Abstract
Listeria monocytogenes of serotype 4b has been implicated in numerous outbreaks of food-borne listeriosis and in ca. 40% of sporadic cases. Strains of this serotype appear to be relatively homogeneous genetically, and molecular markers specific for distinct serotype 4b lineages have not been frequently identified. Here we show that DNA fragments derived from the putative mannitol permease locus of Listeria monocytogenes had an unexpectedly high potential to differentiate among different strains of serotype 4b when used as probes in Southern blotting of EcoRI-digested genomic DNA, yielding four distinct restriction fragment length polymorphism (RFLP) patterns. Strains of two epidemic-associated lineages, including the major epidemic clone implicated in several outbreaks in Europe and North America, had distinct RFLPs which differed from those of all other serotype 4b strains that we screened but which were encountered among strains of serotypes 1/2b and 3b. In addition, three serogroup 4 lineages were found to have unique RFLPs that were not encountered among any other L. monocytogenes strains. One was an unusual lineage of serotype 4b, and the other two were members of the serotype 4a and 4c group. The observed polymorphisms may reflect evolutionary relationships among lineages of L. monocytogenes and may facilitate detection and population genetic analysis of specific lineages.
Invasive illness due to Listeria monocytogenes (listeriosis) has severe symptoms (meningitis, septicemia, and abortions) and is a leading cause of death due to food-borne pathogens in the industrialized world, including the United States (11, 13, 22). The pathogen has also been implicated in outbreaks of febrile gastroenteritis (3, 28).
Numerous investigations have revealed that the species is clonally partitioned along serotypic clusters. Two major genomic divisions have been identified: division I includes serotypes 1/2a, 1/2c, 3a, and 3c, whereas strains of serotypes 1/2b, 3b, and 4b are included in division II (1, 5, 6, 8, 14, 24). In addition, genotyping data suggest the existence of a third, less populous division (26, 35) that consists of strains of serotypes 4a and 4c, derived primarily from animal infections (35). Examination of numerous genes has revealed distinct allelic variants in each division, suggesting a strong linkage disequilibrium maintained by lack of gene flow between divisions.
Strains of just three serotypes, 1/2a, 1/2b, and 4b, constitute more than 95% of human clinical isolates (13, 16). Serotype 4b is of special interest, as it has been implicated not only in ca. 40% of sporadic infections but also in numerous outbreaks of food-borne listeriosis. Earlier major outbreaks in Nova Scotia (coleslaw), California (Jalisco cheese), Switzerland (Vacherin Mont d’s Or cheese), and France (pork tongue in aspic) and numerous other outbreaks in Europe and North America (16, 29) implicated strains of this serotype. More recently, a multistate outbreak in the Unites States, in which contaminated hot dogs were implicated, also involved bacteria of serotype 4b of an apparently novel lineage (2).
Several genotyping data, based on different subtyping tools and approaches, suggest that serotype 4b is genetically rather homogeneous and less diverse than the other serotypes (1/2a and 1/2b) commonly implicated in human illness (5, 14, 21, 23, 27). Furthermore, L. monocytogenes strains implicated in outbreaks have been shown to constitute genetically well-defined clonal lineages (epidemic clones) on the basis of multilocus enzyme electrophoresis and pulsed-field gel electrophoresis (PFGE) (7, 9, 24), but the literature contains only a few descriptions of characterized genetic markers that are unique for epidemic-associated strains. Such descriptions primarily concern the strains of a major epidemic lineage (epidemic clone I [ECI]) which has been implicated in numerous outbreaks in Europe and North America, including the above-mentioned epidemics in Nova Scotia, California, Switzerland. and France. Restriction fragment length polymorphism (RFLP) analysis using DNA probes derived from the ltrB genomic region, which is essential for cold growth of L. monocytogenes, differentiated ECI strains from all other L. monocytogenes strains of serotype 4b that were screened (36). In addition, ECI strains appear to possess a unique DNA modification at GATC sites, which renders their DNA resistant to digestion by Sau3AI (37). Recently, DNA sequences specific to ECI strains have been identified (15), and the genome sequencing of one such strain has been undertaken (www.tigr.org). Genetic markers specific for other lineages of L. monocytogenes serotype 4b, including the strains implicated in the hot dog outbreak in 1998 to 1999 (2), have not yet been described. Such markers would supplement other typing tools (such as PFGE) to facilitate lineage detection and monitoring and would help to elucidate the presently poorly understood evolution of epidemic-associated clonal groups. In this work we describe DNA probes derived from the putative mannitol permease locus of L. monocytogenes which can identify four distinct RFLP patterns within serotype 4b, including RFLPs distinct for epidemic-associated strains.
MATERIALS AND METHODS
Bacterial strains and growth media.
