Abstract
A coastal marine sulfide-oxidizing autotrophic bacterium produces hydrophilic filamentous sulfur as a novel metabolic end product. Phylogenetic analysis placed the organism in the genus Arcobacter in the epsilon subdivision of the Proteobacteria. This motile vibrioid organism can be considered difficult to grow, preferring to grow under microaerophilic conditions in flowing systems in which a sulfide-oxygen gradient has been established. Purified cell cultures were maintained by using this approach. Essentially all 4′,6-diamidino-2-phenylindole dihydrochloride-stained cells in a flowing reactor system hybridized with Arcobacter-specific probes as well as with a probe specific for the sequence obtained from reactor-grown cells. The proposed provisional name for the coastal isolate is “Candidatus Arcobacter sulfidicus.” For cells cultured in a flowing reactor system, the sulfide optimum was higher than and the CO2 fixation activity was as high as or higher than those reported for other sulfur oxidizers, such as Thiomicrospira spp. Cells associated with filamentous sulfur material demonstrated nitrogen fixation capability. No ribulose 1,5-bisphosphate carboxylase/oxygenase could be detected on the basis of radioisotopic activity or by Western blotting techniques, suggesting an alternative pathway of CO2 fixation. The process of microbial filamentous sulfur formation has been documented in a number of marine environments where both sulfide and oxygen are available. Filamentous sulfur formation by “Candidatus Arcobacter sulfidicus” or similar strains may be an ecologically important process, contributing significantly to primary production in such environments.
In the marine environment, hydrogen sulfide is a ubiquitous end product of anaerobic processes of organic matter remineralization (5, 23, 29, 30). At ridge crest sites on the ocean floor, it is produced from the geothermal transformation of sulfate and elemental sulfur leaching via seawater-basaltic rock interaction (26, 40, 62). When brought into contact with the aerobic biosphere, hydrogen sulfide becomes an energy-yielding substrate for chemosynthetic colorless sulfur-oxidizing bacteria. Members of this group include free-living rods or ovoids of the genera Thiobacillus, Thiomonas, Acidiphilium, Thiomicrospira, and Thiovulum (31, 32, 34, 42) as well as the morphologically conspicuous gliding and nongliding filamentous forms of the genera Beggiatoa, Thioploca, and Thiothrix (19, 47, 48, 69). These organisms are characterized by their ability to catalyze the oxidation of sulfide and its chemically and biologically mediated partial oxidation products (polysulfides, Sn2−; elemental sulfur, S0; sulfane monosulfonic acids, HSSnO32−]; and polythionates, SnO62−) (14, 33, 66) coupled to the fixation of carbon dioxide to organic carbon by utilizing the same Calvin-Bassham-Benson cycle enzymes as those used by oxygenic phototrophs.
Because of the differential rates of oxidation of hydrogen sulfide and derived oxidation products, intermediates may substantially accumulate in the environment, most notably, elemental sulfur. Many sulfur oxidizers excrete internal or external elemental sulfur in the form of hydrophilic spherical globules (66) that can serve as an energy reserve should environmental concentrations of hydrogen sulfide substantially decrease (46).
A motile strain of a sulfide-oxidizing bacterium from coastal seawater was described recently as producing a novel metabolic end product in a laboratory culture, namely, rigid irregular sulfur filaments with dimensions of 0.5 to 2.0 μm by 20 to 500 μm (70). Filamentous sulfur was rapidly excreted (∼3 μm min−1) by the vibrioid organism, and the filament became thickened through sulfur deposition by radially attached members of the population. Examination of material collected from diffuse-flow deep-sea hydrothermal vents at 9° N (22) revealed that it was composed largely of filamentous sulfur with a morphology nearly identical to that produced in a laboratory culture system by the coastal strain (70). The rapid and voluminous in situ formation of filamentous sulfur mats at an actively flowing warm water vent, as well as in shipboard reactors inoculated with material from vent collections, was also described recently (71), indicating that this process may occur ubiquitously in many flowing sulfidic environments. In both laboratory and shipboard cultures, vibrioid cells were actively poised within the interface between counteropposing gradients of oxygen and hydrogen sulfide; filament production and presumably active growth are predominantly restricted to these interface regions (70, 71). These vibrioid organisms have adapted to life in high-flow sulfidic environments by the deposition of a product of their energy metabolism, sulfur, into a surface-attached, interlocking mycelial structure that resists turbulence. In this study, we describe the phylogenetic and physiological properties of the coastal vibrioid marine organism first found to produce filamentous sulfur.
MATERIALS AND METHODS
Culture techniques.
Long-term enrichments of the coastal filamentous-sulfur-producing microbe were maintained in continuous-flow reactors containing aerobic, filtered (pore size, 1 μm) coastal seawater pumped at a dilution rate of 0.8 to 1.0 h−1 and enriched with H2S at 400 to 1,200 μM (70, 71). Cells were precleaned and obtained free of sulfur filaments by rapid mixing and then by filtration with 8-μm-pore-size polycarbonate (Poretics) or 10-μm-pore-size nylon (MSI) membranes. A variety of approaches were taken to get cells to grow as individual filament-forming colonies on solid media. Flocculent material obtained directly from the reactors or from precleaned cells was streaked onto natural seawater agar (1.5% Noble agar) plates containing 10 mM Na2S2O3 and onto natural seawater agar plates containing 10 mM neutralized Na2S that were overlaid by a layer of plain natural seawater agar. Plates were streaked with the bacterial suspension and incubated under microaerophilic conditions with an enriched CO2 atmosphere created by using a BBL Campy Pouch system and in Torbal jars with a 5% air-95% N2 atmosphere. Plates were examined for growth over a period of 1 to 4 days.
This organism prefers or requires a liquid phase for filament production to occur. Therefore, autoclaved natural seawater was pumped through a sterile continuous-flow reactor, which was inoculated by filtering an aliquot of precleaned and serially diluted (10−5) cells onto a sterile GF/F glass fiber filter by using a sterile filter unit. This filter was aseptically attached to a glass rod inside the seawater reactor after an appropriate sulfide concentration (approximately 1 mM) had been achieved. Samples were withdrawn over the course of 1 week and examined microscopically. Contamination was assessed by plating on both heterotrophic media (Difco 2216 marine agar and 2216 liquid medium) and autotrophic medium (artificial seawater with 10 mM Na2S2O3) (15).
Heterotrophic growth was tested aerobically on 2216 marine agar and anaerobically on acetate medium with fumarate or nitrate as an alternate electron acceptor by following the procedures of Teske et al. (72).
Microscopy.
Phase-contrast microscopy was used for general observation of cell motility and morphology. Cell number was determined by enumeration of acridine orange-stained cells by epifluorescence microscopy (24). Cell flagellation was determined by observation of negatively stained cells (0.5% uranyl acetate) with a Philips 300 transmission electron microscope. Thin sections were prepared from glutaraldehyde-fixed cells (27) that had been stained with lead citrate and uranyl acetate and poststained with 1% osmium tetroxide and were observed with a Zeiss 10 transmission electron microscope (Zeiss, Oberkochen, Germany).
DNA extraction and 16S rRNA sequencing.
The small-subunit rRNA sequence was determined from reactor-grown cells that were separated from sulfur by filtration and subsequent centrifugation. A determination of cell density was made and the percent contamination in the final cell suspension was estimated by use of colony counts and dilution series on both heterotrophic and autotrophic media as indicated above. The monoculture was shown to contain only ∼0.01% culturable heterotrophic or autotrophic contaminants. Nucleic acids were extracted as described by DeLong (12). Briefly, frozen cell preparations were lysed by resuspension in lysis buffer (50 mM Tris-HCl [pH 8.0], 0.75 M sucrose, 40 mM EDTA, 1 mg of lysozyme ml−1) and incubated for 30 min at 37°C. Subsequently, proteinase K and sodium dodecyl sulfate (SDS) were added to final concentrations of 0.5 mg ml−1 and 1%, respectively, and the lysate was incubated at 55°C for 30 min. Nucleic acids were extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and then with chloroform-isoamyl alcohol (24:1) and were washed several times with sterile deionized water in a microconcentrator (Centricon 100; Amicon).
