Abstract
The objective of our study was to isolate and determine the phylogenetic affiliation of culturable estuarine bacteria capable of catabolizing riverine dissolved organic matter (RDOM) under laboratory conditions. Additions of RDOM consistently promoted the growth of estuarine bacteria in carbon-limited dilution cultures, with seasonal variation in growth rates and yields. At least 42 different taxa were culturable on solid agar media and, according to quantitative DNA-DNA hybridizations, constituted 32 to 89% of the total bacterial number in the enriched treatments. Five species in the Cytophaga-Flexibacter-Bacteroides group and one in the γ-proteobacteria phylogenetic group (Marinomonas sp.) were numerically dominant during the stationary phase of the RDOM-enriched dilution cultures but not in the control cultures. Four of the isolates in Cytophaga-Flexibacter-Bacteroides group were putatively affiliated with the genus Flavobacterium. All dominating isolates were determined to be new species based on comparison to the current databases. The same group of species dominated independently of the season investigated, suggesting a low diversity of bacteria catabolizing RDOM in the estuary. It also suggested a broad tolerance of the dominating species to seasonal variation in hydrography, chemistry, and competition with other species. Taken together, our results suggest that a limited group of bacteria, mainly in the Flavobacterium genus, played an important role in introducing new energy and carbon to the marine system in the northern Baltic Sea.
In this study we sought to identify the group of estuarine bacteria that catabolize riverine dissolved organic matter (RDOM) discharged by rivers into the marine environment in the northern Baltic Sea. Estuarine waters such as the northern Baltic have a large supply of allochthonous material; therefore, carbon produced on land can constitute an important energy source for the food web (5, 18, 23, 29, 33, 37). Bacterial consumption of terrigenous compounds may thereby uncouple bacterial production from phytoplankton production (11, 16) and shift the carbon balance in lakes and estuaries, creating net heterotrophy (14, 18, 30). Therefore, marine bacterial species that catabolize terrigenous compounds may have an important role similar to that of primary producers in releasing new energy and carbon into estuarine environments.
To our knowledge, no studies aiming to identify what marine bacterial species that catabolize allochthonous (riverine or terrestrial) substrates have been reported. Studies of the in situ community show a great diversity of bacterial species present in estuarine environments. In the northern Baltic during the productive season the in situ bacterial community in the photic zone showed a large phylogenetic diversity covering α-, β-, and γ-Proteobacteria and the Cytophaga-Flexibacter-Bacteroides (CFB) group, except during the spring, when five species within the CFB group were common (26, 27). In the Columbia River, its estuary, and adjacent coastal ocean, Crump et al. found that the in situ bacterial community was mainly affiliated with the class Proteobacteria, while the CFB group (particularly the genus Cytophaga) was related to particle-attached bacteria (7).
The discharge of allochthonous dissolved organic matter (DOM) by rivers is also accompanied by a discharge of lotic (river) bacteria that could be successful immigrants into the estuarine environment. It was determined that in the Elbe River β-Proteobacteria numerically dominated growth on organic aggregates, followed by Cytophaga-Flavobacterium (4). In two Japanese rivers the planktonic lotic bacteria were dominated by the Cytophaga-Flavobacterium, Burkholderia-Pseudomonas, and Alcaligenes groups (17). These studies show that a wide range of species occur in, or are introduced to, the estuarine environment, but no study has specifically addressed the question of which bacterial phyla catabolize allochthonous RDOM or how general this capability is.
Our experiment was designed to promote the growth of estuarine bacteria catabolizing substrates in the RDOM pool and compared to a control without RDOM. In order to account for trace amounts of estuarine DOM accompanying the inoculum, the control treatment contained the same concentration of estuarine DOM as did the positive treatment. Experiments were performed during three different seasons to investigate whether a seasonal variation in the diversity of RDOM-catabolizing species occurred. In this study we focused on the population of bacteria that was culturable by initial isolation on two different media. The isolates were taxonomically determined by 16S ribosomal DNA (rDNA) sequencing, and their abundance in the cultures was determined by quantitative DNA-DNA hybridization.
MATERIALS AND METHODS
Study area.
The studied estuary and Öre river are situated in the northern Baltic (station B3, 63°29.98′N, 19°49.14′E [38]). Experiments were conducted during three different seasons (sampling and start on 26 March, 25 May, and 18 November 1998). Allochthonous dissolved organic matter was obtained from the Öre River water 1 km upstream from the river mouth (20-liter samples). Estuarine water was obtained from the national monitoring station B3 by mixing 10 liters of water from a depth of 4 m with 10 liters from 20 m.
