Abstract
Sphaerotilus natans, a filamentous bacterium that causes bulking in activated sludge processes, can assume two distinct morphologies, depending on the substrate concentration for growth; in substrate-rich media it grows as single rod-shaped cells, whereas in substrate-limited media it grows as filaments. To identify genes responsible for sheath formation, we carried out transposon Tn5 mutagenesis. Of the approximately 20,000 mutants obtained, 7 did not form sheathed structures. Sequencing of the Tn5-flanking regions showed that five of the seven Tn5 insertions converged at the same open reading frame, designated sthA. The deduced amino acids encoded by sthA were found to be homologous to glycosyltransferase, which is known to be involved in linking sugars to lipid carriers during bacterial exopolysaccharide biosynthesis. Disruption of the gene of the wild-type strain by inserting a kanamycin resistance gene cassette also resulted in sheathless growth under either type of nutrient condition. These findings indicate that sthA is a crucial component responsible for sheath formation.
Activated sludge systems are used worldwide for wastewater treatment. One of the major operational problems of these systems is excessive growth of filamentous bacteria (5, 9, 25). This results in poor settlement of activated sludge flocs, a problem that is commonly referred to as bulking. For years, Sphaerotilus natans has been considered a primary organism that causes bulking, and a number of studies have been carried out with this species (3–7, 16, 18, 21, 22). Although other types of filamentous bacteria are also known to be involved in bulking (4, 5, 8, 10), Jenkins et al. reported that S. natans was the dominant filamentous organism in 12% of 525 bulking and foaming sludge samples in the United States (9). Thus, S. natans is still a noteworthy causative agent of sludge bulking.
S. natans is characterized by a sheathed structure in which long chains of rod-shaped cells are enclosed (7, 12, 24). However, under some culture conditions, this organism grows as individual cells without forming a sheath. Since only the filamentous growth causes bulking of activated sludge, the growth conditions which determine the cell form have been studied. Gaudy and Wolfe reported that S. natans grew as single cells in the presence of 0.5% glucose and 0.5% peptone but grew as filaments in the presence of 0.1% glucose and 0.1% peptone (6). However, neither additional studies of sheath formation nor the pathway of sheath biosynthesis has been described; hence, nothing is known about the direct trigger that regulates expression of the genes for sheath biosynthesis.
To date, the only available information is the chemical structure of the sheath of S. natans. The sheath was initially found to be a complex composed of polysaccharide, protein, and lipid, with the polysaccharide component consisting of glucose, hexosamine, and various other sugars (18). A more recent study showed that the sheath of S. natans is composed of polysaccharide and protein but not lipid, with the polysaccharide component consisting of glucose and N-acetylgalactosamine at a 1:4 molar ratio and the protein component consisting primarily of glycine and cysteine (22). These reports suggested that synthesis and the subsequent assembly of the sugar and amino acid components of the sheath occur in multiple steps. At present, however, little is known about the synthetic pathway and genes responsible for sheath formation.
In this study, in order to examine the pathway of sheath formation and to find a way to prevent bulking, we analyzed the gene that is crucial for sheath formation by obtaining sheath-deficient transposon mutants of S. natans.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used are listed in Table 1. S. natans IAM 12068 was obtained from the Institute of Molecular and Cellular Biosciences, University of Tokyo. This strain was routinely maintained in 0.1% (wt/vol) nutrient broth (NB) (Difco) plates. A rifampin-resistant S. natans mutant was obtained by successively transferring the wild-type strain to 0.1% NB agar plates supplemented with rifampin (20 μg ml−1). This mutant, designated S. natans S-1, was used as the recipient for transposon mutagenesis as described below. Escherichia coli strains were cultured at 37°C in L broth (LB) or on LB agar plates (19). When necessary, the antibiotics ampicillin (50 μg m1−1), kanamycin (50 μg m1−1), and rifampin (20 μg m1−1) were used.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Pertinent feature(s) | Sourcea |
|---|---|---|
| Sphaerotilus natans strains | ||
| IAM 12068 | Wild type | IAM |
| S-1 | Rifr derivative of IAM 12068 | This study |
| TM1 to TM5 | Tn5-induced sheath-deficient mutant (sthA::Tn5) of S-1, Rifr Kmr | This study |
| TM6 | Tn5-induced sheath-deficient mutant of S-1, Rifr Kmr | This study |
| TM7 | Tn5-induced sheath-deficient mutant of S-1, Rifr Kmr | This study |
| SKM01 | S-1 sthA::Kmr Rifr | This study |
| SGK01 | S-1 sthA-gfp::Kmr Rifr | This study |
| Escherichia coli strains | ||
| DH5α | hsdR17 endA1 recA1 gyrA1 thi relA1 supE44 φ80dlacZδM15 δ(lacZ-argF)U169 | BRL |
| S17-1 | recA pro thi hsdR−M+ chr::RP4-2 | DSMZ |
| Plasmids | ||
| pUC118 | Cloning vector, Apr | Takara Shuzo |
| pT7-Blue | Cloning vector for PCR product, Apr | Novagen |
| pSUP5011 | pBR325 derivative::Tn5-mob, Apr Kmr | DSMZ |
| pUC4K | Source of Kmr cassette | Amersham |
| pUTminiTn5gfp | gfp reporter on mini-Tn5, source of gfp, Apr Tcr | ATCC |
| pSTH10 | Used for disruption of sthA, mob+ Apr Kmr | This study |
| pSTH16 | Used for monitoring sthA expression in vivo, mob+gfp+ Apr Kmr | This study |
IAM, Institute of Applied Microbiology; BRL, Bethesda Research Laboratories; DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen; ATCC, American Type Culture Collection.
