Abstract
Dimeric hemoglobin (VHb) from the bacterium Vitreoscilla sp. strain C1 displays 30 to 53% sequence identity with the heme-binding domain of flavohemoglobins (flavoHbs) and exhibits the presence of potential sites for the interaction with its FAD/NADH reductase partner. The intersubunit contact region of VHb indicates a small interface between two monomers of the homodimer, suggesting that the VHb dimers may dissociate easily. Gel filtration chromatography of VHb exhibited a 25 to 30% monomeric population of VHb, at a low protein concentration (0.05 mg/ml), whereas dimeric VHb remained dominant at a high protein concentration (10 mg/ml). The structural characteristics of VHb suggest that the flavoreductase can also associate and interact with VHb in a manner analogous to flavoHbs and could yield a flavo-VHb complex. To unravel the functional relevance of the VHb-reductase association, the reductase domain of flavoHb from Ralstonia eutropha (formerly Alcaligenes eutrophus) was genetically engineered to generate a VHb-reductase chimera (VHb-R). The physiological implications of VHb and VHb-R were studied in an hmp mutant of Escherichia coli, incapable of producing any flavoHb. Cellular respiration the of the hmp mutant was instantaneously inhibited in the presence of 10 μM nitric oxide (NO) but remained insensitive to NO inhibition when these cells produced VHb-R. In addition, E. coli overproducing VHb-R exhibited NO consumption activity that was two to three times slower in cells overexpressing only VHb and totally undetectable in the control cells. A purified preparation of VHb-R exhibited a three- to fourfold-higher NADH-dependent NO uptake activity than that of VHb alone. Overproduction of VHb-R in the hmp mutant of E. coli conferred relief from the toxicity of sodium nitroprusside, whereas VHb alone provided only partial benefit under similar condition, suggesting that the association of VHb with reductase improves its capability to relieve the deleterious effect of nitrosative stress. Based on these results, it has been proposed that the unique structural features of VHb may allow it to acquire two functional states in vivo, namely, a single-domain homodimer that may participate in facilitated oxygen transfer or a two-domain heterodimer in association with its partner reductase that may be involved in modulating the cellular response under different environmental conditions. Due to this inherent structural flexibility, it may perform multiple functions in the cellular metabolism of its host. Separation of the oxidoreductase domain from VHb may thus provide a physiological advantage to its host.
Flavohemoglobins (flavoHbs), resulting from the integration of an N-terminal heme-binding domain with a flavin-binding reductase domain, have been identified in a number of bacteria and yeast (4, 5, 12, 20, 38, 43). These proteins have been shown to play an important role in the metabolism of oxygen and nitrogenous compounds (3, 6, 29, 35, 36). In addition to their putative role in oxygen sensing, diffusion, and utilization, flavoHbs have also been proposed to function in protection of microorganisms from oxidative and nitrosative stress (14, 17, 30, 31). The first evidence that flavoHbs may be involved in NO metabolism came when a substantial increase in hmp gene transcription in Escherichia coli was observed in the presence of NO and nitrosating agents (35). Subsequently, it has been demonstrated that E. coli flavoHb functions as a NO dioxygenase that participates in S-nitrosothiol and NO metabolism and protects the bacterium against NO toxicity (17). Supporting evidence for flavoHb interaction with NO came from the enhanced sensitivity of hmp mutants of E. coli (31) and Salmonella enterica serovar Typhimurium (6) to NO and nitrosating agents and further from the demonstration that HMP has NO dioxygenase activity forming nitrate (14, 17). The induction of flavoHbs under microaerophilic conditions might be beneficial for bacteria to survive NO killing since the cellular level of NO dioxygenase decreases when the oxygen is limiting (13, 14, 34). These observations suggest that one of the prime functions of these microbial flavoHbs is to provide protection from the toxicity of NO generation.
Hemoglobin from the gram-negative bacterium, Vitreoscilla sp., is a homodimeric globin devoid of any covalently linked flavin domain (7, 24, 44). Dimeric hemoglobin (VHb), however, is closely related to the bacterial and yeast flavoHb heme-binding domain, sharing 43 to 56% sequence homology (41). The evolutionary and functional significance of VHb in existing as single-domain hemoglobin and its implication in Vitreoscilla cellular metabolism remains unclear. In the strictly aerobic bacterium Vitreoscilla sp., expression of the vgb gene is strongly induced by a decrease in oxygen level (8, 9, 26). Extensive biochemical and genetic studies on VHb have suggested that it facilitates oxygen delivery to the respiratory apparatus of its host (45) and thus enhances its growth metabolism (25, 27) or that it may act as an alternative terminal oxidase itself (10). The discovery of NADH-dependent flavin reductase in Vitreoscilla sp. (15, 21) and the extensive sequence and structural similarity of VHb with the heme-binding domain of flavoHb suggests that the heme and the flavin domains might have separated in this bacterium. Interestingly, the flavoreductase (NADH-methemoglobin reductase) of Vitreoscilla usually copurifies along with VHb (15, 45), indicating its close association with VHb. The cellular level of NADH-methemoglobin reductase increases during the growth cycle, parallel to the increase in VHb that accompanies the falling level of dissolved oxygen (8). Under a variety of growth conditions with various oxygen concentrations, the ratio of VHb to reductase remains fairly constant. These observations led us to assume that a significant proportion of VHb may form the transient VHb-reductase chimera (VHb-R) in vivo, analogous to the flavoHbs and may perform functions other than facilitation of oxygen transfer. Thus, the existence of VHb as a single-domain globin may provide it flexibility to attain two different functional states, either as a homodimer or as a heterodimer with reductase that may be functionally relevant in the cellular metabolism of its host.
In the present work, the reductase domain of flavoHb (FHP) from Ralstonia eutropha (4) was appended at the C terminus of VHb to create the VHb-R chimera very similar to the flavoHbs of bacteria and yeast. The functional implications of the VHb-reductase association was studied under various environmental stress conditions utilizing a recombinant E. coli mutant lacking flavoHb. Our results provide new insight into the functional role of this unique bacterial hemoglobin.
MATERIALS AND METHODS
Strains, plasmids, and culture condition.