Treatments.
All polycarbonate bottles, filters, filtration equipment, etc., used during the sample processing were carefully washed with 10% HCl and rinsed with Milli-Q water (Millipore). The following treatments were carried out in duplicate 4-liter bottles. (i) For the +RDOM+I treatment, the enrichment cultures, providing for growth of estuarine bacteria on RDOM, were made from a 3-liter sterile river water sample serially filtered three times through 0.2-μm (pore-size) Gelman Supor filters. The samples were adjusted to the salinity of the estuary (4 practical salinity units [psu]; International Association for the Physical Sciences of the Ocean) with artificial brackish water medium (100× [37]). Finally, 300 ml of estuarine inoculum was added. The inoculum was obtained by gently filtering 2 liters of freshly collected estuarine water three times through a 0.6-μm polycarbonate filter (MSI). This procedure results in a minimum of interference from bactivorous flagellates and bacteria from the river water (37). For the −RDOM+I treatment, the main control was a dilution culture without river water, accounting for the growth of estuarine bacteria on diluted estuarine DOM originating from the inoculum. Three liters of brackish water medium (salinity, 4 psu) was mixed with an estuarine inoculum (300 ml) prepared as described above. (iii) +RDOM−I, an additional control culture without inoculum, was included in order to monitor the potential of freshwater bacteria to escape the serial filtration and grow under estuarine conditions. Three liters of sterile filtered river water was adjusted to the salinity of the estuary (4 psu) with brackish water medium (100×) as in the treatments above. No estuarine water inoculum was added to this treatment.
Inorganic nutrients (6 μM NaNO3 and 0.6 μM Na2PO4, final concentration) were added to all treatments to give an enrichment which approximates the inorganic nutrient concentration at site B3 during unstratified winter conditions. Nutrient values from national monitoring programs at adjacent dates for both the estuary and river showed that the final phosphate concentrations in all treatments and at all seasons after these additions were similar (average, 0.75 μM; coefficient of variation [CV], ±12%). The same was true for nitrate (average, 10.5 μM; CV, ±31%). Ammonium, nitrite, and nutrients of the estuarine inoculum (due to 10-fold dilution) were minor components of the nutrient pools. Major salts, macronutrients, and estuarine DOM were thereby similar in treatment to +RDOM+I and −RDOM+I, the major difference being the large pool of RDOM in the +RDOM+I treatment.
Incubation conditions.
All experimental bottles were incubated with gentle shaking (50 rpm) in the dark at an in situ temperature for approximately 2 weeks (377 to 408 h) until the bacterial community reached stationary phase.
Bacterial numbers.
The total number of bacteria (TNB) was estimated by staining the cells with acridine orange. Subsamples of 20 ml were collected and preserved with formaldehyde (final concentration, 4%) on each day from each flask. Then, 1 to 5 ml of the preserved samples was filtered onto a black 0.2-μm (pore-size) polycarbonate Poretics filter and mounted in oil immersion on a microscope slide (15). Cells were counted in an epifluorescence microscope (Zeiss Universal) with a Neofluar 100× oil immersion objective lens and a 1.25× ocular lens. At least 30 fields and 200 cells were counted for each sample. Cell sizes were determined by image analysis fluorescence microscopy by using a cooled, slow-scan charge-coupled device camera (C4742 Hamamatsu [3]).
The numbers of nucleoid-containing bacterial cells (NucC) were estimated by the DAPI (4′,6′-diamidino-2-phenylindole) staining-destaining protocol described by Zweifel et al. (40). Samples (10 ml) were incubated for 1 h with DAPI solution and Triton X-100 in the dark at 5°C (no formaldehyde preservation). After incubation, 2 to 10 ml of the sample was filtered onto black 0.2-μm (pore-size) polycarbonate filters (Poretics) and counted as described above.
Isolation of bacteria.