Listeria strains were grown at 35°C in brain heart infusion broth (Difco) or on tryptic soy agar supplemented with 0.7% yeast extract (Difco) and were preserved at −70°C in brain heart infusion broth.Escherichia coli strains were grown in Luria-Bertani broth at 35°C. When appropriate, ampicillin was used at 100 μg/ml.
General DNA manipulations and analyses.
Unless otherwise indicated, standard molecular procedures (4) were used for extraction, digestion, and other manipulations of DNA. Plasmids were purified with Wizard miniprep columns (Promega). Southern blotting was performed as described previously (20) with DNA probes labeled with digoxigenin (Genius kit; Roche). The probe for the ltrB region was pST1, which has been previously described (36). Determination of DNA resistance or sensitivity to Sau3AI digestion was as described previously (37). For PCR we used synthetic oligonucleotides (Biosynthesis) and either Taq polymerase (Promega) or Red Taq (Sigma) under previously described conditions (20). To clone PCR fragments, the desired fragment was purified from agarose gels using Geneclean (Bio101) and cloned into pCR1000 (TA cloning; Invitrogen), following the suggestions of the vendors. Sequences of the cloned fragments were determined and analyzed as described previously (25). Restriction enzymes were purchased from MBI Fermentas or from Promega.
Probe constructions.
A genomic region was originally identified in the serotype 1/2a strain 1/2a3 (33) and first used as a probe in Southern blotting in the course of an unrelated research project (33). The region was pursued further in this study because of unusual results obtained with Southern blots of genomic DNAs from different strains. PCR primers HLT28F (5′ TTCTTGGTGGTATGACAGGAAC 3′) and HLT28R (5′ TCACGACCACATCGCATTCGG 3′) derived from the sequence of this region yielded a 464-bp fragment (fragment 28.5) which was cloned in pCR1000, yielding plasmid p28.5. When used as a probe, fragment 28.5 hybridized with a ca. 3.8-kb EcoRI fragment of strain 1/2a3. To clone this larger fragment, the ca. 3.8-kb region of an agarose gel (0.8%) of EcoRI-digested DNA of strain 1/2a3 was excised, and the DNA was purified with Geneclean. The purified fragments were ligated with EcoRI-digested plasmid pGEM-9Zf (Promega) and introduced into E. coli DH5α by transformation. White colonies were selected and inoculated in separate wells of two 96-well plates (200 μl of Luria-Bertani broth with ampicillin in each well). Portions of 150 μl from each of 12 wells were combined in one Eppendorf tube, and plasmid extractions from the mixtures were done with Wizard minipreps. The resulting plasmid preparations were screened by PCR using primers HLT28F2 and HLT28R2 (primer sequences are shown above). Three of the mixtures yielded PCR products of the appropriate size. The 12 cultures composing one of the positive mixtures were grown individually, and their plasmids were screened again by PCR using primers HLT28F2 and HLT28R2. The plasmid from one of the cultures (designated p1-4) produced the expected PCR product, and the plasmid was confirmed to harbor the expected ca. 3.8-kb EcoRI fragment by Southern blotting using fragment 28.5 as a probe. The cloned ca. 3.8-kb fragment was designated probe 1-4. To prepare probes, p28.5 and p1-4 were digested with EcoRI and the cloned fragments were purified from gels using Geneclean and labeled.
Nucleotide sequence analysis.
Routine DNA analysis tools (reference 25 and references therein) were used. Sequence databases included the recently released genome sequence data for L. monocytogenes strain EGD (serotype 1/2a) and for Listeria innocua (http://bioweb.pasteur.fr/cgi-bin/listeria/blast.cgi), as well as the genome sequence data for L. monocytogenes serotype 4b (www.tigr.org).
Nucleotide sequence accession number.
The nucleotide sequence data determined in this study have been deposited in the GenBank database under accession no. AF299095.
RESULTS
The putative mannitol permease gene (mtlA) differentiates between ECI and other serotype 4b strains when used as a probe in Southern blots.
During our search for putative serotype-specific genes that may be involved in teichoic acid glycosylation of L. monocytogenes serotype 1/2a (33), candidate gene fragments were used as probes in Southern blots against a panel of strains representing different known lineages and serotypes. The use of one of these fragments as a probe identified unexpected RFLPs in the panel of strains examined. Although this fragment was subsequently found not to be serotype specific and not to be in a locus involved in teichoic acid glycosylation, we pursued its characterization because of the promising RFLP results. The fragment was sequenced, and a 464-bp internal portion (fragment 28.5) was amplified by PCR and cloned. BLAST analysis of the sequence of this fragment using the recently released (and as of this writing nonannotated) genomic databases of L. monocytogenes serotype 4b (www.tigr.org) and L. monocytogenes strain EGD (serotype 1/2a) (http://bioweb.pasteur.fr/cgi-bin/listeria/blast.cgi) revealed highly similar sequences in both genomic databases, although the identity was higher with strain EGD (97%) than with serotype 4b (94%). This may not be surprising, since the strain which we used, 1/2a3, was of serotype 1/2a, the same as EGD. L. innocua also harbored a homologous sequence (92% identity). The corresponding sequences in the databases of serotype 4b and of strain EGD were 93% identical.