E. coli strains DH5α and JM109 were utilized routinely for the cloning and expression of recombinant genes. HMP lacking E. coli, RB9060 (Δhmp), and its parent strain, YCM10, were generously provided by Alex Ninfa (University of Michigan). HMP overexpressing strain RSC521, carrying recombinant plasmid pPL341, was a kind gift from N. E. Dixon (The Australian National University). Plasmid pUC8:16 (7) was utilized for the expression and production of VHb in E. coli. Recombinant plasmid pGE276 (4), overproducing FHP from R. eutropha, was utilized for retrieving the flavoreductase domain. E. coli strains carrying different recombinant plasmids were grown in Luria-Bertani medium supplemented with 100 μg of ampicillin/ml at 37°C at 200 rpm. Restriction and modification enzymes were obtained from New England Biolabs (Beverly, Mass.).
Gel filtration analysis of VHb.
Size exclusion chromatography of VHb was performed at room temperature on a Superdex-75 column by using the Pharmacia FPLC System. A total of 100 μl of protein dialyzed in 0.05 M phosphate buffer (pH 7.2) was applied on the column in different concentration ranges. The column was preequilibrated with 150 mM phosphate buffer (pH 7.2) before the protein sample was applied. The absorption at 280 and at 410 nm was measured to monitor the elution profile of proteins and VHb, respectively. The molecular weight of the eluted protein was estimated by comparing it with standard molecular weight markers.
Integration of flavoreductase domain of FHP at the C terminus of VHb and construction of VHb-R.
Construction of the VHb-R chimera was done by in-frame fusion of the flavoreductase module of FHP at the carboxy terminus of VHb. The reductase domain of fhp gene encoding flavoHb of R. eutropha was retrieved from the fhp gene present on the recombinant plasmid pGE276 (4) after PCR amplification. Oligonucleotide primers corresponding to the reductase domain of FHP and the VHb gene were designed on the basis of their known oligonucleotide sequences (4, 24) and were as follows: 1, 5′-AGAATTCGGGGTTAAAAGTATTTGTGATTTGATCACAATTA-3′; 2, 5′-ACCCGGGTTCAACTGCTTGAGCGTACAAATC-3′; 3, 5′-ACCCGGGGGCTGGAAAGGGTGGCGCACTTTTGTG-3′; and 4, 5′-GGGATCCGGAACGACCGGTCCCTGACCCAGAAAACA-3′.
The vgb gene was amplified by PCR with oligonucleotides 1 and 2, which resulted in the incorporation of a SmaI site at the C terminus of the gene just after the coding region (thus, the translation termination codon was omitted from the VHb). The fhp reductase domain was separately amplified by using oligonucleotides 3 and 4, and thus the SmaI site was introduced into the gene at the N terminus of the start site of the coding region of the FHP reductase domain. The two PCR-amplified products were cloned separately on the plasmid vector pBluescript KS(+), and the authenticity of the genes were confirmed through nucleotide sequence analysis of the cloned genes. The reductase gene was then appended in frame at the C terminus of the vgb gene by using the SmaI site. The nucleotide sequence of the junction point of the vgb and the fhp reductase genes is given in Fig. 3. The recombinant plasmid pVHb-R, encoding the VHb and the flavoreductase fusion product, VHb-R, under the vgb gene promoter, was used to express chimeric VHb protein in E. coli.
FIG. 3.
Characteristics of VHb-R chimera. (A) Schematic representation of VHb-R. Amino acid residues from 1 to 146 of VHb were fused with the reductase domain of FHP from R. eutropha (amino acid residues 147 to 402). The linker region is derived from the FHP, and its sequence at the junction of VHb and the reductase domain of VHb-R is shown. Since VHb is one residue shorter than the heme-binding domain of FHP, the residues following VHb have been numbered in continuation of VHb (i.e., amino acid residues 147 to 402 here refer to amino acid residues 148 to 403 in FHP). (B) Expression of VHb-R in E. coli. The recombinant plasmids pUC8:16 and pVHb-R were transformed in E. coli DH5α for the expression of VHb and VHb-R, respectively, and the cells carrying these plasmids were grown at 37°C at 100 rpm for 12 h for the expression of VHb and VHb-R. E. coli RSC521 carrying recombinant plasmid pPL341 for the constitutive expression of hmp gene was grown at 37°C for 10 h at 180 rpm. The total cellular protein content (25 to 30 μg) of recombinant E. coli was resolved by SDS–12.5% PAGE, and protein bands were visualized after Coomassie blue staining. M, molecular weight markers. Lane 1, E. coli cell lysate (control); lane 2, cell lysate containing VHb; lane 3, cell lysate containing HMP (44 kDa); lane 4, cell lysate containing VHb-R. (C) Western blot showing cross-reactivity of anti-VHb antibodies with VHb-R. Samples were run on a 12.5% gel prior to Western blotting. Lane 1, VHb-R; lane 2, HMP; lane 3, VHb; lane 4, control.
Comparison of intersubunit contacts in FHP and VHb.
Coordinates of VHb and FHP were retrieved from the Protein Data Bank (1) with the codes 1VHb and 1CQX, respectively. These coordinates were used to construct the model of the VHb-reductase interaction by using program “O” (23). Amino acid residues involved in the interdomain interaction were identified by calculating the accessible surface area of the reductase and VHb individually by using standard computer programs (19, 42) and then compared with those of the intersubunit contact region of FHP.
Determination of NO consumption activity of E. coli carrying VHb and VHb-R.
A Clark-type NO electrode (World Precision Instruments), immersed in a stirred glass vial, was used to measure NO uptake by bacteria or cell extract. A saturated solution of nitric oxide (NO) was prepared in a vessel thoroughly flushed with nitrogen gas according to the procedure described by Poole et al. (35). The container was sealed under nitrogen, and a saturated solution of NO was taken out by using a Hamilton syringe. The concentration of saturated solution of NO was 1.8 mM at room temperature. The nitric oxide dioxygenase activity of isolated VHb and VHb-R was measured in 2 ml of 50 mM potassium phosphate buffer (pH 7.8) containing 1 μM NO and 100 μM NADH as described earlier (13, 14, 30). The cellular NO consumption was measured in a 2-ml reaction mixture in 50 mM phosphate buffer with 10 mM glucose, 20 U of glucose oxidase, and 150 U of catalase (bovine liver catalase; Sigma). Different amounts of cells or cell extract were injected along with 1 μM NO, and the rate of NO consumption was determined from the depletion of NO from the reaction mixture.