When bacterial growth reached stationary phase (usually after 2 weeks) the bacterial abundance was ca. 1 × 106 to 2 × 106 cells ml−1. Culturable bacteria were isolated and counted by spreading samples of 0.1 ml on triplicate plates with ZoBell agar medium and a nutrient-poorer AC medium, with allochthonous carbon as the sole energy and carbon sources, respectively. The latter had an allochthonous carbon as the sole carbon source (see below). To maximize CFU on ZoBell plates dilutions of 10×, 100×, or 1,000× were made, except for the experiment in March. Plates were incubated in the dark at 15°C until no more colonies appeared (usually ca. 5 to 15 days). All different colony morphology types observed were picked in triplicates. Purification of isolates was performed by serially streaking a single-cell colony three times onto new agar plates. The purified isolate was grown in ZoBell medium (10 ml) in the dark at 15°C (1 to 2 days). After the purity of the isolate was confirmed by one more plating it was regrown in liquid ZoBell medium, and 0.8 ml of cell suspension was mixed with 50% glycerol (0.2 ml) and stored at −80°C.
AC medium was prepared by concentrating 20 liters of river water to 180 ml by using tangential ultrafiltration (10,000 Da). This resulted in a dissolved organic carbon (DOC) concentration of 60 mM. Artificial brackish water medium (100× [37]) was added to give a final salinity of 4 psu. PO43− (2.8 mM final concentration) and NO3− (28 mM final concentration) were added to match the carbon content in the concentrated river water to barely allow visible colony formation on the plates. The volume was then adjusted to 200 ml. The medium was added (1.5% final concentration) to Bacto-Agar, and the mixture was autoclaved for 30 min at 121°C.
DOC.
Samples for the determination of DOC were filtered through a 0.2-μm (pore-size) Gelman Supor filter by using a sterile syringe (Plastipak) and filter holders (Millipore) within 4 h of sampling according to the method of Norrman (24). DOC was measured by the high-temperature catalytic oxidation method with a Shimadzu TOC-5000 instrument with platinum-coated Al2O3 granulates as a catalyst. The instrument has been subjected to international intercalibration (25). The detection limit of the DOC method was 8 μM. Milli-Q (Millipore) water was routinely used as an analytical background in our analyses and did not contain detectable levels of DOC (<8 μM). The contribution of DOC from Milli-Q water in the treatments can therefore be regarded as negligible.
16S rDNA sequencing.
A colony of each isolate was resuspended in liquid ZoBell medium and incubated at 15°C for 2 to 10 days. A 10-ml sample of each culture was centrifuged at 1,000 × g for 15 min. The pellet was resuspended in Tris-EDTA (TE) buffer (pH 8.0), cells were lysed with lysozyme (final concentration, 1 mg ml−1; 1 h at 37°C) and proteinase K (final concentration, 100 μg ml−1; 1 h at 37°C), followed by CTAB (cetyltrimethylammonium bromide)-NaCl (final concentration, 1%) treatment and chloroform-phenol extraction. DNA was precipitated in ethanol and resuspended in TE buffer (pH 8.0) or Milli-Q water.
16S rDNA from isolated bacteria was amplified by using Taq polymerase (Boehringer Mannheim). Bacterial universal primers 27F (3′-AGAGTTTGATCATGGCTCAG-5′) and 1492R (3′-TACGGYTACCTTGTTACGACTT-5′) were used for amplification (19). The PCR product was purified with PCR Kleen Spin Columns (Bio-Rad).
Nucleotide sequences were determined from the purified 16S rDNA by automated sequencing with an ABI PRISM II Dye Terminator Cycle Sequencing kit (Perkin-Elmer) with the primers 27F (3′-AGAGTTTGATCATGGCTCAG-5′) and 800R (3′-CCAGGGTATCTAATCC-5′), which gives an ∼800-bp rDNA fragment (19, 39).
Phylogenetic analysis.
The similarity of 16S rDNA sequences of isolates was determined by using the BLAST database search (1). Modified current empirical criteria were used to classify sequences into taxonomic units (see, for example, references 8, 9, and 13) because rDNA similarities are not standardized. If >97% similarity to any sequence in the database was found, the species name of the homologous sequence was used. Otherwise “new” sequences were classified to the genus level at 95 to 97% similarity. Bacteria at a similarity of <95% were affiliated to the class or group level.
