Abstract
A Lactobacillus group-specific PCR primer, S-G-Lab-0677-a-A-17, was developed to selectively amplify 16S ribosomal DNA (rDNA) from lactobacilli and related lactic acid bacteria, including members of the genera Leuconostoc, Pediococcus, and Weissella. Amplicons generated by PCR from a variety of gastrointestinal (GI) tract samples, including those originating from feces and cecum, resulted predominantly in Lactobacillus-like sequences, of which ca. 28% were most similar to the 16S rDNA of Lactobacillus ruminis. Moreover, four sequences of Leuconostoc species were retrieved that, so far, have only been detected in environments other than the GI tract, such as fermented food products. The validity of the primer was further demonstrated by using Lactobacillus-specific PCR and denaturing gradient gel electrophoresis (DGGE) of the 16S rDNA amplicons of fecal and cecal origin from different age groups. The stability of the GI-tract bacterial community in different age groups over various time periods was studied. The Lactobacillus community in three adults over a 2-year period showed variation in composition and stability depending on the individual, while successional change of the Lactobacillus community was observed during the first 5 months of an infant’s life. Furthermore, the specific PCR and DGGE approach was tested to study the retention in fecal samples of a Lactobacillus strain administered during a clinical trial. In conclusion, the combination of specific PCR and DGGE analysis of 16S rDNA amplicons allows the diversity of important groups of bacteria that are present in low numbers in specific ecosystems to be characterized, such as the lactobacilli in the human GI tract.
The human gastrointestinal (GI) tract consists of different habitats, in which the entire colon is occupied by mostly obligately anaerobic bacteria (29). The activity of these commensal bacteria in the GI tract has a major impact on the characteristics of the host. The microbiota-associated roles include protection against pathogens, development of the immune system, and positive effects on colonic health and host nutrition (6, 14). Although the diversity of the gut microbiota has been investigated extensively by anaerobic culture techniques (7, 27), it is receiving renewed interest due to the development and application of molecular techniques, especially those based on the 16S and 23S rRNA genes (45, 47, 48).
Fluorescent in situ hybridization and group-specific hybridization approaches targeting rRNA in combination with advanced microscopy have indicated that the majority of the GI-tract microbial community is not accounted for by cultivation (19, 40, 42, 48). Phylogenetic analysis of rRNA genes, amplified by PCR, has been used as a rapid and efficient strategy to investigate the biodiversity of intestinal bacteria and revealed many novel species (42, 49). Furthermore, fingerprinting methods, such as denaturing or temperature gradient gel electrophoresis (DGGE or TGGE, respectively) of rRNA or ribosomal DNA (rDNA) amplicons, that allow the rapid evaluation of composition and activity of bacteria over time in complex ecosystems (29) have been applied to the human intestine (47, 49, 50). It was observed that each individual harbors a unique dominant bacterial community that is relatively stable in time (49). However, all of these molecular approaches, including a recent study on the bifidobacterial community in human feces (37), focus on the numerically dominant bacteria and do not address the diversity of other important species that are present in low numbers in the GI tract. These include lactic acid bacteria, such as Lactobacillus spp., that have been shown to constitute less than 1% of the total bacterial community (40). The genus Lactobacillus contains a diverse assemblage of gram-positive, catalase-negative, nonsporulating, rod-shaped organisms and includes more than 25 species (4, 10). They inhabit a wide variety of habitats, including the GI tracts of animals and phytosphere, and are traditionally used in the manufacture of fermented foods and, more recently, in functional foods (24, 45). Based on plate counts it was found that Lactobacillus strains are present in the GI tract of 70% of humans that consume a Western-like diet but are not detectable in the remainder of subjects (7, 43). The number of Lactobacillus cells in neonates was found to be in the range of 105 CFU/g of feces, while in infants of 1 month and older the counts ranged from 106 to 108 CFU/g of feces (20).
The primary aim of the present study was to develop a PCR primer targeting the 16S rRNA gene that is highly specific for the genus Lactobacillus. There have been numerous reports on the development of oligonucleotide probes or PCR primers that target specific Lactobacillus species (3, 5, 12, 13, 30, 31, 32) or groups of Lactobacillus species (41). Since, the Lactobacillus genus lacks a clear monophyletic origin and is highly related to other lactic acid bacteria, the design of a Lactobacillus genus-specific probe or primer is challenging. The developed primer was used to analyze the diversity of members of this group in GI-tract samples in space and time. Sequence analysis of the PCR amplicons generated with this primer showed selective amplification of Lactobacillus-like 16S rDNA sequences, some of which have not yet been shown to be present in the GI tract. Furthermore, we demonstrate that application of the primer in combination with DGGE of the 16S rDNA amplicons allows the diversity and development of these bacterial groups to be monitored, as well as their response to the additional dietary intake of lactobacilli within the human GI tract.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Reference microorganisms, their sources, and the growth media for each strain used in this study are indicated in Table 1. Escherichia coli transformants were grown overnight with shaking at 250 rpm in Luria-Bertani broth (35). Lactobacillus strains were grown in tubes containing 10 ml of Lactobacillus MRS broth (Difco Laboratories, Detroit, Mich.) and 0.3 g of cysteine hydrochloride liter−1. The remaining strains were grown in either Luria-Bertani or Wilkins West broth that contains (per liter) 33 g of Wilkins-Chalgren anaerobe broth (Oxoid, Basingstoke, United Kingdom), 2 g of glucose, 4 g of arginine HCl, 5 ml of 5% Tween 80, and 0.3 g of cysteine hydrochloride, and the pH was adjusted to 7.0 with HCl. All bacteria were incubated at 37°C. The strict anaerobes, i.e., the Clostridium, Bacteroides, and Peptostreptococcus species, were handled and cultured in an anaerobic glove box with a constant atmosphere of 96% N2 and 4% H2 and incubated in an anaerobic jar by using Anaerocult A (Merck Microbiologie, Darmstadt, Germany) to ensure an oxygen-free environment. Prior to inoculation of the remaining species, the tubes containing media were boiled for 20 min and cooled to room temperature to remove oxygen from the medium. After inoculation, which was performed on the bench, tubes were rapidly sealed with a paraffin plug to ensure anoxic conditions.
TABLE 1.