Purification of VHb-R.
E. coli cells carrying the recombinant plasmid, pVHb-R, were grown overnight at 37°C at 150 rpm. Cells were harvested after centrifugation and disrupted in a French pressure cell. The lysate was clarified by centrifugation at 16,000 × g. The cell extract was then applied to a DEAE-Sepharose CL-6B (Sigma) column and washed with five column volumes of 20 mM potassium phosphate buffer (pH 7.6) and 50 mM KCl. VHb-R was eluted with linear gradient of 50 to 150 mM KCl, and fractions exhibiting a 410/280-nm ratio larger than 0.06 were pooled and concentrated. VHb-R was further purified as a 44-kDa protein by size exclusion chromatography, which resulted in a >90% pure protein preparation.
Determination of cell respiration in the presence of NO.
Oxygen consumption by cells or cell extract was measured polarographically with a Yellow Springs Instrument model 55 oxygen monitor at 37°C in 3 ml of air-saturated 0.1 M potassium phosphate buffer (pH 7.5). The electrode was calibrated with air-equilibrated water and upon adding sodium dithionite to achieve anoxia. Then, 1 ml of cell culture was concentrated by centrifugation at 14,000 rpm (the number of cells/milliliter was calculated simultaneously by plating on LB medium) and washed twice with 0.1 M potassium phosphate buffer (pH 7.5), and the resulting pellet was added quantitatively to 3 ml of air-saturated buffer. The change in the oxygen concentration of the buffer containing the cells was monitored for 5 min. NO was injected with a Hamilton syringe in the test sample to observe the effect of NO on cell respiration.
Determination of sensitivity of E. coli to sodium nitroprusside.
Wild-type E. coli cells were transformed with the recombinant plasmids pUC8:16 and pVHb-R, encoding VHb and VHb-R, respectively. The expression levels of VHb and VHb-R were 10 and 8% of the total cellular protein, respectively, as determined by densitometry of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Control and recombinant E. coli cells carrying VHb or VHb-R were grown aerobically in LB medium upto an optical density at 600 nm of 0.6. The cells were treated with different concentrations of sodium nitroprusside for 60 min and then serially diluted and plated on LB medium to check the survival of cells. Results were expressed as the percentage of viable cells present in the control culture without any treatment.
RESULTS
Sequence comparison and structural analysis of the intersubunit contact region of VHb and FHP.
The globin domains of flavoHbs discovered thus far exhibit remarkable sequence identity with the VHb (41). At present, the three-dimensional structure of only the R. eutropha flavoHb (FHP) is available (11). VHb exhibits 53% sequence identity and close structural analogy with the three-dimensional structure of the globin module of FHP. Structural and sequence comparisons of VHb with the globin segment of R. eutropha FHP suggests that functional regions important for the interactions with flavoreductase is also conserved in VHb (Fig. 1). In FHP, interaction between the heme- and flavin-binding domains mainly occurs through H and CE helices. A salt bridge is formed between the heme- and FAD-binding domains of FHP through Lys84, which is a conserved residue in other flavoHbs (41). VHb also exhibits the presence of lysine (Lys84) at this position (Fig. 1), indicating that it may interact with its partner flavoreductase in an analogous manner. Additionally, the H helix of FHP extends into the flavin domain and remains in the close vicinity of the FAD-binding region (11). In VHb, the corresponding region of the H helix (Glu116 to Gln143) is also extended (41) and may function similarly. Another functionally important segment of R. eutropha FHP is the CE region of its heme domain. It adopts an extended conformation that generates contact and hydrogen bonding with the adenine portion of FAD through the residues His47 and Gln48. Analogous residues in VHb are Arg47 and Gln48 (Fig. 1). The glutamine residue at position 48 is fully conserved in flavoHbs discovered so far, indicating a key role for this residue both in VHb and in flavoHbs. The higher flexibility of the CE region in VHb further suggests that it may be important for the interaction with FAD-binding protein in a way that is very similar to that of the FHP of R. eutropha. Comparison of intersubunit contact region of FHP with that of corresponding regions of VHb indicates that the majority of side chain interactions between the globin and the reductase module in FHP may be more or less the same in VHb on its association with a reductase partner (Fig. 1). In order to understand the precise nature of subunit interaction of VHb, structural features of the VHb interface were inspected in the VHb three-dimensional structure model generated by using coordinates of VHb available from the protein data bank (accession codes 1VHb and 2VHb). One of the prominent features of the VHb structure has been a very small subunit interface between its two monomers. The accessible surface area for VHb dimerization was found to be ca. 905 Å2 by using different computer programs (19, 42). Although this value was relatively higher than the previously reported value (41), it is nonetheless small compared to typical buried accessible areas for other dimeric proteins (22, 32). The small amount of intersubunit contact suggests a weak association of the two monomers of VHb. Interestingly, comparison of the VHb interface with that of globin and the flavoreductase of FHP indicated a partial overlap of the interface involved in the dimerization of the VHb and FHP globin-flavoreductase interaction. Thus, association of VHb with reductase possibly disrupts the VHb homodimeric structure that may be facilitated by small intersubunit contacts in the VHb homodimer.
FIG. 1.
Sequence alignment of globin domains of FHP and VHb. Identical residues are marked (✽), and functionally similar amino acid residues are indicated by a period. Amino acid residues of FHP interacting with the FAD-binding domain and the corrsponding region of VHb are shaded. Amino acid residues of VHb participating in homodimeric assembly of VHb and overlapping with the FAD-binding domain of FHP are indicated in boldface.
Pattern of homodimeric assembly of VHb.
Detail comparison and analysis of structural features of the VHb interface suggested that the VHb homodimer may dissociate easily. Since the cellular concentration of VHb is induced more than 50-fold when cells are exposed to oxygen limitation (2, 9, 26), we checked whether the cellular concentration of the VHb affects its conformational state and self-association properties. The relative levels of monomers and homodimers in the total VHb population were examined at different protein concentrations by using gel filtration chromatography. Figure 2A shows gel filtration elution profiles of the VHb at high and low protein concentrations. Under identical experimental conditions, at protein concentrations of 10 mg/ml, the majority of the VHb eluted with an apparent size of 29 kDa, indicating the dimeric state of VHb. When VHb was analyzed at a lower protein concentration (0.05 mg/ml), the elution profile exhibited the appearance of another peak corresponding to 15 kDa, along with the dimeric species of VHb (Fig. 2B) that constituted 25 to 30% of total protein. The presence of a significant level of monomeric VHb at a lower protein concentration indicated that the intersubunit contact of VHb may not be very strong and can easily disrupt under specific physiological conditions.