A 1.5-kb region of the 16S rRNA gene was directly amplified from the purified nucleic acids in a 30-cycle PCR at an annealing temperature of 40°C with primers broadly specific for the Bacteria rRNA gene (8F, 5′-AGAGTTTGATCMTGGC-3′, and 1492R, 5′-TACCTTGTTACGACTT-3′) (8). PCR products were purified by using a QIAquick Spin PCR purification kit (Qiagen, Inc., Chatsworth, Calif.) as described by the manufacturer. A Taq Dyedeoxy Terminator cycle sequencing kit (Applied Biosytems, Foster City, Calif.) was used to directly sequence the PCR products, according to the protocol provided by the manufacturer, with sequencing primers described previously (8) and an Applied Biosystems 373S DNA sequencer.
Phylogenetic analysis.
The 16S rRNA gene sequence of the sulfide-oxidizing organism was checked by comparing it to the predicted bacterial secondary structure and by using the CHECK_CHIMERA program available from the Ribosomal Database Project (38). The sequence and related sequences, as determined by a BLAST search (2), were loaded into the 16S rRNA sequence database of the Technical University of Munich (Munich, Germany) by using the ARB program package (http://www.mikro.biologie.tu-muenchen.de). The sequences were aligned automatically by using ARB_ALIGN and then were checked and corrected manually, considering the secondary structure of the rRNA molecule. The sequences of the sulfide-oxidizing bacterium and representatives of several genera from five subdivisions of the Proteobacteria were then imported into PAUP, version 4.0b8 (Sinauer Associates, Sunderland, Mass.), for phylogenetic analysis. Only positions that were conserved in 50% of the 16S rRNAs of the ɛ-Proteobacteria were considered for the analysis.
Models for use in minimum-evolution and maximum-likelihood searches were chosen by using the likelihood ratio test in the Modeltest program, version 3.04 (51). The chosen substitution model corresponded to the GTR+I+G model (six classes of substitutions and unequal base frequencies [general time reversible {GTR}]), the proportion of sites invariant [I], and the evolutionary rate of the remaining positions of sites varying according to a gamma distribution [G]). The specific model parameters are available upon request. 16S rRNA trees were inferred by using distance analysis with minimum evolution as the optimum criterion and the distance measure set to maximum likelihood, parsimony analysis, and maximum-likelihood analysis. Heuristic searches under the minimum-evolution and maximum-parsimony criteria were performed by using 1,000 random-addition replicates with Tree-bisection-reconnection (TBR) branch swapping. Bootstrapping of minimum-evolution and maximum-parsimony analyses was done with 1,000 bootstrap replicates of 10 random-addition replicates each. Heuristic searches under the maximum-likelihood criterion were performed by using five random-addition replicates with TBR branch swapping. The phylogenetic trees were edited with the TREEVIEW for Macintosh program, version 1.6.5 (50).
Oligonucleotide probes.
The rRNA-targeted oligonucleotides used (probe nomenclature according to Alm et al. [1]; given in parentheses) were as follows: ARC94 (S-G-Arc-0094-a-A-18) and ARC1430 (S-G-1430-Arc-a-A-18) (65), which target the genus Arcobacter; EUB338 (S-D-Bact-0338-a-A-18) (3), which is specific for Bacteria; and CARCS219 (S-S-Carcs-0219-a-A-18) (5′-ATAGGCCGATCGAAGAGC-3′), which targets the sequence of the sulfide oxidizer (to be designated “Candidatus Arcobacter sulfidicus”). In addition, a nonsense probe complementary to EUB338 (NON338) was used to account for background fluorescence due to nonspecific binding. Probe CARCS219 was designed by using the tool PROBE_DESIGN as implemented in the ARB software package. The probe specificity was evaluated with the PROBE_MATCH tool of the ARB package against the rRNA database of the Technical University of Munich. The probe was further checked against sequences in the Ribosomal Database Project by using the CHECK_PROBE program and against GenBank sequences by using the BLAST algorithm. Probe CARCS219 had three or more mismatches with all sequences in the different databases. Arcobacter strain KT0913, isolated from North Sea bacterioplankton (16), served as a negative control for probe CARCS219 and as a positive control for probes ARC94 and ARC1430. Oligonucleotides were synthesized and fluorescently labeled with 5,5′-disulfo-1,1-(γ-carbopentynyl)-3,3,3′,3′-tetramethylindolocarbocyanin-N-hydroxy-succinimide ester (Cy-3) or with 5 (6)-carboxyfluorescein-N-hydroxysuccinimide ester (Fluos) at the 5′ end at ThermoHybaid GmbH, Interactiva Division (Ulm, Germany).
In situ hybridization.
Samples were taken from the reactor and fixed for 3 h with paraformaldehyde as previously described (3). The samples were stored in a 1:1 mixture of phosphate-buffered saline and 96% ethanol at −20°C. The fixed samples were applied to a well of gelatin-coated glass slides (3), air dried, and sequentially dehydrated in 50, 80, and 96% ethanol for 3 min each. For hybridization, the fluorescently labeled probe was mixed 1:10 with hybridization buffer (0.9 M NaCl, 20% formamide, 20 mM Tris-HCl [pH 8], 0.01% SDS), and 10 μl of the mixture was added to a well. The final probe concentration was 5 ng μl−1. Hybridization was performed at 46°C for 90 min by using a chamber equilibrated with hybridization buffer. Hybridization was followed by a stringent washing step for 15 min with prewarmed washing buffer (20 mM Tris-HCl, 225 mM NaCl, 0.01% SDS) at 48°C. Washing buffer was removed by rinsing the slides with distilled water. The slides were air dried, stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; 1 μg ml−1) for 5 min in the dark, and finally rinsed again with distilled water. The slides were mounted in Vectashield (Vector Laboratories Inc., Burlingame, Calif.) to avoid bleaching and were examined with an Axiophot microscope (Zeiss) with filter sets specific for DAPI, Fluos, and Cy-3. Pictures were taken with an Axiocam video camera (Zeiss) and analyzed with Axiovision software.
Carbon dioxide fixation experiments.
Cells were tested for their ability to incorporate CO2 under batch and continuous-flow culture conditions. Samples were removed from the enrichment reactor and tested in batch cultures as (i) bulk sulfur filaments with associated cells, (ii) cells disassociated from sulfur filaments by mixing but unfiltered, or (iii) precleaned cells obtained by filtration. In certain instances, H2S was removed by preaeration and known quantities of H2S were added prior to incubation of samples. The level of added isotope (NaH14CO3) was in the range of 0.043 to 0.062 μCi of NaHCO3 μmol−1 (i.e., 0.095 to 0.136 μCi ml−1). To determine the level of incorporated carbon, aliquots were filtered at various time points and assayed by previously described procedures (73). The CO2 fixation data were subjected to a nonlinear fit to the following equation:
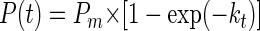 |
In this equation, P(t) is the fixation observed at time t, Pm is the asymptote, and k is the exponential rate constant. See the figures for the best-fit curves through the data. By taking advantage of the fact that as t approaches 0, the exponential function P(t) approaches the linear function Pmkt, initial rates were computed as the slope, Pmk, by using the best-fit values for Pm and k.
Incorporation of CO2 by the organism, under flowing conditions versus batch conditions, was conducted by use of a presterilized continuous-flow reactor fed from a carboy of sterile seawater containing NaH14CO3 (0.047 μCi μmol−1; 0.104 μCi ml−1). A precleaned and diluted cell suspension was filtered onto a sterile GF/F filter (see “Culture Techniques” above) attached to a sterile glass rod and was placed in the H2S-enriched seawater-containing reactor. The reactor received an initial level of isotope to make the culture equivalent in specific activity to the inflowing seawater. This culture was allowed to grow for 48 h prior to sampling for carbon fixation levels as described above. Filamentous sulfur and associated cells were harvested after 48 h of growth on the GF/F filter in the flowing-seawater reactor and assayed for carbon fixation levels. The sulfide concentration in the reactor ranged from 1,090 to 1,370 μM over the course of incubation.
Nitrogen fixation assays.
