Abstract
A new animal model, the streptomycin-treated mini-pig, was developed in order to allow colonization of defined strains of Enterococcus faecalis in numbers sufficient to study plasmid transfer. Transfer of the pheromone-inducible pCF10 plasmid between streptomycin-resistant strains of E. faecalis OG1 was investigated in the model. The plasmid encodes resistance to tetracycline. Numbers of recipient, donor, and transconjugant bacteria were monitored by selective plating of fecal samples, and transconjugants were subsequently verified by PCR. After being ingested by the mini-pigs, the recipient strain persisted in the intestine at levels between 106 and 107 CFU per g of feces throughout the experiment. The donor strain, which carried different resistance markers but was otherwise chromosomally isogenic to the recipient strain, was given to the pigs 3 weeks after the recipient strain. The donor cells were initially present in high numbers (106 CFU per g) in feces, but they did not persist in the intestine at detectable levels. Immediately after introduction of the donor bacteria, transconjugant cells appeared and persisted in fecal samples at levels between 103 and 104 CFU per g until the end of the experiment. These observations showed that even in the absence of selective tetracycline pressure, plasmid pCF10 was transferred from ingested E. faecalis cells to other E. faecalis organisms already present in the intestinal environment and that the plasmid subsequently persisted in the intestine.
During the last few decades, enterococci have been identified as an important cause of nosocomial infections (26, 30, 36). Even though these organisms are a natural part of the intestinal flora of most mammals (13) and are generally of relatively low virulence (32), some enterococcal infections are severe and difficult to treat because they are resistant to a large variety of antimicrobial agents (31, 39).
Recently, a lot of focus has been put on vancomycin-resistant strains of Enterococcus faecium (previously Streptococcus faecium), since vancomycin is important for treatment of human infections (4, 18, 27, 29). Also, studies of the mechanisms responsible for spread of tetracycline resistance are of importance, since tetracycline-resistant enterococci are very commonly found in high numbers in the intestinal tract of humans, as well as of pigs and broilers used for human consumption (2). Large amounts of tetracycline are used for treatment of infections in Danish farm animals (9), and even very low concentrations of this antibiotic have been suggested to have an important influence on the spread and establishment of resistance genes in the animal intestine (T. R. Licht, C. Struve, B. B. Christensen, R. L. Poulsen, S. Molin, and K. A. Krogfelt, submitted for publication).
A factor promoting high occurrence of resistance to antimicrobial agents among enterococci could be the remarkably efficient mechanisms for transfer of resistance genes associated with these organisms. Enterococci are known to harbor transferable genetic elements, conjugative transposons, which have an unusually broad host range and can even be transferred between gram-negative and gram-positive bacterial cells (7). In addition to this, Enterococcus faecalis (previously Streptococcus faecalis) can carry a variety of so-called pheromone-inducible plasmids. The transfer of such plasmids is initiated in response to a small signal peptide (the pheromone), which available recipient bacteria constitutively excrete (for reviews, see references 8, 11, and 41). The transfer is highly efficient in liquid suspension and can reach frequencies of more than 1 transconjugant per donor in 2-h matings (12). Only very few studies have addressed the impact of this unique mechanism of transfer on plasmid exchange between intestinal bacteria in vivo (20, 25).
Even though pheromones are hydrophobic and susceptible to proteases, recent results from our laboratories indicate that these peptides do have an effect on plasmid transfer in intestinal mucus (23). The present study investigated the transfer of the pheromone-inducible pCF10 plasmid (5) between E. faecalis strains colonizing the intestinal tract. This plasmid encodes resistance to tetracycline. The resistance determinant tet(M) is present on the Tn916-like conjugative transposon Tn925, which is stably integrated into the plasmid DNA (6). In order to measure the transfer of pCF10 in the intestinal tract of animals used for human consumption, we developed a streptomycin-treated mini-pig model based on the streptomycin-treated mouse model (19, 34, 35), which has previously been used for in vivo studies of plasmid transfer between gram-negative bacteria (22).
MATERIALS AND METHODS
Bacterial strains and growth media.
In all animal experiments, the streptomycin- and spectinomycin-resistant strain E. faecalis OG1SS (15) was used as recipient strain. The strain used as donor was E. faecalis OG1RFS, which is a spontaneously streptomycin-resistant mutant of E. faecalis OG1RF (10) and is therefore also resistant to rifampin and fusidin. The donor strain carried the pheromone-inducible, conjugative plasmid pCF10 (5), which encodes resistance to tetracycline. For verification of the donor capacity exhibited by transconjugants from fecal samples, E. faecalis OG1RF (without pCF10) was used as recipient. Fluent cultures of the strains were grown in brain heart infusion medium (Oxoid) or Todd-Hewitt broth (Difco Laboratories), and strains carrying the pCF10 plasmid were grown in the presence of tetracycline (10 μg/ml).
