Abstract
A promoter-fusion study with a Tn 5-based promoter probe vector had earlier found that the hutU gene which encodes the enzyme urocanase for the histidine utilization pathway is upregulated at a lower temperature (4° C) in the Antarctic psychrotrophic bacterium Pseudomonas syringae. To examine the characteristics of the urocanase gene and its promoter elements from the psychrotroph, the complete hutU and its upstream region from P. syringae were cloned, sequenced, and analyzed in the present study. Northern blot and primer extension analyses suggested that the hutU gene is inducible upon a downshift of temperature (22 to 4°C) and that there is more than one transcription initiation site. One of the initiation sites was specific to the cells grown at 4°C, which was different from the common initiation sites observed at both 4 and 22°C. Although no typical promoter consensus sequences were observed in the flanking region of the transcription initiation sites, there was a characteristic CAAAA sequence at the −10 position of the promoters. Additionally, the location of the transcription and translation initiation sites suggested that the hutU mRNA contains a long 5′-untranslated region, a characteristic feature of many cold-inducible genes of mesophilic bacteria. A comparison of deduced amino acid sequences of urocanase from various bacteria, including the mesophilic and psychrotrophic Pseudomonas spp., suggests that there is a high degree of similarity between the enzymes. The enzyme sequence contains a signature motif (GXGX2GX10G) of the Rossmann fold for dinucleotide (NAD+) binding and two conserved cysteine residues in and around the active site. The psychrotrophic enzyme, however, has an extended N-terminal end.
Antarctic bacteria provide a useful model system for studying cold adaptation (15, 17, 31, 36). These organisms are generally represented by the psychrotrophs and psychrophiles, which have the ability to grow at 0°C. They can transcribe at this lower temperature both in vitro and in vivo (31). However, nothing much is known about the nature of promoter and regulatory elements from these bacteria or about the mechanism of transcription at lower temperatures. Most of the transcriptional studies thus far have been carried out with only mesophilic bacteria, and the RNA polymerase from these bacteria, including Escherichia coli, cannot transcribe at 0°C. A recent study from our laboratory has demonstrated that the RNA polymerase of the Antarctic psychrotrophic bacterium Pseudomonas syringae can transcribe at 0°C. The polymerase from the bacterium was not only active at the low temperature but also could transcribe in vitro preferentially the cold-inducible gene of E. coli cspA from a supercoiled template (43). However, absolutely no information is available with regard to the characteristics of promoter sequence, such as the −10 and −35 elements from the bacterium for such low-temperature-specific transcription. Neither is any information available for the in vivo recognition of promoter sequences by RNA polymerase from the cold-adapted P. syringae. Therefore, we initially attempted to identify the genes from the Antarctic P. syringae that are upregulated at low temperature, with the help of Tn 5-mediated random genomic fusions of a promoter-less reporter gene, lacZ (23). One of the fusions that produced at least 10- to 14-fold more β-galactosidase at a low temperature (4°C) was identified by cloning and sequencing of ca. 450 bp of DNA sequence proximal to the Tn 5 insertion site. The fusion was in the hutU gene, which encodes an enzyme, urocanase, of the histidine utilization pathway of bacteria (13, 23). A direct assay of urocanase activity from the P. syringae and a few more Antarctic Pseudomonas species and comparison of it with that of the mesophilic Pseudomonas putida suggested that the hutU gene is upregulated in the psychrotrophs but not in the mesophile. Therefore, it appeared to us that the hutU gene might be a useful model for investigating the mechanism of gene regulation at low temperatures in the Antarctic bacterium. Accordingly, we cloned and sequenced the DNA encompassing the hutU gene and its upstream and downstream regions and identified different open reading frames (ORFs) in the region. We also examined transcripts from bacterial cells grown at low (4°C) and high (22°C) temperatures by Northern and primer extension analyses, and we identified the transcription start sites and other putative regulatory elements of the hutU gene. Additionally, we compare here the deduced amino acid sequences of the urocanase from the psychrotrophic P. syringae and other bacteria, including the mesophilic P. putida, in order to examine the possible amino acid substitutions due to a low-temperature adaptation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Antarctic psychrotrophic bacterium P. syringae Lz4W, which grew Soptimally at 22°C, was isolated, identified, and maintained as reported earlier (42). Routinely, the culture was grown on Antarctic bacterial medium, which contains 0.5% peptone (wt/vol) and 0.2% (wt/vol) yeast extract, at room temperature (22°C) or at a cold temperature (4°C) when needed. The E. coli cells were grown at 37°C in Luria-Bertani (LB) medium and maintained on LB agar plates.
DNA manipulation techniques and cloning of hutU gene.
The bacterial genomic DNA was prepared as described previously (32). Isolation of plasmid DNA, restriction endonuclease digestion, ligation, transformation, and agarose gel electrophoretic separation of DNA were carried out as described by Sambrook et al. (39).
For cloning of the hutU gene from P. syringae, a 522-bp DNA fragment from the plasmid pF43, which contained the hutU proximal region of the promoter fusion clone F43 (23), was used as a probe. Initially, a 2.4-kbp PstI DNA fragment containing the hutU gene was cloned in pUC19 (ph180). Subsequently, three more overlapping fragments containing the upstream 3.5-kbp EcoRI-KpnI fragment (phUp) and the downstream 2.54-kbp SalI and 1.2-kbp PstI fragments (ph20 and ph39, respectively) were cloned. A total of 6.578 kbp were thus cloned spanning the region (Fig. 1A).
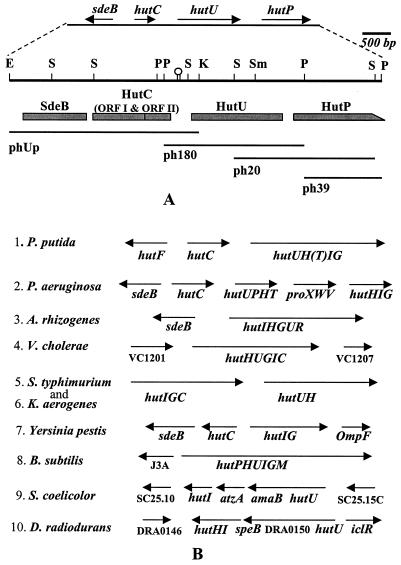
FIG. 1.