DNA techniques.
Standard genetic techniques (i.e., restriction enzyme digestion, ligation, plasmid isolation, and transformation of DNA into E. coli) were carried out as described by Sambrook et al. (19). Restriction enzymes and T4 DNA ligase were purchased from Takara Shuzo, Kyoto, Japan. Nucleotide sequencing was performed by the dideoxy chain termination method with a Bigdye terminator cycle sequencing Ready Reaction kit and a model 310 automated DNA sequencer (PE Applied Biosystems).
Transposon mutagenesis.
Transposon mutagenesis with Tn5 was accomplished through biparental mating as follows. Exponentially grown E. coli S17-1(pSUP5011) cells and S. natans S-1 cells grown at 30°C in 1.0% NB to the stationary phase were harvested and resuspended in minimal volumes of saline (0.9% NaCl). The two suspensions were mixed and dispensed onto a sterile nitrocellulose filter on a 1.0% NB agar plate, which was then incubated overnight at 30°C. The cells were then washed with a minimal volume of saline, plated onto 0.1% NB agar plates containing rifampin and kanamycin, and incubated for 3 to 4 days at 30°C. Sheath-deficient mutants were obtained by visually selecting colonies with a smooth morphology.
Southern blot hybridization.
Chromosomal DNA was prepared by the procedure of Murray and Thompson (13). DNA digested with a restriction enzyme was separated by agarose gel electrophoresis and transferred onto a nylon membrane (Hybond N+; Amersham Pharmacia Biotech). Hybridization was carried out at 55°C with an AlkPhos direct labeling and detection kit (Amersham Pharmacia Biotech) by using a 1.8-kb BamHI fragment of pSUP5011 as a Tn5-specific probe according to the manufacturer’s instructions. The membrane was then washed at the same temperature.
Cloning of Tn5-inserted regions from the sheath-deficient mutants.
Chromosomal DNA was digested with SacI and ligated into the SacI site of pUC118. Transformants were selected on LB agar plates containing kanamycin.
Disruption of the sthA gene.
The fact that sthA is essential for sheath formation was confirmed by disrupting the gene by insertion of the kanamycin resistance (Kmr) gene. The plasmid used for double-crossover integration was constructed as follows. A 2.3-kb DNA fragment situated entirely within the sthA gene was amplified by PCR by using primers 5′-TGACGCAGTTGGTACAAGTC-3′ (upstream region of sthA) and 5′-AGATCCTTCAGGCGGATGCT-3′ (downstream region of sthA) and PyroBEST DNA polymerase according to the instructions of the supplier (Takara Shuzo). The thermal cycling protocol consisted of an initial denaturation step at 95°C for 1 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 2 min. Adenine was added to the 3′ terminus of the PCR product by using Taq polymerase (Takara Shuzo), and the product was cloned into pT7Blue by TA cloning. A unique AscI site within sthA in the PCR product was used as the insertion site of the Kmr gene cassette as follows. A 1.2-kb HincII fragment carrying Kmr prepared from pUC4K was inserted into the AscI site (blunted by T4 DNA polymerase) of sthA in the same orientation without producing a polar mutation. The 1.8-kb BamHI fragment carrying the mob site from pSUP5011 was then inserted at the BamHI site of the construct to generate pSTH10. E. coli S17-1 was then transformed by pSTH10. Plasmid pSTH10 was introduced into S. natans S-1 by conjugation with E. coli S17-1 harboring pSTH10 as described above (transposon mutagenesis). The transconjugant was selected on 0.1% NB agar plates supplemented with rifampin and kanamycin. Kmr insertion into sthA was confirmed by Southern blot hybridization by using the Kmr gene cassette as the probe and PCR amplification performed with the primers mentioned above.