FIG. 2.
Gel filtration elution profile of VHb with a linear NaCl gradient at 10 (A) or 0.05 (B) mg/ml.
Integration of the reductase domain of FHP with VHb and characterization of the VHb-R chimera.
Looking into the possible interaction of VHb with flavoreductase in vivo, we attempted to understand the physiological role of this association. Since primary and the three-dimensional structure of VHb shares maximum identity with the N-terminal, heme-binding domain of FHP and since both the globin- and flavin-binding reductase domains of FHP fold independently (11), we genetically engineered the flavin-binding oxidoreductase module of FHP at the C-terminal end of the VHb to generate the VHb-R chimera (Fig. 3A). This fusion protein was overexpressed in E. coli under the oxygen-regulated promoter of VHb as a 44-kDa protein (Fig. 3B). Expression of the VHb-R chimera conferred a pinkish tinge to the recombinant cells, indicating that the heme-binding VHb module of this fusion protein is functional. VHb-specific antibodies raised against native VHb cross-reacted with VHb-R (Fig. 3C), suggesting that the globin module of VHb-R is similar to VHb. CO difference spectra of cell lysate carrying VHb-R exhibited a characteristic Soret peak at 419 nm, as has been observed in the case of native VHb (data not shown). The CO difference spectrum of control E. coli cells did not display these characteristics, indicating that the recorded CO-binding activity is due to overexpression of VHb-R and is not an artifact of E. coli flavoHb expression. The emission spectrum (excited at 460 nm) of VHb-R showed its peak at 532, which was absent in native VHb, indicating that the VHb-R carries a flavin cofactor native unlike that of VHb. These observations suggest that VHb-R has independently folded into distinct heme- and flavin-binding domains in a way that is very similar to that of flavoHbs.
Overproduction of VHb-R in an hmp null mutant of E. coli provides NO consumption capability.
A NO detoxification function for flavoHbs with associated reductases has been suggested by the identification of the efficient nitric oxide dioxygenase activity in E. coli with flavoHb (13, 36). In order to check that VHb and VHb-R exhibit this characteristic, vgb and vgbr genes encoding VHb and VHb-R, respectively, were overexpressed in an hmp null mutant of E. coli (RB 9060), and their NO consumption activity was monitored. Whole cells or cell extract of E. coli 9060 displayed no measurable NO consumption when tested alone. However, a slow but measurable NO utilization was observed by whole cells and cell extract of wild-type E. coli cells (Table 1). When E. coli cells carrying VHb were examined, a distinct NO consumption activity was observed in these cells. VHb-R-carrying cells exhibited an approximately twofold-higher NO utilization ability than did cells carrying only VHb. SDS-PAGE analysis of E. coli carrying VHb and VHb-R indicated that the expression levels of these two proteins were ca. 10 and 8% of the total cellular protein, respectively. These results suggested that the association of the reductase domain with VHb results in a functional protein capable of catalytically consuming NO. The slower-NO-utilizing capability of VHb may be due to partial association of alternative flavoreductase (e.g., cytochrome P450 reductase) with VHb in vivo during overexpression of VHb, where a significant proportion of VHb remains in a dimeric state.
TABLE 1.
Nitric oxide (NO) utilization of recombinant E. coli carrying VHb and VHb-R
| E. coli strain | NO consumptiona in:
|
|
|---|---|---|
| Cell culture (nmol/min/108 cells) | Cell extract (nmol/min/mg of protein) | |
| YCM10 (hmp+) | 6 (2.4) | 30 (6.5) |
| RB9060 (Δhmp) | ND | ND |
| DH5α | 6.8 (2.5) | 36.5 (5) |
| DH5α (VHb) | 15.6 (3.5) | 118 (16.2) |
| DH5α (VHb-R) | 33 (5.1) | 255 (24.2) |
The standard deviation is given in parantheses. For cell culture, cells grown for 8 to 10 h were suspended in 2 ml of oxygen-saturated reaction buffer and exposed to NO (1 μM) as described in Materials and Methods. For the cell extract, cells were harvested from cells grown 10 to 12 h by centrifugation (14,000 rpm) and then lysed in 50 mM potassium phosphate buffer (pH 7.8) by sonication.
The presence of VHb-R protects cellular respiration from NO toxicity.
One of the prime cellular targets for the toxic effect of NO and related reactive nitrogen species may be respiratory enzymes (40); therefore, the oxygen uptake capabilities of recombinant E. coli carrying VHb, VHb-R, and HMP were compared in the presence or absence of NO. In the absence of NO, the respiration of wild-type and hmp mutant E. coli strains remained more or less similar, as has been observed previously by Stevanin et al. (40), whereas cells carrying VHb and VHb-R exhibited a nearly 20 to 30% higher oxygen uptake rate than the wild-type cells. The injection of NO solution into a respiring cell suspension of E. coli immediately inhibited oxygen uptake. To test that the VHb-reductase association confers protection from the NO-mediated toxicity on cellular respiration, the respiration rate of E. coli carrying VHb and VHb-R was determined and compared with that of E. coli cells overexpressing HMP (Table 2). Under aerobic conditions, the oxygen uptake by an hmp mutant of E. coli was completely blocked in the presence of 10 μM NO, whereas wild-type E. coli exhibited a ca. 40% inhibition in oxygen uptake compared to that of the control cells. The result shows that even the basal level of HMP provided a protective effect against NO toxicity.
TABLE 2.