Selective plating from fecal samples and intestinal contents were done on the following media: reinforced clostridial agar (Oxoid) for total aerobic and anaerobic microflora, MacConkey agar no. 3 (Oxoid) for selection of Escherichia coli, Bacteroides bile esculin agar (Becton Dickinson) for selection of the Bacteroides group, and Slanetz and Bartley medium (Oxoid) for selection of enterococci. In addition, brain heart infusion agar (Oxoid) was used for selection and for further analysis of isolated bacteria. Brain heart infusion agar and Slanetz and Bartley medium were supplemented with the appropriate antibiotics as described in Results. Recipient, donor, and transconjugant bacteria were selected on Slanetz and Bartley plates containing, respectively, (i) streptomycin and spectinomycin; (ii) streptomycin, rifampin, and fusidin; and (iii) streptomycin, spectinomycin, and tetracycline. Since transconjugant cells also grew on the plates selective for recipient bacteria, the “real” recipient numbers were calculated by subtracting the numbers of transconjugant colonies from the numbers of colonies appearing on the plates selective for recipients. For verification of the donor capacity of transconjugants formed in vivo, secondary transconjugants formed in broth were selected on plates containing rifampin, fusidin, and tetracycline. Antibiotics were purchased from Sigma and used at the following concentrations: streptomycin, 1,000 μg/ml; spectinomycin, 500 μg/ml; rifampin, 25 μg/ml; fusidin, 25 μg/ml; tetracycline, 10 μg/ml.
Bacteroides bile esculin agar and half of the reinforced clostridial agar plates were incubated under anaerobic conditions (Anoxymat, HART, BioLab) at 37°C for 2 days. All other media were incubated aerobically for 2 days at 37°C, with the exception of MacConkey agar plates, which were incubated for 1 day only.
Animals.
Three female Göttingen mini-pigs (Ellegaard, Dalmose, Denmark) were used for these experiments. The animals were caged individually in special isolators and treated according to the recommendations given by the breeder (www.minipigs.com), who also delivered their feed (Special Diets Services, Ellegaard). Drinking water was given ad libitum. At arrival, the mini-pigs were 7 to 8 weeks old and weighed approximately 4 kg. They were allowed to adapt to the new environments for a minimum period of 1 week before any sampling was done.
Streptomycin treatment and colonization of mini-pigs.
Fecal sampling from the three animals was carried out regularly throughout the experiment. Sampling was done with an interval of 1 to 3 days. On day 22 measured from the first day of sampling, the three mini-pigs were given 5 g of streptomycin sulfate (Sigma) per liter of drinking water. After this point, all drinking water given to the animals throughout the experiment contained streptomycin (5 g per liter) and was freshly prepared every morning. One day after the onset of streptomycin treatment (day 23), 25 ml of an overnight culture containing 2.5 × 108 CFU per ml of the recipient strain, E. faecalis OG1SS, was mixed in the morning feed for each of the animals. Three weeks after the recipient inoculation (day 45), they similarly received another 25-ml dose containing 4.7 × 108 CFU/ml of the donor strain, E. faecalis OG1RFS(pCF10). The three mini-pigs were euthanatized as described below at day 72, 74, and 79, respectively.
Euthanatization and sampling from intestinal contents.
Prior to euthanatization, the animals were anesthetized by intramuscular injection of 50 mg of ketaminol/ml (0.4 ml per kg of body weight) and 20 mg of narcoxyl/ml (0.1 ml per kg). Both of these chemicals were purchased from Veterinaria AG, Zurich, Switzerland. Subsequently, an intraperitoneal injection (1 ml per kg) as well as an intravenous injection (0.5 ml per kg) of 200-mg/ml pentobarbital (Pharmacy of the Royal Veterinary and Agricultural University, Denmark) was given to the animals. The animals were bled, and samples were taken from the contents of the stomach, duodenum, jejunum, ileum, cecum, colon, and rectum. Immediately thereafter, the samples were analyzed as described below.
Analysis of fecal samples and intestinal contents.
Samples from the mini-pigs were diluted 10-fold (vol/wt) in maximum recovery dilution buffer (Oxoid). The dilution was homogenized (120 s, maximal speed) using a laboratory blender (Stomacher 400, Seward). From the homogenized sample, further dilutions were made and plated on the appropriate selective agar plates. The plates were incubated as described above, and numbers of CFU per gram of feces were estimated.
Verification of transconjugant and donor bacteria by PCR.
In the period after day 45 of the experiment, three colonies per animal per day were isolated from the fecal platings on Slanetz and Bartley medium containing streptomycin, spectinomycin, and tetracycline, which supported growth of putative transconjugant colonies. In the period from day 0 until day 22, 10 of the background colonies, which in this period only occasionally showed up on this medium, were collected. In addition, all colonies appearing on Slanetz and Bartley medium containing streptomycin, rifampin, and fusidin, which supported growth of donor colonies, were isolated. The colonies were grown in liquid medium and frozen as glycerol cultures for later PCR analysis.
PCR was carried out with three different primer sets. The first primer set verified the presence of the tetracycline resistance determinant tet(M). This verification procedure has previously been described (1). The second primer set placed tet(M) on the Tn916-like conjugative transposon, Tn925, which is known to be integrated on pCF10 (6). The primers used for this purpose were Tn916-2, 5′-CTAGATTGCGTCCAA-3′, and ReversetetM, 5′-TTGTTAGAGCCATATCTTAG-3′ (Y. Agerso, L. B. Jensen, M. Givskov, and M. C. Roberts, submitted for publication).