Further analysis of the nucleotide sequence of fragment 28.5 identified a partial open reading frame with significant sequence identity to mtlA, encoding mannitol permease of Bacillus stearothermophilus (15) (60% over 560 nucleotides [nt]), E. coli (19) (56% over 557 nt), Staphylococcus carnosus (12) (67% over 193 nt), and numerous other bacteria. The deduced gene product was also found to have significant similarity to mannitol permeases of the bacteria named above, with identities of 43% (145 amino acids [aa]), 34% (187 aa), and 37% (149 aa) with the mannitol permeases of B. stearothermophilus, E. coli, and S. carnosus, respectively. TBLASTX analysis of the contig harboring the entire putative mtlA gene (contig 22, nt 33967 to 36524 in the L. monocytogenes 4b genomic sequence database [www.tigr.org]) showed that the deduced amino acid sequence from fragment 28.5 had 98% identity over 185 aa with an internal fragment of the putative MtlA of serotype 4b.
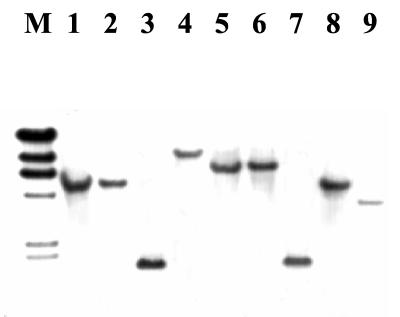
Use of the cloned fragment 28.5 as a probe (probe 28.5) in Southern blots of EcoRI-digested genomic DNA revealed several distinct RFLPs among serotype 4b strains. Interestingly, one hybridizing band of ca. 1.8 kb was identified in strains of the major epidemic clone (ECI) (Fig. 1, lanes 3 and 7), whereas strains from an unrelated epidemic lineage (ECII) had a different hybridizing band, of ca. 5 kb (Fig. 1, lanes 1, 2, and 8). A still different band, of ca. 6.4 kb, was produced with strains which were of apparent sporadic origin, i.e., not associated with any known epidemics (Fig. 1, lanes 5 and 6). One strain (strain 267), which on the basis of other evidence (Z. Lan, R. Y. Kanenaka, and S. Kathariou, unpublished data) represents an unusual genotype, also had a unique hybridizing band, of ca. 9.4 kb (Fig. 1, lane 4).
FIG. 1.
Southern blot of EcoRI-digested genomic DNAs from L. monocytogenes strains with probe 28.5. Lane M, λ molecular size markers (fragment sizes [from top to bottom] are 23, 9.4, 6.6, 4.4, 2.3, and 2.0 kb). Lanes: 1, 2, and 8, serotype 4b strains LM20, LM21, and LM50 (clinical isolates from an epidemic unrelated to ECI and referred to here as ECII), respectively; 3 and 7, serotype 4b ECI strains LM265 and F2381 (Jalisco outbreak), respectively; 4, serotype 4b strain LM267; 5 and 6, serotype 4b sporadic strains LM266 and 4b1, respectively; 9, serotype 1/2a strain 1/2a3 (33).
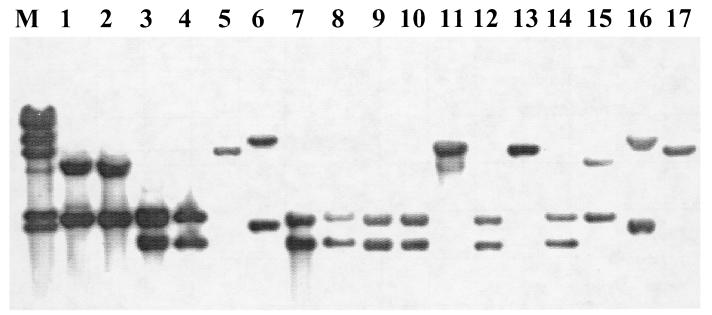
Four distinct RFLP patterns, designated RFLP patterns I to IV, were also observed in serotype 4b strains when the larger (ca. 3.8-kb) genomic fragment was used as a probe (probe 1-4) with EcoRI-digested DNAs (Fig. 2 and Table 1). Pattern I (one band of ca. 6.4 kb) was obtained with sporadic isolates (Fig. 2, lanes 5, 11, 13, and 17), whereas strains of ECI yielded RFLP pattern II, consisting of the previously observed ca. 1.8-kb fragment plus an additional hybridizing fragment of ca. 2.3 kb (Fig. 2, lanes 3, 4, 7 to 10, 12, and 14). The new, ca. 2.3-kb fragment was also observed when the strains of the other epidemic lineage (ECII) were probed, resulting in RFLP pattern III, with two bands of ca. 5 and 2.3 kb for these strains (Fig. 2, lanes 1, 2, and 15). Strain 267, which, as mentioned above, represents an unusual serotype 4b genotype, yielded RFLP pattern IV, consisting of the original ca. 9.4-kb fragment obtained with probe 28.5 and an additional fragment of ca. 2.1 kb (Fig. 2, lane 6). Interestingly, two other isolates from our collection, which on the basis of other genetic studies also appeared to have unusual genotypes similar to those of strain 267, hybridized identically to strain 267 with this probe (Fig. 2, lanes 6 and 16, and Table 1).