Physical map and the organization of the hutU region of P. syringae. (A) A 6.578-kbp DNA from the hutU region of P. syringae was sequenced and analyzed by cloning the region on four overlapping DNA restriction fragments (phUp, ph180, ph20, and ph39) shown below the physical map. Only the major restriction enzyme recognition sites (E, EcoRI; K, KpnI; P, PstI; S, SalI; Sm, SmaI) are indicated on the physical map. The genes (sdeB, hutC, hutU, and hutP) and their directions of transcription are shown at the top. The putative ORFs, such as SdeB, HutC ORFI and ORFII, HutU, and HutP, are shown as shaded boxes below the physical map. The incomplete ORF of HutP is indicated as a box with a slope at the C-terminal end. A hairpin structure between HutC and HutU indicates the location of a dyad structure in the DNA sequence. (B) The organization of the genes in the hutU region of a few bacteria identified either by earlier genetic studies or by recent genome sequence analyses is shown for comparison. The directions of transcription are indicated above the genes by arrows. The genes encoding various enzymes involved in histidine utilization (hut) are abbreviated as hutH (for histidase), hutU (for urocanase), hutI (for imidazolone propionate hydrolase), hutF (for FIGLUase), hutG (for formylglutamate amidohydrolase), hutT (for inducible histidine/urocanate transporter), hutP (putative transporter with similarity to purine-cytosine permease), hutC orfI (putative repressor of hut operon), and hutC orfII (3′ downstream ORF of hutC). The unknown ORFs of the hutU region in some bacteria are indicated directly by their ORF identification numbers (e.g., VC1201, VC1207, J3A, SC25.10, SC25.15C, and DRA0146).
DNA sequence analysis.
Nucleotide sequence determinations were carried out on an automated DNA sequencer (ABI model 377), by using double-stranded plasmid DNAs as a template and the ABI PRISM Dye terminator cycle sequencing method (Perkin-Elmer). The BLAST programs (3, 4) were employed for DNA sequence homology search in the NCBI GenBank sequence database (http://www.ncbi.Snlm.gov/). PCGENE programs were also used for various DNA and amino acid sequence analyses. Secondary structure prediction analysis of protein was carried out by using the Predict-Protein PHD mail server at EMBL, Heidelberg (predict-Help@ EMBL-Heidelberg.DE), Germany, which uses combined evolutionary information and neural networks for structural predictions (34, 35).
The nucleotide sequence reported here has been submitted to GenBank under accession number AF326719.
Isolation of RNA and Northern analysis.
RNA was isolated by the hot phenol method (1) from the bacterial cells (optical density at 600 nm of ∼0.5) grown at 22 and 4°C. For the isolation of RNA from cold-shocked cells, P. syringae was initially grown at 22°C and then shifted to 4°C and incubated for various time periods before the isolation. Northern hybridization was carried out as described earlier (32) but with the hutU- and hutP-specific probes. Densitometer scanning (Molecular Dynamics) of the autoradiograms was carried out for estimating the fold induction of RNA.
Primer extension analysis.
Primer extension analyses were carried out by two methods on the total RNA isolated from 4 and 22°C grown cells. In the first method (39), a32P-end-labeled primer (5′-GGGTAGTTGAAGTCACGGGTAAC-3′) corresponding to the complementary sequence of the putative N-terminal end of the HutU-ORF (−7 to +16 nucleotides with respect to the GTG initiation codon) of P. syringae was extended in the presence of Moloney murine leukemia virus reverse transcriptase (Pharmacia). In the second method, the unlabeled primer was extended in the presence of [α-32P]ATP and Moloney murine leukemia virus reverse transcriptase as described earlier (8). Both methods produced similar results, except that the primer-extended products were sharper with less background on the autoradiograms obtained by the second method. DNA sequencing reactions were also carried out with the same primer on the double-stranded DNA template (ph180) with a Sequenase 2.0 Kit (U.S. Biochemicals) and run in parallel with the primer extended products on an 8 M urea-6% polyacrylamide gel for mapping the transcriptional start points as described previously (39).
Enzyme assays.
Urocanase activity was assayed spectrophotometrically by the method of George and Phillips (14). β -Galactosidase was assayed by the method of Miller (28). Proteins were estimated by the method of Bradford (10).
RESULTS
Cloning and sequence analysis of the hutU region from P. syringae.
A 522-bp Tn 5-proximal sequence of the hutU promoter fusion clone (F43) of P. syringae (23) was used as a probe to clone the whole of the hutU gene and its upstream and downstream sequences, as described in Materials and Methods. The reported region (6.578 kbp) was cloned on three overlapping DNA fragments (Fig. 1A). The DNA sequences of the region were determined (accession no. AF326719) and analyzed by a BLAST search (3, 4). Four complete and one incomplete ORFs were identified which had homology with the hypothetical s deB gene homologue of Yersinia pestis (accession no. AL031866); the hutC (ORFI), hutC (ORFII), and hutU genes of the hut operon of P. putida (2, 13, 19); and the gene yxlA homologue for purine cytosine permease of Bacillus subtilis (accession no. E70081), respectively (Fig. 1). At the level of amino acids, the SdeB homologue (456 amino acids [aa]) was 68% similar to the Y. pestis sequence, and the HutC ORFI (249 aa), HutC ORFII (192 aa), and HutU (565 aa) were 91, 63, and 93% similar, respectively, to the P. putida sequences. The ORF of the last gene (yxlA homologue) was incomplete (433 aa versus the complete 457 aa of YxlA), as shown in Fig. 1A. The direction of transcription of the sdeB was opposite to the direction of the other genes. When this gene organization was compared with the organization of the hut operon of the mesophilic P. putida (19), it was observed that the relative positions of hutC (orfI), hutC (orfII), and hutU in these two bacteria were identical. The difference was noticed in the hutU downstream region of P. syringae, which was occupied by a homologue to the gene for purine-cytosine permease (henceforth referred to as hutP) of B. subtilis and yeast, instead of the hutH (encoding histidase) found in P. putida (Fig. 1B). Electronically submitted data (accession no. AF032970) reveal that a homologue of hutT (encoding an inducible urocanate transporter) is present in the downstream of hutH in P. putida, which was not earlier identified by the genetic study (19, 20). The genome sequence data of Pseudomonas aeruginosa (http://www.pseudomonas.com) reveals that the downstream region of the hutU gene in the bacterium (Fig. 1B) contains the homologues for both hutP (gene for purine-cytosine permease) and hutT (gene for urocanate transporter). The importance and functional significance of the occurrence of a purine-cytosine permease homologue in the hut operon of P. syringae are not clear at present. However, from a comparative analysis of the genes surrounding the hutU region of different bacteria (Fig. 1B), it is apparent that the organization of the genes in the hutU region is quite divergent among various bacteria.