Isolates having >97% 16S rDNA similarity to each other were further evaluated by genomic DNA-DNA hybridization. If they had >70% DNA-DNA similarity they were classified as the same species (36). Taxonomic names were based on the classification proposed by Bergey’s Manual of Systematic Bacteriology (2). The sequences were aligned by using the secondary structure of the small subunit rRNA. A phylogenetic tree was constructed by using comparisons between distance (Kimura’s two-parameter model) and the maximum parsimony method. Comparison between different optimality criterion trees was done by use of a Shimodaira-Kasegawa test to find the best tree based on a maximum-likelihood optimality criterion. The permutation test was used to evaluate the significance of the tree, and the jackknife method was used to estimate the significance of the branches.
Screening for abundant species.
Reverse-hybridization experiments were conducted to screen for dominating species in the cultures. The community DNAs from all seasons and treatments were radioactively labeled with a random priming kit (High Prime DNA Labeling Kit; Boehringer Mannheim) and [α-32P]dATP (3,000 to 6,000 Ci mmol−1; Amersham Pharmacia Biotech). The labeled community DNA from each treatment and season was then hybridized toward cells of all isolated strains (312 strains), applied in duplicates onto hybridization membranes at the estimated saturation level of the probe DNA to allow a quantitative comparison (ca. 109 cells per dot). Cells of isolated strains were blotted and treated as described below for preparing dilution series. Hybridization conditions and the detection method were as specified below.
Abundance of culturable populations.
On the basis of the reverse hybridization, isolates showing a strong dot blot hybridization signal were quantified on the basis of the total DNA-DNA hybridization protocol, as originally described by Pinhassi et al. (27). Chromosomal DNA of an isolate (extracted as described in the 16S rDNA sequencing section above) was labeled by using a random priming kit (High Prime DNA Labeling Kit) and [α-32P]dATP (3,000 to 6,000 Ci/mmol; Amersham Pharmacia Biotech). Labeled DNA was purified from free nucleotides with spin columns (MicroSpin S-300 HR; Amersham Pharmacia Biotech) and used as a probe.
A standard curve of bacterial abundance versus signal intensity from the dot blot hybridization of each bacterial strain was prepared and hybridized together with the unknown samples from the cultures in all cases. Bacterial counts were obtained by direct microscopic enumeration as described above. A dilution series was then made by suspending bacteria in autoclaved, 0.2-μm-filtered brackish water. Duplicate samples from 1 × 104 to 5 × 106 cells per dot were applied onto hybridization membranes (Hybond-N+; Amersham) by using a blotting apparatus (Gibco-BRL). Cells were lysed as described below for community DNA.
Bacterial cells from experimental water (volumes of 10 or 20 ml) were filtered onto the same membranes as the bacterial standards. The samples were lysed in the blotting apparatus by covering the slots with 100 μl of 0.5 M NaOH, followed by incubation for 5 min. The solution was filtered dry, and the slot was covered with 100 μl of 1 M Tris-HCl (pH 7.4) for 5 min. Finally, the dot was covered with 100 μl of 1.5 M NaCl-0.5 M Tris-HCl (pH 7.4), and the solution was filtered dry. The DNA was linked to the membrane by optimal cross-linking (1,200 mJ cm−2, 14 s). Membranes were prehybridized in a solution consisting of 10% dextran sulfate, 1% sodium dodecyl sulfate (SDS), and 100 μg of salmon sperm DNA per ml for at least 2 h at 69°C in a hybridization incubator (Robbins Scientific Model 400). Denatured probe was added to the hybridization tube (Robbins Scientific). The membranes with community DNA and a standard curve of each isolate were hybridized in the same hybridization tube overnight at 69°C. The membranes were washed with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate)-0.1% SDS solution at 74°C for 45 min and in 2× SSC at room temperature for 45 min. Washed membranes were exposed on a PhosphorImager (Molecular Dynamics) for detection of the hybridization signal. The relationship between the hybridization signal and the number of bacteria was obtained from the slope of the standard curve for each isolate.
Nucleotide sequence accession numbers.
Sequences reported have been submitted to GenBank under accession numbers AF321008 to AF321049. The designation GOBB3 used for isolate names originates from the Swedish national monitoring station B3 in the Gulf of Bothnia.
RESULTS
Bacterial growth in the treatments.