FIG. 2.
Southern blot of EcoRI-digested genomic DNAs from L. monocytogenes serotype 4b strains with probe 1-4. Lane M, λ molecular size markers, as described in the legend for Fig. 1. RFLP pattern I, lanes 5, 11, 13, and 17 (presumably sporadic strains of serotype 4b, not associated with ECI or ECII); RFLP pattern II, ECI strains from Hawaii (lanes 3 and 4), Jalisco (lanes 7, 8, 9, and 12), and Nova Scotia (lanes 10 and 14); RFLP pattern III, ECII strains LM 20 (lane 1), LM 21 (lane 2), and LM 50, (lane 15); RFLP pattern IV, strains 267 (lane 6) and A2 (lane 16) (both serotype 4b and of unusual genotype).
TABLE 1.
RFLP patterns in different Listeria strains determined with probe 1–4
| RFLP pattern | Fragments (kb) hybridizing with probe 2–4 | Strains (no. screened)
|
||
|---|---|---|---|---|
|
L. monocytogenes
|
Other Listeria speciesa | |||
| Serotype 4b | Other serotypes | |||
| I | 6.4 | Sporadicb (9) | 4d (1) | L. innocua (12) |
| II | 2.3, 1.8 | ECI (15) | 1/2b (4) | |
| 4e (1) | ||||
| III | 5.0, 2.3 | ECII (4) | 1/2b (6) | L. innocua (3)c |
| 3b (1) | ||||
| IV | 9.4, 2.1 | B2, A2, 267 (3) | ||
| V | 2.0, 1.8 | 4a (1) | ||
| 4a or 4c (2) | ||||
| VI | 2.8, 1.8 | 4a or 4c (1) | ||
| VII | 3.8 | 1/2a (3) | ||
| 3a (1) | ||||
| 3c (1) | ||||
| 1/2c (1) | ||||
| VIII | 8.0, 2.6 | “4”d (1) | ||
L. ivanovii, L. seeligeri, L. welshimeri, and L. grayi did not hybridize with the probe.
Serotype 4b strains not associated with a known epidemic (presumably of sporadic origin).
Distinct L. innocua lineage (lineage I) with serotype 4b-like teichoic acid composition (18).
Unusual clinical isolate of L. monocytogenes, with an atypical serotypic designation.
Southern blots of HindIII-digested DNAs with the pST1 probe derived from the ltrB locus, which is essential for cold tolerance of L. monocytogenes (36), confirmed that the strains of RFLP pattern II had the epidemic-specific RFLP (data not shown). In addition, the genomic DNAs of these strains were resistant to Sau3AI digestion (data not shown), suggesting modification of cytosines at the GATC sites, as described earlier for this epidemic clone (37).
On the basis of PFGE and other genotyping data, strains of the rare serotypes 4d and 4e cannot be distinguished from those of serotype 4b (8, 20, 25). Similar findings were obtained with probes 28.5 and 1-4. The two available strains of serotype 4d and 4e (both type strains, originating at the American Type Culture Collection [ATCC]) hybridized with probe 1-4, yielding RFLP patterns I and II, respectively (Table 1).
Serotype 1/2b strains have two distinct RFLP patterns, which are also produced by epidemic-associated strains of serotype 4b.
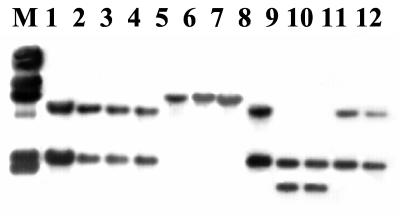
Serotypes 1/2b and 3b are the two other constituents of the major genomic division (division II) which includes serotype 4b strains. Two distinct RFLP patterns were observed in strains of serotype 1/2b. Interestingly, the RFLP patterns were identical to patterns II and III, which were found in epidemic-associated strains of serotype 4b but not in other strains of that serotype. In serotype 1/2b, RFLP patterns II and III were obtained in four and six strains, respectively. The single available serotype 3b strain had RFLP pattern III as well. The representative serotype 1/2b RFLPs are shown in Fig. 3 (pattern II in lanes 9 and 10 and pattern III in lanes 8, 11, and 12).