Bacterial strains, their sources, and media used for their cultivation
| Species | Sourcea | Mediumb | PCR productc
|
|
|---|---|---|---|---|
| 0.7 kb | 0.4 kb | |||
| Bacteroides fragilis | MMB S16 | WW | − | ND |
| Bacteroides thetaiotaomicron | MMB S20 | WW | − | ND |
| Bifidobacterium lactis Bb12 | VTT | WW | − | ND |
| Butyrivibrio fibrisolvens-like clone A11 | LB | − | ND | |
| Clostridium beijerinckii | MMB 3318 | WW | − | ND |
| Clostridium bifermentans | NIZO B529 | WW | − | ND |
| Clostridium celerescens-like clone A54 | LB | − | ND | |
| Coprococcus eutactus-like clone A19 | LB | − | ND | |
| Enterococcus faecalis | DSM 20478 | WW | − | ND |
| Escherichia coli JM109 | Promega | LB | − | ND |
| Eubacterium biforme-like clone F44 | LB | ND | − | |
| Eubacterium halii-like clone A07 | LB | − | − | |
| Eubacterium rectale-like clone A13 | LB | − | − | |
| Fusobacterium prausnitzii-like clone A03 | LB | − | ND | |
| Fusobacterium prausnitzii-like clone A10 | LB | − | ND | |
| Lactococcus lactis subsp. lactis | NIZO B17 | MRS | − | ND |
| Lactobacillus acidophilus | VTT E96276 | MRS | + | + |
| Lactobacillus brevis | VTT E91458 | MRS | + | + |
| Lactobacillus buchneri | VTT E93445 | MRS | + | + |
| Lactobacillus curvatus | VTT E90391 | MRS | + | + |
| Lactobacillus fermentum | VTT E93489 | MRS | + | + |
| Lactobacillus helveticus | ATTC 15009 | MRS | + | + |
| Lactobacillus helveticus | NCIMB 8652 | MRS | + | + |
| Lactobacillus paracasei F19 | VTT | MRS | + | + |
| Lactobacillus plantarum | VTT E79098 | MRS | + | + |
| Lactobacillus reuteri | VTT E92142 | MRS | + | + |
| Lactobacillus rhamnosus LGG | ATCC 53103 | MRS | + | + |
| Lactobacillus ruminis | VTT E91470 | MRS | + | + |
| Lactobacillus salivarius subsp. salicinius | DSM 20554 | MRS | + | + |
| Leuconostoc sp. | NIZO B179 | WW | + | + |
| Pediococcus sp. | FM 0001 | MRS | + | + |
| Peptostreptococcus anaerobius | MMB 2828 | WW | − | ND |
| Roseburia cecicola-like clone A22 | LB | − | ND | |
| Ruminococcus obeum-like clone A20 | LB | − | ND | |
| Streptococcus thermophilus | NIZO B119 | WW | − | ND |
| Vagococcus fluvialis | DSM 5731 | WW | − | ND |
| Weissella kandleri | DSM 20593 | WW | + | ND |
MMB, Department of Medical Microbiology, University of Groningen, 9713 GZ Groningen, The Netherlands. VTT, VTT Culture Collection, FIN-02044, Finland. NIZO, Netherlands Institute for Dairy Research, 6710 BA Ede, The Netherlands. DSM, German Collection for Microorganisms, 3300 Braunschweig-Stöckheim, Germany. ATCC, American Type Culture Collection, Rockville, Md. NCIMB, National Collection of Industrial, Food, and Marine Bacteria, Aberdeen, AB24 3RY, Scotland, United Kingdom. FM, Food Microbiology, Wageningen University, 6703 HD Wageningen, The Netherlands.
WW, Wilkins West; LB, Luria-Bertani broth; MRS, Lactobacillus MRS broth.
The formation of specific PCR products with a size of 0.7 kb by using the primer set Bact-0011f and Lab-0677r or of 0.4 kb by using the primer set Lab-0159f and Uni-0515r at an annealing temperature of 66°C is also indicated (+, present; −, absent). ND, not determined.
Recovery, preparation, and storage of fecal and cecal samples.
Fecal samples were provided by healthy volunteers of both sexes, and of different ages, in spatula containers and were stored at −20°C until further use. Informed consent was obtained from the parents of children. One healthy full-term baby boy, vaginally delivered and breast-fed, participated in this study. The infant was monitored over time and was breast-fed until day 17, received the first formula food after 2 weeks, and then received progressively more formula. Solid food was given after 3 months. Besides these non-trial-affiliated volunteers, fecal samples were provided by individuals that were involved in a trial wherein subjects were fed with fermented products containing the probiotic Lactobacillus paracasei F19 (15, 24). Upon use samples were thawed on ice-water and mixed, after which they were homogenized in 0.05 M ice-cold potassium phosphate buffer (pH 7.0) at a ratio of 3 g of feces (wet weight) per 50 ml (49). Cecal samples from healthy adults were collected as described previously (8, 22, 23).
DNA isolation.
Bacterial DNA from fecal and cecal samples was isolated according to the method of Zoetendal et al. (49). DNA from 5-ml bacterial cultures was isolated by centrifugation of the cells (5 min, 1,500 × g); the cells were then resuspended in 1 ml of Tris-EDTA buffer (pH 8.0) (35). The cell suspension was further treated as described above by bead beating, phenol-chloroform extraction, and ethanol precipitation (49).
Primer design and PCR conditions.
All primers used in this study are listed in Table 2. 16S rDNA sequences of phylogenetically related species were retrieved from GenBank (www.ncbi.nlm.nih.gov) and used to perform multiple alignments by using CLUSTAL W (46). A potential target site starting at position 677 was selected and, based on this site, a 17-mer PCR primer was designed and designated S-G-Lab-0677-a-A-17 (Lab-0677r) according to the OPD nomenclature (1). A second primer, S-G-Lab-0159-a-S-20 (Lab-0159f), was designed based on another conserved site starting at position 158 targeting Lactobacillus and Enterococcus genera (11). The specificity of the primers was screened by submitting the sequence to the Check Probe program of Ribosomal Database Project (RDP; www.cme.msu.edu/RDP) (21) and the ARB software package (O. Strunck and W. Ludwig [http://www.mikro.biologie.tu-muenchen.de/pub/ARB/documentation/arb.ps]). The genomic DNA of relevant reference strains and predominant GI-tract species was used as the template in PCR experiments to validate the Lab-0677r and Lab-0159f primer specificities (Table 1).
TABLE 2.
Primers used in this study
| Primera | Primer sequence (5′-3′) | Source or reference |
|---|---|---|
| S-D-Bact-0011-a-S-17 | AGA GTT TGA T (C/T) (A/C) TGG CTC AG | 16 |
| S-D-Bact-0124-a-S-27 | GGA CGG GTG AGT AAC ACG | 18 |
| S-D-Lab-0158-a-S-20 | TGG AAA CAG (A/G) TG CTA ATA CC | 11 |
| S-G-Lab-0159-a-S-20 | GGA AAC AG (A/G) TGC TAA TAC CG | This study |
| S-*-Univ-0515-a-A-24 | ATC GTA TTA CCG CGG CTG CTG GCA | 18 |
| S-G-Lab-0677-a-A-17 | CAC CGC TAC ACA TGG AG | This study |
| S-G-Lacb-0722-a-A-25 | (C/T) CA CCG CTA CAC ATG (A/G) AG TTC CAC T | 39 |
| GC-clamp | CGC CGG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG G | 29 |
| T7 | TAA TAC GAC TCA CTA TAG G | Promega |
| Sp6 | GAT TTA GGT GAC ACT ATA G | Promega |
Apart from the GC clamp and primers T7 and Spb, the nomenclature is according to Alm et al. (2).
Primers were synthesized commercially by MWG Biotech AG, Ebersberg, Germany. Bacterial primer S-D-Bact-0011-a-S-17 (Bact-0011f), formerly named 27f (18), was paired with the designed reverse primer Lab-0677r, while primer Lab-0159f was paired with the universal reverse primer S-*-Univ-0515-b-A-24 with a GC clamp (Uni-0515-GCr). The GC clamp attached at the 5′ end of one of the primers creates products suitable for separation by DGGE and/or TGGE. PCR was performed employing the Taq polymerase kit from Life Technologies (Gaithersburg, Md.). PCR mixtures of 50 μl contained 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 3 mM MgCl2, 50 mM concentrations of each deoxynucleoside triphosphate, 1.25 U of Taq polymerase, 10 pmol of each primer, and ca. 250 ng of genomic DNA isolated from pure cultures. Samples were amplified in a PE Applied Biosytems GenAmp PCR system 9700 (Foster City, Calif.) by using the following program: predenaturation at 94°C for 5 min; 35 cycles of denaturation 94°C for 30 s, variable annealing temperature for 20 s, and extension at 68°C for 40 s; and a final extension at 68°C for 7 min.