The third primer set was designed to verify that the transposon is positioned on the pCF10 plasmid. The sequences of these primers were PCF10-39838, 5′-CATGAGCCAAAGAGGC-3′, and Tn916-3-out, 5′-GATAACTAGATTTTTATGCTATTTTTAACT A-3′. The template sequence information used for design of the third primer set was kindly provided by Gary M. Dunny, University of Minnesota.
Test of donor capacity exhibited by transconjugants formed in vivo.
From each of the three animals, two of the transconjugant colonies collected for PCR verification as described above were also tested for their ability to transfer tetracycline resistance to E. faecalis OG1RF. Liquid overnight cultures of the transconjugant (now donor) strains were washed, and 100-μl volumes of each of these cultures were mixed with 100-μl volumes of a washed overnight culture of E. faecalis OG1RF (now recipient). Todd-Hewitt broth was added to a total volume of 1 ml, and mating was carried out at 37°C overnight. Numbers of secondary transconjugants were determined by dilution and plating on selective agar plates. For comparison, a broth mating experiment was carried out in a similar way using donor and recipient E. faecalis strains identical to those fed to the mini-pigs.
PFGE of colonized recipient bacteria.
Throughout the colonization period beginning at day 23 of the experiment, one colony per animal per day was isolated from the fecal platings on Slanetz and Bartley medium containing streptomycin and spectinomycin, which supported growth of putative recipient colonies (E. faecalis OG1SS). These colonies were grown in liquid medium and frozen as glycerol cultures for pulsed-field gel electrophoresis (PFGE) analysis in order to investigate whether genetic rearrangements occurred in the E. faecalis OG1SS cells as a result of adaptation to the intestinal environment. PFGE analysis was carried out as previously described (17).
RESULTS
Streptomycin treatment and introduction of E. faecalis OG1.
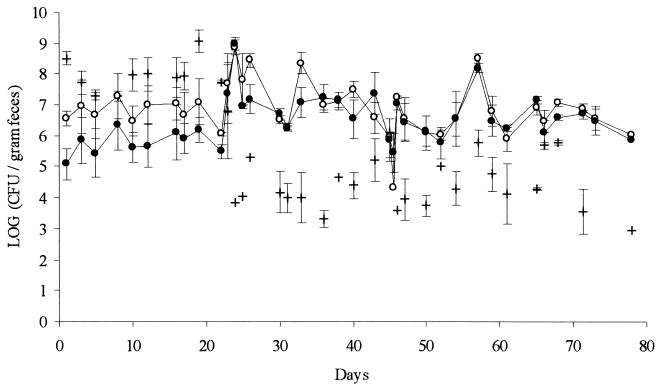
Three mini-pigs were treated with streptomycin and dosed with E. faecalis OG1 as described in Materials and Methods. Total levels of aerobic and anaerobic culturable bacteria remained between 109 and 1010 CFU per g of fecal material during the entire experiment (data not shown). However, the composition of the flora changed as shown in Fig. 1. Until the onset of streptomycin treatment (day 22), the amount of streptomycin-resistant enterococci was about 1 decade lower than the total amount of enterococci, suggesting that up to 90% of the intestinal enterococci could be removed by streptomycin. This would leave a free niche for colonization of the introduced, streptomycin-resistant recipient strain. During streptomycin treatment and after introduction of the recipient strain (day 23), practically all enterococci recovered from feces were resistant to streptomycin. The amount of fecal enterococci fluctuated between 106 and 108 CFU per g of feces. In general, the fluctuations in enterococcal numbers seemed to occur in a parallel manner and also when compared to the recipient numbers, which are shown in Fig. 2. When looking at data from the three animals separately, this parallelism was even clearer, suggesting that the fluctuations could be explained mainly by fluctuations in the amount of dry weight per gram of fresh feces. Immediately after the onset of streptomycin treatment, the fecal levels of gram-negative aerobes, presumably mainly E. coli, dropped about three decades (Fig. 1). At the same time, anaerobic counts of the Bacteroides fragilis group dropped from about 108 down to 105 to 106 CFU per g (for clarity, these data are not included in Fig. 1).
FIG. 1.
Concentration in fecal samples of total enterococci (open circles), streptomycin-resistant enterococci (closed circles), and E. coli (+). From day 22 of the experiment, the animals were treated with streptomycin. At days 23 and 45, respectively, the recipient and donor strains of E. faecalis OG1 were introduced. The figure shows the average values from three mini-pigs. Error bars represent standard errors of the means.
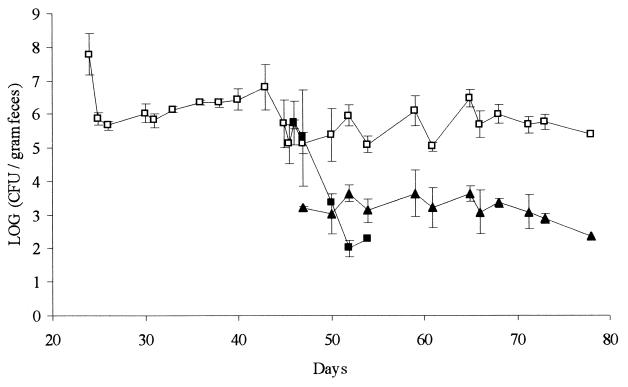
FIG. 2.