FIG. 3.
Southern blot of EcoRI-digested genomic DNAs from L. monocytogenes serotype 4b strains with probe 1-4. Lane M, λ molecular size markers, as described in the legend for Fig. 1. Lanes: 1, RFLP pattern III of L. monocytogenes strain LM 50 (ECII); 2 to 4, RFLP pattern III of L. innocua strains F8735, F8596, and F7833 (lineage I, described in reference 18), respectively; 5 to 7, RFLP pattern I of L. innocua strains 6a, LM 248, and LM 250, respectively; 8, 11, and 12, RFLP pattern III of L. monocytogenes strains LM 249, F4242, and F4236 (serotype 1/2b), respectively; 9 and 10, RFLP pattern II of L. monocytogenes strains F4245 and F4260 (also serotype 1/2b), respectively.
Strains of the other major genetic division of L. monocytogenes (division I, consisting of serotypes 1/2a, 1/2c, 3a, and 3c) yielded one hybridizing band of ca. 3.8 kb with either probe (e.g., Fig. 1, lane 9). Thus, a single RFLP pattern (pattern VII) was identified in this division.
The RFLP patterns obtained with the different serotypes of L. monocytogenes are summarized in Table 1.
Among other listeriae, only L. innocua harbors sequences with detectable homology to probes 28.5 and 1-4 and yields RFLP patterns also encountered within serotype 4b.
L. innocua strains hybridized with both probes. With probe 1-4, RFLP pattern I (also obtained with non-epidemic L. monocytogenes serotype 4b) was produced by the type strain (serotype 6a) and several other strains. Examples of this RFLP pattern are shown in Fig. 3, lanes 5, 6, and 7. Interestingly, pattern III, which was obtained with certain epidemic-associated L. monocytogenes serotype 4b (ECII) strains and with some of the serotype 1/2b strains, was also observed with three strains of L. innocua when probe 1-4 was used (Fig. 3, lanes 2 to 4). It is noteworthy that these L. innocua strains represent a unique lineage, which is indistinguishable from L. monocytogenes serotype 4b in terms of teichoic acid composition and the presence of serotype 4b-specific genes (18).
No signals were observed in Southern blots with DNAs from L. seeligeri (three strains) or L. ivanovii, L. welshimeri, and L. grayi (one strain each).
The hybridization results with L. innocua and other listeriae, using probe 1-4, are summarized in Table 1.
Probe 1-4 can differentiate strains of serotypes 4a and 4c from other strains, including other isolates of serogroup 4 (serotypes 4b, 4d, and 4e).
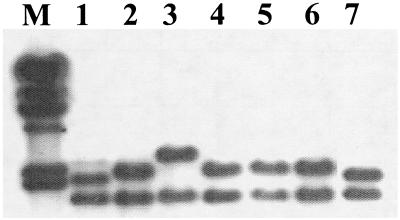
With the small probe 28.5, strains of serotypes 4a and 4c reacted similarly to ECI strains, producing one hybridizing fragment of ca. 1.8 kb (data not shown). When the larger probe 1-4 was used, however, these strains yielded two unique RFLP patterns, V and VI, not seen among any other listeriae (Table 1). Pattern V was produced by the type strain of serotype 4a and two clinical isolates (one animal and one human). The hybridizing bands from the latter strains are shown in Fig. 4 (lanes 1 and 7). The top hybridizing band of these strains was slightly but consistently smaller than the corresponding band of ECI strains (Fig. 4, lanes 2, 4, 5, and 6). Pattern VI was encountered in only one strain, a human clinical isolate (Fig. 4, lane 3).
FIG. 4.
Southern blot of EcoRI-digested genomic DNAs from L. monocytogenes serotype 4b strains with probe 1-4. Lane M, λ molecular size markers, as described in the legend for Fig. 1. Lanes 1 and 7, RFLP pattern V of strains OLM 8 and OLM 151 (isolates from animal and human illness, respectively, of serotype 4a or 4c); lanes 2, 4, 5, and6, RFLP pattern II of ECI strains OLM10, OLM 65, OLM 97, and OLM 116, respectively; lane 3, RFLP pattern VI of strain OLM 59 (human clinical isolate, serotype 4a or 4c).
The unique RFLPs of the 4a and 4c strains differentiate them from all other screened strains of L. monocytogenes. A similar statement can be made for the three serotype 4b strains (A2, B2, and 267) with RFLP pattern IV, which was not encountered among any of the other strains which we screened (Table 1). An additional unique RFLP pattern (pattern VIII) was encountered in a serogroup 4 strain of L. monocytogenes which appears to have an atypical antigenic composition and does not conform to the known serogroup 4 serotypes (Table 1).