It is also interesting that the genetic data of P. putida had earlier established the occurrence of the hutF gene (encoding the enzyme formiminoglutamate iminohydrolase [FIGLUase]) in the upstream region of hutC (19, 20). The hutF gene was shown transcribed divergently from hutC, similar to the direction of transcription of the sdeB homologues observed in the genome sequence of many gram-negative bacteria, such as P. syringae, P. aeruginosa, Agrobacterium rhizogenes, and Y. pestis (Fig. 1B). Since the DNA sequence for the hutF gene is not known from P. putida or any other bacterium, it remains to be determined whether the sdeB homologue, which also has similarity with the atrazine chlorohydrolases (AtzA, 23% identity; accession no. U55933.1) and the melamine deaminase (TriA, 23% identity; accession no. AF312304.1) of Pseudomonas species, is the hutF gene of these gram-negative bacteria. In this context, it is interesting that the DNA sequence 108 bp upstream of the hutC region of P. putida (accession no. M33922) contains a beginning of a divergently transcribed ORF (N-terminal 19 aa), which is 93% similar to the first 19 aa of the sedB homologue of P. syringae. It is also important to note that hutF is generally thought to be unique for Pseudomonas species. The two genes, hutF and hutG, encoding FIGLUase and formylglutamate hydrolase, respectively, are involved in the catalytic conversion of formiminoglutamate (FIGLU) to glutamate and formate by a two-step process in pseudomonads. This is in contrast to other bacteria, such as Klebsiella and B. subtilis, where a single enzyme encoded by hutG catalyzes the conversion of FIGLU to glutamate and formamide at the final step of the histidine degradation pathway (19, 26).
Predictive analysis of the amino acid sequence of urocanase from P. syringae.
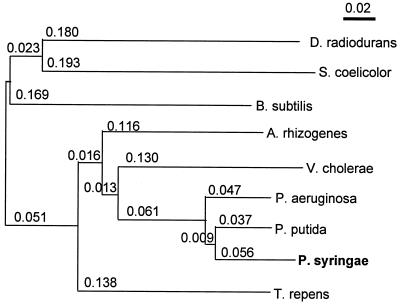
In recent times comparative analyses of proteins from psychrophilic, mesophilic, and thermophilic bacteria have shed new light on the structural adaptability of the enzymes for catalysis at low temperatures and have provided some clues as to their thermal stability (11, 15). Therefore, it was interesting to compare the primary sequence of the urocanase from the psychrotrophic and mesophilic bacteria. Accordingly, the deduced amino acid sequences of the enzyme from the psychrotrophic P. syringae and mesophilic P. putida were compared (Fig. 2). It was observed that the P. syringae enzyme contained an overall identity of ca. 90% (similarity of 93%) to the P. putida enzyme but had an additional 8-aa extension at the N-terminal end (making it 565 aa long). A BLAST search analysis picked up the urocanase homologues from various other bacteria for which the hutU gene sequences are known. An alignment of the deduced amino acid sequences of the enzyme from these bacteria is shown in Fig. 2. Although the enzyme seems to be quite conserved, it fell into two distinct groups of gram-positive (B. subtilis, Streptomyces coelicolor, and Deinococcus radiodurans) and gram-negative (P. syringae, P. putida, P. aeruginosa,Vibrio cholerae, and A. rhizogenes) strains (Fig. 3). The urocanase from the plant (Trifolium repens) was found clustered with the enzymes from gram-negative bacteria.
FIG. 2.
Multiple sequence alignment of the deduced amino acid sequence of urocanase and the predicted secondary structure of the enzyme. The sequences of the ShutU genes (the numbers in parentheses are GenBank accession numbers) include P. putida (M33923), P. aeruginosa (AE004922), A. rhizogenes (AB039932), T. repens (P53385), V. cholerae (AE004200), Y. pestis (AL031866), B. subtilis (P25503), S. coelicolor A3 (AL354048), and D. radiodurans (AE001862). The CLUSTAL X (1.8) Program was used for the alignment. The amino acids have been shown in single letter code. The characters (“✽” and “ : ”) indicate the positions of perfectly conserved amino acids and substitution by similar amino acids, respectively. A period indicates that the position is conserved in most organisms. The highly conserved active site of the enzyme (FQGLPARICW) is shown within a box. Apart from the conserved cysteine within the active-site box, a second conserved cysteine (C-363 of P. syringae corresponding to C-355 of the mesophilic P. putida) that is important for enzyme activity (25) has also been marked. The predicted secondary structure shown above the aligned sequences is based on PHDs (profile-fed neural network system) developed by Rost et al. (34, 35) and found at predict-Help@EMBL-Heidelberg.DE. The helices have been shown as cylinders, the β-sheets are indicated as block arrows, and the other structures, including the loops and coils, appear as thick straight lines. The Rossman fold for dinucleotide binding sequence GXSGX2G-X10-G (where X is a nonspecific amino acid) has been marked below the aligned sequences. A β-sheet of“SLNIE” following the helix of the Rossman fold is also highly conserved, which contains an acidic residue (E or D) at the 3′ end of the sheet that is generally specific for NAD+. Also note that a predicted short N-terminal helix (aa residue 10 or 13) is present only in P. syringae among the compared organisms.
FIG. 3.
Relationship among bacterial urocanases. Similarities between the enzymes have been shown as an unrooted neighbor-joining tree, giving all branch lengths (indicated by numerical values). The tree has been drawn by a neighbor-joining plot of the CLUSTAL X (version 1.8) program. The urocanase sequences of gram-negative bacteria (e.g., P. syringae, P. putida, P. aeruginosa, V. cholerae, and A. rhizogenes) and gram-positive bacteria (e.g., B. subtilis, S. coelicolor, and D. radiodurans) fall into two distinct clusters and may have diverged early in evolution. The only sequence of possible plant origin (33), from T. repens, is closer to the sequence of gram-negative bacteria.
From the compositional analyses of the amino acids of urocanase it was apparent that the contents of isoleucine plus leucine (13.1%), the aromatic amino acids (7.2%), and proline (4.4%), which are generally known to change in the psychrophilic enzymes (15), were similar in P. syringae and mesophilic P. putida (13.28, 7.5, and 4.4%, respectively). A slight increase in the content of serine and threonine (10.4%) was, however, noticed in P. syringae when it was compared with the enzymes from P. putida and other organisms (7.98 to 9.5%). The arginine content (5.3%), which is known to stabilize the helices, was also marginally lower in P. syringae than in the P. putida enzyme (5.9%). The P. syringae enzyme also contained eight cysteines compared to the seven cysteines of P. putida and P. aeruginosa. Six of these cysteines are conserved among them and are located in equivalent positions, including the two important cysteines C-410 and C-354 (C-419 and C-363 in P. syringae), which were shown to be involved in catalysis and substrate binding, respectively, in the enzyme from P. putida (25). Interestingly, the radiotolerant bacterium D. radiodurans contains only these two conserved cysteines in the enzyme.