Concentration in fecal samples of recipient E. faecalis OG1SS (open squares), donor E. faecalis OG1RFS(pCF10) (closed squares), and transconjugant E. faecalis OG1SS(pCF10) (closed triangles). At days 23 and 45, respectively, the recipient and donor strains were introduced. The figure shows the average values from three mini-pigs. Error bars represent standard errors of the means.
The total amount of indigenous tetracycline-resistant bacteria in all three animals as measured by plating on brain heart infusion agar supplemented with tetracycline was about 108 to 109 CFU per g of fecal material and was largely unaffected by streptomycin treatment. The level of indigenous, tetracycline-resistant enterococci was just above 104 CFU per g but dropped below the detection limit in all three animals immediately after streptomycin treatment and introduction of the recipient strain, probably because the tetracycline-sensitive recipient bacteria replaced most of the indigenous enterococci (data not shown).
Transfer of plasmid pCF10 to colonized recipient bacteria.
Before day 23, there was generally no growth of bacteria from feces on the plates selective for the recipient strain, although colonies occasionally occurred in numbers close to the detection limit of 50 CFU per g of fecal material. The recipient strain colonized all three animals at levels between 106 and 107 CFU per g of feces (Fig. 2). No genetic rearrangements of the recipient cells, which could be detected by PFGE analysis, occurred during the colonization period (data not shown).
Prior to day 45, where the donor strain was ingested by the animals no growth was observed after plating of fecal samples on medium selective for this strain. In all three animals, the donor bacteria were initially detectable in high numbers (106 CFU per g of feces) but were subsequently rapidly cleared from the intestine (Fig. 2). At day 57, the donor cells were no longer detectable in fecal samples from any of the three mini-pigs. (detection limit, 50 CFU/g of feces). All of the donor cell colonies isolated from fecal samples were subsequently transferred to plates containing tetracycline in order to verify that they still carried the pCF10 plasmid. Furthermore, it was verified that all enterococci which were selected on plates containing rifampin were also resistant to fusidin, and vice versa, in order to make sure that the donor cells did not loose any of these resistances during passage through the gastrointestinal (GI) tract.
Transconjugant cells were detectable in fecal samples 1 day after donor inoculation. Before this day, there was generally no growth of bacteria from feces on the plates selective for transconjugant cells, although colonies occasionally occurred in numbers close to the detection limit of 50 CFU per g of fecal material. The transconjugant bacteria were present in fecal samples at levels between 103 and 104 CFU per g (Fig. 2). Unlike the donor cells, transconjugant cells persisted in the intestine at these levels throughout the experiment. In vitro broth mating experiments confirmed that the donor capacity of transconjugants isolated from feces was not significantly different from that of the original donor strain.
PCR verification of transconjugant and donor bacteria.
PCR analysis was carried out using three different primer sets in order to verify (i) the presence of the tet(M) resistance determinant, (ii) that the tet(M) gene was located on a Tn916-like transposon, and (iii) that the transposon was located on plasmid pCF10. The first two primer sets gave a positive PCR for all of the analyzed putative transconjugant cells (i.e., 3 colonies per animal per day). However, they also gave positive reactions from 10 background strains which were isolated prior to day 22 on medium similar to that used for selection of transconjugant cells. The third primer set gave a positive reaction for all of the analyzed transconjugant colonies and a negative reaction for all of the background strains (data not shown). All colonies isolated on the medium supporting growth of the donor strain gave a positive reaction with all three primer sets.
Bacterial flora in different segments of the digestive tract.
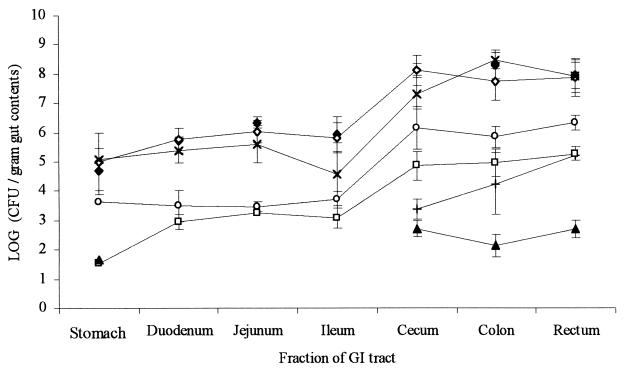
Immediately after euthanatization of the mini-pigs, part of the contents were removed from a number of different segments of the GI tract, and the bacterial flora of the contents was analyzed (Fig. 3). As expected, the concentration of all investigated microorganisms was higher in the large intestine than in the small intestine. The proportion of the total flora constituted by enterococci was more or less the same throughout the GI tract (Fig. 3), and all of the detected enterococci were resistant to streptomycin (data not shown). In all investigated parts of the intestine, a very significant proportion of the total aerobic bacteria was resistant to tetracycline (Fig. 3). Nevertheless, the total numbers of tetracycline-resistant enterococci (data not shown) were identical to the numbers of enterococci resistant to streptomycin, spectinomycin, and tetracycline, suggesting that they were all the result of conjugative pCF10 transfer between the strains introduced during the experiment. The recipient strain was not seen to colonize preferentially in a certain part of the GI tract but was distributed in the intestine in a similar way as the rest of the enterococcal population. Recipient bacteria constituted up to 20% of the total enterococci (Fig. 3).