DISCUSSION
Numerous genotyping data suggest that L. monocytogenes is partitioned in two major divisions that correlate with flagellar (H) antigen types (31) and the corresponding serotypic designations (division I with serotypes 1/2b, 3b, and 4b and division II with serotypes 1/a, 1/c, 3a, and 3c) (1, 5, 6, 8, 14, 24). Strains of serotypes 4a and 4c appear to constitute a third division (26, 35).
Although division-specific genetic variation has been identified in many genes, including those involved in virulence (26, 34, 35), there has been a scarcity of well-defined genetic markers for specific lineages within the major divisions. The lack of lineage-specific genetic markers has been an issue especially for serotype 4b, which appears to be genetically more homogeneous than serogroup 1/2 (5, 14, 21, 23, 27) and which, in addition, is often implicated in food-borne epidemics. The gene probes described here may therefore be of special interest as subtyping tools for this serotype.
The major epidemic clone of L. monocytogenes, ECI, yielded a distinct RFLP pattern (pattern II), whereas another epidemic cluster of strains, ECII, had a different RFLP pattern (pattern III). Patterns II and III were not found among any of the other serotype 4b strains which we screened but were obtained for strains of serotypes 1/2b and 3b. This may suggest a common origin of these epidemic serotype 4b lineages and lineages of serotypes 1/2b and 3b, at least in the probed genomic region. The genetic background of ECI and ECII remains poorly understood at this time.
RFLP pattern II was also obtained with the single available serotype 4e strain (the type strain from the ATCC) (Table 1). This was not surprising to us, because on the basis of both ltrB RFLP and Sau3AI digestion resistance, this strain is indistinguishable from serotype 4b ECI strains (36, 37). In addition, the single available serotype 4d strain (also the type strain from the ATCC) had RFLP pattern I, identical to that of nonepidemic serotype 4b strains (Table 1). None of the genotyping tools that we and others have used, including detection of serotype-specific genes (20, 25) and PFGE (8), has been able to differentiate between serotype 4b and serotype 4d or 4e. Furthermore, serotype 4d and 4e strains also reacted with monoclonal antibodies which reacted with serotype 4b but no other serotypes of L. monocytogenes (17).
The nonpathogenic species L. innocua, which is considered to be the one most closely related genetically and bacteriologically to L. monocytogenes (30), had two RFLPs, both of which were encountered in serotype 4b as well. It is noteworthy that L. innocua lineage I shared the same RFLP pattern (pattern III) as the epidemic strains of L. monocytogenes ECII. This lineage of L. innocua is unique in that its teichoic acid-associated surface antigens are identical to those of L. monocytogenes 4b and in harboring serotype 4b-specific teichoic acid glycosylation genes (18), facts which may be of relevance in terms of the origin of this lineage. Other L. innocua strains had pattern I, which is seen in non-epidemic L. monocytogenes serotype 4b strains. The presence of apparently common RFLP patterns between L. monocytogenes serotype 4b and L. innocua is of potential evolutionary interest.
We identified a surprisingly high number of RFLP patterns in serogroup 4 (e.g., patterns IV, V, VI, and VIII [Table 1]) which were specific and unique to single lineages, suggesting that the RFLPs arose in a specific lineage. Pattern IV was found in only three L. monocytogenes serotype 4b strains in our collection which were unusual in terms of PFGE pattern and additional genetic markers (Lan et al., unpublished data). One of these strains was a clinical isolate from 1999, whereas the other two were environmental strains isolated earlier (1995) from a produce storage facility. These strains may represent a unique clone not commonly seen within serotype 4b but apparently capable of causing human illness. The strains of patterns V and VI were of serotypes 4a and 4c (including the 4a type strain). It is worth noting that the use of the large probe was essential in identifying the unique RFLP patterns of these strains, as patterns were identical (1.8-kb band) when the small probe 28.5 was used. Strains of serotypes 4a and 4c are genetically quite distinct from other L. monocytogenes strains and represent a distinct lineage (26, 35). In addition, strains of serotypes 4a and 4c had attenuated virulence in mice, although they were still capable of inducing a protective immune response (10, 32). In our laboratory, these strains have been found to have unique RFLPs with probes derived from a serotype-specific DNA region (20). Serotypes 4a and 4c are rarely found in food or environmental samples and are also rare in human infection but constituted 8 to 25% of animal listeriosis isolates in one study (35). This may be a genetically distinct, animal-adapted lineage, although we must note that of the four 4a and 4c strains that we examined, two were of human clinical origin.