RNA extraction and quantification of sthA mRNA by real-time quantitative RT-PCR.
Filamentous S. natans cells (strain S-1) were grown in 0.1% NB and harvested at the exponential phase. Single cells of the same strain were grown on a 1.0% NB agar plate for 10 h and harvested. Total RNAs were isolated by using ISOGEN-LS according to the instructions of the supplier (Nippon Gene, Tokyo, Japan). DNA contamination was then eliminated with RNase-free DNase I (Promega, Madison, Wis.), after which each isolated RNA was quantified with a RiboGreen RNA quantification kit (Molecular Probes, Eugene, Oreg.) and stored at −80°C until it was used.
Reverse transcription (RT)-PCR was carried by using a two-step protocol. A 500-ng portion of total RNA and 20 pmol of antisense primer sthAREV (5′-ATGCGGGTCTTGCGGATGAA-3′; positions 1093 to 1084 of the predicted coding region) were added to an RTG RT-PCR kit (Amersham Pharmacia Biochem) to obtain a final volume of 50 μl. Although this kit is ordinarily used for one-step RT-PCR, in this case only the RT reaction was carried out. Samples were incubated for 30 min at 42°C and then heated at 95°C for 5 min to inactivate the reverse transcriptase. Aliquots (1 μl) of the RT reaction mixture were then added to a LightCycler FastStart DNA Master SYBR Green I kit (Roche Diagnostics) containing 0.5 μM primer sthAFOR (5′-ATCGGCACCGCAATCTCGGT-3′; positions 220 to 240 of the predicted coding region) and 0.5 μM primer sthAREV. The thermal cycling protocol consisted of an initial denaturation step at 95°C for 10 min, followed by 55 cycles of denaturation at 94°C for 15 s, annealing at 60°C for 5 s, and extension at 72°C for 40 s. The PCR and monitoring of the PCR products were carried out with the LightCycler apparatus (Roche Diagnostics). Samples containing known amounts of an sthA fragment were used to generate a standard curve from which mRNA levels were determined.
In vivo monitoring of sthA expression with the GFP gene.
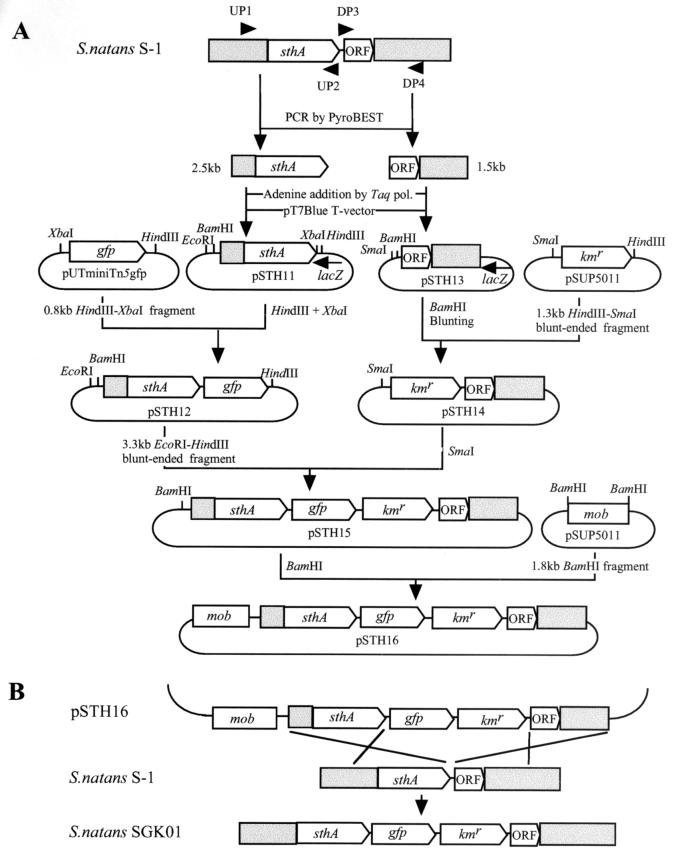
To examine expression of sthA in living cells, the green fluorescent protein (GFP) gene was inserted downstream of sthA in the chromosome in order to be transcribed by readthrough from sthA. To do this, plasmid pSTH16 was constructed as shown schematically in Fig. 1A. This construct was introduced into S. natans S-1 by conjugation with E. coli S17-1(pSTH16), as described above, and the transconjugants were selected on 0.1% NB agar plates supplemented with rifampin and kanamycin (Fig. 1B). The double-crossover event at the desired position was verified by Southern blot hybridization by using the 1.3-kb Kmr cassette as the probe and PCR amplification performed with primers UP1 and DP4 (Fig. 1). GFP fluorescence was detected with an Olympus AX-80TR microscope equipped with a high-pressure mercury bulb and a fluorescence mirror unit (U-MWIBA/GFP; Olympus Optical, Tokyo, Japan).