Predictive structural analyses of the urocanase sequence from P. syringae exhibited some interesting features (Fig. 2). The enzyme contained a conserved β-α-β structural motif of the dinucleotide binding Rossman fold (GXGX2G-X10-G/A) for NAD+ binding at the 182- to 198-aa region. A ψ BLAST analysis suggested that this NAD binding region has structural homology with the similar region of the glutamate dehydrogenase, including an acidic amino acid residue (E-205 in the case of P. syringae urocanase) at the end of the β-strand (201SLNIE205) of the β-α-β motif (7). The acidic amino acid residue commonly forms the hydrogen bonds to the adenine ribose hydroxyls and is generally thought to be an indicator of NAD+ specificity as opposed to the NADP+ specificity (6, 7). An implication of the identification of the Rossman fold in urocanase would be that, although the loop is similar to that of other NAD+ binding proteins, it has to be juxtaposed within a tightly held tertiary or quaternary structural fold of the protein. This is because the exogenously added NAD could not be incorporated into the enzyme in vitro, nor could the coenzyme be dissociated from the protein without irreversible denaturation of the protein (33). The present predictive analysis (Fig. 2) also suggests that the P. syringae enzyme contains at the N-terminal end a short α-helix that is absent in other bacteria.
Temperature-dependent expression of hutU in P. syringae.
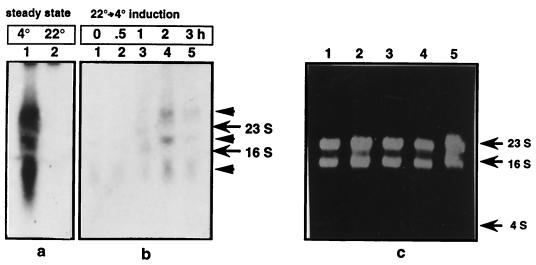
Northern analyses were carried out to examine the expression of the hutU gene in P. syringae at low (4°C) and high (22°C) temperatures. It was observed that, during the steady-state growth of the bacterium, the amount of transcripts produced at 4°C from the hutU gene and hut operon was ca. 20-fold higher than the amount present at 22°C. Three transcripts (ca. 7.1, 2.1, and 1.0 kb) hybridized to the probe of the 2.4-kbp PstI fragment containing both hutU and hutP genes (Fig. 4a). The hutP-specific probe, however, hybridized only to the 7.1-kb transcript (data not shown). Thus, it would appear that the 7.1-kb transcript might represent the polycistronic mRNA, whereas the 2.1- and 1.0-kb transcripts represent the processed and/or degraded product from the operon. The 2.1-kb transcript can potentially encode the full-length hutU gene and therefore might be physiologically important.
FIG. 4.
Northern analysis of transcripts from the hutU region in P. syringae. The RNAs were isolated either from the cells exponentially growing at 4 and 22°C or from “cold-shocked” cells after a shift of the culture from 22 to 4°C at different time points (0, 0.5, 1, 2, and 3 h) after that shift. The transcripts from the hutU region are shown at a steady-state level (a) and during the cold-shock induction (b). (c) The RNA loading control (10 μg) used in the study is shown in a ethidium bromide-stained gel. The positions of the 23S and 16S rRNAs and of the 4S RNA are marked by arrows. The positive signals due to the hybridization of a 32P-labeled 2.4-kbp DNA fragment probe spanning all of hutU and part of hutP (derived from ph180) are marked by arrowheads.
Interestingly, upon a temperature downshift of the bacterium from 22 to 4°C, a “cold-shock” response was noticed because the amount of mRNAs from the hut operon increased only about a maximum of two- to threefold after the shift (Fig. 4b). The amount of the mRNAs was at a maximum by 2 h after the shift and decreased subsequently. Thus, it appears that the hut operon might have cold-responsive regulatory elements that are known to occur in cold-inducible genes of mesophilic bacteria (12, 29).
Transcription start site and promoter region of the hutU gene of P. syringae.
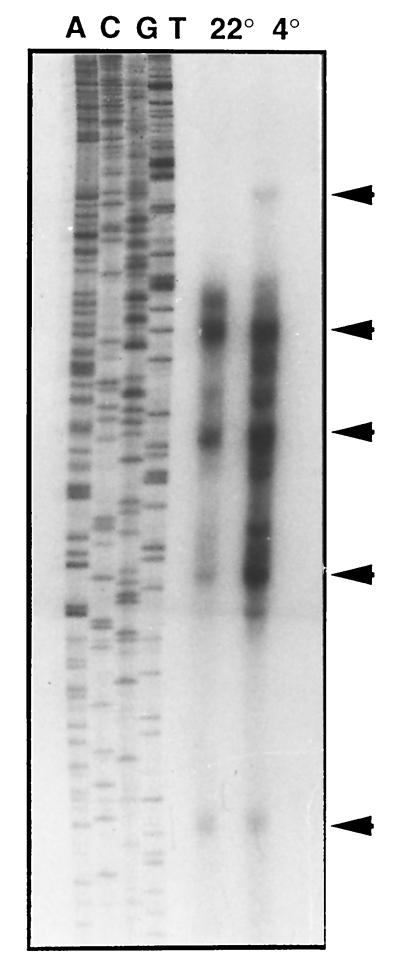
Primer extension analyses were carried out to locate the transcription start sites of the hutU gene, with the RNAs isolated from cells of P. syringae grown at low (4°C) and high (22°C) temperatures (Fig. 5). Among several primer-extended products, the longest product (topmost arrow in Fig. 5) was repeatedly observed only with the RNAs isolated from the cells grown at 4°C. Most of the other extended products were common in cells grown at both both low and high temperatures. Based on the longest primer-extended products, the transcription start sites for the low temperature (4°C) and the common site for both low and high temperatures (4 and 22° C) were located in the 5′ region of the hutU gene (Fig. A). The rest of the smaller extended products of the primer extension reactions (Fig. 5) could either represent the true transcription start sites or the processed or degraded ends of the mRNAs.