Effect of NO addition on cellular respiration of E. coli
| E. coli strain | Oxygen uptake (μmol/min/108 cells)a in:
|
||
|---|---|---|---|
| Control | NO (10 μM) | NO (20 μM) | |
| YCM10 (hmp+) | 7.5 (1.4) | 4.3 (0.9) | 0.5 (0.1) |
| RB9060 (Δhmp) | 6.8 (1.3) | 0.3 (0.05) | 0.0 |
| DH5α | 7.1 (1.6) | 4.1 (1.1) | 0.8 (0.2) |
| DH5α (VHb)b | 9.8 (1.5) | 8.1 (1.6) | 3.6 (1.1) |
| DH5α (VHb-R)b | 9.1 (1.6) | 9.9 (1.5) | 8.6 (1.4) |
| RSC521 (HMP)b | 7.9 (1.1) | 7.3 (1.4) | 6.9 (1.1) |
Prior to oxygen uptake measurements, the 0.1 M potassium phosphate buffer was stirred vigorously for 30 min to be air saturated (DO = 250 μM). One mililiter of cell culture (the cell number was simultaneously determined by plating on LB medium) was concentrated by centrifugation at 14,000 rpm for 10 min and added quantitatively to 4 ml of air-saturated buffer. The change in the oxygen concentration of the buffer containing the cells was recorded with or without NO addition. Data are the means of four to five individual measurements. Values in parentheses are the standard deviations.
Recombinant plasmids pUC8:16 and pVHb-R were transformed in E. coli DH5α for the overexpression of VHb and VHb-R, respectively. E. coli RSC521, carrying plasmid pPL341, was utilized for the constitutive expression of HMP. Densitometric scanning of cellular proteins of recombinant E. coli, separated by SDS-PAGE, indicated that VHb, VHb-R, and HMP constituted 10, 8, and 6% of the total cellular proteins, respectively.
These results are in line with the previous observations made by Stevanin et al. (40). When 10 μM NO was added to a cell suspension of E. coli cells overexpressing VHb and VHb-R, there was no significant effect on oxygen uptake of these cells (Table 2), indicating that the presence of both VHb and VHb-R provide protection to the cellular respiration of E. coli from NO toxicity. However, when NO was added at a higher concentration (20 μM), VHb-carrying E. coli exhibited partial inhibition of cellular respiration (56% of the control cells), whereas the respiratory activity of VHb-R-producing cells was not significantly different than that of cells not exposed to NO. E. coli cells overproducing HMP were more or less insensitive to 10 μM NO exposure. These results show that the presence of VHb and VHb-R, like HMP, provides protective effect against a toxic level of NO and that the level of protection provided by VHb-R was higher than that provided by VHb alone.
Functional characteristics of purified VHb-R.
Since the results presented above indicated that both VHb and VHb-R protect the cellular respiration of recombinant E. coli from NO toxicity, a logical next step was to test the interaction of NO with the purified preparation of these proteins. The addition of 3.5 μM NO to the purified preparation of VHb-R resulted in the immediate depletion of NO in the presence of 0.1 mM NADH, resulting in the complete removal of NO within 15 s. Compared to the VHb-R, the purified preparation of VHb exhibited a distinctly lower rate of NO removal and depleted NO from the reaction mixture in ca. 50 s under similar experimental conditions (Fig. 4). VHb-R exhibited a nearly twofold increase in NO uptake compared to that of VHb alone. It is quite possible that the protein preparation of VHb carries some minor flavoreductase contamination since it has been observed earlier that a flavoreductase (NADH-methemoglobin reductase) usually copurifies along with VHb (15) and thus may be contributing to the slow NO uptake via VHb. Alternatively, the association of native Vitreoscilla reductase may be essential for achieving its full potential for the NO consumption activity. In the absence of VHb-R or VHb, the addition of NO elicited a very slow removal of NO that may be due to the nonenzymatic reaction of NO with oxygen. These results reveal that the ability of VHb to interact with NO is greatly enhanced in association with its partner reductase.
FIG. 4.
NO consumption activity of purified VHb and VHb-R. NO consumption in purified VHb and VHb-R protein preparation was determined from the rate of NO removal after the addition of 3 μM NO in 2 ml of 50 mM potassium phosphate buffer (pH 7.8) containing 100 μM NADH (solid line), buffer containing VHb (dotted line), or buffer containing VHb-R (dashed line). The heme contents of VHb and VHb-R in the sample were 0.95 and 0.68 mol, respectively. NO signal was recorded as arbitrary units according to the published procedure (16). One unit of NO signal represents 0.1 μM NO.
The presence of VHb and VHb-R improves growth and cell viability under nitrosative stress conditions.
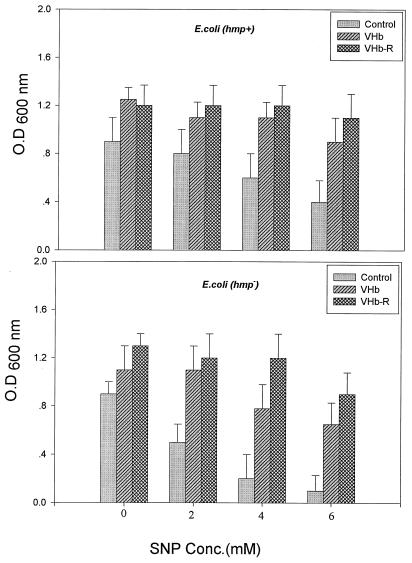
We next addressed the question of whether the VHb-R association could elicit a similar response in vivo and provide protection to its host cells from nitrosative stress. We therefore studied the physiological response of recombinant E. coli carrying VHb or VHb-R in the presence of the nitrosating agent SNP. Wild-type E. coli exhibited >60% cell survival after treatment with 2 mM SNP. Under similar conditions, only 30 to 35% of the cells were able to survive in the case of the hmp mutant, suggesting an enhanced sensitivity of E. coli toward SNP in the absence of flavoHb. VHb- and VHb-R-overexpressing cells of wild-type and hmp mutant E. coli were more or less insensitive to 2 mM SNP. This result was further supported by the growth profile of E. coli carrying VHb and VHb-R in the presence of SNP (Fig. 5). The growth rate and cell density of both wild-type and hmp mutant E. coli strains was enhanced when VHb was overexpressed in these cells. The growth pattern of VHb-R carrying cells in 2, 4, and 6 mM SNP was more or less similar to that of the control cells that were not exposed to this nitrosative agent. In contrast, control cells not having any hemoglobin-like protein exhibited ca. 40 and 60% growth inhibition in the presence of 4 and 6 mM SNP, respectively. Taken together, these data show that both VHb and VHb-R relieve the cytotoxic effect of SNP. It is quite possible that a part of VHb actually associates with flavoreductase of E. coli in vivo and functions like a flavoHb which, in turn, may provide the protective effect since a basal level of HMP in E. coli is sufficient to confer protection against nitrosative stress (40). The presence of VHb-R provides significantly higher cell survival than VHb, suggesting that the association of flavoreductase with VHb enhances its capability to cope better with the toxic effect of a nitrosative stress response.