PCR to investigate the general or Lactobacillus-specific GI-tract bacterial community by DGGE was performed with the following 16S rDNA primer combinations: (i) S-D-Bact-0124-a-S-27 with a GC clamp (Bact-0124-GCf) and Univ-515r or (ii) Lab-0159f and Uni-0515-GCr. Nested PCR was performed with these primers on previously generated products from amplification with Bact-0011f and Lab-0677r. The cycling program consisted of 94°C for 5 min; 35 cycles of 94°C for 30 s, 56°C for 20 s, and 68°C for 40 min; and finally 7 min at 68°C. PCR products that were used as templates in nested PCRs were purified with the Qiaquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
DGGE analysis of PCR amplicons.
PCR products generated with primers Bact-0124f-GC and Univ-0515r or primers Lab-0159f and Univ-0515r-GC were separated by DGGE according to the specifications of Muyzer et al. (29) by using the Dcode system (Bio-Rad Laboratories, Hercules, Calif.) with the following modifications. Polyacrylamide gels (dimensions, 200 by 200 by 1 mm) consisted of 8% (vol/vol) polyacrylamide (37.5:1 acrylamide-bisacrylamide) and 0.5× TAE (35). Denaturing acrylamide of 100% was defined as 7 M urea and 40% formamide. The gels were poured from the top by using a gradient maker and a pump (Econopump; Bio-Rad Laboratories, Hercules, Calif.) set at a speed of 4.5 ml/min, and gradients of 30 to 60% were used for the separation of the generated amplicons. Before polymerization of the denaturing gel (28-ml gradient volume), a 7.5-ml stacking gel without denaturing chemicals was added, and the appropriate comb was subsequently inserted. Electrophoresis was performed for 16 h at 85 V in a 0.5× TAE buffer at a constant temperature of 60°C. Gels were stained with AgNO3 according to the method of Sanguinetti et al. (36).
Cloning of the PCR-amplified products.
PCR amplicons were purified with the Qiaquick PCR purification kit according to the manufacturer’s instructions. PCR products were cloned into E. coli JM109 by using the Promega pGEM-T vector system (Promega, Madison, Wis.). PCR was performed on cell lysates of ampicillin-resistant transformants by using pGEM-T specific primers T7 and Sp6 to confirm the size of the inserts. To establish the diversity within the group of 25 selected clones of each origin, amplicons of the correct size were subjected to restriction fragment length polymorphism analysis by using the restriction enzyme MspI. Plasmids containing a unique insert of the appropriate size or corresponding to a band in the community fingerprint were purified by the QIAprep spin miniprep kit (Qiagen, Hilden, Germany) and were subjected to DNA sequence analysis.
Sequence analysis.
One microgram of purified pGEM plasmid was used for sequence analysis of the cloned 16S rDNA fragments. Sequencing reactions were performed with the Sequenase (T7) sequencing kit (Amersham Life Sciences, Slough, United Kingdom) according to the manufacturer’s specifications by using the IRD-800 5′-prime end-labeled primers T7 and Sp6. Sequences were automatically analyzed on a LI-COR DNA sequencer 4000L (Lincoln, Nebr.) and corrected manually.
Phylogenetic placement.
Phylogenetic analysis was performed by using the ARB software package. Sequences were aligned and a rooted neighbor-joining tree (E. coli positions 31 to 648) was constructed by using Bacillus subtilis as an outgroup species.
Nucleotide sequence accession numbers.
Thirty-nine 0.7-kb and nine 0.4-kb sequences of the 16S rDNA determined in this study were deposited with the GenBank database under accession numbers AF335874 to AF335918 and AF368382 to AF368390, respectively.
RESULTS
Development and evaluation of a Lactobacillus group-specific primer.
Based on the alignment of the complete 16S rRNA sequences of various members of the three recognized Lactobacillus subgroups (4) and those of several other related lactic acid bacteria, a potential PCR primer binding site was identified starting at E. coli position 677 of the 16S rDNA (Table 3). Primer S-G-Lab-0677-a-A-17, complementary to this part of the 16S rRNA, was designed taking into consideration (i) the nucleotide mismatches with related species, (ii) the complementarity with the target at the 3′ end, possibly ending with a C; (iii) a G+C content of >50%, and (iv) a size of ca. 20 nucleotides. Sequence comparison by using the Check Probe, ARB, and BLAST programs confirmed that the targeted region was conserved only among all of the 16S rRNA sequences of Lactobacillus, Leuconostoc, Pediococcus, Weissella, and Aerococcus species present in the Ribosomal Database Project and GenBank, except for L. maltaromicus, L. vitulinus, L. catenaformis, and L. helveticus. The first three of the latter lactobacilli are more related to Clostridium and Carnobacterium spp. than to the genus Lactobacillus (38). Their exclusion may have only limited consequences since these particular Lactobacillus spp., as well as members of the Pediococcus, Weissella, and Aerococcus species, are generally not indigenous to the human GI tract. Moreover, specific PCR products were obtained with DNA from two strains of L. helveticus, suggesting that the mismatches found in the database with the primer are most likely the result of sequencing errors (see below). The terminal 3′ end of the primer was also complementary to Eubacterium biforme and Eubacterium cylindroides 16S rDNA sequences. Most importantly, the primer had one mismatch at the 3′ end with 16S rDNA sequences of several other related species, including some bacteria commonly found in large numbers in the GI tract, such as Clostridium, Eubacterium, Bacteroides, and Bifidobacterium spp., that should prevent annealing (Table 3).
TABLE 3.
Alignment of primer S-G-Lab-0677-a-A-17 with target sequences of 16S rDNA (region 677 to 693 of E. coli numbering) from Lactobacillus and other species
| Primer or species (no. of species found)a | Sequence |
|---|---|
| S-G-Lab-0677-a-A-17 reverse primer | 5′-C ACC GCT ACA CAT GGA G-3′ |
| All Lactobacillus spp. subgroupsb (94) | GTA AGG TGG CGA TGT GTA CCT CAA G |
| All Leuconostoc spp. (18) | GTA AGG TGG CGA TGT GTA CCT CAA G |
| All Weissella spp. (16) | GTA AGG TGG CGA TGT GTA CCT CAA G |
| All Pediococcus spp. (12) | GTA AAG TGG CGA TGT GTA CCT CAA G |
| Bacteroides fragilis | GTA AAG TGG CGA TGT GGTGCT TAA G |
| Bacteroides vulgatus | GTA AAG TGG CGA TGT GGTGCT TAA G |
| Bifidobacterium bifidum | GTA AGG TGG CAA TGT GGC CCT TAA G |
| Clostridium coccoides | GTA AAG TGG CGA TGT GAT CCT TAA G |
| Clostridium perfringens | GTA AAG TGG CGA TGT GAT CCT TAA G |
| Clostridium ramosum | GTA AAG TGG CGA TGT GTA CCT TAA G |
| Enterococcus faecalis | GTA AAG TGG CGA TGT GTA CCT TAA G |
| Eubacterium biforme | GTA AAA TGG CGA TGT GTA CCT CAA G |
| Eubacterium rectale | GTA AAG TGG CGA TGT GAT CCT TAA G |
| Lactococcus lactis | GTA AAG TGG CGA TGT GTA CCT TAA G |
| Staphylococcus aureus | GTA AAG TGG CGA TGT GTA CCT TAA G |
| Streptococcus thermophilus | GTA AAG TGG CGA TGT GTA CCT TAA G |
| Vagococcus fluvialis | GTA AAG TGG CGA TGT GTA CCT TAA G |
The number of species found in the database is indicated in parentheses. The mismatches with the primer sequence are indicated in boldface and are underlined.