FIG. 5.
Primer extension analysis of hutU transcripts. The RNAs prepared from cells of P. syringae grown at 22 and 4°C were used for primer extension reactions in the presence of Moloney murine leukemia virus reverse transcriptase and [α-32P]ATP, as described in Materials and Methods. The products were analyzed on a sequencing gel (8 M urea-6% polyacrylamide gel electrophoresis) in parallel with the sequencing reaction products (i.e., A, C, G, and T) carried out with the same primer. The major primer-extended products are marked by arrowheads. Note that the two longest primer-extended products were considered for analyzing the promoters in the present study (see Fig. 6). The 4°C specific longest product was found repeatedly absent in the 22°C RNA specific reactions. The shortest product (the lowermost arrow) was compatible with a putative ς 54-specific promoter located in the upstream region of the hutU gene.
The low-temperature (4°C) specific transcript starts with a G, which is 219 nucleotides upstream of the putative translation initiation codon GTG of the HutU ORF. The common transcription start site for both low and high temperatures is located 39 nucleotides downstream from the former start site (Fig. 6A). Thus, the transcripts from the hut operon seem to have a long 5′-untranslated region (5′ -UTR) sequence that is a characteristic of the cold-shock genes in mesophilic bacteria, including E. coli (12). Upon further examination of the DNA sequences around the transcription start sites, it was observed that the −10 region has a characteristic CAAAA sequence at both temperatures. The −35 sequences in the promoter region of the 4°C specific and the 4 and 22°C common transcripts were TGTTAC and CCTGCG, respectively. Interestingly, the promoter region of the 4°C specific transcript had a second CAAAA sequence at the −15 position; the significance of this, if any, is not yet clear.
FIG. 6.
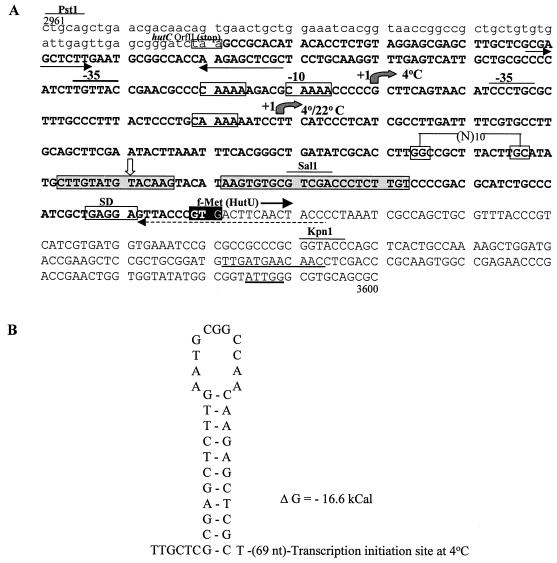
(A) DNA sequence from the upstream region of hutU of P. syringae containing the promoter elements and other putative regulatory sequences. The dashed arrow shown below the sequence at beginning of the hutU-specific ORF was used for mapping the transcription start sites. The temperature (4°C and 4 or 22°C) specific transcription start sites have been indicated by shaded arrows. The characteristic CAAAA sequence in the promoters is shown within the boxes. The putative ς 54-specific promoter sequence [GG-(N10)-GC] and the corresponding transcription start site (vertical open arrow) are marked on the sequence. The putative repressor HutC-binding sequence (CTTGTATGTACAAG) and CAP binding sequence (AAGTGTGCGTCGACCCTCTTGT) have been marked by shaded boxes. The translation initiation codon (GTG), the Shine-Dalgarno (SD) sequence, a putative cold box-like sequence, and a cold-shock protein binding sequence (ATTGG) downstream of the translational initiation site of hutU are marked. The sequence of the 3′ end of orfII of hutC are underlined and in lowercase. The nucleotide numbers refer to the number in the DNA sequence of the region from P. syringae (accession no. AF326719). (B) Putative regulatory sequence with the potential hairpin structure in the promoter region of hutU. The location of the hairpin is marked by two opposing arrows below the sequence in panel A.
The sequence at the upstream of the GTG translational start codon of the hutU gene of P. syringae contained a putative HutC repressor binding motif (CTTGTATGTACAAG), which is slightly different from the sequence observed in P. putida and Klebsiella aerogenes (2, 40). Interestingly, in the latter organisms this sequence overlaps with the −35 element of the promoter region, in contrast to the case in P. syringae, which contains the sequence within the 5′ -UTR of the gene (Fig. 6A). It was also observed that a putative nitrogen assimilatory cofactor-binding sequence [ATA-(N6)-TAT] is located overlapped with the HutC-binding motif (41). A putative catabolite activator protein (CAP) binding sequence was also noticed in the downstream of transcription initiation site. The sequence, AAGTGTGC(N6)CCTCTTGT, is located at the 35 nucleotides upstream of the GTG translation inititation codon of the hutU ORF. In K. aerogenes a similar sequence was observed centered around nucleotide −81.5 of the promoter region of the hutUH gene cluster (30). Since the CAP binding sequence occurs downstream of the transcription initiation site of P. syringae, the catabolite repressor in this case could act in theory as a transcriptional roadblock for RNA polymerase. It was also noticed that a putative conserved“cold-box”-like sequence (TTGATGAACAACC), which occurs at the 5′ end of the 5′-UTR of the cold-inducible genes of E. coli, is located 123 nucleotides downstream of the translation initiation codon of HutU in P. syringae (Fig. 6A). Since the location of the cold box is at variance with E. coli, any functional significance of the element in P. syringae remains to be determined.
A putative nitrogen regulatory ςN promoter element [GG-(N10)-GC] was also noticed 81 nucleotides upstream of the translation start site of the HutU ORF of P. syringae. This is interesting for the observation that the shortest primer-extended product (lowest arrow in Fig. 5) is located svery close to the expected position of the transcription start site (at the −13 position rather than at the expected −12 position of the initiation site) of the above ςN promoter (Fig. 6A). Since histidine is utilized as a source of both carbon and nitrogen in bacteria, the hut operon is likely to be regulated by both vegetative (ς70 family) and ς N-specific promoters (27), and therefore the occurrence of the ς N-promoter might be physiologically important.
Expression of hutU gene in E. coli.