FIG. 1.
Schematic representation of a method for monitoring sthA expression in vivo. (A) Construction of pSTH16, which was used to insert gfp and the Kmr cassette downstream of sthA by double-crossover integration. A 2.5-kb fragment containing sthA (open box) and its upstream region (shaded box) was amplified from genomic DNA by using PyroBEST polymerase and primers UP1 (5′-ATGAGCGATACCCACTTCGGCTTC-3′) and UP2 (5′-TGTGGCGTGCACGACATGTTCGAT-3′). Adenine was added to the 3′ termini of the PCR products by using Taq polymerase, and the products were cloned into pT7Blue by the TA cloning procedure. A plasmid in which the orientation of the insert was opposite that of lacZ was then selected and designated pSTH11. A 0.8-kb XbaI-HindIII fragment, including promoterless gfp, was prepared from pUTminiTn5gfp and ligated into pSTH11 which had been digested with XbaI and HindIII to generate pSTH12. A 1.5-kb fragment that included the region downstream of the sthA gene (ORF and shaded boxes) was amplified from genomic DNA by using PyroBEST polymerase with primers DP3 (5′-TGAAGGCCTGCGGGACGACAGAAGC-3′) and DP4 (5′-TGCTCGTCGATGAAGCGGCTGGCCT-3′), and the PCR product was cloned into pT7Blue as described above. A plasmid in which the orientation of the insert was opposite that of lacZ was selected and designated pSTH13. A 1.3-kb HindIII-SmaI fragment carrying Kmr prepared from pSUP5011 was blunted and ligated into pSTH13 which had been digested with BamHI and blunted in order to generate pSTH14. pSTH12 was digested with EcoRI and HindIII, and the 3.3-kb EcoRI-HindIII fragment containing sthA-gfp of pSTH12 was blunted and ligated into the SmaI site of pSTH14. A plasmid in which the insert was oriented properly was selected and designated pSTH15. Finally, a 1.8-kb BamHI fragment carrying the mob site prepared from pSUP5011 was inserted into the BamHI site of pSTH15, yielding pSTH16, in which gfp and the Kmr cassette were situated between sthA and its downstream ORF, without disrupting either gene. pol., polymerase. (B) Strategy for insertion of gfp and the Kmr cassette between sthA and its downstream ORF.
Scanning electron microscopy.
Single cells of strain S-1 and an sthA disruptant (sthA::Tn5, strain TM1) grown on 1.0% NB agar plates were harvested and fixed with 2% glutaraldehyde for 1 h at room temperature, rinsed with phosphate buffer, and postfixed with 1% osmium tetroxide overnight at 4°C. The cells were then dehydrated by using a graded series of ethanol solutions, critical point dried, and coated with platinum. Samples were observed with a model S-4500 field emission scanning electron microscope (Hitachi, Tokyo, Japan).
Nucleotide sequence accession numbers.
The nucleotide sequence data determined in this study have been deposited in the DDBJ/GenBank/EMBL nucleotide sequence database under accession numbers AB050638 to AB050640.
RESULTS
Morphological traits of S. natans IAM 12068.
A previous study showed that S. natans can have two distinct morphologies, depending upon the substrate concentration (6). We found that the strain used in this study formed rough colonies comprised of filamentous cells when it was grown on 0.1% NB agar plates, whereas it formed smooth colonies comprised mostly of single cells when it was grown on 1.0% NB agar plates, although some of the cells formed filaments, particularly after prolonged incubation. These morphological traits were highly reproducible; therefore, all mutation experiments and subsequent verification were performed under these conditions.