FIG. 3.
Concentration in various intestinal segments of total aerobic microorganisms (closed diamonds), total anaerobic microorganisms (open diamonds), total tetracycline-resistant aerobic microorganisms (×), E. coli (+), total enterococci (open circles), recipient bacteria (open squares), and transconjugant bacteria (closed triangles). The figure shows the average values from three mini-pigs, measured at the day of euthanatization. Error bars represent standard errors of the means.
DISCUSSION
The pheromone-inducible plasmid pCF10, which encodes tetracycline resistance, is known to transfer extremely efficiently between E. faecalis cells in liquid medium in vitro (3, 12). We have investigated the transfer of this plasmid between isogenic strains of E. faecalis OG1 colonizing the intestinal tract of streptomycin-treated mini-pigs in vivo.
Pilot experiments carried out in our lab had shown that the intestine of conventional mini-pigs, carrying an undisturbed intestinal microflora, after oral ingestion of E. faecalis OG1 was not colonized with this strain at levels which would make it possible to detect plasmid transfer (data not shown). We therefore developed a streptomycin-treated mini-pig model for this purpose. Streptomycin treatment has previously been used to break down the colonization barrier (40) in the mouse intestinal flora, thereby allowing various streptomycin-resistant gram-negative bacteria to colonize the intestinal tract (19, 34, 35). The MIC of streptomycin for enterococci is approximately 1,024 μg/ml, while that for the intestinal gram-negative organisms E. coli and Salmonella sp. is only approximately 16 μg/ml (9). Nevertheless, 5 g/liter of streptomycin sulfate in the drinking water of the animals, which was used in the mouse model for colonization of various gram-negative organisms (14, 24, 33), was seen to be sufficient to enable also stable colonization of our streptomycin-resistant E. faecalis OG1SS recipient strain in the pig model. The recipient strain colonized at a density of 106 to 107 cells per g of feces and constituted around 20% of the total enterococci (Fig. 1, 2, and 3). In the conventional mini-pigs, the amount of indigenous, tetracycline-resistant enterococci was between 104 and 105 CFU per g of feces. PCR analysis showed that 10 out of 10 of these strains carried the tet(M) gene on a Tn916-like transposon similar to the Tn925 element that is present on pCF10 (6). Consistent with this, a recent report shows that a considerable part of the indigenous tetracycline resistance in the intestinal flora of swine is indeed encoded on transferable elements (16). However, the indigenous tetracycline-resistant enterococci were replaced by the tetracycline-sensitive recipient strain immediately after the onset of streptomycin treatment and oral inoculation of the recipient cells. This observation made it unlikely that the recipient would pick up tetracycline resistance from transferable elements present in the indigenous flora, and as expected it did not happen during the 3 weeks where the recipient was allowed to establish in the intestine prior to introduction of the donor strain.
The E. faecalis OG1RFS(pCF10) donor strain, which is streptomycin resistant and chromosomally isogenic with the recipient except for the resistance markers, was not able to persist in the intestine. Even though it was detectable in high numbers directly after inoculation, the amount of donor cells in fecal samples dropped rapidly, and 10 days later no donor cells could be detected in feces from any of the three animals (Fig. 2). We suggest two explanations for this observation. One is that the recipient cells had already been adapted to the intestinal environment and therefore prevented establishment of the donor bacteria, which would be competing for the same niche. This has been seen for isogenic lactococci as well as for isogenic enterococci in germ-free rats (21, 37), but it is inconsistent with observations of isogenic E. coli strains in streptomycin-treated mice (22). PFGE analysis showed no detectable adaptation of the recipient organisms during colonization (data not shown). However, this does not exclude that a phenotypic adaptation occurred. An alternative reason for the inability of the donor bacteria to persist could be that they form clumps when they enter the intestinal environment. The recipient cells present in the intestine continuously excrete cCF10 pheromones (8), which are therefore likely to be present in the intestinal contents when the donor cells come in. Pheromones induce the expression of an adhesion substance on the donor cell surfaces which causes clumping of the E. faecalis cells (11), and the formation of clumps has in other cases been shown to give bacterial cells a disadvantage in colonizing the intestine (14).
Donor cell colonies which were isolated from fecal samples by selection for resistance to rifampin and fusidin were subsequently tested for resistance to tetracycline, and it was confirmed by PCR that all of the donor colonies still carried the pCF10 plasmid. This indicates that the pCF10 plasmid did not segregate in the intestinal environment but was stably maintained in the donor bacteria. However, the rapid elimination of these bacteria from the intestine suggests that only minimal donor cell proliferation occurred, and this minimizes the probability for plasmid segregation.
One to 2 days after inoculation of the donor strain, 103 to 104 transconjugant cells, E. faecalis OG1SS/pCF10, were detected per gram of fecal sample from all three animals by selection for resistance to streptomycin, spectinomycin, and tetracycline. The transconjugant bacteria persisted at these levels throughout the remaining 3 weeks of the experiment (Fig. 2). PCR analysis confirmed that the transconjugant cells carried the pCF10 plasmid and that the conjugative transposon Tn925, which is known to be stably integrated into this plasmid (6), was indeed still present on pCF10. Plasmid pCF10 was not found in the indigenous, tetracycline-resistant bacteria, which were isolated from feces prior to introduction of the donor strain. We therefore conclude that the observed transconjugant colonies were a result of conjugative mating between the donor and recipient enterococci, which had been fed to the pigs.