Since the hutU gene of P. syringae contains some common features, including a long 5′-UTR of cold-inducible genes observed in E. coli, it was interesting to examine the temperature-dependent expression of the gene in the mesophilic bacterium. The expression of the hutU gene in E. coli was examined by transforming the bacterium with a multicopy plasmid (ph180) that contained a 2.6-kbp DNA fragment with the structural gene for urocanase and its upstream regulatory sequences from P. syringae. The cell extracts prepared from the E. coli grown at 37°C exhibited a modest activity of the enzyme (8.3 μmol min−1 mg of protein−1). However, the extracts prepared from the cells grown at a lower temperature (15°C) did not contain any detectable urocanase activity. Thus, it appears that the hutU gene of P. syringae is not expressed in E. coli at the lower growth temperature. Whether the lack of expression is due to the absence of cognate regulatory sequences and factors for transcription or to the inability of translation of the hutU mRNA at the lower temperature in E. coli remains unknown.
DISCUSSION
The present study was undertaken to determine and analyze the DNA sequences of the hutU gene and its promoter region in order to identify, if possible, the putative regulatory cis elements for the regulation of the hut operon in the psychrotrophic bacterium P. syringae. Additionally, it was thought that an analysis of the deduced amino acid sequence of the enzyme urocanase (the hutU-encoded product) from P. syringae, in comparison with the enzyme from mesophilic P. putida, might provide the clue to the nature of substitution of amino acids in a cold active enzyme. An earlier study had demonstrated that a urocanase from the psychrophilic P. putida A.3.12 could retain, at 0°C, 30% of its maximum activity seen at 30°C (21). The urocanase in P. putida is a homodimer that contains tightly bound NAD+ in each subunit, where an intact NAD+ is essential for its catalytic reaction. Mechanistically, the enzyme is unique for the fact that the NAD+ in this case does not function as a simple redox reagent but as an electrophile for the catalytic addition of water to the urocanate for its conversion into imidazolone propionic acid (7).
Analysis of the genetic organization and promoter region of the hutU in P. syringae.
The organization of the genes in the hut operon is known to be variable among gram-positive and gram-negative bacteria. The present study indicates that the organization of the hut operon is also variable within the Pseudomonas species (Fig. 1B). The analysis also indicates that the nature of the permease gene within the hut operon is variable. For example, Antarctic P. syringae has the hutP gene encoding permease belonging to transporter class (TC) 2.A.39.1 (family NCS1), which is different from hutT encoding a transporter of a different class (TC 2A.1.6.4, family MFS) observed in P. putida (38). The pathogenic P. aeruginosa has both hutP and hutT within the hut gene cluster. The published genetic study of P. putida had earlier failed to locate any permease or transporter gene in the operon (19, 20). A recent study of Sinorhizobium meliloti has demonstrated that the histidine-degrading hutH gene is linked to a histidine transporter of the ATP-binding cassette type (9). Thus, there seems to be a random recruitment of the histidine transporters during the microbial evolution of the histidine degradation pathway.
The regulation of the hut operon, which had mainly been studied in three gram-negative bacteria, including Salmonella sp., K. aerogenes, and P. putida, and one gram-positive bacterium (B. subtilis), had also been found to be variable (16, 26). For example, in B. subtilis histidine was found to be the main inducer of the operon, while in P. putida urocanase was found to be the inducer. Similarly, a positive activator, HutP, was observed to be the main regulator of the operon in B. subtilis, while the repressor HutC was found to be the main negative regulator of the operon in P. putida. The regulation of the operon is, however, complex in both systems for it is subjected to both carbon catabolite repression and nitrogen metabolite regulation in both of the organisms. The temperature-dependent expression of the operon in the cold-adapted bacterium P. syringae might further add to the complexity of the regulation of the operon.
In order to investigate the mechanism of temperature-dependent regulation of the operon, the promoter region of hutU, i.e., the first gene of the operon in P. syringae, has been identified here by primer extension analysis. It appears that the mRNA for the hutU gene has a long 5′-UTR that is a characteristic feature of many cold-inducible genes of mesophilic bacteria, including E. coli. It also appears that the promoter region of the hutU of P. syringae contains various putative cis-acting regulatory elements that have been characterized earlier in mesophilic bacteria. However, the locations of these elements are at variance with the known positions. For example, the repressor HutC-binding site, which is known to overlap either with the −35 sequence (e.g., in P. putida) or with the region between the −10 and −35 sequences (e.g., in K. aerogenes) of the hut promoters, has been found at the +143 base (with respect to the 4°C specific +1 site) and at the +95 base (with respect to the 4 or 22°C specific common +1 site) region of the 5′-UTR of the hutU gene of P. syringae (Fig. 6A). Similarly, a putative binding sequence [ATA-(N6)-TAT] for the positive activator NAC (for nitrogen assimilation control) protein (41) is located downstream of the transcription sites (data not shown). Interestingly, the HutC- and NAC binding sites mentioned above overlap each other in P. syringae, the significance of which is not yet clear. As pointed out above, the CAP binding sequence is also located downstream of the transcription initiation sites of the operon in the cold-adapted bacterium (Fig. 6A).
The identification of a unique sequence, CAAAA, at the −10 site of the hutU promoter of P. syringae is interesting. This sequence is probably important for the initiation of transcription at both low and high temperatures (e.g., 4 and 22°C). An extra CAAAA sequence that is observed a half turn away (5 bp) from the DNA helix in the region of the 4°C specific transcription start site might also be important for increased expression of the gene at lower temperatures. The occurrence of a putative ςN (ς54) specific promoter sequence, GG-(N10)-GC, and a corresponding primer-extended product of RNA suggests that the operon might be transcribed by RNA polymerase containing both vegetative sigma and nitrogen regulatory sigma factors, depending upon the environmental signals. The observation of five or six primer-extended products of RNA from the hutU gene might also be a reflection of the complex regulatory process of the operon.
It is also interesting that the putative cold-box sequence observed in the 5′-UTR of the cold-inducible genes of mesophilic E. coli is present in the coding region of hutU mRNA of P. syringae (Fig. 6). Whether this sequence has any role in P. syringae at lower temperatures is a matter of conjecture. Recently, it has been shown in E. coli that the Y-box sequence (5′-ATTGG-3′ or its inverted repeat 5′-CCAAT-3′) might be important for the regulation of cold-inducible genes by an antitermination mechanism (5). In the psychrotrophic Yersinia enterocolitica, the induction of the pnp gene encoding polynucleotide phosphorylase at low temperatures has also been shown to be regulated by the Y-box sequence located 230 bp upstream of the translation start site (18). The promoter region of the low-temperature-inducible gene icdII for isocitrate dehydrogenase in the psychrophilic bacterium Vibrio sp. strain AEB1 also contains a CCAAT sequence 2 bp upstream of the −35 site of the promoter that is essential for low-temperature induction (37). The −10 and −35 sequences of the promoter (TAACTA and TTATAG, respectively) in the bacterium were, however, not novel in any sense compared to other housekeeping genes. The analyses of the present study failed to show any Y-box sequence in the proper context of the promoter, although a 5′-ATTGG-3′ sequence is observed 185 bp downstream of the GTG initiation codon of HutU (Fig. 6A).