Includes all Lactobacillus species in GenBank, except Lactobacillus maltaromicus, L. catenaformis, and L. vitulinus.
The primer was experimentally tested by performing PCR on genomic DNA isolated from a range of Lactobacillus species, related lactic acid and other bacteria, as well as cloned 16S rDNA sequences that have been demonstrated to be present in large numbers in the GI tract (49). The optimal annealing temperature was empirically determined by raising it in steps of 2°C from 56 to 68°C, by using genomic DNA of 27 bacterial species and plasmid DNA from nine clones (Table 1). The optimum was found to be 66°C and, after a maximum of 35 cycles, products were obtained with DNA from all tested Lactobacillus, Leuconostoc, and Weissella spp., whereas the 16S rDNA of all other species was not amplified (Table 1).
Biodiversity of the Lactobacillus genus evaluated by Lab-0677r.
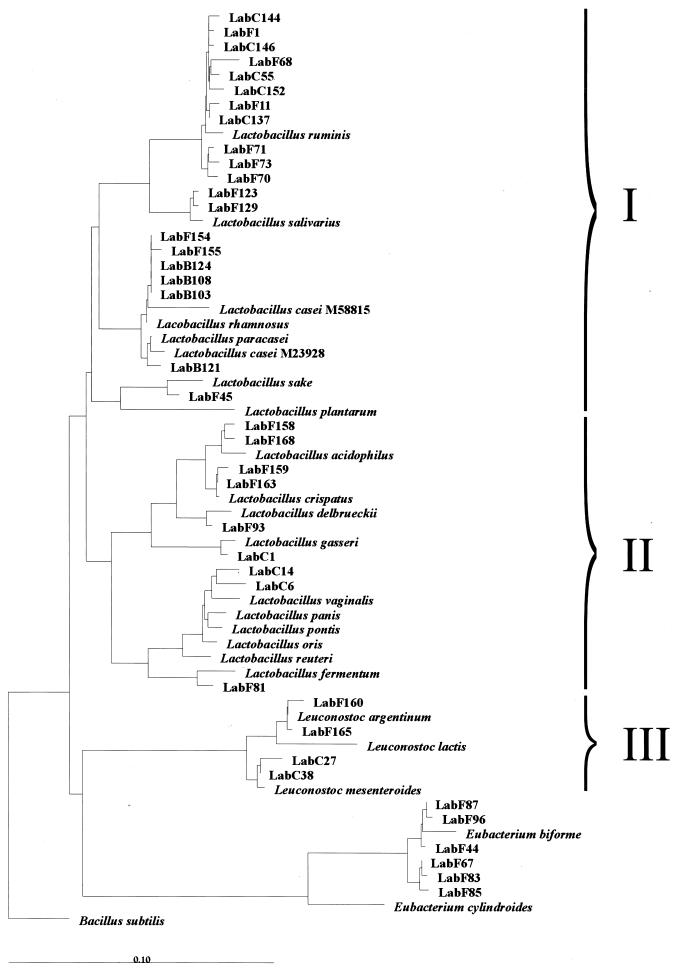
The diversity of members of the Lactobacillus genus was investigated by using primer S-G-Lab-0677-a-A-17 in combination with primer Bact-0011f on bacterial DNA isolated from fecal and other intestinal samples that generated 0.7-kb PCR products. Sequence analysis of unique clones of fecal origin from five healthy adults and an infant, as well as samples taken from the cecal chyme during intestinal intubation in four healthy volunteers, resulted in sequences with significant identity (97 to 99%) to the 16S rDNA of known Lactobacillus spp. (Table 4). Among all 39 sequences retrieved, L. ruminis-like sequences were the most frequently encountered in both fecal and cecal samples. Leuconostoc-like sequences that showed similarity to Leuconostoc mesenteroides and Leuconostoc argentinum were also found in these samples. Eubacterium biforme-like sequences were found in the feces of three adults (Table 4). Phylogenetic analysis clearly showed that sequences were retrieved from all three subgroups of the genus Lactobacillus (Fig. 1). Most representatives were positioned in the Lactobacillus casei-Pediococcus subgroup (I), with the majority being most similar to L. ruminis. Sequences retrieved from the infant feces grouped with L. salivarius, L. rhamnosus, and L. paracasei. Some sequences grouped within the L. delbrueckii subgroup (II), while the Leuconostoc paramesenteroides subgroup (III) was represented by sequences that were similar to Leuconostoc mesenteroides and Leuconostoc argentinum. Overall, the sequence results confirmed the validity of the developed primer.
TABLE 4.
Clones with the percentage of identity to known sequences in GenBank and the RDP database that were retrieved from intestinal samples of different origins by using the primer sets Bact-0011f and Lab-0677r (A) and Lab-0159f and Uni-0515r (B)
| Primer set | Origin | Individual | Speciesa | No. of clones | Clone(s) (% similarity) |
|---|---|---|---|---|---|
| A | Fecal, adult | A | Lactobacillus sakei | 1 | F45 (98) |
| Eubacterium biforme | 1 | F44 (98) | |||
| B | Lactobacillus ruminis | 4 | F68 (98), F70 (98), F71 (98), F73 (98) | ||
| Eubacterium biforme | 1 | F67 (97) | |||
| C | Lactobacillus delbrueckii | 1 | F93 (98) | ||
| Lactobacillus fermentum | 1 | F81 (99) | |||
| Eubacterium biforme | 4 | F83 (97), F85 (97), F87 (98), F96 (98) | |||
| D | Lactobacillus ruminis | 2 | F1 (99), F11 (98) | ||
| E | Lactobacillus acidophilus | 2 | F158 (98), F168 (98) | ||
| Lactobacillus crispatus | 2 | F159 (99), F163 (99) | |||
| Lactobacillus rhamnosus | 2 | F154 (99), F155 (97) | |||
| Leuconostoc argentinum | 2 | F160 (98), F165 (99) | |||
| Fecal, infant | F | Lactobacillus casei | 1 | B121 (99) | |
| Lactobacillus salivarius | 2 | B123 (99), B129 (99) | |||
| Lactobacillus rhamnosus | 3 | B103 (99), B108 (99), B124 (99) | |||
| Cecal, adult | G | Lactobacillus ruminis | 4 | S137 (99), S144 (98), S146 (99), S152 (98) | |
| H | Lactobacillus gasseri | 1 | S1 (99) | ||
| Lactobacillus vaginalis | 2 | S6 (97), S14 (98) | |||
| I | Leuconostoc mesenteroides | 2 | S27 (98), S38 (99) | ||
| J | Lactobacillus ruminis | 1 | S55 (98) | ||
| Totalb | 39 | ||||
| B | Fecal adult | K | Lactobacillus gasseri | 1 | F706 (99) |
| Lactobacillus casei | 1 | F703 (98) | |||
| Lactobacillus paracasei | 1 | F723 (99) | |||
| L | Lactobacillus ruminis | 1 | F748 (98) | ||
| Lactobacillus ruminis | 1 | F754 (98) | |||
| Lactobacillus sakei | 1 | F749 (98) | |||
| Lactobacillus casei | 1 | F747 (98) | |||
| M | Lactobacillus ruminis | 1 | F761 (99) | ||
| Lactobacillus ruminis | 1 | F763 (98) |
Species were named according to their closest relative.