The genomic region from which these probes were derived remains to be characterized. Our sequence analysis results suggest that this is a putative mannitol permease locus. Preliminary biochemical characterization of a mutant with an insertion mutation in the putative mtlA gene suggests that the mutant was incapable of mannitol uptake (H. L. Tran, S. Kathariou, and R. Hutkins, unpublished findings). In B. stearothermophilus and S. carnosus, mtlA is the first gene in an operon that contains additional genes involved in mannitol metabolism (12, 15). Such mannitol metabolism genes are expected to be identified when the annotated L. monocytogenes genome sequence data are released.
In conclusion, the probes described here have molecular subtyping potential in identifying several distinct lineages of L. monocytogenes of serogroup 4. These and similar probes that may become identified in the future may also be used as tools to elucidate the evolutionary relationships among different lineages (including epidemic-associated lineages) of L. monocytogenes as well as between L. monocytogenes and the closely related nonpathogenic species L. innocua.
Acknowledgments
This work was partially supported by U.S. Department of Agriculture Competitive Research Initiative AAFS grant 99-35201-8183 and by International Life Sciences Institute-North America.
We thank the investigators who have provided strains used in this work and listed in Table 1 of reference 36. We also thank Rebecca Kanenaka, Nancy Freitag, and Wen Lin for providing strains. We thank Zheng Lan, Jianying Li, Edward Lanwermeyer, and all other members of our laboratory for valuable feedback and support throughout the course of this work.
REFERENCES
- 1.Aarts, H. J., L. E. Hakemulder, and A. M. Van Hoef. 1999. Genomic typing of Listeria monocytogenes strains by automated laser fluorescence analysis of amplified fragment length polymorphism fingerprint patterns. Int. J. Food Microbiol. 49:95–102. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1999. Update: multistate outbreak of listeriosis—United States, 1998–1999. Morb. Mortal. Wkly. Rep. 47:1117–1118. [PubMed] [Google Scholar]
- 3.Aureli, P., G. C. Fiorucci, D. Caroli, G. Marchiaro, O. Novara, L. Leone, and S. Salmaso. 2000. An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria monocytogenes. N. Engl. J. Med. 342:1236–1241. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. D. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bibb, W. F., B. Schwartz, B. G. Gellin, B. D. Plikaytis, and R. E. Weaver. 1989. Analysis of Listeria monocytogenes by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Int. J. Food Microbiol. 8:233–239. [DOI] [PubMed] [Google Scholar]
- 6.Bibb, W. F., B. G. Gellin, R. Weaver, B. Schwartz, B. D. Plikaytis, M. W. Reeves, R. W. Pinner, and C. V. Broome. 1990. Analysis of clinical and food-borne isolates of Listeria monocytogenes in the United States by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Appl. Environ. Microbiol. 56:2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boerlin, P., and J. C. Piffaretti. 1991. Typing of human, animal, food, and environmental isolates of Listeria monocytogenes by multilocus enzyme electrophoresis. Appl. Environ. Microbiol. 57:1624–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60:2584–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchrieser, C., R. Brosch, B. Catimel, and J. Rocourt. 1993. Pulsed-field gel electrophoresis applied for comparing Listeria monocytogenes strains involved in outbreaks. Can. J. Microbiol. 39:395–401. [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty, T., F. Ebel, J. Wehland, J. Dufrenne, and S. Notermans. 1994. Naturally occurring virulence-attenuated isolates of Listeria monocytogenes capable of inducing long term protection against infection by virulent strains of homologous and heterologous serotypes. FEMS Immunol. Med. Microbiol. 10:1–9. [DOI] [PubMed] [Google Scholar]
- 11.Farber, J. M., and P. L. Peterkin. 1991.Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, R., and W. Hengstenberg. 1992. Mannitol-specific enzyme II of the phosphoenolpyruvate-dependent phosphotransferase system of Staphylococcus carnosus. Sequence and expression in Escherichia coli and structural comparison with the enzyme II mannitol of Escherichia coli. Eur. J. Biochem. 204:963–969. [DOI] [PubMed] [Google Scholar]
- 13.Gellin, B. G., and C. V. Broome. 1989. Listeriosis. JAMA 261:1313–1320. [PubMed] [Google Scholar]
- 14.Graves, L., M., B. Swaminathan, M. W. Reeves, S. B. Hunter, R. E. Weaver, B. D. Plikaytis, and A. Schuchat. 1994. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J. Clin. Microbiol. 32:2936–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henstra, S. A., B. Tolner, B., R. H. ten Hoeve Duurkens, W. N. Konings, and G. T. Robillard. 1996. Cloning, expression, and isolation of the mannitol transport protein from the thermophilic bacterium Bacillus stearothermophilus. J. Bacteriol. 178:5586–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Herd, M., and C. Kocks. 2001. Gene fragments distinguishing an epidemic-associated strain from a virulent prototype strain of Listeria monocytogenes belong to a distinct functional subset of genes and partially cross-hybridize with other Listeria species. Infect. Immun. 69:3972–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kathariou, S. 2000. Pathogenesis determinants of Listeria monocytogenes, p.295–314. In J. W. Cary, J. E. Linz, and D. Bhatnagar (ed.), Microbial foodborne diseases. Technomics Publishing Co., Inc., Lancaster, Pa.