Isolation of sheath-deficient mutants.
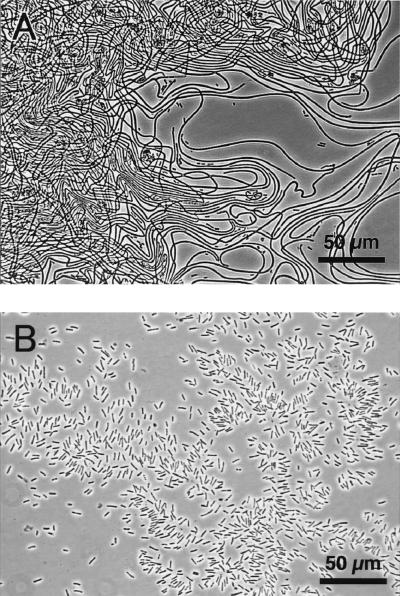
Rifampin-resistant strain S-1, which was derived from S. natans IAM 12068, was used for all mutation experiments. The shape of strain S-1 cells was identical to the shape of the wild-type strain cells, and the characteristic NB concentration-dependent morphological features were also identical. When Tn5 transposon mutagenesis was used, approximately 20,000 kanamycin-resistant mutants were obtained. Seven of these mutants (TM1 to TM7) had smooth colony morphology on 0.1% NB agar plates. Microscopic observation revealed that all of the cells were rod shaped and did not form sheathed structures. The inability to form sheaths was confirmed by culturing the mutants in 0.1% NB (strain S-1 formed sheaths in the same medium). Without exception, these mutants grew continuously as single cells without forming sheaths (Fig. 2); no revertants (sheath-forming cells) were observed.
FIG. 2.
Photomicrographs of S. natans cells. (A) Parental strain S-1 cells; (B) sheath-deficient mutant TM1 cells. Both strains were cultured in 0.1% NB.
Cloning and sequencing of Tn5-containing DNA fragments from sheath-deficient mutants.
The Tn5-containing genomic region was identified by Southern blot analysis with a Tn5-specific probe. The restriction profiles of KpnI, EcoRI, and SacI were identical for the genomes of five of the mutants (TM1 to TM5), suggesting that in these cases the Tn5 insert was located in the same specific region (data not shown). By contrast, the Tn5 insertions in the remaining two mutants (TM6 and TM7) were found to be located elsewhere.
To identify the gene disrupted by the Tn5 insertion, SacI fragments harboring Tn5 were cloned into pUC118, and their Tn5-flanking regions were sequenced. Consistent with the Southern blot analysis results, DNA sequencing revealed that five of the insertions occurred in the same open reading frame (ORF), designated sthA, which consisted of 1,407 bp encoding a 469-amino-acid protein (accession number AB050638). In addition, a putative Shine-Dalgarno sequence (AAGG) was found 9 bp upstream from the ATG codon.
One of the two remaining Tn5 insertions, TM6 (accession number AB050640), was situated in a region homologous to genes encoding the putative glycosyltransferases in the database (e.g., WbpT in Pseudomonas aeruginosa [28% amino acid similarity], which is thought to be involved in exopolysaccharide [EPS]) synthesis [1]). The other Tn5 insertion, TM7 (accession number AB050639), was situated in a region homologous to genes encoding the putative RNA helicases or RNA polymerase-associated proteins (e.g., HepA in E. coli [43% amino acid similarity] [14]). Southern blot analyses suggested that these genes were located some distance from sthA and thus were not clustered with sthA on the genome (data not shown). In subsequent experiments we focused on the sthA gene.
Inactivation of sthA.
Whether disrupting sthA blocked sheath formation was investigated by inactivating sthA by insertion of a Kmr cassette. The cassette was inserted at a unique AscI site in sthA, and successful insertion was confirmed by Southern blot hybridization and PCR amplification. The disruptant constructed (SKM01) grew as smooth colonies on 0.1% NB agar plates and as single cells in 0.1% NB and never formed a sheathed structure. This result, together with the results of the Tn5 mutagenesis experiments, strongly indicates that sthA is essential for sheath formation.
Analysis of sthA transcription.