For determination of nitrogen fixation activity, filamentous material was used directly from the continuous-growth reactor and assayed for ethylene production by the acetylene reduction technique (converted to relative units of nitrogen) (13, 59, 60).
Rubisco assays.
Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) activity in cell extracts was measured at 30°C by the procedure of Beudeker et al. (6) as modified by Nelson and Jannasch (47). An extract from spinach was used as the positive control. Protein was determined for the cell extracts by the Coomassie brilliant blue dye binding technique (7) with a Bio-Rad protein assay kit. Ovalbumin served as the standard.
Reactor-grown cells were filtered free of sulfur and screened for the expression of form I (large-subunit) or form II Rubisco by Western blotting. The cell pellets were homogenized and boiled for 5 min in SDS-polyacrylamide gel electrophoresis sample buffer by published protocols (4). Approximately 20 to 25 μg of total protein, as estimated by the Bradford method (7), was loaded per sample and separated on a 10% acrylamide gel. Proteins were transferred from the gel and immobilized on a polyvinylidene difluoride membrane (Millipore) by electroblotting. Blots were incubated with antisera against large-subunit form I Rubisco from spinach and with form II-specific antisera raised against Rhodospirillum rubrum Rubisco (55). Blots were developed by using an alkaline phosphatase-conjugated 2° antibody system according to the manufacturer’s instructions (Bio-Rad). The assay was repeated five times with three different cell pellets and increasing concentrations of total protein.
Chemical analyses.
Because the produced sulfur filaments were hydrophilic, they were analyzed for the possible presence of a coating of protein or carbohydrate. Reactor-grown filamentous material was harvested, stirred, and allowed to settle. Samples of this material were taken for chemical assays, dry-weight determinations, and direct cell counts. The following chemical assays were performed. Carbohydrate was determined by the anthrone reaction (76) with glucose and starch as standards. Protein was determined for undigested samples and for samples digested in 0.1 and 1.0 N NaOH for 3 h at 60°C by using a Bio-Rad protein assay kit. Aliquots for dry-weight determinations were filtered onto preweighed 0.45-μm-pore-size filters (Millipore) and dried to a constant weight at 50°C. These weights were corrected for retained seawater salts by using control filters. Other aliquots of the filaments were appropriately diluted in sterile seawater, and acridine orange direct counts were obtained to determine cell density. Triplicate subsamples were used for all assays, weight determinations, and cell counts.
The filaments were examined microscopically for the presence of any visible coating. First, filaments were monitored microscopically for any residual material as the sulfur was slowly dissolved by wicking 95% ethanol through the field of view. Second, a microscopic examination for a protein coating was made with fluorescein isothiocyanate by the procedure of Crissman and Steinkamp (10) as modified by Olson et al. (49).
The particulate organic carbon (POC) content of reactor-grown filamentous material was determined by using a Perkin-Elmer 2400 C/N analyzer. The sulfur content of samples washed with distilled water and air dried was determined by using the benzene method (28, 48). Elemental sulfur (sublimed powder; Mallinckrodt) served as the standard. Sulfide in the reactor seawater or in radioactivity experiments was determined by the method of Cline (9).
Nucleotide sequence accession number.
The 16S rRNA sequence of “Candidatus Arcobacter sulfidicus” has been deposited in GenBank under accession number AY035822.
RESULTS
Culturing.
The only method that uses solid medium and that supported the growth and formation of filament-producing colonies was that of sulfide-containing agar plates possessing a thin film of surface moisture. These were streaked with reactor material or with precleaned and serially diluted cells and incubated under microaerophilic conditions in the BBL Campy Pouch system. Diffuse white colonies formed on the aqueous surface film of these plates and increased in size over a 48-h period. When the Campy Pouches were opened and examined, the colonies were seen to contain actively motile vibrios and accompanying sulfur filaments. Colony formation did not occur on similarly prepared sulfide-containing agar plates incubated in a Torbal jar under 5% air-95% N2 and not enriched with CO2. The organism did not grow on thiosulfate-containing agar plates under any conditions.
Growth of purified cells was achieved with a presterilized seawater reactor which was maintained with sterile H2S-enriched seawater at flow rates and sulfide concentrations similar to those used in the enrichment reactor. Cells inoculated from serial dilutions of freshly filtered reactor material became established in this system and continued to grow over a test period of 1 week or longer. The woolly appearance of the material was somewhat reduced under these conditions, but the cells were actively motile and free of contaminants, as determined by plating on both heterotrophic (Difco 2216) and autotrophic(10 mM Na2S2O3) media and by microscopic morphological examination (only motile vibrioid cells were seen). This purified culture system was used for certain radioisotopic growth experiments.
No growth of precleaned cells from sulfur filaments occurred heterotrophically on plates of 2216 marine agar incubated aerobically or in agar shake tubes containing acetate and incubated anaerobically with nitrate or fumarate as an alternate electron acceptor.
Microscopy.
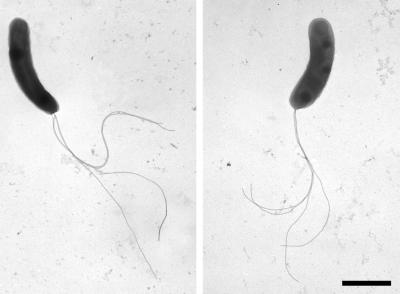
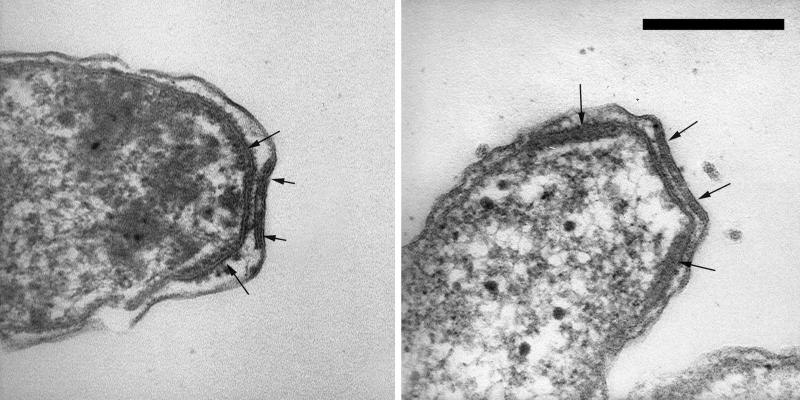
The cells responsible for the production and excretion of the filamentous sulfur were gram-negative, highly motile vibrios (approximately 0.5 by 2.0 μm) with four polar flagella (Fig. 1). The organism possessed a multilaminar polar organelle (Fig. 2) morphologically similar to that previously described for members of the genus Campylobacter (formally Vibrio fetus) (54). In microscopic examination of freshly sampled sulfur filaments, vibrioid cell motility was the only morphology observed. The organisms possessed the ability to attach and detach from solid surfaces (e.g., glass slide or existing sulfur threads) as conditions of their preferred oxygen-sulfide gradient changed.
FIG. 1.
Transmission electron micrographs of negatively stained cells of the filamentous-sulfur-producing microbe “Candidatus Arcobacter sulfidicus” showing four polar flagella. Bar, 1 μm.
FIG. 2.
Thin-section transmission electron micrographs showing the multilaminar polar membrane structure (arrows) found in the filamentous-sulfur-producing microbe “Candidatus Arcobacter sulfidicus.” Bar, 0.2 μm.
16S rRNA sequencing and phylogenetic analysis.
PCR amplification performed with the Bacteria 16S rRNA primers produced a single band of the expected size (about 1,500 bp) on an agarose gel. Direct sequencing of this PCR product yielded a single unambiguous sequence. The 16S rRNA sequence obtained here did not show any chimeric artifacts, as established by the CHECK_CHIMERA program from the Ribosomal Database Project. To eliminate misincorporation errors inherited during direct sequencing of PCR products, secondary structure models (75) were used to examine the nature and position of the sequence variation. The secondary structure of this sequence showed that all of the substitutions, relative to the Escherichia coli sequence, were restricted to highly variable regions of the 16S rRNA sequence, were also largely compensated for by the corresponding base substitutions in the complementary stem region, and therefore did not disturb the highly conserved secondary structure. All signature nucleotides distinguishing the delta subdivision of the Proteobacteria from the epsilon subdivision of the Proteobacteria (21) were also present in the sulfide-oxidizing strain sequence.