After euthanatization of the three mini-pigs, the bacterial flora in different segments of the GI tract was analyzed (Fig. 3). A significant part of the total bacterial population in all parts of the digestive tract was resistant to tetracycline, but all of the tetracycline-resistant enterococci had a resistance pattern similar to that of the transconjugant bacteria. A recent report indicates that resistance to tetracycline is more common among the E. coli colonizing the ileum and cecum of swine than among the E. coli found in colon and rectum and that the upper gastrointestinal tract thus may act as a reservoir for resistant E. coli organisms (28). However, in the present study, where a large part of the E. coli organisms had been removed by streptomycin treatment, the fraction of total bacteria resistant to tetracycline was practically the same in all of the cecal, colonic, and rectal segments investigated (Fig 3).
Since the levels of transconjugant bacteria initially observed after donor cell inoculation did not increase with time (Fig. 2), it seems that the majority of transfer events took place between the originally ingested donor cells and the colonized recipient organisms. Assuming that pCF10 did not segregate in the transconjugant cells, the result of continuous secondary transfer events between transconjugant and recipient bacteria would be that more and more of the available recipient cells turned into transconjugant cells over time. This did clearly not happen, and the amount of transconjugant bacteria created by secondary transfer must therefore be negligible compared to that created by proliferation of primary transconjugant cells (Fig. 2). In vitro broth mating experiments showed that the donor capacity of transconjugants collected from fecal samples was not reduced compared to that of the original donor strain (data not shown). However, the kinetics of gene transfer and the absence of secondary transfer in the intestinal environment has previously been reported and discussed for conjugation between intestinal E. coli organisms (22). As it was suggested for E. coli, the lack of secondary transfer between E. faecalis in the intestinal environment might be due to an absence of mixing in the colonic mucus layer, which prevents transconjugant bacteria from getting in contact with new potential recipient organisms. This would also explain why the transconjugant cells did not have the putative disadvantage of clumping, described above for the donor cells. We speculate that since transconjugant cells are already distributed in the intestinal contents and mucus layer when they are formed, they may have a better ability to persist than newly inoculated donor bacteria, which have not yet established in the intestinal mucus at the time when formation of aggregation substance is induced.
The amount of transconjugant cells detected in feces was similar to what has been observed for the transfer of R1drd19 between E. coli bacteria (22), even though the initial levels of donor and recipient E. faecalis cells in the present experiment was one to two decades lower than that reported in the E. coli transfer experiment. This suggests that the pCF10 transfer mechanism may be slightly more efficient in the intestinal environment than transfer mediated by flexible pili, such as those encoded by R1drd19.
We must emphasize that the results presented here do not show whether in vivo transfer of pCF10 occurred as a result of pheromone induction or by some other (as yet unrecognized) mechanism. However, recent experiments from our laboratories suggest that the presence of pheromones induces transfer of pCF10 in intestinal mucus from gnotobiotic rats (23). These experiments also show that the plasmid transfer rate coefficient as defined by Simonsen and coworkers (38) is only approximately 10-fold lower for pCF10 in mucus than in broth, which means that transfer occurs quite efficiently in mucus in spite of the high viscosity of this medium (23). Another report shows that transfer of plasmids belonging to the pheromone-responsive pAM group occurs at high frequencies in the intestinal tract of Syrian hamsters (20). These observations strongly support the findings reported in the present paper and indicate that transfer of pheromone-inducible plasmids between Enterococcus cells occurs in the gastrointestinal tract of many mammals.
Certain antibiotics are known to induce an aggregation response in E. faecalis cells carrying pheromone-inducible pAD1 (43), and one might therefore speculate that the presence of streptomycin in the intestinal environment could cause increased transfer of pCF10. However, we find this very unlikely, since the reported aggregation response induced by antibiotics does not result in increased plasmid transfer. Furthermore, streptomycin does not induce aggregation of pAD1-carrying cells, and antibiotics inducing aggregation of pAD1-carrying cells do not induce aggregation of bacteria carrying pCF10 (43).
The observations presented here confirm that the transient passing of cells carrying conjugative plasmids represents a potential source of spread of resistance genes to the indigenous intestinal flora (22). The fact that the transconjugant E. faecalis formed by the initial mating events was able to persist in the intestine even in the absence of selective tetracycline pressure emphasizes the risk which is associated with the extensive use of antibiotics for domestic animals (42). We speculate that if animals already carrying pCF10-encoded tetracycline resistance in their intestinal flora are exposed to tetracycline, the vertical spread of this plasmid in the intestinal microflora will be even more pronounced. When such animals are subsequently used for human consumption, there also might be a considerable risk for spread of tetracycline resistance among the human intestinal microorganisms, including potentially pathogenic bacteria. Experiments addressing the role of tetracycline as well as of pheromones in the intestinal environment are currently being carried out in our laboratories.
Acknowledgments
This work was financed by a grant from The Directorate for Food, Fisheries and Agro Business under the Danish Ministry of Food, Agriculture and Fisheries.