FIG. 5.
Effects of sodium nitroprusside on growth profile of recombinant E. coli carrying VHb and VHb-R. Growth of wild-type and hmp mutant E. coli strains carrying recombinant plasmids pUC8:16 and pVHb-R encoding VHb and VHb-R, respectively, was initiated in phosphate-buffered LB medium in the presence of the indicated concentration of SNP with 1% inoculum of overnight seed culture. Cultures were grown at 37°C for 12 h at 180 rpm, and the final cell optical density (O.D.) of each culture was recorded at 600 nm. Data are presented as averages of results of four individual experiments.
DISCUSSION
The primary structure of the VHb is closely related to the N-terminal heme-binding domain of bacterial and yeast flavoHbs. VHb lacks a C-terminal reductase module and exists as single domain hemoglobin. A flavin containing reductase that cofractionates with VHb has been isolated and purified from the Vitreoscilla (15, 21), suggesting that a noncovalent assembly of these proteins in vivo could yield a functional two chain hybrid very similar to flavoHbs. Evolutionary implications of the separation of heme and reductase domains in Vitreoscilla cellular metabolism, however, remain unclear.
In the present study, we have demonstrated that the association of a flavoreductase module with VHb may generate a flavoHb-like hybrid protein which may interact with nitric oxide and help to relieve the deleterious cellular response under nitrosative stress. The structural features of VHb provide strong evidence for the VHb-reductase association. This is supported by the fact that the functional residues of the heme-binding domain of flavoHb, which is involved in the interaction with the flavoreductase, are also conserved in VHb. Furthermore, the subunit interaction of dimeric VHb is relatively weak and can easily dissociate in order to interact with its partner flavoreductase. Since the interface of the VHb dimer partially overlaps with the functional region of VHb presumed to be involved in the interaction with flavoreductase (Fig. 1), the association of flavoreductase with VHb may disrupt the dimeric state of VHb due to weak subunit interaction of the VHb dimer. The appearance of monomeric VHb at a low protein concentration supports the structural prediction that the VHb homodimeric assembly may be relatively weak. It may be functionally relevant for the VHb since the weak subunit interaction of the VHb may allow it to dissociate easily into a monomer and interact with the reductase to form a flavo-VHb-like structure.
If VHb associates with its flavoreductase partner in a manner very similar to that of the flavoHbs, then what could be the evolutionary implications of separation of the heme- and flavin-binding domains? VHb is hyperinduced under hypoxic condition in its native host Vitreoscilla, and the recombinant host, E. coli. Based on its oxygen-binding properties (45) and oxygen-dependent biosynthesis (2, 8), it has been proposed that the function of the VHb is to facilitate oxygen flux to the vigorously respiring membranes of Vitreoscilla sp., which is an obligate aerobe usually found in a hypoxic habitat (37). Genetic engineering of VHb in several industrial microbial hosts (25, 27, 28), animal cell lines (33), and transgenic plants (18) enhances their oxygen-utlizing capability and results in higher productivity. A major fraction (40 to 50%) of cellular VHb remains concentrated near the respiratory membranes and exhibits strong affinity for the membrane-binding and cytochrome bo encapsulated into the proteoliposomes (39). These properties are consistent with its putative role of sequestering oxygen from the environment and feeding it to the respiratory terminal oxidases (45), thereby facilitating respiration under hypoxic conditions.
Recent research has revealed a special role in nitric oxide metabolism or detoxification for hemoglobins associated with flavoreductases (3, 14, 30, 31, 35). Both the VHb and the VHb-R chimera are capable of utilizing NO, protect cellular respiration from NO toxicity, and relieve the nitrosative stress of E. coli. However, integration of the reductase domain with the VHb provides an enhanced capability for NO consumption and exhibits better protection from nitrosative stress. Since the NO uptake capability of purified VHb is significantly lower than that of the VHb-R, it can be assumed that the partial protection provided by VHb may possibly be due to its interaction with the E. coli reductase in vivo. It is quite plausible that dimeric VHb may solely participate in maintaining cellular oxygen flux and facilitating oxygen transfer, whereas the VHb-flavoreductase association may be involved in an additional function(s) by modulating the cellular response toward reactive nitrogen species, which are common products of organic and decaying reactions of hypoxic habitat, e.g., stagnant ponds, rotten vegetables, and cowdung, etc., where Vitreoscilla species are usually found (37). Although a flavoreductase has been found usually associated with VHb (15, 21), how VHb interacts with flavoreductase in vivo is not known as yet and remains to be determined. However, the present study demonstrates that the association of the reductase with the VHb improves its capability for NO consumption and the detoxification of nitrosative stress. FlavoHbs have been identified in a number of prokaryotes and eukaryotes, including E. coli (43), Bacillus subtilis (29), Erwinia spp. (12), and serovar Typhimurium (6). Although the functions of all the flavoHbs have not been established as yet, HMPs from yeast, E. coli, and serovar Typhimurium have been shown to protect cells from NO toxicity (6, 14, 30, 40). Accordingly, NO detoxification is probably an evolutionarily conserved function of hemoglobin-flavoreductase hybrid proteins. Whether VHb-reductase association has any functional implications in the cellular metabolism of Vitreoscilla is not yet known and remains to be elucidated.