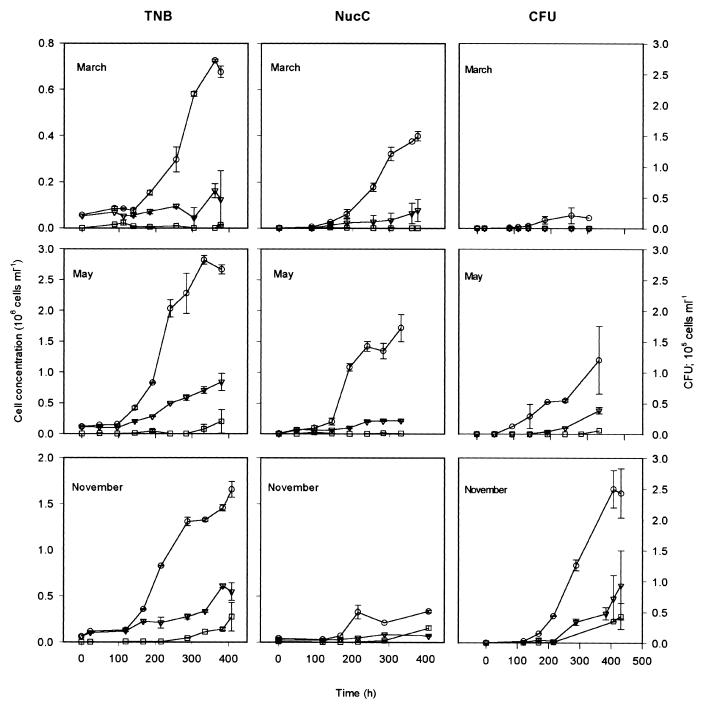
RDOM consistently promoted the growth of estuarine bacteria at all seasons, with a higher growth rate and yield than in the controls. A marked increase in the TNB, in total number of cells with a stainable nucleus (NucC), and in CFU (on ZoBell plates) was observed in the treatment with RDOM and estuarine inoculum (+RDOM+I) at all seasons. A smaller increase in cell numbers was also observed in treatments without RDOM (−RDOM+I), probably as a result of growth on estuarine DOM from the inoculum. Growth in the treatment with RDOM but without inoculum (+RDOM−I) was barely detectable in March and May (Fig. 1). However, in the November experiment there was a small increase in TNB, NucC, and CFU in the +RDOM−I treatment with estuarine salt concentration due to freshwater bacteria escaping the filtering procedure. All of these cell growth parameters had a lag period of 100 to 120 h in all treatments and an exponential growth phase and, after 250 to 300 h, the cultures reached stationary phase. Higher end values of TNB, NucC, and CFU were consistently observed with the +RDOM+I treatments compared to the −RDOM+I treatments. The growth rate in the log phase was usually two times higher with the +RDOM+I treatment than with the −RDOM+I control, but the differences in CFU between treatments were less clear due to higher replicate variance.
FIG. 1.
Growth curve based on TNB values (left panels) in three different seasons (March, May, and November). The middle panels show the growth of bacteria with a stainable nucleus (NucC) during the three different seasons. The right panels show the growth curve based on CFU on ZoBell agar plates in three different seasons. The bars represent the range between the lowest and highest values of the replicate treatments. Symbols: ○, +RDOM+I; ▵, −RDOM+I; □, +RDOM−I.
The bacterial biomass produced matched the DOC consumed well, with a growth efficiency of 5 to 16%. The latter was close to the average growth efficiency from a 3-year field study reported for the same type of dilution cultures by Wikner et al. (37). Due to the moderate nutrient additions made to avoid extreme levels, the DOC consumption was, however, slightly lower than was found in that study. The DOC consumption observed was therefore sufficient to promote the bacterial carbon production in the cultures.
Seasonal variation in the +RDOM+I treatment.
The increase in cells until the plateau phase (ΔN) and NucC varied markedly between seasons. The ΔN was highest in May and lowest in March. The growth rate of TNB (μTNB) was lower in March than in May and November (analysis of covariance linearity test, P = 0.04). The specific growth rate based on NucC counts (μNucC) was similar (P > 0.05) in all experiments.
Seasonal variation with the −RDOM+I treatment.
The growth rate of TNB was clearly highest in the May experiment (Fig. 1). The specific growth rate based on NucC counts (μNucC) was not different (P > 0.05) in March, May, and November. Also, the ΔN was higher in the May experiment than in the rest of the experiments, wherein the TNB and NucC reached 0.77 × 106 and 0.213 × 106 cells ml−1, respectively.
Seasonal variation with the +RDOM−I treatment.