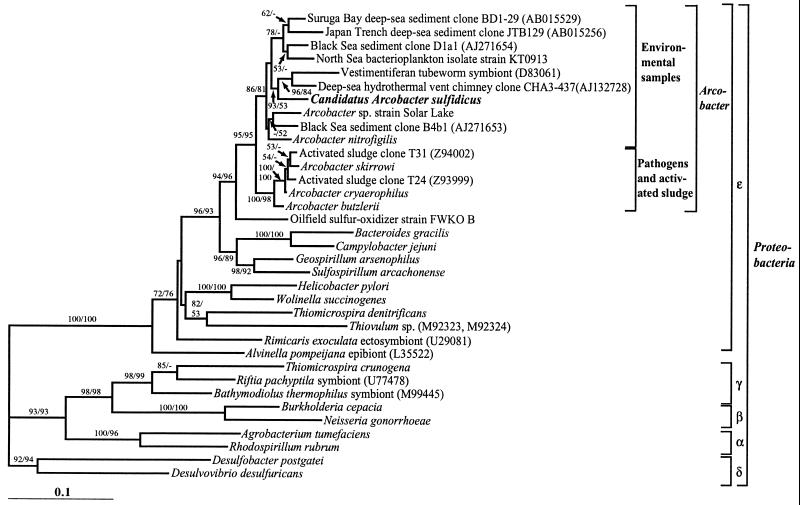
Tree construction analyses performed with distance, parsimony, and maximum-likelihood methods unambiguously placed the filamentous-sulfur-producing strain sequence in the genus Arcobacter in the epsilon subdivision of the Proteobacteria (Fig. 3). The 16S rDNA sequence of the strain exhibited 93 to 94% identity with those of all other known Arcobacter spp., with the uncultured deep-sea eubacterium CHA3-437 (GenBank AJ132728) sequence being its closest relative. The phylogenetic analysis revealed further that the genus Arcobacter can be divided into two main clusters, i.e., a cluster containing the pathogens and sequences from uncultured activated-sludge microbes and an environmental cluster exclusively containing sequences of cultured and uncultured organisms of marine or saline origins, including the filamentous-sulfur-producing strain.
FIG. 3.
16S rRNA-based phylogenetic tree showing the relationship of the filamentous-sulfur-producing microbe “Candidatus Arcobacter sulfidicus” to selected cultured and environmental Proteobacteria sequences. The tree was constructed by using distance analysis in PAUP*, version 4.0b8, with minimum evolution as the optimal criterion and the distance measure set to maximum likelihood. Trees constructed with other reconstruction algorithms (parsimony and maximum likelihood) resulted in the same overall topology. The numbers at the nodes are bootstrap confidence values expressed as percentages of 1,000 bootstrap replications. Bootstrap values from both distance (first number) and parsimony (second number) analyses are depicted. Bootstrap values of less than 50% are not shown. The scale bar represents 0.1 estimated change per nucleotide.
In situ hybridization.
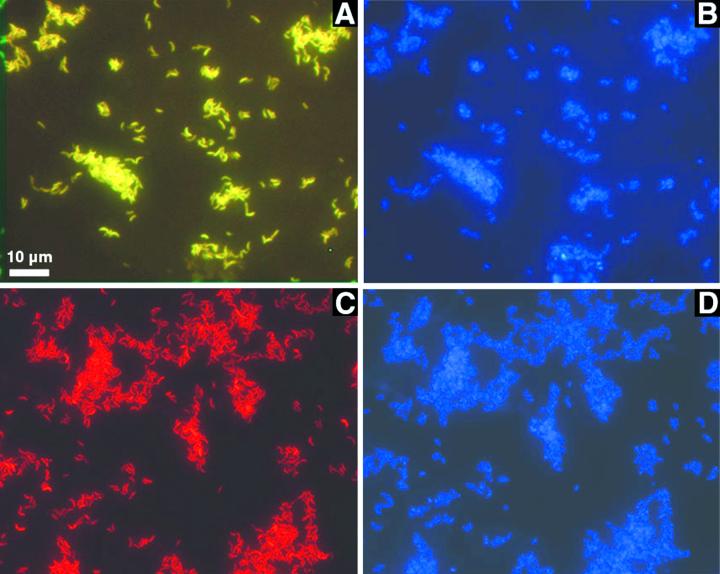
The source of the sequence was further tested by fluorescent in situ hybridization by using oligonucleotide probes specific for the genus Arcobacter and for the sequence of the sulfur-oxidizing bacterium. All cells that could be stained with DAPI (Fig. 4B and D) hybridized with both Arcobacter-specific probes (ARC94 and ARC1430) (Fig. 4A) and hybridized with the species-specific probe CARCS219 (Fig. 4C) as well as the universal Bacteria probe EUB338 (data not shown). No hybridization was found with probe NON338, which was used to detect nonspecific binding of the label. Cells of Arcobacter strain KTO913 hybridized with the genus-specific probes ARC94 and ARC1430 but not with CARCS219, indicating the species specificity of the probe (data not shown). Interestingly, the sequence of Arcobacter strain KTO913 has one mismatch with the sequence of probe ARC1430, whereas the sulfide-oxidizing strain has none. When probe ARC1430 was used at a higher stringency by increasing the formamide concentration to 30% (compared to 20% in the normal protocol), cells of Arcobacter strain KTO913 did not hybridize anymore, while cells of the sulfide-oxidizing strain still gave a clear hybridization signal. These data confirm the hypothesis that the sequence amplified was the sequence of the dominant organism in the reactor. In addition, the simultaneous hybridization with two different labeled probes targeting opposite ends of the 16S rDNA demonstrates further that the sequence obtained belongs to just one organism and rules out the possibility of a chimeric sequence.
FIG. 4.
Whole-cell fluorescent in situ hybridization and DAPI staining of reactor-grown “Candidatus Arcobacter sulfidicus” cells. (A) Simultaneous hybridization with the arcobacter-specific probes ARC94 (Cy-3 labeled, red) and ARC1430 (fluorescein labeled, green) showing yellow cells as a composite image. (B) Same field as in panel A stained with DAPI. (C) Hybridization with probe CARCS219 (Cy-3 labeled, red), specific for “Candidatus Arcobacter sulfidicus.” (D) Same field as in panel C stained with DAPI.
Carbon dioxide fixation experiments.
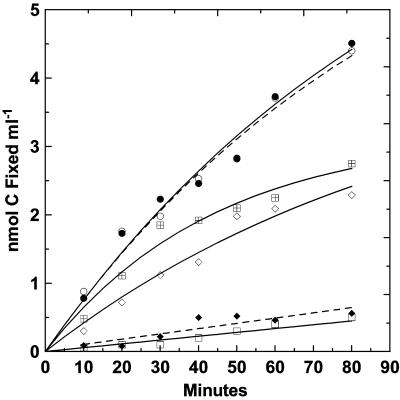
Cells were able to fix CO2 under batch and continuous-flow culture conditions. Carbon fixation by cells disassociated from sulfur filaments and incubated in batch cultures at various initial concentrations of sulfide is shown in Fig. 5. The initial rate of CO2 fixation peaked at 0.077 nmol of C ml−1 min−1 for sulfide concentrations of 460 and 1,230 μM. Fixation was incrementally lower, 0.066 nmol of C ml−1 min−1, at 160 μM sulfide and rapidly decreased as the electron donor was consumed during the incubation. After 60 min, the rate dropped to that which was observed in the control without added sulfide (i.e., 0.006 nmol of C ml−1 min−1), suggesting that sulfide was totally consumed by this time. At 90 min, the readdition of sulfide (350 μM) to the test culture originally containing 460 μM sulfide resulted in an immediate increase to approximately the initial fixation rate (data not shown). Sulfide inhibition occurred at 2,460 μM, as reflected by the approximate twofold reduction in initial carbon fixation rate to 0.042 nmol of C ml−1 min−1. Minimal fixation occurred above the sulfide-free control rate when cultures were amended with pasteurized (80°C, 10 min) sulfur filaments. Based on the time-zero cell count of 5.8 × 106 cells ml−1, the initial fixation rate was 0.80 × 10−9 μmol of C cell−1 h−1 in the 1,230 μM sulfide tube.