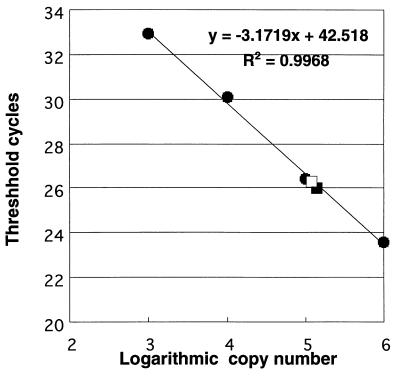
To determine whether sthA is expressed only by filamentous cells, we first attempted to detect the gene transcript by Northern blot hybridization. However, all of our attempts were unsuccessful, most likely because the mRNA was either too unstable or too long to be transferred onto a nylon membrane. Therefore, we performed quantitative RT-PCR to determine the level of sthA transcription using strain S-1. As shown in Fig. 3, mRNAs encoding the sthA gene product were found in both single cells (grown on 1.0% NB agar plates) and filamentous cells (grown in 0.1% NB); in each case, approximately 1.0 × 105 copies per 10 ng of total RNA were detected, suggesting that the sthA gene is expressed at the same level in both morphotypes of cells.
FIG. 3.
Real-time quantitative RT-PCR assay for evaluating sthA transcription. A standard curve was generated by plotting the threshold cycle (the cycle at which the fluorescent signal became detectable above the background signal) as a function of the log of the template copy number. sthA mRNA levels in filamentous cells (□) and single cells (▪) were calculated from the standard curve.
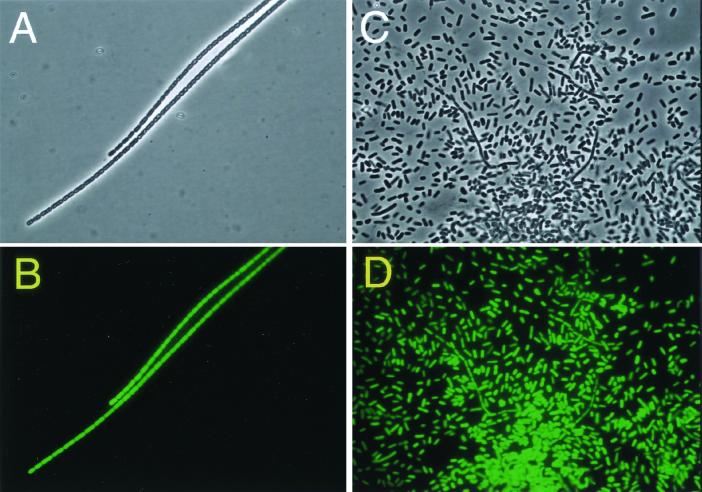
To confirm this finding, we examined expression of sthA in living cells using the GFP gene as a reporter gene. The morphology of the mutant constructed (SGK01) was identical to that of the parental strain (S-1), and the ability to alter its morphology by using different NB concentrations was retained. The mutant was found to express GFP in both single and filamentous cells (Fig. 4), confirming that sthA was expressed regardless of the cell morphology.
FIG. 4.
Expression of GFP in filamentous and single cells. Filamentous cells (A and B) were obtained by growing the mutant with gfp::Kmr inserted (SGK01) in 0.1% NB, and single cells (C and D) were obtained by culturing the same mutant on 1.0% NB agar plates. (A and C) Phase-contrast micrographs; (B and D) GFP fluorescence emitted by the cells in panels A and C, respectively.
Microscopic observation of single cells of the parental strain and the sthA-deficient mutant.
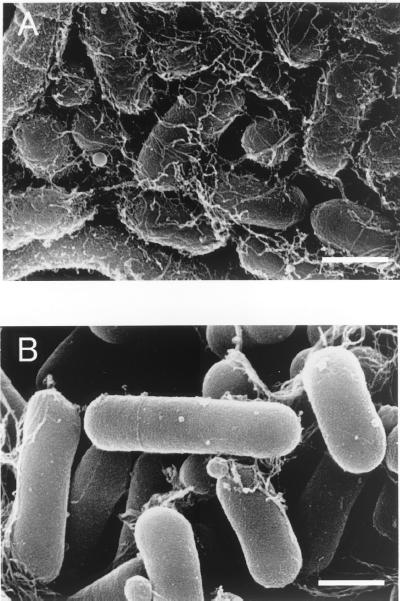
Scanning electron microscopy revealed that the single-cell morphotype of parental strain S-1 (grown on a 1.0% NB agar plate) produced much larger amounts of extracellular matrix-like materials than an sthA-deficient mutant (TM1) produced (Fig. 5).