In order to identify any other putative regulatory elements in the promoter region of hutU, the potential secondary structural elements were also examined. One such structure with a dyad symmetry (ΔG = −16.6 kcal) is located 70 nucleotides upstream of the transcription start site. Such a hairpin loop structure can potentially function as a transcription stop signal for upstream hutC or as a regulatory element for transcription of the hutU gene.
Analysis of the urocanase sequence of P. syringae.
The deduced amino acid sequence of the urocanase from P. syringae, compared with the sequences of the enzyme homologues from other mesophilic bacteria, including K. aerogenes, A. rhizogenes, B. subtilis,S. coelicolor, and D. radiodurans, did not show any obvious preference for any specific substitution of amino acids in the protein due to the low-temperature adaptation. The active site of the enzyme in these bacteria (Fig. 2) has an almost identical sequence, FQGLPARICW, including the essential cysteine of the mesophilic P. putida (25). The two conserved cysteines (C-410 and C-354 of P. putida) are present in all of them. All of these enzymes have also a distinct signature motif (GXGX2GX10G) of the Rossmann sfold, including an equivalent acidic residue at the end of the β-strand of the α/β-fold (Fig. 2). However, multiple alignment of the amino acid sequence suggests that the urocanase has two distinct diverged branches that might be related to the phylogenetic origin of gram-negative and gram-positive bacteria (Fig. 3).
The cold sensitivity of many cold-labile enzymes, including NAD+-specific glutamate dehydrogenase, from various bacteria has been ascribed to the ready dissociation of the monomeric subunits at a lower temperature as a result of weakening hydrophobic bonds (22). Since the hydrophobic bonds stabilize the quaternary structures of proteins and since the enzymes from cold-adapted bacteria have in general reduced hydrophobic interactions to acquire flexibility for functioning at lower temperatures, it might be suicidal for cold active dimeric and oligomeric proteins to adopt a similar strategy. This could be one of the reasons why there are not many substitutions at the level of primary sequence in the dimeric enzyme urocanase from mesophilic P. putida and psychrotrophic P. syringae. In fact, a recent study shows that the hydrophobic character of the homodimeric enzyme malate dehydrogenase of the psychrophilic bacterium Aquaspirillium arcticum is similar to that of the enzyme from the thermophilic bacterium Thermus flavus (24). However, three major differences were noticed in the psychrophilic enzyme that were implicated in the efficient catalysis at lower temperature. The differences include (i) an increased relative flexibility at the active-site region of the enzyme; (ii) favorable surface charge distribution, such as more positive potential around the negatively charged substrate (oxaloacetate) binding site and decreased negative potential around the cofactor NADH binding site; and (iii) reduced intersubunit ion pairs and decreased buried surface area in the dimeric interface of the enzyme (24). A similar structure-function study on the urocanase of P. syringae would be useful to substantiate the generality of these findings.
In conclusion, the present study shows that the −10 and −35 characteristics of the promoter of the hutU gene that is upregulated at low temperatures in P. syringae are unique. The occurrence of multiple A’s in CAAAA might be important for low-temperature melting of the promoter. The present study also reflects the possible complexity and uniqueness of regulation in the operon of P. syringae due to the presence of many putative regulatory cis elements that are located downstream rather than in the usual location upstream of the transcription start site. The present study also identifies two distinct clusters of urocanase sequences among the bacteria that might be related to their origin or lineage. The identification of a Rossmann fold for NAD+ binding is also important for future modeling of the enzyme since this site is presumed to be different from other NAD-requiring enzymes for its inaccessibility without denaturation of the urocanase.
Acknowledgments
A part of this research was funded by a grant to M.K.R. from the Indian Department of Science and Technology (DST). K.L.J. was a CSIR postdoctoral fellow during this work.
We thank N. Nagesh for DNA sequencing, M. K. Basu and Pratik Jagtap for computer analyses, and K. Regha for the DNA sequence of the hutC region of P. syringae.
REFERENCES
- 1.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for the functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905–11910. [PubMed] [Google Scholar]
- 2.Allison, S. L., and A. T. Phillips. 1990. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Pseudomonas putida. J. Bacteriol. 172:5470–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Jhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae, W., B. Xia, M. Inouye, and K. Severinov. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. USA 97:7784–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, P. J., K. L. Britton, P. C. Engel, G. W. Farrants, K. S. Lilley, D. W. Rice, and T. J. Stillman. 1992. Subunit assembly and active site location in the structure of glutamate dehydrogenase. Proteins 12:75–86. [DOI] [PubMed] [Google Scholar]
- 7.Baker, P. J., K. L. Britton, D. W. Rice, A. Rob, and T. J. Stillman. 1992. Structural consequences of sequence patterns in the finger-print region of the nucleotide binding fold. Implications for nucleotide specificity. J. Mol. Biol. 228:662–671. [DOI] [PubMed] [Google Scholar]
- 8.Balas, D., E. Fernandez-Moreira, and A. de La Campa. 1998. Molecular characterization of the gene encoding the DNA gyrase A subunit of Streptococcus pneumoniae. J. Bacteriol. 180:2854–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boncompagni, E., L. Dupont, T. Mignot, M.Østeräs, A. Lambert, M.-C. Poggi, and D. L. Rudulier. 2000. Characterization of a Sinorhizobium meliloti ATP-binding cassette histidine transporter also involved in betaine and proline uptake. J. Bacteriol. 182:3717–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- 11.Di Fraia, R., V. Wilquet, M. A. Ciardiello, V. Carratore, A. Antignani, L. Camardella, N. Glansdorff, and G. Di Prisco. 2000. NADP+-dependent glutamate dehydrogenase in the Antarctic psychrotrophic bacterium Psychrobacter sp. TAD1: characterization, protein and DNA sequence, and relationship to other glutamate dehydrogenases. Eur. J. Biochem. 267:121–131. [DOI] [PubMed] [Google Scholar]