FIG. 5.
Carbon dioxide fixation by “Candidatus Arcobacter sulfidicus” cells harvested free of sulfur filaments from an actively growing reactor enrichment. Incubation was carried out with batch cultures in the presence of various initial hydrogen sulfide concentrations: □, no sulfide; ⋄, 160 μM; •, 460 μM; ○, 1,230 μM; [  ], 2,460 μM. ⋄, pasteurized sulfur filaments.
], 2,460 μM. ⋄, pasteurized sulfur filaments.
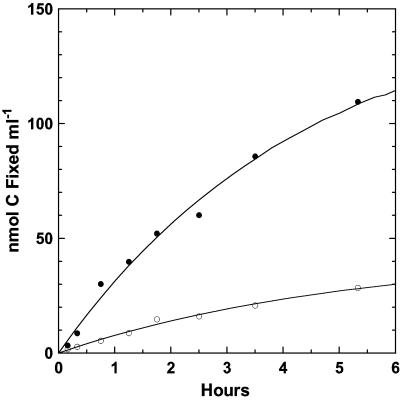
In order to determine where the majority of carbon fixation was occurring, a comparison was made between reactor-grown filaments with associated cells and filament-free seawater containing disassociated cells (Fig. 6), both at an initial concentration of 1,500 μM sulfide. The latter had a cell density 1.5 times higher than that used in the previous incubation experiment (Fig. 5) and had a higher initial fixation rate of 0.14 nmol of C ml−1 min−1. Based upon an initial population of 8.66 × 106 cells ml−1, the initial CO2 fixation rate in the filament-free sample was 0.97 × 10−9 μmol of C cell−1 h−1, a rate comparable to the rate noted above for dissociated cells. The initial fixation rate in the reactor filamentous material (0.54 nmol of C ml−1 min−1), on the other hand, was fourfold higher, indicating that a large number of cells remained associated with the filaments (microscopically verified), even after intense agitation to disassociate cells from the sulfur filaments.
FIG. 6.
Carbon dioxide fixation by “Candidatus Arcobacter sulfidicus.” Reactor-grown filaments with associated cells • were compared with filament-free seawater obtained from the reactor and containing cells dissociated from the filamentous matrix ○ (i.e., reactor filaments were harvested, stirred rapidly for 5 min, and allowed to settle for 5 min before the seawater above was collected).
A carbon dioxide fixation experiment conducted with a purified cell suspension growing in a continuous-flow reactor for 48 h resulted in the highest measured level of carbon fixation on both per-volume and per-cell bases. Direct counts of the cell numbers on both the intact filamentous sulfur (after making appropriate dilutions for counting) and the cells present in a filament-free 10-μm filtrate of this material were obtained at 48 h. Based on these counts, there was good agreement in the level of carbon fixation over 48 h between intact filaments with associated cells and filtered cells. The values were 4.8 × 10−8 μmol of C fixed cell−1 (0.364 μmol of C fixed ml−1 at 7.6 × 106 cells ml−1) for the bulk filaments and 5.0 × 10−8 μmol of C fixed cell−1 (0.184 μmol of C ml−1 at 3.56 × 106 cells ml−1) for the 10-μm filtered cells. Based on the total amount of fixed carbon over 48 h, the population demonstrated a fixation rate of 1.04 × 10−9 μmol of C cell−1 h−1 for the whole incubation period; comparable yet slightly lower rates were measured in batch culture, based on their initial slope. This high and sustained level of CO2 fixation illustrates the importance of a dynamic supply of both oxygen and sulfide to support optimal autotrophic growth.
Nitrogen fixation.
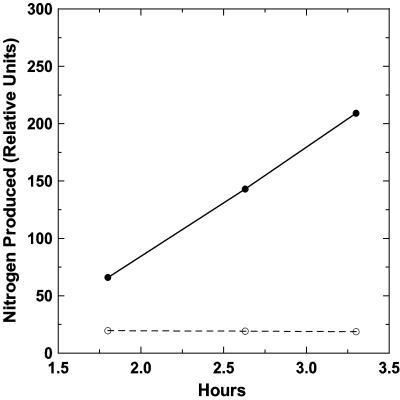
Filamentous material harvested from a flowing reactor enrichment culture exhibited a highly positive rate of N2 fixation, as shown by acetylene reduction (Fig. 7). Sterilized seawater from the reactor from which the filamentous material had been removed showed no acetylene reduction activity.
FIG. 7.
Relative rate of nitrogen fixation, as assayed by acetylene reduction. Filamentous material freshly harvested from a reactor • was compared with seawater obtained from the reactor and from which the filamentous material had been removed ○ (0.45-μm filtration).
Rubisco activity.
Rubisco activity was not detected in cell extracts of cells freshly harvested from the enrichment reactor. Microscopic examination showed that cell breakage had been accomplished, yielding a cell extract with a protein content of 600 μg ml−1. The assay procedure functioned normally, as confirmed by using spinach as a positive control.
Neither form I nor form II Rubisco was detected in cell extracts by Western blotting with antisera specific to each form, although positive controls for both form I and form II Rubisco proteins were detected and estimated to be 55 and 52 kDa, respectively (data not shown).
Chemical analyses.
Examination of reactor-grown filamentous material for protein or carbohydrate coatings responsible for its hydrophilic properties indicated that little to no organic material coated or would be available to coat the sulfur filaments. The dry weight of samples of filamentous reactor material (i.e., sulfur and associated cells) corrected for sea salts was 1,220 μg ml−1, and the samples contained 1.65 × 108 cells ml−1, as determined by direct counting. The measured POC was 20.4 μg of C ml−1. Of the total dry weight per milliliter, the POC amounted to only 1.67%, with the remaining 98.3% being sulfur. The anthrone determination for the reactor material yielded an average of 2.5 μg of carbohydrate ml−1 (range, 1.8 to 3.4; n = 3), and the protein determination indicated an average of 4.0 μg ml−1 in the 1.0 N NaOH digest (values in undigested material or in the 0.1 N NaOH digest were lower). The total of these two organic components alone (6.5 μg) accounts for 32% of the measured 20.4 μg of organic carbon in the sample. By empirical calculations (cell mass, 50%, as dry weight; 20% dry weight as carbon), the cell fraction in this test should contain about 9 μg of carbon, but it must be noted that these types of calculations can be in error by up to a factor of two. The analyses indicate that at least one-third (and probably as much as one-half, considering lipids and nucleic acids) of the organic carbon present (i.e., 1.67%, as dry weight) can be accounted for by the carbohydrate and protein of the cellular fraction. Thus, a small amount (e.g., 10 μg) of carbon-containing material may be left to coat the large amount of sulfur filaments, or it may actually be structurally incorporated and act as an amphiphile in the sulfur moiety.
No residual structure or materials were microscopically visible when sulfur threads were dissolved with ethanol. Additionally, there was no indication of any specific protein coating of the sulfur filaments by fluorescein isothiocyanate staining. Control orthorhombic elemental sulfur and filamentous sulfur threads demonstrated the same low level of background yellow fluorescence. Additional sulfur benzene assays, done on a dry-weight basis with filamentous sulfur and rhombic sublimed sulfur as the control, showed the filamentous material to be equivalent to 100% sulfur.
DISCUSSION
In recent years, especially due to the advent of molecular methods, Arcobacter spp. have been described from a variety of habitats, including activated sludge (65), aborted fetuses of farm animals (74), salt marsh sediments (39, 74), Wadden Sea sediments (35), North Sea bacterioplankton (16), in association with vestimentiferan tube worms (45), a deep-sea hydrothermal vent chimney (V. Cilia and D. Prieur, unpublished data; uncultured eubacterium CHA3-437), hypersaline cyanobacterial mats from Solar Lake (Sinai) (72) and, most recently, from the redox interface of the Cariaco Basin (37). In the latter two habitats, the Arcobacter cells may be exposed to oxic-anoxic fluctuations and significant sulfide levels (approximately 20 to 200 μM), albeit lower than the optimum demonstrated for the sulfide oxidizer in the present study.