- 17.Kathariou, S., C. Mizumoto, R. D. Allen, A. K. Fok, and A. A. Benedict. 1994. Monoclonal antibodies with a high degree of specificity for Listeria monocytogenes serotype 4b. Appl. Environ. Microbiol. 60:3548–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan, Z., F. Fiedler, and S. Kathariou. 2000. A sheep in wolf’s clothing: Listeria innocua strains with teichoic acid-associated surface antigens and genes characteristic of Listeria monocytogenes serogroup 4. J. Bacteriol. 182:6161–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, C. A., and M. H. Saier, Jr. 1983. Mannitol-specific enzyme II of the bacterial phosphotransferase system. III. The nucleotide sequence of the permease gene. J. Biol. Chem. 258:10761–10767. [PubMed] [Google Scholar]
- 20.Lei, X.-H., N. Promadej, and S. Kathariou. 1997. DNA fragments from regions involved in surface antigen expression specially identify Listeria monocytogenes serovar 4 and a subset thereof: cluster IIB (serotype 4b, 4d, and 4e). Appl. Environ. Microbiol. 63:1077–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margolles, A., B. Mayo and, C. G. de los Reyes-Gavilan. 1998. Polymorphism of Listeria monocytogenes and Listeria innocua strains isolated from short-ripened cheeses. J. Appl. Microbiol. 84:255–262. [DOI] [PubMed] [Google Scholar]
- 22.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O‘Donoghue, K., K. Bowker, J. McLauchlin, D. S. Reeves, P. M. Bennett, and A. P. MacGowan. 1995. Typing of Listeria monocytogenes by random amplified polymorphic DNA (RAPD) analysis. Int. J. Food Microbiol. 27:245–252. [DOI] [PubMed] [Google Scholar]
- 24.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Promadej, N., F. Fiedler, P. Cossart, S. Dramsi, and S. Kathariou. 1999. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serotype-specific gene. J. Bacteriol. 181:418–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053–2061. [DOI] [PubMed] [Google Scholar]
- 27.Ridley, A. M. 1995. Evaluation of a restriction fragment length polymorphism typing method for Listeria monocytogenes. Res. Microbiol. 146:21–34. [DOI] [PubMed] [Google Scholar]
- 28.Schlech, W. F., III. 1997. Listeria gastroenteritis—old syndrome, new pathogen. N. Engl. J. Med. 336:130–132. [DOI] [PubMed] [Google Scholar]
- 29.Schuchat, A., B. Swaminathan, and C. V. Broome. 1991. Epidemiology of human listeriosis. Clin. Microbiol. Rev. 4:169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seeliger, H. P. 1981. Nonpathogenic listeriae:L. innocua sp. n. Zentbl. Bakteriol. Mikrobiol. Hyg. A 249:487–493. [PubMed] [Google Scholar]
- 31.Seeliger, H. P., and K. Hoehne. 1979. Serotypes of Listeria monocytogenes and related species. Methods Microbiol. 13:31–49. [Google Scholar]
- 32.Sokolovic, Z., S. Schuller, J. Bohne, A. Baur, U. Rdest, C. Dickneite, T. Nichterlein, and W. Goebel. 1996. Differences in virulence and in expression of PrfA and PrfA-regulated virulence genes of Listeria monocytogenes strains belonging to serogroup 4. Infect. Immun. 64:4008–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran, H. L., F. Fiedler, D. A. Hodgson, and S. Kathariou. 1999. Transposon-induced mutations in two loci of Listeria monocytogenes serotype 1/2a result in phage resistance and lack of N-acetylglucosamine in the teichoic acid of the cell wall. Appl. Environ. Microbiol. 65:4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vines, A., M. W. Reeves, S. Hunter, and B. Swaminathan. 1992. Restriction fragment length polymorphism in four virulence-associated genes of Listeria monocytogenes. Res. Microbiol. 143:281–294. [DOI] [PubMed] [Google Scholar]
- 35.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, P. L., and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng, W., and S. Kathariou. 1995. Differentiation of epidemic-associated strains of Listeria monocytogenes by restriction fragment length polymorphism in a gene region essential for growth at low temperature (4°C). Appl. Environ. Microbiol. 61:4310–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng, W., and S. Kathariou. 1997. Host-mediated modification of Sau3AI restriction in Listeria monocytogenes: prevalence in epidemic-associated strains. Appl. Environ. Microbiol. 63:3085–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]