The total number of clones used for phylogenetic analysis.
FIG. 1.
Phylogenetic tree based upon the neighbor-joining method of partial 16S rDNA sequences (E. coli positions 31 to 648) from clones derived by PCR with the primer Bact-0011f and the specific primer Lab-0677r. Suffixes: F, feces; C, cecal chyme; B, baby. Reference sequences are included which were found to be the closest relatives of the retrieved clones. Three subgroups are indicated: I, L. casei-Pediococcus group; II, L. delbrueckii group; III, Leuconostoc paramesenteroides group. B. subtilis is used as an outgroup. The scale bar represents (calculated) distance. The origin of the clones is presented in Table 4.
DGGE analysis of Lactobacillus amplicons.
To facilitate rapid monitoring of Lactobacillus-specific sequences, use was made of sequence-specific separation of PCR amplicons by DGGE that requires having the GC clamp attached to one of the primers. However, a low product yield was observed when either of the primers, Bact-0011f or Lab-0677r, was equipped with a GC clamp, and therefore we opted for the use of a nested-PCR approach. This involved a first round of PCR with primers Bact-0011f and Lab-0677r amplifying the Lactobacillus-like community, followed by a second PCR with established GC-containing universal primers. To prevent amplification of residual genomic DNA or low-yield aspecific amplicons formed in the first PCR, only samples yielding sufficient product (15 ng/μl) were used for a second round of PCR with primers Bact-0124-GCf and Uni-0515r.
Succession of Lactobacillus spp. after birth.
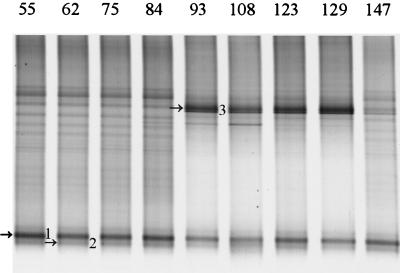
The intestinal microbial community of infants is developing and therefore highly unstable (20; C. Favier, unpublished data). A study of the development of the Lactobacillus-like community in babies was performed by PCR with primers Bact-0011f and Lab-0677r on fecal DNA from baby F from delivery (day 1) to 5 months later (day 147) at regular intervals. No PCR products were obtained up to day 55, indicating that the template was absent or that the amount of template was too low to be detected. The PCR amplicons obtained from days 55 to 147 were used for the nested PCR with primers Bact-0124-GCf and Uni-0515r, and the resulting 0.4-kb fragments were separated in a 30 to 60% DGGE gel (Fig. 2). Two amplicons with different intensity were present from day 55 throughout the experiment, and the sequences of these 0.4-kb fragments were identical to clones earlier retrieved from the infant, i.e., B103 (L. rhamnosus) and B121 (L. casei) (Table 4). Successional change was observed with amplicon 3, which was identical to clone B123 (L. salivarius), that appeared on day 93 and faded between days 129 and 147.
FIG. 2.
Development of the Lactobacillus-like community over time in an infant. DGGE analysis of amplicons generated by nested PCR from fecal samples taken on day 55 up to day 147. Fragments that are indicated by arrows and numbers were identified by the Lab-clone library of the infant as described in Table 4. The origin of the fragments and the corresponding clone are indicated as follows: 1, L. rhamnosus (B103); 2, L. casei (B121); and 3, L. salivarius (B123).
Effect of consumption of lactobacilli on the Lactobacillus sp. diversity in children.
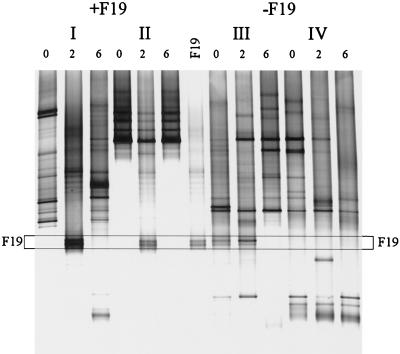
The fate of orally administered L. paracasei F19 in a clinical trial with children was investigated with primer Lab-0677r by using nested PCR and DGGE. Fecal samples from children between 10 and 18 months of age that were given 1010 CFU of L. paracasei F19 twice a day for 3 weeks or a placebo for the same time period were analyzed in a blind fashion. Although L. paracasei F19 was not detected in the dominant bacterial community of these young children (47), it could be detected as a double band when the Lab-0677-specific PCR-DGGE approach was used (Fig. 3). This analysis also revealed that in some children the specific community is also unstable. This instability of the Lactobacillus-like community was particularly evident in two individuals and appeared independent of the L. paracasei F19 administration. In individual I the community was changing throughout the experiment, but despite this change strain F19 could be detected during the administration period. In individual III, the double band of strain F19 or a Lactobacillus species with the same melting behavior was present even before the trial started and was enhanced during the experiment. However, after the cessation of placebo-administration (Fig. 3, individual III) it disappeared. In a subject where the Lactobacillus-like community appeared to be stable (Fig. 3, individual II), the administration of L. paracasei F19 had a temporary impact on the composition but it returned to its original composition after cessation of the strain administration.
FIG. 3.
Effect of orally administered L. paracasei F19 on the Lactobacillus-like community over time. DGGE analysis of amplicons generated in a nested PCR with primers Bact-0124-GCf and Uni-0515r. Children were administered L. paracasei F19 (I and II) or placebo (III and IV) for 4 weeks. Fecal samples were taken before the experiment (lanes 0), after 2 weeks of administration (lanes 2), and 2 weeks after administration had ceased (lanes 6). The PCR amplicon for L. paracasei F19 was concomitantly separated. The fragments in the profiles corresponding to F19 are indicated by the box.
Lactobacillus-like community development in time and space in adults.