Based on the proposal (43) and the recommendation for implementation (44) of a procaryotic category Candidatus, we are submitting the provisional name “Candidatus Arcobacter sulfidicus” for the sulfide-oxidizing, filamentous-sulfur-producing vibrioid organism described in this study. This status is intended for difficult-to-grow or presently uncultivable procaryotes for which genetic information allows formal assignment within established genera. There are at present cleaned and frozen purified cell suspensions from which new enrichments can be generated, but no transferable pure cultures of “Candidatus Arcobacter sulfidicus” are maintained. While production of this filamentous-sulfur-producing organism was achieved in purified cultures under certain conditions of this study, as indicated microscopically and by the absence of contaminants on plates, it should presently be considered a difficult-to-cultivate organism. This suggestion is based on the growth apparatus and conditions required and not on the nutritional aspects of cultivation. The morphological and physiological characteristics of “Candidatus Arcobacter sulfidicus,” such as cell shape, motility, location of flagella, Gram reaction, microaerophilic preference, mesophilic temperature preference, nitrogen fixation (as in Arcobacter nitrofigilis), and habitat, are characteristic for members of the genus Arcobacter (11, 61, 74). Furthermore, cells from the reactor hybridized with both probes specific for the genus Arcobacter. The finding that all DAPI-stained cells hybridized to probe CARCS219, which is complementary to a unique region of the 16S RNA sequence of “Candidatus Arcobacter sulfidicus,” indicates that the enrichment conditions led to the establishment of essentially a monoculture of one species.
The coastal “Candidatus Arcobacter sulfidicus” of this study possesses a multilaminar polar membrane morphologically similar to that described for members of the genus Campylobacter (64) and detailed for V. fetus (i.e., Campylobacter fetus) (54). As the genus Arcobacter was only recently created in 1991 for two species of aerotolerant campylobacters (74), the occurrence of this polar organelle in reclassified members of this genus has not been examined thus far. The function of this membranous organelle in Campylobacter is at present unknown. In “Candidatus Arcobacter sulfidicus,” it may play a role in the excretion of the sulfur filaments, analogous to the polar organelle referred to as the Faûré-Fremiet-Rouiller organelle (17) in the sulfide-oxidizing bacterium Thiovulum, which is believed to play a role in the excretion of slime threads (77). Perhaps the greatest divergence of “Candidatus Arcobacter sulfidicus” from other members of the genus Arcobacter is its mode of nutrition. While phylogenetically related procaryotes can have diverse physiologies, the chemoautotrophic mode of growth in this strain versus the chemoorganotrophic capability of other arcobacters is a significant and at present unexplained divergence. An interesting comparison, however, is the recently described sulfide oxidizer FWKO B from oil field brine (20). It is closely related to the Arcobacter branch (Fig. 3), but it is not a member of this genus, based on base positions in the 16S rRNA that are shared among all arcobacters but that are different in strain FWKO B. However, the metabolism of FWKO B is similar to that of “Candidatus Arcobacter sulfidicus” in that it is chemoautotrophic and can utilize sulfide but not thiosulfate as an energy source.
While the autotrophic potential of the organism was clearly demonstrated, the pathway for the fixation of CO2 may not be that of the Calvin-Bassham-Benson pathway. Rubisco activity could not be demonstrated by the procedures used, nor was there any detection of form I or form II Rubisco by Western blotting. Microorganisms fixing CO2 by the Calvin cycle have isotopic fractionation values of bulk cell material relative to the dissolved inorganic carbon source on the order of a Δδ13C value of −20 to −26‰ (18, 36, 57) but may be somewhat more enriched in 13C, depending on the form of Rubisco present (55). As a preliminary determination of what alternative pathway of CO2 fixation to test for, we measured the isotopic discrimination values (Δδ13C) for cells grown in two reactors (average value for duplicate samples of 8-μm filtered cells from each reactor). We obtained from the two reactors Δδ13C values of −12.5‰ and −13.5‰, corrected for the δ13C of the source seawater dissolved inorganic carbon. This preliminary measurement indicates that other pathways of autotrophic CO2 fixation need to be investigated for this organism, particularly the reductive tricarboxylic acid pathway, which has been demonstrated in anaerobic and microaerophilic bacteria and archaea (Δδ13C, −8‰ to −12‰) (18), or the 3-hydroxypropionate pathway identified for Chloroflexus (Δδ13C, −13.7‰) and certain archaea (25, 41). These alternative pathways of CO2 fixation are presently being investigated for “Candidatus Arcobacter sulfidicus” by using diagnostic enzyme assays (18, 68).
A comparison of the level of CO2 fixation can be made for autotrophic organisms grown on sulfide or thiosulfate and for “Candidatus Arcobacter sulfidicus” growing on sulfide in the sterile continuous-flow system. For Thiomicrospira sp. strain L-12, Ruby and Jannasch (56) reported a fixation rate in the presence of 100 to 300 μM sulfide of 0.7 × 10−9 μmol of C cell−1 h−1. For other autotrophic Thiomicrospira strains (58) grown on thiosulfate, the fixation rate was reported to be in the range of 3.1 to 6.1 μg of HCO3 fixed per 106 cells after 23 h of growth. The highest level in this range of fixation converts to 0.44 × 10−9 μmol of C cell−1 h−1. These rates compare well with our data of 0.80 to 1.04 × 10−9 μmol of C cell−1 h−1 derived from batch-culture and continuous-flow reactor experiments and indicate that sulfide-oxidizing “Candidatus Arcobacter sulfidicus” has a CO2 fixation capability equivalent to or better than that of traditional sulfur-oxidizing microbes.
It was believed that for the amount and rapidity of biomass produced, the cell population should likely be capable of fixing nitrogen. Nitrogen fixation rates were very high for the sulfide-oxidizing enrichment sampled from an actively growing reactor population. Nitrogen fixation was previously described for at least one species of Arcobacter, i.e., A. nitrofigilis (74), formerly Campylobacter nitrofigilis (39).
What confers the hydrophilicity to the filamentous sulfur threads produced by the organism is unclear. In contrast to the common, room-temperature-stable form of solid orthorhombic S8 elemental sulfur, typical microbially produced intracellular sulfur and extracellular sulfur are hydrophilic in nature and are spherical or ellipsoidal globules with a liquid-like character (66). Analysis by ion chromatography has shown that sulfur globules appear to be membranous structures substantially composed of long-chain (average, S30) amphiphilic polythionates or polysulfides that have formed into membranous unilamellar or multilamellar vesicles surrounding an aqueous interior (66, 67). Furthermore, there is the potential for organic molecules to be present at the terminal ends of sulfur chains, as reported by Prange et al. (52) for phototrophic sulfur bacteria. Our present evidence, based upon bulk chemical analysis of the sulfur filaments and fluorometric techniques for visualizing protein, suggests that the excreted filamentous sulfur threads of “Candidatus Arcobacter sulfidicus” are entirely sulfur. There were no visible signs of an organic coating on the filaments, although evidence at present does not preclude the presence of an organic or inorganic amphiphile that is integrated into the structure of the sulfur filaments in a manner similar to that found in sulfur globules. We are currently conducting instrumental analyses of the composition of the sulfur filaments and what might confer the hydrophilicity to these microbially produced products.