Considering the beneficial effect of VHb on growth and product formation in several heterologous systems (18, 25, 27, 33), its unique structural characteristics (41), and the conservation of functional residues in the VHb important for the interaction with the reductase, it could be argued that the VHb may have a functional advantage in remaining as a single domain. Unusual structural characteristics of VHb may allow it to remain in two functional states in vivo and perform multiple functions in the cellular metabolism of its natural host, Vitreoscilla sp., which is an obligate aerobe but thrives in an oxygen-poor environment and is unable to ferment sugars. Its metabolism is mainly limited to aerobic oxidation of amino acids (37). The capability of the VHb to remain in more than one form, therefore, may give it flexibility to face the environmental stress of its natural habitat and to cope with the oxygen limitation simultaneously to carry on its aerobic metabolism. In the homodimeric state, due to its unusual distal site conformation, it can bind oxygen and facilitate oxygen metabolism within the cellular environment whereas, due to its unique structural characteristics, it can dissociate under physiological conditions, pairs easily with the reductase domain, and attains a flavoHb-like structure. Experiments are under way to study the kinetics of interaction of the VHb with the NADH-methemoglobin reductase isolated from Vitreoscilla.
It is assumed that ancestral hemoglobins appeared in bacteria at a time when the earth’s atmosphere was anoxic and NO was abundant in that primordial atmosphere (16). Thus, the primary function of ancestral hemoglobins must have been the detoxification of atmospheric NO. The origin of oxygenic photosynthetic cyanobacteria and free oxygen in the atmosphere (1.8 billions years ago) resulted in an aerobic atmosphere and aerobic metabolism. The presence of a limiting amount of atmospheric oxygen probably caused ancient hemoglobins to evolve into oxygen storage and transport proteins. It is interesting that species related to Vitreoscilla have been previously known as colorless cyanobacteria (37). The separation and interaction of heme- and flavin-binding domains, the close similarity of flavoHbs, and the oxygen-binding characteristics of VHb suggest that Vitreoscilla single-domain hemoglobin might represent an interesting link between ancient NO-metabolizing flavoHbs and oxygen-transporting hemoglobins.
Acknowledgments
We thank B. Friedrich for providing plasmid pGE276 and Alex Ninfa for the hmp mutant of E. coli.
Financial support was provided by C.S.I.R (R.K. and P.R.) and the Indian Department of Science and Technology.
REFERENCES
- 1.Berman, H. M., J. Westbrook, Z. Feng, G. Gililand, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boerman, S., and D. A. Webster. 1982. Control of heme content in Vitreoscilla by oxygen. J. Gen. Appl. Microbiol. 28:35–43. [Google Scholar]
- 3.Buisson, N., and R. Labbe-Bois. 1998. Flavohemoglobin expression and function in Saccharomyces cerevisiae. No relationship with respiration and complex response to oxidative stress. J. Biol. Chem. 273:9527–9533. [DOI] [PubMed] [Google Scholar]
- 4.Cramm, R., R. A. Siddiqui, and B. Friedrich. 1994. Primary sequence and evidence for a physiological function of the flavohemoprotein of Alcaligenes eutrophus. J. Biol. Chem. 269:7349–7354. [PubMed] [Google Scholar]
- 5.Crawford, M. J., D. R. Sherman, and D. E. Goldberg. 1995. Regulation of Saccharomyces cervisiae flavohemoglobin expression. J. Biol. Chem. 270:6997–7003. [DOI] [PubMed] [Google Scholar]
- 6.Crawford, M. J., and D. E. Goldberg. 1998. Regulation of the Salmonella typhimurium flavohemoglobin gene. A new pathway for bacterial gene expression in response to nitric oxide. J. Biol. Chem. 273:34028–34032. [DOI] [PubMed] [Google Scholar]
- 7.Dikshit, K. L., and D. A. Webster. 1988. Cloning, characterization and expression of the bacterial globin gene from Vitreoscilla in Escherichia coli. Gene 70:377–386. [DOI] [PubMed] [Google Scholar]
- 8.Dikshit, K. L., D. Spaulding, A. Braun, and D. A. Webster. 1989. Oxygen inhibition of globin gene transcription and bacterial haemoglobin synthesis in Vitreoscilla. J. Gen. Microbiol. 135:2601–2609. [DOI] [PubMed] [Google Scholar]
- 9.Dikshit, K. L., R. P. Dikshit, and D. A. Webster. 1990. Study of Vitreoscilla globin (vgb) gene expression and promoter activity in E. coli through transcriptional fusion. Nucleic Acids Res. 18:4149–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dikshit, R. P., K. L. Dikshit, Y. Liu, and D. A. Webster. 1992. The bacterial hemoglobin from Vitreoscilla can support the aerobic growth of Escherichia coli lacking terminal oxidases. Arch. Biochem. Biophys. 293:241–245. [DOI] [PubMed] [Google Scholar]
- 11.Ermler, U., R. A. Siddiqui, R. Cramm, and B. Friedrich. 1995. Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 Å resolution. EMBO J. 14:6067–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favey, S., G. Labesse, V. Vouille, and M. Boccara. 1995. Flavohemoprotein HMPX: a new pathogenicity determinant in Erwinia crysanthemi strain 3937. Microbiology 141:863–871. [DOI] [PubMed] [Google Scholar]
- 13.Gardner, A. M., L. A. Martin, and P. R. Gardner. 2000. Steady state and transient kinetics of Escherichia coli nitric oxide dioxygenase (flavohemoglobin). The B10 tyrosine hydroxyl is essential for dioxygen binding and catalysis. J. Biol. Chem. 275:12581–12589. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, P. R., A. M. Gardner, L. A. Martin, and A. L. Salzman. 1998. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. USA 95:10378–10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzales-Prevatt, V., and D. A. Webster. 1980. Purification and properties of NADH-cytochrome o reductase from Vitreoscilla. J. Biol. Chem. 255:1478–1482. [PubMed] [Google Scholar]
- 16.Hardison, R. 1999. The evolution of hemoglobin. Am. Scientist 87:126–137. [Google Scholar]
- 17.Hausladen, A., A. J. Gow, and J. S. Stamler. 1998. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. USA 95:14100–14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmberg, N., G. Lilius, J. E. Bailey, and L. Bulow. 1997. Transgenic tobacco expressing Vitreoscilla hemoglobin exhibits enhanced growth and altered metabolite production. Nat. Biotechnol. 15:244–247. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard, S. J., and J. M. Thornton. 1993. NACCESS computer program. Department of Biochemistry and Molecular Biology, University College of London, London, England.