There was no significant growth during experiments in March and May with the +RDOM−I treatments, suggesting growth of contaminating freshwater strains to be negligible (low ΔN, Fig. 1). In the November experiment both TNB and NucC showed a significant increase in ΔN, where the growth rate and yield of NucC were comparable to those obtained with the −RDOM+I treatment.
CFU numbers.
Like the TNB and NucC, the CFU counts increased with time with the +RDOM+I and −RDOM+I treatments for all tested seasons and also with the +RDOM−I treatment in the November experiment. The increase in CFU was generally greater with the +RDOM+I treatment in all seasons (Fig. 1).
CFU versus TNB and NucC.
The fraction of CFU relative to the TNB and NucC did not change much over time in the cultures. Generally CFU constituted a low proportion of the TNB and NucC, being <10% of the TNB in most cases and <25% of the NucC. The proportion of culturable bacteria was usually higher with the +RDOM+I treatment compared to the −RDOM+I treatment.
Genotype characterization of the isolated strains.
A total of 94 isolates were subjected to 16S rDNA sequencing. This took into account that similar colony morphologies, appearing in different seasons, could be different species. The selection of strains also encompassed the possibility that replicates of a given colony morphology could be different species. Originally, isolates were grouped subjectively into 23 different colony morphology types. They were distinguished on agar plates by color, surface, size, and other morphological characteristics (data not shown). The 16S rDNA sequencing resulted in 42 different identified taxa of culturable bacteria in the experimental bottles.
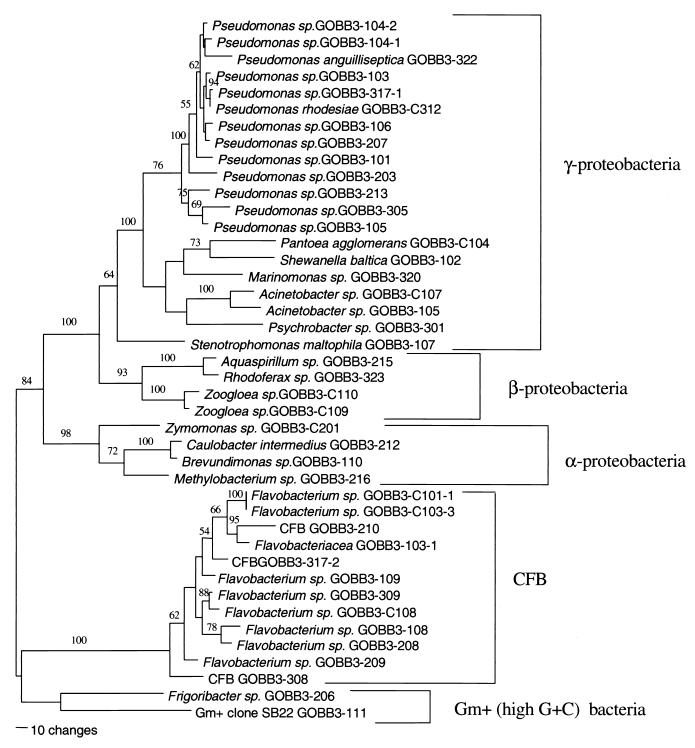
Most colony-forming bacteria on the applied solid media belonged to the γ-Proteobacteria subclass. The second most common class on the plate surface was the CFB group with several novel taxa. Two strains belonged to gram-positive bacteria (high G+C content) in the Firmicutes class. The α- and β-proteobacterium subclasses were represented by four and five taxa, respectively (Fig. 2). The species composition of dominating bacteria on the plate surface was thus clearly different from that measured in the liquid cultures (see below).
FIG. 2.
Phylogenetic relationships between isolated strains from all seasons with the +RDOM+I and −RDOM+I treatments. The maximum parsimony tree was used for selecting the best-fitting tree. The first-order jackknife values above 50% are shown, and the length of the lines corresponds to the number of base pair differences between species. The presented tree was significant (P = 0.01, permutation test): tree length, 1,865; consistency index, 0.481; retention index, 0.769.
Screening for abundant populations.
Reverse DNA-DNA hybridization was used to perform a rapid screening for most abundant strains. This analysis showed that 30 strains out of 312 dots (two to three replicates of all found colony morphologies) were detectable. These 30 strains were chosen for further quantification of abundant species in the late-exponential-growth phase by quantitative DNA-DNA hybridization. Isolates selected for the probe preparation came from different treatments and seasons with no apparent systematic distribution.