We thank Anne Ørngreen, DVFA, and Grethe Østergaard, DVFA, for excellent assistance with animal experiments, Inge M. Hansen, DVL, for excellent technical assistance, and Anette M. Kühle Hammerum, DVL, for helpful advice and discussions. Gary M. Dunny, University of Minnesota, is acknowledged for kindly providing the DNA sequence information used for PCR analysis.
REFERENCES
- 1.Aarestrup, F. M., Y. Agerso, P. Ahrens, J. C. O. Jorgensen, M. Madsen, and L. B. Jensen. 2000. Antimicrobial susceptibility of resistance genes in staphylococci from poultry. Vet. Microbiol. 74:353–364. [DOI] [PubMed] [Google Scholar]
- 2.Aarestrup, F. M., Y. Agerso, P. Gerner-Smidt, M. Madsen, and L. B. Jensen. 2000. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 37:127–137. [DOI] [PubMed] [Google Scholar]
- 3.Andrup, L., and K. Andersen. 1999. A comparison of the kinetics of plasmid transfer in the conjugation systems encoded by the F plasmid from Escherichia coli and plasmid pCF10 from Enterococcus faecalis. Microbiology 145:2001–2009. [DOI] [PubMed] [Google Scholar]
- 4.Boyce, J. M., S. M. Opal, J. W. Chow, M. J. Zervos, G. Potter-Bynoe, C. B. Sherman, R. L. Romulo, S. Fortna, and A. A. Medeiros. 1994. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistance. J. Clin. Microbiol. 32:1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie, P. J., and G. M. Dunny. 1986. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid 15:230–241. [DOI] [PubMed] [Google Scholar]
- 6.Christie, P. J., R. Z. Korman, S. A. Zahler, J. C. Adsit, and G. M. Dunny. 1987. Two conjugation systems associated with Streptococcus faecalis plasmid pCF10: identification of a conjugative transposon that transfers between S. faecalis and Bacillus subtilis. J. Bacteriol. 169:2529–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clewell, D. B., S. E. Flannagan, and D. B. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229–236. [DOI] [PubMed] [Google Scholar]
- 8.Clewell, D. B., and K. E. Weaver. 1989. Sex pheromones and plasmid transfer in Enterococcus faecalis. Plasmid 21:175–184. [DOI] [PubMed] [Google Scholar]
- 9.Danish Integrated Antimicrobial Resistance Monitoring and Research Programme. 1999. DANMAP 99—consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Danish Veterinary Laboratory, Copenhagen, Denmark.
- 10.Dunny, G. M., and D. B. Clewell. 1975. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J. Bacteriol. 124:784–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunny, G. M., B. A. B. Leonard, and P. J. Hedberg. 1995. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J. Bacteriol. 177:871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunny, G. M., M. Yuhasz, and E. Ehrenfeld. 1982. Genetic and physiological analysis of conjugation in Streptococcus faecalis. J. Bacteriol. 151:855–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Facklam, R. R., D. F. Sahm, and L. M. Teixeira. 1999. Enterococcus, p.297–305. In P. R. Murray (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 14.Favre-Bonté, S., T. R. Licht, C. Forestier, and K. A. Krogfelt. 1999. Klebsiella pneumoniae capsule expression is necessary for colonization of the large intestine of streptomycin-treated mice. Infect. Immun. 67:6152–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haack, B. J., and R. E. Andrews, Jr. 2000. Isolation of Tn916-like conjugal elements from swine lot effluent. Can. J. Microbiol. 46:542–549. [DOI] [PubMed] [Google Scholar]
- 17.Hammerum, A. M., V. Fussing, F. M. Aarestrup, and H. C. Wegener. 2000. Characterization of vancomycin-resistant and vancomycin-susceptible Enterococcus faecium isolates from humans, chickens and pigs by RiboPrinting and pulsed-field gel electrophoresis. J. Antimicrob. Chemother. 45:677–680. [DOI] [PubMed] [Google Scholar]
- 18.Handwerger, S., B. Raucher, D. Altarac, J. Monka, S. Marchione, K. V. Singh, B. E. Murray, J. Wolff, and B. Walters. 1993. Nosocomial outbreak due to Enterococcus faecium highly resistant to vancomycin, penicillin, and gentamicin. Clin. Infect. Dis. 16:750–755. [DOI] [PubMed] [Google Scholar]
- 19.Hentges, D. J., J. U. Que, S. W. Casey, and A. J. Stein. 1984. The influence of streptomycin on colonization resistance in mice. Microecol. Ther. 14:53–62. [Google Scholar]
- 20.Huycke, M. M., M. S. Gilmore, B. D. Jett, and J. L. Booth. 1992. Transfer of pheromone-inducible plasmids between Enterococcus faecalis in the Syrian hamster gastrointestinal tract. J. Infect. Dis. 166:1188–1191. [DOI] [PubMed] [Google Scholar]
- 21.Jacobsen, B. L., M. Scou, A. M. Hammerum, and L. B. Jensen. 1999. Horizontal transfer of the satA gene encoding streptogramin A resistance between isogenic Enterococcus faecium strains in the gastrointestinal tract of gnotobiotic rats. Microb. Ecol. Health Dis. 11:241–247. [Google Scholar]