- 20.Iijima, M., H. Shimizu, Y. Tanaka, and H. Urushihara. 2000. Identification and characterization of two flavohemoglobin genes in Dictyostelium discoideum. Cell Struct. Funct. 25:47–55. [DOI] [PubMed] [Google Scholar]
- 21.Jacob, W., D. A. Webster, and P. M. Kroneck. 1992. NADH-dependent methmoglobin reductase from the obligate aerobe Vitreoscilla: improved method of purification and reexamination of prosthetic groups. Arch. Biochem. Biophys. 292:29–33. [DOI] [PubMed] [Google Scholar]
- 22.Janin, J., S. Miller, and C. Chothia. 1988. Surface, subunit interface, and interior of oligomeric proteins. J. Mol. Biol. 204:155–164. [DOI] [PubMed] [Google Scholar]
- 23.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A47:110–119. [DOI] [PubMed] [Google Scholar]
- 24.Joshi, M., S. Mande, and K. L. Dikshit. 1998. Hemoglobin biosynthesis in Vitreoscilla stercoraria DW: cloning, expression, and characterization of a new homolog of a bacterial globin gene. Appl. Environ. Microbiol. 64:2220–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khosla, C., and J. E. Bailey. 1988. Heterologous expression of a bacterial haemoglobin improves the growth properties of recombinant Escherichia coli. Nature 331:633–635. [DOI] [PubMed] [Google Scholar]
- 26.Khosla, C., and J. E. Bailey. 1989. Characterization of the oxygen-dependent promoter of the Vitreoscilla hemoglobin gene in Escherichia coli. J. Bacteriol. 171:5995–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khosla, C., J. E. Curtis, J. DeModena, U. Rinas, and J. E. Bailey. 1990. Expression of intracellular hemoglobin improves protein synthesis in oxygen-limited Escherichia coli. Bio/Technology 8:849–853. [DOI] [PubMed] [Google Scholar]
- 28.Khosravi, M., D. A. Webster, and B. C. Stark. 1990. Presence of the bacterial hemoglobin gene improves α-amylase production of a recombinant Escherichia coli strain. Plasmid 24:190–194. [DOI] [PubMed] [Google Scholar]
- 29.LaCelle, M., M. Kumano, K. Kurita, K. Yamane, P. Zuber, and M. M. Nakano. 1996. Oxygen-controlled regulation of the flavohemoglobin gene in Bacillus subtilis. J. Bacteriol. 178:3803–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, L., M. Zeng, A. Hausladen, J. Heitman, and J. S. Stamler. 2000. Protection from nitrosative stress by yeast flavohemoglobin. Proc. Natl. Acad. Sci. USA 97:4672–4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Membrillo-Hernandez, J., M. D. Coopamah, M. F. Anjum, T. M. Stevanin, A. Kelly, M. N. Hughes, and R. K. Poole. 1999. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser” and paraquat and is essential for transcriptional responses to oxidative stress. J. Biol. Chem. 274:748–754. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell, D. T., G. B. K. Itto, and M. L. Hackert. 1995. Structural analysis of monomeric hemichrime and dimeric cyanomet hemoglobins from Caudina arenicola. J. Mol. Biol. 251:421–431. [DOI] [PubMed] [Google Scholar]
- 33.Pendse, G. J., and J. E. Bailey. 1994. Effect of Vitreoscilla hemoglobin expression in growth and specific tissue plasminogen activator productivity in recombinant Chinese hamster ovary cells. Biotechnol. Bioeng. 44:1367–1370. [DOI] [PubMed] [Google Scholar]
- 34.Mills, C. E., S. Sedelnikova, B. Soballe, M. N. Hughes, and R. K. Poole. 2001. Escherichia coli flavohemoglobin (HMP) with equistoichiometric FAD and haem contents has a low affinity for dioxygen in the absence or presence of nitric oxide. Biochem. J. 353:207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poole, R. K., M. F. Anjum, J. Membrillo-Hernandez, S. O. Kim, M. N. Hughes, and V. Stewart. 1996. Nitric oxide, nitrite and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 178:5487–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole, R. K., and M. N. Hughes. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36:775–783. [DOI] [PubMed] [Google Scholar]
- 37.Pringsheim, E. G. 1951. The Vitreoscillaceae: a family of colourless, gliding, filamentous organisms. J. Gen. Microbiol. 5:124–149. [DOI] [PubMed] [Google Scholar]
- 38.Probst, I., G. Wolf, and H. G. Schlegel. 1979. An oxygen binding flavohemoprotein from Alcaligenes eutrophus. Biochim. Biophys. Acta 576:471–478. [DOI] [PubMed] [Google Scholar]
- 39.Ramandeep, K. W. Hwang, M. Raje, K. J. Kim, B. C. Stark, K. L. Dikshit, and D. A. Webster. 2001. Vitreoscilla hemoglobin: intracellular localization and binding to membranes. J. Biol. Chem. 276:24781–24789. [DOI] [PubMed] [Google Scholar]
- 40.Stevanin, T. M., N. Ioannidis, C. E. Mills, S. O. Kim, M. N. Hughes, and R. K. Poole. 2000. Flavohemoglobin HMP affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo′ or bd′ from nitric oxide. J. Biol. Chem. 275:35868–35875. [DOI] [PubMed] [Google Scholar]
- 41.Taricone, C., A. Galizzi, A. Coda, P. Ascenzi, and M. Bolognesi. 1997. Unusual structure of the oxygen binding site in the dimeric bacterial hemoglobin from Vitreoscilla sp. Structure 5:497–507. [DOI] [PubMed]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasudevan, S. G., W. I. Armarego, D. C. Shaw, P. E. Lilley, N. E. Dixon, and R. K. Poole. 1991. Isolation and nucleotide sequence of hmp gene that encodes a hemoglobin-like protein in E. coli K-12. Mol. Gen. Genet. 226:49–58. [DOI] [PubMed] [Google Scholar]
- 44.Wakabayashi, S., H. Matsubara, and D. A. Webster. 1986. Primary sequence of a dimeric bacterial hemoglobin from Vitreoscilla. Nature 322:481–483. [DOI] [PubMed] [Google Scholar]
- 45.Webster, D. A. 1987. Structure and function of bacterial hemoglobin and related proteins, p.245–265. In G. L. Eichorn and L. G. Magill (ed.), Advances in inorganic biochemistry, vol. 7. Elsevier, New York, N.Y. [PubMed]