Dominating strains.
Selected and quantified populations by dot blot DNA-DNA hybridization constituted 32 to 89% of the TNB in the +RDOM+I treatment, depending on the season investigated. The diversity in this treatment was significantly different from that in the controls but was not different between the seasons. The significance of the dispersions was tested by using the multivariate randomization test with the grouping factors RDOM (levels +/−), seasons, and duplicates (P < 0.05 [22]).
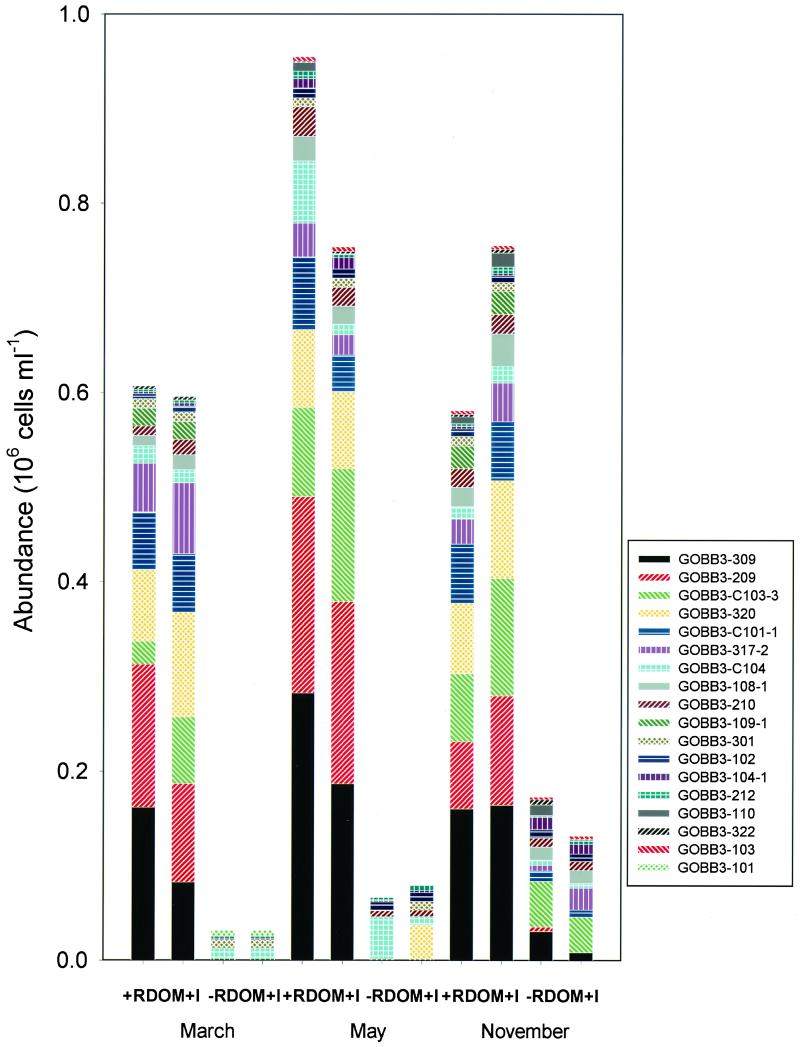
A small group of five to six populations numerically dominated in the +RDOM+I treatment and accounted for 10 to 86% of the quantified bacterial numbers depending on the season (Table 1 and Fig. 3). They were significantly higher in the presence of RDOM than in the control without RDOM (P < 0.05, nonparametric Mann-Whitney test). This group of potential RDOM-catabolizing bacterioplankton were all nondescribed taxa, most of them belonging to the CFB group and the Flavobacterium genus. One of the dominating isolates in the CFB group was not similar (similarity, <95%) to any genus according to the available sequence information in databases. The only dominating isolate outside the CFB class was a Marinomonas sp. strain GOBB3-320 in the γ-Proteobacteria subclass. The same strains dominated during all seasons and in both replicate cultures of the +RDOM+I treatment (Fig. 3). Generally, the species diversity was similar in duplicate bottles, although −RDOM+I treatment replicates were more variable than with the +RDOM+I and +RDOM−I treatments.
TABLE 1.