- 22.Licht, T. R., B. B. Christensen, K. A. Krogfelt, and S. Molin. 1999. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 145:2615–2622. [DOI] [PubMed] [Google Scholar]
- 23.Licht, T. R., A. M. Hammerum, L. B. Jensen, and B. L. Jacobsen. Effect of pheromone induction on transfer of the Enterococcus faecalis plasmid pCF10 in intestinal mucus ex vivo. FEMS Microbiol. Lett., in press. [DOI] [PubMed]
- 24.Licht, T. R., K. A. Krogfelt, P. S. Cohen, L. K. Poulsen, J. Urbance, and S. Molin. 1996. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect. Immun. 64:3811–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcinek, H., R. Wirth, A. Muscholl-Silberhorn, and M. Gauer. 1998. Enterococcus faecalis gene transfer under natural conditions in municipal sewage water treatment plants. Appl. Environ. Microbiol. 64:626–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moellering, R. C. 1992. Emergence of enterococcus as a significant pathogen. Clin. Infect. Dis. 14:1173–1178. [DOI] [PubMed] [Google Scholar]
- 27.Montecalvo, M. A., H. Horowitz, C. Gedris, C. Carbonaro, F. C. Tenover, A. Issah, P. Cook, and G. P. Wormser. 1994. Outbreak of vancomycin-, ampicillin-, and aminoglycoside-resistant Enterococcus faecium bacteremia in an adult oncology unit. Antimicrob. Agents Chemother. 38:1363–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moro, M. H., G. W. Beran, R. W. Griffith, and L. J. Hoffman. 2000. Effects of heat stress on the antimicrobial drug resistance of Escherichia coli of the intestinal flora of swine. J. Appl. Microbiol. 88:836–844. [DOI] [PubMed] [Google Scholar]
- 29.Morris, J. G., D. K. Shay, J. N. Hebden, R. J. McCarter, B. E. Perdue, W. Jarvis, J. A. Johnson, T. C. Dowling, L. B. Polish, and R. S. Schwalbe. 1995. Enterococci resistant to multiple antimicrobial agents, including vancomycin: establishment of endemicity in a university medical center. Ann. Intern. Med. 123:250–259. [DOI] [PubMed] [Google Scholar]
- 30.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray, B. E. 1991. New aspects of antimicrobial resistance and the resulting therapeutic dilemmas. J. Infect. Dis. 163:1184–1194. [PubMed] [Google Scholar]
- 32.Murray, B. E. 1992. Enterococci, p. 1415–1421. In S. L. Gorback, J. G. Bartlett, and N. R. Blacklow (ed.), Infectious diseases. The W. B. Saunders Co., Philadelphia, Pa.
- 33.Poulsen, L. K., T. R. Licht, C. Rang, K. A. Krogfelt, and S. Molin. 1995. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J. Bacteriol. 177:5840–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Que, J. U., S. W. Casey, and D. J. Hentges. 1986. Factors responsible for increased susceptibility of mice to intestinal colonization after treatment with streptomycin. Infect. Immun. 53:116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Que, J. U., and D. J. Hentges. 1985. Effect of streptomycin administration on colonization resistance to Salmonella typhimurium in mice. Infect. Immun. 48:169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaberg, D. R., D. H. Culver, and R. P. Gayes. 1991. Major trends in the microbial etiology of nosocomial infection. Am. J. Med. 91:72–76. [DOI] [PubMed] [Google Scholar]
- 37.Schlundt, J., P. Saadbye, B. Lohmann, B. L. Jacobsen, and E. M. Nielsen. 1994. Conjugal transfer of plasmid DNA between Lactococcus lactis strains and distribution of transconjugants in the digestive tract of gnotobiotic rats. Microb. Ecol. Health Dis. 7:59–69. [Google Scholar]
- 38.Simonsen, L., D. M. Gordon, F. M. Stewart, and B. R. Levin. 1990. Estimating the rate of plasmid transfer: an end-point method. J. Gen. Microbiol. 136:2319–2325. [DOI] [PubMed] [Google Scholar]
- 39.Spera, R. V., and B. F. Farber. 1994. Mulitidrug resistant Enterococcus faecium. An untreatable nosocomial pathogen. Drugs 48:678–688. [DOI] [PubMed] [Google Scholar]
- 40.van der Waij, D., J. M. Berghuis-De Vries, and J. E. C. Lekkerkerk-van der Wees. 1971. Colonization resistance of the digestive tract in conventional and antibiotic-treated animals. J. Hyg. 69:405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wirth, R. 1994. The sex pheromone system of Enterococcus faecalis. More than just a plasmid-collection mechanism? Eur. J. Biochem. 222:235–246. [DOI] [PubMed] [Google Scholar]
- 42.Witte, W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996–997. [DOI] [PubMed] [Google Scholar]
- 43.Wu, K., F. Y. An, and D. B. Clewell. 1999. Enterococcus faecalis pheromone-responding plasmid pAD1 gives rise to an aggregation (clumping) response when cells are exposed to subinhibitory concentrations of chloramphenicol, erythromycin, or tetracycline. Plasmid 41:82–88. [DOI] [PubMed] [Google Scholar]