FIG. 5.
Scanning electron micrographs of single S. natans cells. (A) Parental strain S-1 cells; (B) sthA-deficient mutant TM1 cells. Both strains were grown on 1.0% NB agar plates. Bars, 1 μm.
DISCUSSION
The results of Tn5 mutagenesis and a subsequent gene disruption analysis made it clear that the sthA gene is crucial for sheath construction by S. natans. The putative amino acid sequence indicated that the sthA gene product is homologous to glycosyltransferases involved in bacterial EPS biosynthesis, including GumD from Xanthomonas campestris (31.6% amino acid identity) (2), EpsD from Lactococcus lactis (35.0% identity) (23), and ExoY from Sinorhizobium meliloti (37% identity) (17). These enzymes catalyze the transfer of sugar to undecaprenol phosphate, which acts as a sugar carrier in the membrane (15, 26). Considering that disrupting sthA by insertion of either Tn5 or a Kmr cassette blocked formation of the sheathed structure, the data strongly suggest that sthA encodes a similar enzyme catalyzing the glycosyl transfer essential for sheath formation.
Interestingly, transcriptional analyses based on quantitative RT-PCR or expression of the gfp reporter gene showed that sthA is transcribed in both filamentous and single cells. It has been reported that S. natans produces EPS at the outermost layer of both sheathed and single cells (6, 21). Our scanning electron microscopy results revealed that the single-cell morphotype of the parental strain produced extracellular matrix-like material, which appeared to be EPS, whereas the sthA-deficient mutant produced far less extracellular material. Disruption of sthA did not completely suppress extracellular polymer production, but it resulted in an obvious difference between the wild-type and sthA-deficient cells.
Our results strongly indicate that sthA is involved in both sheath synthesis and EPS synthesis. Moreover, genetic analysis of its downstream region revealed that sthA is followed by an ORF homologous to genes encoding EPS-exporting proteins (unpublished results). Thus, sheath formation and EPS formation very likely share common synthetic processes involved in the formation of extracellular materials.
As mentioned above, we also obtained two other transposon insertions that yielded sheathless mutants. Without doubt, sheath formation and EPS formation involve a number of synthetic steps and hence a number of genes, virtually none of which have been characterized. Still, the fact that five of the seven sheathless mutants were found to have disrupted sthA genes suggests that the sthA gene product is a crucial component of the mechanism responsible for sheath biosynthesis and subsequent assembly.
The filamentous form of S. natans is often observed in activated sludges, and a certain number of filaments are thought to be required for proper floc formation in order to retain the settleability of sludges (11, 20). However, excessive growth of the filamentous form of S. natans results in poor settling of sludge flocs (bulking). One effective way to prevent bulking would be to inhibit filamentous growth of this bacterium. If we could inhibit expression of the sthA gene or the activity of the sthA gene product with chemicals, we could temporarily inhibit the excess growth of filaments and thus prevent bulking when the size of the S. natans population approaches a bulking level. If similar genes are present in other filamentous bacteria, this method could be widely applicable for preventing bulking. In preliminary experiments, we found that Leptothrix discophora, which is phylogenetically affiliated with S. natans, also harbors sthA and its downstream genes (H. Yoshihara, T. Suzuki, and Y. Kamagata, unpublished data). Inhibition of filament formation is a new approach for preventing bulking. To better understand the pathway of filament formation, we should determine the in situ function of the products of sthA and its downstream genes, and such a study is under way.
REFERENCES
- 1.Belanger, M., L. L. Burrows, and J. S. Lam. 1999. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology 145:3505–3521. [DOI] [PubMed] [Google Scholar]
- 2.Chou, F. L., H. C. Chou, Y. S. Lin, B. Y. Yang, N. T. Lin, S. F. Weng, and Y. H. Tseng. 1997. The Xanthomonas campestris gumD gene required for synthesis of xanthan gum is involved in normal pigmentation and virulence in causing black rot. Biochem. Biophys. Res. Commun. 233:265–269. [DOI] [PubMed] [Google Scholar]
- 3.Dondero, N. C., R. A. Phillips, and H. Heukelekian. 1961. Isolation and preservation of culture of Sphaerotilus. Appl. Microbiol. 9:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eikelboom, D. H. 1975. Filamentous organisms observed in active sludge. Water Res. 9:365–388. [Google Scholar]
- 5.Eikelboom, D. H., and H. J. J. van Buijsen. 1981. Microscopic sludge investigation manual. Report A94a. IMG-TNO, Delft, The Netherlands.