- 12.Fang, L., Y. Hou, and M. Inouye. 1998. Role of cold-box region in the 5′ untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J. Bacteriol. 180:90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fessenmaier, M., R. Frank, J. Retey, and C. Schubert. 1991. Cloning and sequencing the urocanase gene (hutU) from Pseudomonas putida. FEBS Lett. 286:55–57. [DOI] [PubMed] [Google Scholar]
- 14.George, D. J., and A. T. Phillips. 1970. Identification of α-ketobutyrate as the prosthetic group of urocanase from Pseudomonas putida. J. Biol. Chem. 245:528–537. [PubMed] [Google Scholar]
- 15.Gerday, C., M. Aittaleb, M. Bentahir, J. Chessa, P. Claverie, T. Collins, S. D’Amico, J. Dumont, G. Garsoux, D. Georlette, A. Hoyoux, T. Lonhienne, M. Meuwis, and G. Feller. 2000. Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol. 18:103–107. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg, R. B., and B. Magasanik. 1985. Gene order of the histidine utilization (hut) operons in Klebsiella aerogenes. J. Bacteriol. 122:1025–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gounot, A. M. 1991. Bacterial life at low temperature: physiological aspects and biotechnological implications. J. Appl. Bacteriol. 71:386–397. [DOI] [PubMed] [Google Scholar]
- 18.Goverde, R. L., J. H. Veld, J. G. Kusters, and F. R. Mooi. 1998. The psychrotrophic bacterium Yersinia enterocolitica requires expression of pnp, the gene for polynucleotide phosphorylase, for growth at low temperature (5°C). Mol. Microbiol. 28:555–569. [DOI] [PubMed] [Google Scholar]
- 19.Hu, L., and A. T. Phillips. 1988. Organization and multiple regulation of histidine utilization genes in Pseudomonas putida. J. Bacteriol. 170:4272–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, L., S. Allison, and A. T. Phillips. 1989. Identification of multiple repressor recognition sites in the hut system of Pseudomonas putida. J. Bacteriol. 171:4189–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hug, D. H., and J. K. Hunter. 1974. Effect of temperature on urocanase from a psychrophile, Pseudomonas putida. Biochemistry 13:1427–1431. [DOI] [PubMed] [Google Scholar]
- 22.Jahns, T., and H. Kaltwasser. 1993. Properties of the cold-labile NAD+-specific glutamate dehydrogenase from Bacillus cereus DSM31. J. Gen. Microbiol. 139:775–780. [DOI] [PubMed] [Google Scholar]
- 23.Kannan, K., K. L. Janiyani, S. Shivaji, and M. K. Ray. 1998. Histidine utilization operon (hut) is upregulated at low temperature in the Antarctic psychrotrophic bacterium Pseudomonas syringae. FEMS Microbiol. Lett. 161:7–14. [DOI] [PubMed] [Google Scholar]
- 24.Kim, S., K. Y. Hwang, S. H. Kim, H. C. Sung, Y. S. Han, and Y. Cho. 1999. Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum. J. Biol. Chem. 274:11761–11767. [DOI] [PubMed] [Google Scholar]
- 25.Lenz, M., and J. Retey. 1993. Cloning, expression and mutational analysis of the urocanase gene (hutU) from Pseudomonas putida. Eur. J. Biochem. 217:429–434. [DOI] [PubMed] [Google Scholar]
- 26.Magasanik, B. 1978. Regulation in the hut system, p.373–387. In J. Miller and W. Reznikoff (ed.), The operon. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Merrick, M. J. 1993. In a class of its own: the RNA polymerase sigma factor ς54 (ς N). Mol. Microbiol. 10:903–909. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Mitta, M., L. Fang, and M. Inouye. 1997. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol. Microbiol. 26:321–335. [DOI] [PubMed] [Google Scholar]
- 30.Osuna, R., B. K. Janes, and R. A. Bender. 1994. Roles of catabolite activator protein sites centered at −81.5 and −41.5 in the activation of the Klebsiella aerogenes histidine utilization operon hutUH. J. Bacteriol. 176:5513–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray, M. K., G. Seshu Kumar, K. Janiyani, K. Kannan, P. Jagtap, M. K. Basu, and S. Shivaji. 1998. Adaptation to low temperature and regulation of gene expression in Antarctic psychrotrophic bacteria. J. Biosci. 23:423–435. [Google Scholar]
- 32.Ray, M. K., T. Sitaramamma, S. Ghandhi, and S. Shivaji. 1994. Occurrence and expression of cspA, a cold shock gene, in Antarctic psychrotrophic bacteria. FEMS Microbiol. Lett. S 116:55–60. [DOI] [PubMed] [Google Scholar]
- 33.Retey, J. 1994. The urocanase story: a novel role of NAD+ as electrophile. Arch. Biochem. Biophys. 314:1–16. [DOI] [PubMed] [Google Scholar]
- 34.Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structures. Proteins 19:55–72. [DOI] [PubMed] [Google Scholar]
- 35.Rost, B., C. Sander, and R. Schneider. 1994. PHD: an automatic mail server for protein secondary structure prediction. Comput. Appl. Biol. Sci. 10:53–60. [DOI] [PubMed] [Google Scholar]
- 36.Russel, N. J. 1990. Cold adaptation of microorganisms. Philos. Trans. R. Soc. London SB326:595–611. [DOI] [PubMed] [Google Scholar]
- 37.Sahara, T., M. Suzuki, J. Tsuruha, Y. Takada, and N. Fukunaga. 1999. cis-Acting elements responsible for low-temperature-inducible expression of the gene coding for the thermolabile isocitrate dehydrogenase isozyme of a psychrophilic bacterium, Vibrio sp. strain ABE-1. J. Bacteriol. 181:2602–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Schwacha, A., and R. A. Bender. 1990. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Klebsiella aerogenes. J. Bacteriol. 172:5477–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwacha, A., and R. A. Bender. 1993. The product of the Klebsiella aerogenes nac (nitrogen assimilation control) gene is sufficient for activation of the hut operons and repression of the gdh operon. J. Bacteriol. 175:2116–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shivaji, S., N. S. Rao, L. Saisree, V. Sheth, G. S. N. Reddy, and P. M. Bhargava. 1989. Isolation and identification of Pseudomonas spp. from Schirmacher Oasis, Antarctica. Appl. Environ. Microbiol. 55:767–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uma, S., R. S. Jadhav, G. Seshu Kuamr, S. Shivaji, and M. K. Ray. 1999. A RNA polymerase with transcriptional activity at 0°C from the Antarctic bacterium Pseudomonas syringae. FEBS Lett. 453:313–317. [DOI] [PubMed] [Google Scholar]