The metabolic capability of filamentous sulfur formation is not limited to coastally derived “Candidatus Arcobacter sulfidicus,” as it has now been documented to occur in other sulfidic environments. Filamentous sulfur was first compared with and found to be identical to material collected from an earlier flocculent discharge at a 9°N hydrothermal vent on the East Pacific Rise (EPR) (22, 70). The process of microbial filamentous sulfur formation was recently documented as occurring in situ at 9°N EPR deep-sea hydrothermal vents as well as in shipboard experiments by using inocula collected from these sites (71). Visual and microscopic observations of filamentous sulfur have also been made for extensive emission discharges at deep-sea vent sites on the CoAxial segment of the Juan de Fuca Ridge (53), for surface sediments of Guaymas Basin (71; unpublished data), and for Aegean Sea shallow marine vents (63). In the last example, the dominant organism was shown by in situ hybridization to belong to the genus Arcobacter (63). All of these results indicate that this process of microbial filamentous sulfur formation is an ecologically important process and perhaps is ubiquitous in marine environments where sulfide and oxygen are available at the necessary concentrations and gradients. Since “Candidatus Arcobacter sulfidicus” is retrieved from a coastal habitat, it would be significant to know whether and to what extent similar organisms are involved in filamentous sulfur formation in distinct and different sulfidic environments. In addition, it will also be interesting to determine whether filamentous sulfur formation is widely distributed among different bacteria or is restricted to Arcobacter spp.
Acknowledgments
This work was supported by National Science Foundation grant IBN-9630054 and by a postdoctoral fellowship awarded to S. M. Sievert from the Woods Hole Oceanographic Institution.
We gratefully acknowledge Michael Atkins and Andrew McArthur for help with the phylogenetic analyses and Heike Eilers for providing cells of Arcobacter strain KT0913. We thank Kathleen Scott for conducting the seawater δ13C analyses and Marshall Otter for conducting the cell carbon δ13C analyses. We also thank Andreas Teske for helpful discussions and advice.
Footnotes
Contribution 10519 of the Woods Hole Oceanographic Institution.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997., Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. I. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Battersby, N. S., S. J. Malcolm, C. M. Brown, and S. O. Stanley. 1985. Sulphate reduction in oxic and sub-oxic North-East Atlantic sediments. FEMS Microbiol. Ecol. 31:225–228. [Google Scholar]
- 6.Beudeker, R. F., G. C. Cannon, J. G. Kuenen, and J. M. Shively. 1980. Relations between d-ribulose-1,5-bisphosphate carboxylase, carboxysomes and CO2 fixing capacity in the obligate chemolithotroph Thiobacillus neapolitanus grown under different limitations in the chemostat. Arch. Microbiol. 124:185–189. [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- 8.Buchholz-Cleven, B. E. E., B. Rattunde, and K. L. Straub. 1997. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst. Appl. Microbiol. 20:301–309. [Google Scholar]
- 9.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454–458. [Google Scholar]
- 10.Crissman, H. A., and J. A. Steinkamp. 1982. Rapid, one-step staining procedures for analysis of cellular DNA and protein by single and dual laser flow cytometry. Cytometry 3:84–90. [DOI] [PubMed] [Google Scholar]
- 11.de Boer, E., J. Tilburg, D. Woodward, H. Lior, and W. Johnson. 1996. A selective medium for the isolation of Arcobacter from meats. Lett. Appl. Microbiol. 23:64–66. [DOI] [PubMed] [Google Scholar]
- 12.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 85:5685–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dilworth, M. J. 1966. Acetylene reduction by nitrogen-fixing preparations from Clostridium pasteurianum. Biochim. Biophys. Acta 127:285–294. [DOI] [PubMed] [Google Scholar]
- 14.Dugan, P. 1991. Microbial conversions of sulfur and their potential for bioprocessing of fossil fuels. Resour. Conserv. Recycl. 5:101–125. [Google Scholar]
- 15.Eberhard, C., C. O. Wirsen, and H. W. Jannasch. 1995. Oxidation of polymetal sulfides by chemolithoautotrophic bacteria from deep-sea hydrothermal vents. Geomicrobiol. J. 13:145–164. [Google Scholar]
- 16.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faûré-Fremiet, E., and C. Rouiller. 1958. Etude au microscope électronique d’une bactérie sulfureuse, Thiovulum majus Hinze. Exp. Cell Res. 14:29–46. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs, G. 1989. Alternative pathways of autotrophic CO2 fixation, p.365–382. In H. G. Schlegel and B. Bowien (ed.), Autotrophic bacteria. Science Tech Publishers, Madison, Wis.
- 19.Gallardo, V. A. 1977. Large benthic microbial communities in sulphide biota under Peru-Chile subsurface countercurrent. Nature 268:331–332. [Google Scholar]
- 20.Gevertz, D., A. J. Telang, G. Voordouw, and G. E. Jenneman. 2000. Isolation and characterization of strains CVO and FWKO B, two novel nitrate-reducing, sulfide-oxidizing bacteria isolated from oil field brine. Appl. Environ. Microbiol. 66:2491–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haddad, A., F. Camacho, P. Durand, and S. C. Cary. 1995. Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaeate Alvinella pompejana. Appl. Environ. Microbiol. 61:1679–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haymon, R. M., D. J. Fornari, K. L. Von Damm, M. D. Lilley, M. R. Perfit, J. M. Edmond, W. C. Shanks III, R. A. Lutz, J. M. Grebmeier, S. Carbotte, D. Wright, E. McLaughlin, M. Smith, N. Beedle, and E. Olson. 1993. Volcanic eruption of the mid-ocean ridge along the East Pacific Rise crest at 9°45–52′N: direct submersible observations of seafloor phenomena associated with an eruption event in April, 1991. Earth Planet. Sci. Lett. 119:85–101. [Google Scholar]
- 23.Hines, M. E., M. J. Spencer, J. B. Tugel, W. B. Lyons, and G. E. Jones. 1985. Seasonal sulfate reduction and iron mobilization in estuarine sediments, p.222–229. In D. E. Caldwell, J. A. Brierley, and C. L. Brierley (ed.), Planetary ecology. Van Nostrand Reinhold Co., New York, N.Y.
- 24.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holo, H., and R. Sirevåg. 1986. Autotrophic growth and CO2 fixation of Chloroflexus aurantiacus Arch. Microbiol. 145:173–180. [Google Scholar]
- 26.Jannasch, H. W., and M. J. Mottl. 1985. Geomicrobiology of deep-sea hydrothermal vents. Science 229:717–725. [DOI] [PubMed] [Google Scholar]
- 27.Jannasch, H. W., and C. O. Wirsen. 1981. Morphological survey of microbial mats near deep-sea thermal vents. Appl. Environ. Microbiol. 41:528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javor, B. J., D. B. Wilmot, and R. D. Vetter. 1990. pH-Dependent metabolism of thiosulfate and sulfur globules in the chemolithotrophic marine bacterium Thiomicrospira crunogena. Arch. Microbiol. 154:231–238. [Google Scholar]
- 29.Jørgensen, B. B., L. X. Zawacki, and H. W. Jannasch. 1990. Thermophilic bacterial sulfate reduction in deep-sea sediments at the Guaymas Basin hydrothermal vent site (Gulf of California). Deep Sea Res. 37:695–710. [Google Scholar]
- 30.Jørgensen, B. B. 1982. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature 296:643–645. [Google Scholar]
- 31.Kishimoto, N., Y. Kosako, N. Wakao, T. Tano, and A. Hiraishi. 1995. Transfer of Acidiphilium facilis and Acidiphilium aminolytica to the genus Acidocella gen. nov., and emendation of the genus Acidiphilium. Syst. Appl. Microbiol. 18:85–91. [Google Scholar]
- 32.Kuenen, J. G., and O. H. Tuovinen. 1981. The genera Thiobacillus and Thiomicrospira, p.1023–1036. In M. P. Starr, H. Stolz, H. G. Truper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes. Springer-Verlag, Berlin, Germany.
- 33.Kuenen, J. G. 1975. Colourless sulfur bacteria and their role in the sulfur cycle. Plant Soil. 43:49–76. [Google Scholar]
- 34.LaRivière, J. W. M. 1963. Cultivation and properties of Thiovulum majus Hinze, p.61–72. In C. H. Oppenheimer (ed.), Symposium on marine microbiology. Charles C Thomas, Publisher, Springfield, Ill.