- 6.Gaudy, E., and R. S. Wolfe. 1961. Factors affecting filamentous growth of Sphaerotilus natans. Appl. Microbiol. 10:1962–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoeniger, J. F., H. D. Tauschel, and J. L. Stokes. 1973. The fine structure of Sphaerotilus natans. Can. J. Microbiol. 19:309–313. [DOI] [PubMed] [Google Scholar]
- 8.Howarth, R., R. F. Unz, E. M. Seviour, R. J. Seviour, L. L. Blackall, R. W. Pickup, J. G. Jones, J. Yaguchi, and I. M. Head. 1999. Phylogenetic relationships of filamentous sulfur bacteria (Thiothrix spp. and Eikelboom type 021N bacteria) isolated from wastewater-treatment plants and description of Thiothrix eikelboomii sp. nov., Thiothrix unzii sp. nov., Thiothrix fructosivorans sp. nov. and Thiothrix defluvii sp. nov. Int. J. Syst. Bacteriol. 49:1817–1827. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins, D., M. G. Richard, and G. T. Daigger. 1993. Manual on the causes and control of activated sludge bulking and forming, 2nd ed. Lewis Publishers, Chelsea, Mich.
- 10.Kanagawa, T., Y. Kamagata, S. Aruga, T. Kohno, M. Horn, and M. Wagner. 2000. Phylogenetic analysis of and oligonucleotide probe development for Eikelboom type 021N filamentous bacteria isolated from bulking activated sludge. Appl. Environ. Microbiol. 66:5043–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lao, A. O., P. F. Strom, and D. Jenkins. 1984. Growth kinetics of Sphaerotilus natans and a floc former in pure and dual continuous culture. J. Water Pollut. Control Fed. 56:41–51. [Google Scholar]
- 12.Mulder, E. G. 1989. Sheathed bacteria, p. 1994–2008. In J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holt (ed.), Bergey’s manual of systematic bacteriology, vol. 3. Williams and Wilkins, Baltimore, Md.
- 13.Murray, M. G., and W. F. Thompson. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muzzin, O., E. A. Campbell, L. Xia, E. Severinova, S. A. Darst, and K. Severinov. 1998. Disruption of Escherichia coli HepA, an RNA polymerase-associated protein, causes UV sensitivity. J. Biol. Chem. 273:15157–15161. [DOI] [PubMed] [Google Scholar]
- 15.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495–503. [DOI] [PubMed] [Google Scholar]
- 16.Rensink, J. H. 1974. New approach to preventing bulking sludge. J. Water Pollut. Control Fed. 46:1888–1894. [Google Scholar]
- 17.Reuber, T. L., and G. C. Walker. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269–280. [DOI] [PubMed] [Google Scholar]
- 18.Romano, A. H., and J. P. Peloquin. 1963. Composition of the sheath of Sphaerotilus natans. J. Bacteriol. 86:252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Sezgin, M., D. Jenkins, and D. S. Parker. 1978. A unified theory of filamentous activated sludge bulking. J. Water Pollut. Control Fed. 50:362–381. [Google Scholar]
- 21.Takada, M., K. Iohara, S. Shinmaru, I. Suzuki, and J. Koizumi. 2000. Purification and properties of an enzyme capable of degrading the sheath of Sphaerotilus natans. Appl. Environ. Microbiol. 66:4998–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda, M., F. Nakano, T. Nagase, K. Iohara, and J. Koizumi. 1998. Isolation and chemical composition of the sheath of Sphaerotilus natans. Biosci. Biotechnol. Biochem. 62:1138–1143. [DOI] [PubMed] [Google Scholar]
- 23.van Kranenburg, R., J. D. Marugg, S. van Swam II, N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387–397. [DOI] [PubMed] [Google Scholar]
- 24.van Veen, W. L., E. G. Mulder, and M. H. Deinema. 1978. The Sphaerotilus-Leptothrix group of bacteria. Microbiol. Rev. 42:329–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wanner, J. 1994. Activated sludge bulking and foaming control. Technomic Publishing Company, Lancaster, Pa.
- 26.Whitfield, C., and M. A. Valvano. 1993. Biosynthesis and expression of cell-surface polysaccharides in gram-negative bacteria. Adv. Microb. Physiol. 35:135–246. [DOI] [PubMed] [Google Scholar]