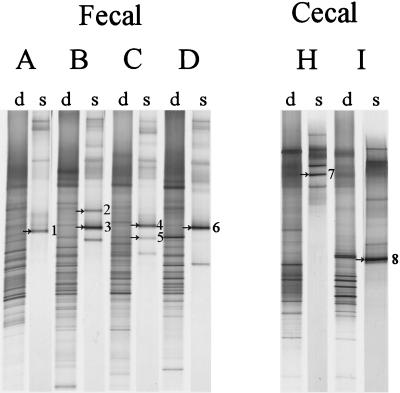
While Lactobacillus and Leuconostoc sequences were readily obtained from adult fecal samples, analysis of the amplicons obtained by nested PCR revealed an overrepresentation of Eubacterium biforme sequences and only small amounts derived from the Lactobacillus group (data not shown). Therefore, another more specific primer Lab-0159f, which targets the Lactobacillus and Enterococcus genera, was used in a nested-PCR strategy with Uni-0515-GCr. Although the Lab-0159f primer resulted in aspecific PCR products when used as a primary PCR primer on fecal samples (data not shown), it was very selective when used in the nested-PCR approach to reamplify amplicons obtained with Bact-0011f and Lab-0677r and circumvented reamplification of Eubacterium biforme-like sequences (Table 1). Comparison of the dominant bacterial community with the Lactobacillus-like community revealed a decreased diversity for the Lactobacillus-like sequences where relatively few bands were present (Fig. 4). Identification of the bands resulted in the retrieval of Lactobacillus- and Leucono stoc-like sequences with identities to known species ranging from 98 to 99%.
FIG. 4.
DGGE analysis of the dominant (d) and Lactobacillus-like community (s) diversity in fecal and cecal samples from six adults (A to D, H, I), after PCR with primers Bact-0124-GCf and Uni-0515r and nested PCR with primers Lab-0159f and Uni-0515-GCr, respectively. The dominant fragments in the Lactobacillus-like patterns indicated by arrows and numbers were identified by clones from the existing clone library of Lactobacillus-like sequences, as described in Table 4. The origin of the fragments and the corresponding clones are indicated as follows: 1, L. sakei (F45); 2, L. ruminis (F68); 3, L. ruminis (F70); 4, L. delbrueckii (F93); 5, L. fermentum (F81); 6, L. ruminis (F1); 7, L. gasseri (S1); and 8, Leuconostoc mesenteroides (S27).
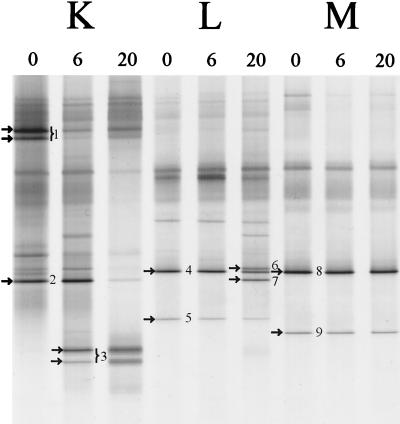
The fecal bacterial community of several healthy adults was monitored over a period of 20 months (Fig. 5). The DGGE patterns of the dominant bacterial community of each individual were specific and stable (data not shown), which is in agreement with previous results (50). The DGGE fingerprints of the dominant Lactobacillus-like community obtained with the nested PCR approach with the specific Lab-0159f primer revealed a variable stability of the Lactobacillus-like community among the different individuals (Fig. 5). The diversity of species increased in both individuals K and L, but individual M appeared to harbor a stable community of lactobacilli. PCR-DGGE is a semiquantitative method in which variation in band-intensities reflects the relative number of specific amplicons. For example, in individual K (Fig. 5) at 6 months the amount of sequences close to L. gasseri (fragment 1) decreased relative to that of the newly emerging sequences resembling L. paracasei (fragment 3). Identification of the dominant 0.4-kb fragments in the three adults resulted in sequences most similar (98 to 99%) to L. casei, L. paracasei, L. gasseri, L. sakei, and L. ruminis. Notably, in adults L and M sequences originating from L. ruminis-like sequences were dominantly present throughout the observed time frame.
FIG. 5.
Monitoring of the Lactobacillus-like community of adults in time. DGGE analysis of amplicons generated by nested PCR with primers Lab-0159f and Uni-0515-GCr, originating from three individuals (K, L, and M) from whom fecal samples were obtained at 0, 6, and 20 months. The dominant fragments in Lactobacillus-like patterns indicated by arrows and numbers were sequenced and compared to known sequences in GenBank, as described in Table 4. The origin of the fragments and the corresponding clones (see Table 4) are indicated as follows: 1, L. gasseri (F706); 2, L. casei (F703); 3, L. paracasei (F723); 4, L. ruminis (F748); 5, L. ruminis (F754); 6, L. sakei (F749); 7, L. casei (F747); 8, L. ruminis (F761); and 9, L. ruminis (F763).
DISCUSSION
This study describes the development of a Lactobacillus group-specific primer that is used to investigate bacterial diversity in the human GI tract. Currently, the Lactobacillus genus constitutes three phylogenetic clusters, the L. delbrueckii group, the L. casei-Pediococcus group, and the distinct Leuconostoc group. It is widely acknowledged that the taxonomy of the genus is unsatisfactory, which is caused by the phylogenetically heterogeneous nature of this large assembly of microorganisms (4, 38). Additionally, the 16S rRNA sequences of this genus are highly similar to other species, including Streptococcus and Enterococcus spp. Nevertheless, a Lactobacillus group-specific probe was developed by Sghir et al. (39) that was used in dot blot hybridization but has only a single mismatch with enterococci, streptococci and staphylococci. Harmsen et al. (11) developed an oligonucleotide probe for fluorescent in situ hybridization that targets the three subgroups and has additional specificity for some Weissella, Vagococcus, and Enterococcus species. Attempts to use the latter two oligonucleotide probes, being the sense version of S-G-Lab-0158-a-A-20 (11) or probe S-G-Lacb-0722-a-A-25 (39) directly, as PCR primers resulted in nonspecific PCR products (data not shown). In general it is not possible to use probes as specific primers since the specific bases of a probe are located centrally, while PCR primers harbor their specificity at the 3′ end, where initial hybridization and elongation take place. New alignments of the 16S rRNA revealed that a reverse primer, S-G-Lab-0677-a-A-17, targeting E. coli positions 677 to 693, would specifically encompass Lactobacillus, Leuconostoc, Weissella, Pediococcus, and Aerococcus. Phylogenetically related genera that are usually present in intestinal samples in reasonably high numbers such as Streptococcus, Enterococcus, Vagococcus, and Staphylococcus were eliminated, as well as more abundant genera such as Bifidobacterium, Bacteroides, Eubacterium, and Clostridium.
The specificity of primer S-G-Lab-0677-a-A-17 was established by performing PCR with this primer on bacterial DNA from pure cultures. This resulted in the formation of specific amplicons for Lactobacillus, Leuconostoc, Pediococcus, and Weissella species. Additionally, the effectiveness of the primer was confirmed during the investigation of the diversity of the Lactobacillus sequences in a variety of intestinal samples amplified with primer Lab-0677r and Bact-0011f. Notably, no PCR products were obtained from fecal samples for three out of eight adult subjects, suggesting that either the numbers were below the detection level or Lactobacillus-like species were completely absent. Samples from five adults of different ages yielded PCR products that were used for cloning and sequencing. Specific PCR products were also readily obtained when cecal material from the proximal colon of another four healthy volunteers was used as a template. Differences between cecal and fecal microbiota have been reported in humans with lactobacilli in reasonably high numbers in the cecum (22, 33).
