseudomonas C12B. Biochem. J. 185:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodgson, K. S., J. W. Fitzgerald, and W. J. Payne. 1974. Chemically defined inducers of alkylsulphatases present in Pseudomonas C12B. Biochem. J. 138:53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodgson, K. S., and G. F. White. 1983. Some microbial enzymes involved in the biodegradation of sulphated surfactants. Top. Enzyme Ferment. Biotechnol. 7:90–155. [Google Scholar]
- 10.George, A. L., and G. F. White. 1999. Optimization of the methylene blue assay for anionic surfactants added to estuarine and marine water. Environ. Toxicol. Chem. 18:2232–2236. [DOI] [PubMed] [Google Scholar]
- 11.Harada, T. 1964. The formation of sulfatases by Pseudomonas aeruginosa. Biochim. Biophys. Acta 81:193–196. [Google Scholar]
- 12.Hauthal, H. G. 1992. Trends in surfactants. Chim. Oggi 10:9–13. [Google Scholar]
- 13.Hayashi, K. 1975. A rapid determination of sodium dodecyl sulfate with methylene blue. Anal. Biochem. 67:503–506. [DOI] [PubMed] [Google Scholar]
- 14.Karsa, D. A. 1987. The industrial applications of surfactants. Special publication 59. Royal Society of Chemistry, London, England.
- 15.Karsa, D. R. (ed.). 1992. Industrial applications of surfactants, vol. III. Special publication 107. Royal Society of Chemistry, Cambridge, England.
- 16.Krauch, M., and W. Kunz. 1964. Organic name reactions. John Wiley & Sons, Inc., New York, N.Y.
- 17.Lillis, V., K. S. Dodgson, and G. F. White. 1983. Initiation of activation of a pre-emergent herbicide by a novel alkylsulfatase of Pseudomonas putida FLA. Appl. Environ. Microbiol. 46:988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:765–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matcham, G. J. W., B. Bartholomew, K. S. Dodgson, W. J. Payne, and J. W. Fitzgerald. 1977. Stereospecificity and complexity of microbial sulphohydrolases involved in the biodegradation of secondary alkylsulphate detergents. FEMS Microbiol. Lett. 1:197–200. [Google Scholar]
- 20.McMurray, J. 1984. Organic chemistry. Brooks/Cole Publishing Company, Monterey, Calif.
- 21.Payne, W. J., and V. E. Feisal. 1963. Bacterial utilization of dodecyl sulfate and dodecylbenzene sulfonate. Appl. Microbiol. 11:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Payne, W. J., J. W. Fitzgerald, and K. S. Dodgson. 1974. Methods for visualization of enzymes in polyacrylamide gels. Appl. Microbiol. 27:154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers, E. M. 1995. Efficacy of the non-staining KOH technique for rapidly determining Gram reactions of food-borne and waterborne bacteria and yeasts. Appl. Environ. Microbiol. 61:3756–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rammler, D. H. 1964. Sulfur metabolism of Aerobacter aerogenes. I. A repressible sulfatase. Biochemistry 3:199–206. [DOI] [PubMed] [Google Scholar]
- 25.Shi, J., P. J. Kloepper-Sams, S. T. Giolando, T. W. Federle, D. J. Versteeg, and S. E. Belanger. 2000. Biodegradable high solubility alkyl sulfate surfactants: environmental safety profiles, p.1525–1531. In 5th World Surfactant Congress. Federchimica, Milan, Italy.
- 26.Swisher, R. D. 1987. Surfactant biodegradation, 2nd ed., vol. 18. Marcel Dekker, Inc., New York, N.Y.