- 35.Llobet-Brossa, E., R. Rosselló-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madigan, M. T., R. Takigiku, R. G. Lee, H. Gest, and J. M. Hayes. 1989. Carbon isotope fractionation by thermophilic phototrophic sulfur bacteria: evidence for autotrophic growth in natural populations. Appl. Environ. Microbiol. 55:639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Christoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maidak, B. J., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McClung, C. R., D. G. Patriquin, and R. E. Davis. 1983. Campylobacter nitrofigilis sp. nov., a nitrogen-fixing bacterium associated with roots of Spartina alterniflora. Int. J. Syst. Bacteriol. 33:605–612. [Google Scholar]
- 40.McDuff, R. E., and J. M. Edmond. 1982. On the fate of sulfate during hydrothermal circulation at mid-ocean ridges. Earth Planet. Sci. Lett. 57:117–132. [Google Scholar]
- 41.Menendez, C., Z. Bauer, H. Huber, N. Gad’on, K. Stetter, and G. Fuchs. 1999. Presence of acetyl coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J. Bacteriol. 181:1088–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreira, D., and R. Amils. 1997. Phylogeny of Thiobacillus cuprinus and other mixotrophic thiobacilli: proposal for Thiomonas gen. nov. Int. J. Syst. Bacteriol. 47:522–528. [DOI] [PubMed] [Google Scholar]
- 43.Murray, R. G. E., and K. H. Schleifer. 1994. Taxonomic notes: a proposal for recording the properties of putative taxa of procaryotes. Int. J. Syst. Bacteriol. 44:174–176. [DOI] [PubMed] [Google Scholar]
- 44.Murray, R. G. E., and E. Stackebrandt. 1995. Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int. J. Syst. Bacteriol. 45:186–187. [DOI] [PubMed] [Google Scholar]
- 45.Naganuma, T., C. Kato, H. Hirayama, N. Moriyama, J. Hashimoto, and K. Horikoshi. 1997. Intracellular occurrence of ɛ-proteobacterial 16S rDNA sequences in the vestimentiferan trophosome. J. Oceanogr. 53:193–197. [Google Scholar]
- 46.Nelson, D. C., and R. W. Castenholz. 1981. Use of reduced sulfur compounds by Beggiatoa sp. J. Bacteriol. 147:140–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson, D. C., and H. W. Jannasch. 1983. Chemoautotrophic growth of a marine Beggiatoa in sulfide-gradient cultures. Arch. Microbiol. 136:262–269. [Google Scholar]
- 48.Odintsova, E. V., A. P. Wood, and D. P. Kelly. 1993. Chemolithotrophic growth of Thiothrix ramosa. Arch. Microbiol. 160:152–157. [Google Scholar]
- 49.Olson, R. J., D. Vaulot, and S. W. Chisholm. 1986. Effects of environmental stresses on the cell cycle of two marine phytoplankton species. Plant Physiol. 80:918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358. [DOI] [PubMed] [Google Scholar]
- 51.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818. [DOI] [PubMed] [Google Scholar]
- 52.Prange, A., I. Arzberrger, C. Engemann, H. Modrow, O. Schumann, H. G. Truper, R. Studel, C. Dahl, and J. Hormes. 1999. In situ analysis of sulfur in the sulfur globules of phototrophic sulfur bacteria by X-ray absorption near edge spectroscopy. Biochim. Biophys. Acta 1428:446–454. [DOI] [PubMed] [Google Scholar]
- 53.Rawls, R. L. 1998. Some like it hot. Chem. Eng. News 76:35–39. [Google Scholar]
- 54.Ritchie, A. E., R. F. Keeler, and J. H. Bryner. 1966. Anatomical features of Vibrio fetus: electron microscopic survey. J. Gen. Microbiol. 43:427–438. [DOI] [PubMed] [Google Scholar]
- 55.Robinson, J. J., and C. M. Cavanaugh. 1995. Expression of form I and form II ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo) in chemoautotrophic symbioses: implications for the interpretation of stable carbon isotope ratios. Limnol. Oceanogr. 40:1496–1502. [Google Scholar]
- 56.Ruby, E. G., and H. W. Jannasch. 1982. Physiological characteristics of Thiomicrospira sp. strain L-12 isolated from deep-sea hydrothermal vents. J. Bacteriol. 149:161–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruby, E. G., H. W. Jannasch, and W. G. Deuser. 1987. Fractionation of stable carbon isotopes during chemoautotrophic growth of sulfur-oxidizing bacteria. Appl. Environ. Microbiol. 53:1940–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruby, E. G., C. O. Wirsen, and H. W. Jannasch. 1981. Chemolithotrophic sulfur-oxidizing bacteria from the Galapagos Rift hydrothermal vents. Appl. Environ. Microbiol. 42:317–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schöllhorn, R., and R. H. Burris. 1966. Study of the intermediates of nitrogen fixation. Fed. Proc. 24:710. [Google Scholar]
- 60.Schöllhorn, R., and R. H. Burris. 1967. Acetylene as a competitive inhibitor of N2 fixation. Proc. Natl. Acad. Sci. USA 58:213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schroeder-Tucker, L., I. Wesley, J. A. Kiehlbauch, G. A. Erickson, L. A. Thomas, and D. J. Larsen. 1996. Phenotypic and ribosomal RNA characterization of Arcobacter species isolated from porcine aborted fetuses. J. Vet. Diagn. Investig. 8:186–195. [DOI] [PubMed] [Google Scholar]
- 62.Shanks, W. C., III, J. L. Bischoff, and R. J. Rosenbauer. 1981. Seawater sulfate reduction and sulfur isotope fractionation in basaltic systems: interaction of seawater with fayalite and magnetite at 250–350° C. Geochim. Cosmochim. Acta 45:1977–1995. [Google Scholar]
- 63.Sievert, S. M., T. Brinkhoff, G. Muyzer, W. Ziebis, and J. Kuever. 1999. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 65:3834–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smibert, R. M. 1984. Genus Campylobacter Sebald and Véron 1963, 907AL, p.111–118. In N. R. Kreig and J. G. Holt (ed.), Bergey’s manual of systematic bacteriology. The Williams & Wilkins Co., Baltimore, Md.
- 65.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steudel, R. 1989. On the nature of the “elemental sulfur” (S0) produced by sulfur-oxidizing bacteria—a model for S0 globules, p.289–303. In H. G. Schlegel and B. Bowien (ed.), Autotrophic bacteria. Science Tech Publishers, Madison, Wis.
- 67.Steudel, R., G. Holdt, and T. Gobel. 1989. Ion-pair chromatographic separation of inorganic sulphur anions including polysulfide. J. Chromatogr. 47:442–446. [Google Scholar]
- 68.Strauss, G., and G. Fuchs. 1993. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxpropionate cycle. Eur. J. Biochem. 215:633–643. [DOI] [PubMed] [Google Scholar]
- 69.Strohl, W. R., and J. M. Larkin. 1978. Enumeration, isolation, and characterization of Beggiatoa from freshwater sediments. Appl. Environ. Microbiol. 36:755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor, C. D., and C. O. Wirsen. 1997. Microbiology and ecology of filamentous sulfur formation. Science 277:1483–1485. [Google Scholar]
- 71.Taylor, C. D., C. O. Wirsen, and F. Gaill. 1999. Rapid microbial production of filamentous sulfur mats at hydrothermal vents. Appl. Environ. Microbiol. 65:2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teske, A., P. Sigalevich, Y. Cohen, and G. Muyzer. 1996. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl. Environ. Microbiol. 62:4210–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuttle, J. H., and H. W. Jannasch. 1977. Thiosulfate stimulation of microbial dark assimilation of carbon dioxide in shallow marine environments. Microb. Ecol. 4:9–25. [DOI] [PubMed] [Google Scholar]
- 74.Vandamme, P., E. Falsen, R. Rossau, B. Hoste, P. Segers, R. Tytgat, and J. De Ley. 1991. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. J. Syst. Bacteriol. 41:88–103. [DOI] [PubMed] [Google Scholar]
- 75.Van de Peer, Y., S. Chapelle, and R. De Wachter. 1996. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 24:3381–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Viles, F. J., and L. Silverman. 1949. Determination of starch and cellulose with anthrone. Anal. Chem. 21:950–953. [Google Scholar]
- 77.Wirsen, C. O., and H. W. Jannasch. 1978. Physiological and morphological observations on Thiovulum spp. J. Bacteriol. 136:765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]