Previous culturing studies of the human GI tract reported Lactobacillus species similar to those described here. For example L. fermentum, L. salivarius, and L. casei have been isolated from both the rectum and the oral cavity (1), while L. ruminis, L. paracasei, L. crispatus, L. vaginalis, and L. gasseri were isolated from stools (25, 26, 44). However, none of the retrieved 16S rRNA sequences were identical to those of known Lactobacillus spp., and some showed considerable differences. Unexpectedly, a number of sequences in this study were similar to those of species that are frequently found in fermented food products. L. delbrueckii and L. acidophilus are known to be present in yogurt, while L. sakei is used in industrial meat and sourdough fermentation (9, 28). Members of the Leuconostoc group are also well known for their use in fermented food products and as food spoilage microorganisms. Leuconostoc mesenteroides and Leuconostoc argentinum-like sequences were retrieved from cecal and fecal samples. However, the retrieved intestinal sequences are not identical to those of starter bacteria and therefore may be derived from several indigenous species rather than being of a transitional nature from ingested foods. Mitsuoka (26) reported on the occasional presence of L. ruminis in the GI tract of humans, but L. ruminis-like sequences were found in 6 of the 13 individual samples examined in this study, indicating that it is a common GI-tract species. As mentioned none of the sequences were identical and in two clones (F155 and S6) the similarity to deposited sequences was only 97%. This indicates that, although the Lactobacillus genus is considered culturable, the diversity appears to be still insufficiently described.
As Eubacterium biforme and Eubacterium cylindroides are considered to be common constituents of the adult GI tract (7), this coincidentally prohibits the strict specificity of primer S-G-Lab-0677-a-A-17 for the 16S rDNA of Lactobacillus clusters. Eubacterium-like sequences were found in fecal samples from three of five adult individuals, with similarities ranging from 97 to 98%. The screened material of cecal origin lacked this common constituent, which supports the presence of differences in the specific bacterial communities of the proximal and distal parts of the large intestine as observed from culturing studies (33).
PCR-DGGE with nested PCR in which primers Bact-0124-GCf and Uni-0515r were used resulted in an overrepresentation of Eubacterium biforme-like sequences. This species is apparently present in large amounts in adult fecal samples and hence the use of general bacterial primers in the nested PCR amplified them to large proportions. This almost obscured the Lactobacillus and Leuconostoc bacterial community (data not shown) which was demonstrated to be present by the earlier cloning and sequencing results (Table 4). The use of a second, more general Lactobacillus-targeted PCR primer, Lab-0159f, in a nested approach readily resulted in the retrieval of only Lactobacillus- and Leuconostoc-like sequences which matched the bands found in DGGE analysis (Fig. 2 and 3 and Table 4). This supported the earlier cloning results of this study that L. ruminis is a dominant organism. Culture-based approaches showed that the Lactobacillus-like community in the GI tract at the strain level was highly fluctuating in composition and numbers over time and was different from individual to individual (17, 25). The current culture-independent approach reveals a relatively stable Lactobacillus composition in the three individuals while the relative proportions could vary considerably in time depending on the individual (25). The 16S rDNA is a discriminative gene on the species level, but exact copies of sequences for different strains of the same species can undermine its power for strain detection. Thus, there is a possibility that for some individuals the stability in time of the Lactobacillus-like community as observed in this study may not fully represent the actual situation on the strain level.
In contrast to adults, PCR with primers Bact-0124GCf and Uni-0515r can efficiently be used in babies and infants where Eubacterium biforme strains are either absent or in sufficiently low numbers not to result in a distorted representation of the Lactobacillus community. The combination of specific PCR and DGGE could effectively demonstrate the succession of the different Lactobacillus-like species for the first 5 months of an infant’s life.
Increasing consumer awareness of the link between diet and health has promoted the introduction of lactic acid bacteria, especially lactobacilli, into functional foods such as probiotics that may exert an effect in the human GI tract (24, 34). Beneficial influences of probiotics reported include protection against GI infections and inflammatory bowel disease, antiallergic effects, and other immune-related effects (24). The effect of consumption of probiotics on the composition of the dominant microbiota was investigated for several probiotic feeding trials by using DGGE, which indicated that no extensive changes occurred (47). The present study clearly demonstrates that specific PCR with S-G-Lab-0677-a-A-17, followed by DGGE, allows monitoring of the presence of a L. paracasei strain in clinical trials. In conclusion, the strategy described here that combines specific Lactobacillus PCR with DGGE is widely applicable and elevates this significant group of bacteria, which is often present in low numbers within an ecosystem, from total community obscurity.
Acknowledgments
We thank Joël Doré, Abdelghani Sghir, and Christine Favier for providing us with fecal and cecal samples and DNA from various sources, and we also thank the many volunteers who donated samples. We are grateful to Rangne Fondén for providing samples of the ARLA probiotic trial and Maria Saarela for providing the Lactobacillus VTT strains. We are also grateful to Masja Nierop-Groot and Patrick Verbaarschot for providing us with additional strains. Hermie Harmsen is especially acknowledged for providing many bacterial strains and for helpful discussions on the initial primer design.
This work was supported by grants from the European Community: EC-FAIR-CT96-3035 and EC-FAIR-CT96-1028.
REFERENCES
- 1.Ahrné, S., S. Nobaek, B. Jeppson, I. Adlerberth, A. E. Wold, and G. Molin. 1998. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Appl. Microbiol. 85:88–94. [DOI] [PubMed] [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chagnaud, P., K. Machinis, L. A. Coutte, A. Marecat, and Y. Mercenier. 2001. Rapid PCR-based procedure to identify lactic acid bacteria: application to six common Lactobacillus species. J. Microbiol. Methods 44:139–148. [DOI] [PubMed] [Google Scholar]
- 4.Collins, M. D., U. Rodrigues, M. Aguirre, J. A. E. Farrow, A. Martinez-Murcia, B. A. Philips, A. M. Williams, and S. Wallbanks. 1991. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol. Lett. 77:5–12. [Google Scholar]
- 5.Delley, M., B. Mollet, and H. Hottinger. 1990. A DNA probe for Lactobacillus delbrueckeii. Appl. Environ. Microbiol. 56:1967–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falk, P. G., L. V. Hooper, T. Midtvedt and J. I. Gordon. 1998. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62:1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finegold, S. M., V. L. Sutter, and G. E. Mathisen. 1983. Normal indigenous intestinal flora, p.3–31. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, Inc., New York, N.Y.
- 8.Flourié, B., C. Florent, J. P. Jouany, P. Thivend, F. Etachaud, and J. C. Rambaud. 1986. Colonic metabolism of wheat starch in health of humans. Effects on fecal outputs and clinical symptoms. Gastroenterology 90:111–119. [DOI] [PubMed] [Google Scholar]
- 9.Hammes, W. P., A. Bantleon, and S. Min. 1990. Lactic acid bacteria in meat fermentation. FEMS Microbiol. Rev. 87:165–174. [Google Scholar]
- 10.Hammes, W. P., and R. F. Vogel. 1995. The genus Lactobacillus, p.19–52. In B. J. B. Wood and W. H. Holzapfel (ed.), The genera of lactic acid bacteria. The lactic acid bacteria, vol. 2. Chapman & Hall, Glasgow, United Kingdom.
- 11.Harmsen, H. J. M., P. Elfferich, F. Schut, and G. W. Welling. 1999. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in fecal samples by fluorescent in situ hybridization. Microbiol. Ecol. Health Dis. 11:3–12. [Google Scholar]
- 12.Hensiek, R., G. Krupp, and E. Stackebrandt. 1992. Development of diagnostic oligonucleotide probes for four Lactobacillus species occurring in the intestinal tract. Syst. Appl. Microbiol. 15:123–128. [Google Scholar]
- 13.Hertel, C., W. Ludwig, M. Obst, R. F. Vogel, W. P. Hammes, and K. H. Schleifer. 1991. 23S rRNA-targeted oligonucleotide probes for rapid identification of meat lactobacilli. Syst. Appl. Microbiol. 14:173–177. [Google Scholar]
- 14.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884. [DOI] [PubMed] [Google Scholar]
- 15.Juntunen, M., P. V. Kirjavainen, A. C. Ouwehand, S. J. Salminen, and E. Isolauri. 2001. Adherence of probiotic bacteria to human intestinal mucus in healthy infants and during rotavirus infection. Clin. Diagn. Lab. Immunol. 2:293–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kane, M. D., L. K. Poulsen, and D. Stahl. 1993. Monitoring the enrichment and isolation of sulfate reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura, K., A. L. McCartney, M. A. McConnell, and G. W. Tannock. 1997. Analysis of fecal populations of bifidobacteria and lactobacilli and investigations of the immunological responses of their human hosts to the predominant strains. Appl. Environ. Microbiol. 63:3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p.115–175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 19.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA targeted probe and its application in fecal samples. Appl. Environ. Microbiol. 61:3069–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackie, R. I., A. Sghir, and H. R. Gaskins. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69:1035–1045. [DOI] [PubMed] [Google Scholar]
- 21.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. J. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marteau, P., B. Flourié, C. Cherbut, J. L. Correze, P. Pellier, J. Seylaz, and J. C. Rambaud. 1994. Digestibility and bulking effect of ispaghula husks in healthy humans. Gut 35:1747–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marteau, P., P. Pochart, J. Doré, C. Béra-Maillet, A. Bernalier, and G. Corthier. 2001. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl. Environ. Microbiol. 67:4939–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattila-Sandholm, T., S. Blum, J. K. Collins, R. Crittenden, W. M. de Vos, C. Dunne, R. Fondén, G. Grenov, E. Isolauri, B. Kiely, P. Marteau, L. Morelli, A. Ouwehand, R. Reniero, M. Saarela, S. Salminen, M. Saxelin, E. Schiffrin, F. Shanahan, E. Vaughan, and A. von Wright. 1999. Probiotics: towards demonstrating efficacy. Trends Food Sci. Technol. 10:393–399. [Google Scholar]
- 25.McCartney, A. L., W. Wenzhi, and G. W. Tannock. 1996. Molecular analysis of the composition of the bifidobacterial and Lactobacillus microflora of humans. Appl. Environ. Microbiol. 62:4608–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitsuoka, T. 1992. The human gastrointestinal tract, p.69–114. In B. J. B. Wood (ed.), The lactic acid bacteria, vol. 1. The lactic acid bacteria in health and disease. Elsevier Applied Science, London, United Kingdom.
- 27.Moore, W. E. C., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller, M. R. A., M. A. Ehrmann, and R. F. Vogel. 2000. Multiplex PCR for the detection of Lactobacillus pontis and two related species in a sourdough fermentation. Appl. Environ. Microbiol. 66:2113–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muyzer, G., E. C. de Waal, and G. A. Uitterlinden. 1993. Profiling of complex populations by denaturing gradient gel electrophoresis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osawa, R., K. Kuroiso, S. Goto, and A. Shimizu. 2000. Isolation of tannin-degrading lactobacilli from humans and fermented foods. Appl. Environ. Microbiol. 66:3093–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrick, H. A. R., R. E. Ambrosio, and W. H. Holzapfel. 1988. Isolation of a DNA probe for Lactobacillus curvatus. Appl. Environ. Microbiol. 54:404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pilloud, N., and B. Mollet. 1990. DNA probes for the detection of Lactobacillus helveticus. Syst. Appl. Microbiol. 13:345–349. [Google Scholar]
- 33.Pochart, P., F. Lémann, B. Flourié, P. Pellier, I. Goderel, and J. C. Rambaud. 1993. Pyxigraphic sampling to enumerate methanogens and anaerobes in the right colon of healthy humans. Gastroenterology 108:1281–1285. [DOI] [PubMed] [Google Scholar]
- 34.Salminen, S., E. Isolauri, and E. Salminen. 1996. Clinical uses of probiotics for stabilizing the gut mucosal barrier: successful strains and future challenges. Antonie Leeuwenhoek 70:347–358. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Sanguinetti, C. J., E. Dias Neto, and A. J. G. Simpson. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques 17:915–919. [PubMed] [Google Scholar]
- 37.Satokari, R. M., E. E. Vaughan, A. D. L. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schleifer, K. H., and W. Ludwig. 1995. Phylogenetic relationships of lactic acid bacteria, p.7–17. In B. J. B. Wood and W. H. Holzapfel (ed.), The genera of lactic acid bacteria. The lactic acid bacteria, vol. 2. Chapman & Hall, Glasgow, United Kingdom.
- 39.Sghir, A., D. Antonopoulos, and R. I. Mackie. 1998. Design and evaluation of a Lactobacillus group-specific ribosomal RNA-targeted hybridization probe and its application to the study of intestinal microecology in pigs. Syst. Appl. Microbiol. 21:291–296. [DOI] [PubMed] [Google Scholar]
- 40.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Doré. 2000. Quantification of bacterial groups within the human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson, J. M., V. J. McCracken, H. R. Gaskins, and R. I. Mackie. 2000. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Appl. Environ. Microbiol. 66:4705–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tannock, G. W. 1991. The microecology of lactobacilli inhabiting the gastrointestinal tract. Adv. Microb. Ecol. 11:147–171. [Google Scholar]
- 44.Tannock, G. W., A. Tilsala-Timisjarvi, S. Rodtong, J. Ng, K. Munro, and T. Alatossava. 1999. Identification of Lactobacillus isolates from the gastrointestinal tract, silage, and yoghurt by 16S–23S rRNA gene intergenic spacer region sequence comparisons. Appl. Environ. Microbiol. 65:4264–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tannock, G. W. 1999. Analysis of the intestinal microflora: a renaissance. Antonie Leeuwenhoek 76:265–278. [PubMed] [Google Scholar]
- 46.Thompson, J. D., Higgins, D. G., and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaughan, E. E., H. G. H. J. Heilig, E. G. Zoetendal, R. Satokari, J. K. Collins, A. D. L. Akkermans, and W. M. de Vos. 1999. Molecular approaches to study probiotic bacteria. Trends Food Sci. Technol. 10:400–404. [Google Scholar]
- 48.Vaughan, E. E., F. Schut, H. G. H. J. Heilig, E. G. Zoetendal, W. M. de Vos, and A. D. L. Akkermans. 2000. A molecular view of the intestinal ecosystem. Curr. Issues Intestinal Microbiol. 1:1–12. [PubMed] [Google Scholar]
- 49.Zoetendal, E. G., A. D. L. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zoetendal, E. G., A. D. L. Akkermans, and W. M. de Vos. 2001. Molecular characterisation of microbial communities based on 16S rRNA sequence diversity, p.267–298. In L. Dijkshoorn, K. Towner, and M. Struelens (ed.), New approaches for the generation and analysis of microbial fingerprints. Elsevier, Amsterdam, The Netherlands.