Abundance of bacterial populations as quantified by DNA-DNA hybridizationa
| Treatment and strain | Mean no. of bacteria (104 cells ml−1) ± SEb in:
|
||
|---|---|---|---|
| March | May | November | |
| +RDOM+I | |||
| Flavobacterium sp. strain GOBB3-309 | 12.3 ± 4.0 | 23.5 ± 4.5 | 16.3 ± 0.0 |
| Flavobacterium sp. strain GOBB3-209 | 12.8 ± 2.3 | 2.4 ± 0.1 | 9.3 ± 2.2 |
| Flavobacterium sp. strain GOBB3-C103-3 | 9.3 ± 1.8 | 8.2 ± 0.0 | 8.9 ± 1.5 |
| Marinomonas sp. strain GOBB3-320 | 4.8 ± 2.3 | 11.8 ± 2.3 | 9.8 ± 2.6 |
| Flavobacterium sp. strain GOBB3-C101-1 | 6.2 ± 0.1 | 5.8 ± 1.9 | 6.2 ± 0.1 |
| CFB GOBB3-317-2 | 6.3 ± 1.2 | 2.9 ± 0.7 | 3.3 ± 0.7 |
| −RDOM+I | |||
| Flavobacterium sp. strain GOBB3-309 | 0 | 0.8 ± 0.0 | 5.6 ± 2.4 |
| Flavobacterium sp. strain GOBB3-209 | 0 | 0 | 0 |
| Flavobacterium sp. strain GOBB3-C103-3 | 4.1 ± 0.1 | 0 | 1.9 ± 0.2 |
| Marinomonas sp. strain GOBB3-320 | 0 | 1.3 ± 0.0 | 0.9 ± 0.1 |
| Flavobacterium sp. strain GOBB3-C101-1 | 3.1 ± 0.9 | 0.9 ± 0.0 | 1.8 ± 0.2 |
| CFB GOBB3-317-2 | 0 | 0 | 1.7 ± 0.9 |
The most dominating taxa in the +RDOM+I treatment in comparison to their abundance in the −RDOM+I treatment are shown. Abundance values were statistically different between treatments for all species (P < 0.01, Mann-Whitney test).
Mean value of two replicates.
FIG. 3.
Diversity of isolated strains based on quantitative DNA-DNA hybridization in duplicate treatments. The upward order of patterns in the bars corresponds to the downward order in the legend.
In the control (−RDOM+I), with a diluted inoculum containing only estuarine DOM as a substrate, culturable bacterioplankton constituted 10 to 75% of the TNB. In the control treatment, seasonal differences of species composition and abundance were significant, in contrast to the positive treatment (the multivariate randomization test, P < 0.05). The species in the control were different from those dominating in the +RDOM+I treatment, representing several phyla such as α- and γ-proteobacteria and the CFB group. However, the latter phylum was represented by different species than in the +RDOM+I treatment.
Since the growth in the treatment with RDOM but without inoculum (+RDOM−I) was barely detectable, it was difficult to quantify the bacterial species in these samples. However, the γ-proteobacterium Pantoea agglomerans GOBB3-C104 was found in detectable amounts.
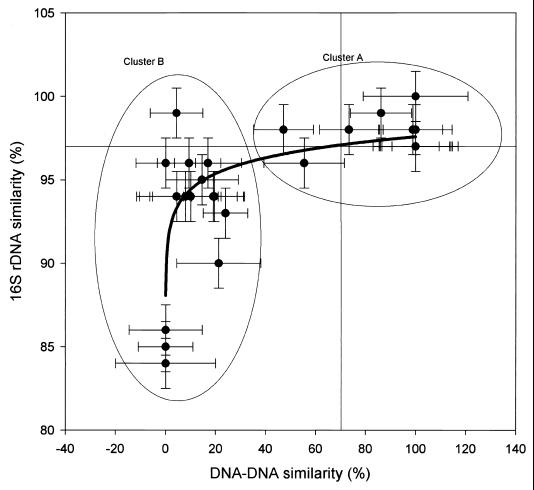
The extent of cross-hybridization of genomic DNA between several strains was analyzed. According to this analysis, the average extent of cross-hybridization among the dominating isolates within the Flavobacterium genus was 12.0% ± 2.2%. By plotting the 16S rDNA similarity versus the DNA-DNA similarity, the tested strains clearly grouped into two separate clusters (Fig. 4), the first including the same species and the second representing different species.
FIG. 4.
Comparison of relationship between 16S rDNA sequence similarities and the DNA-DNA similarities. The curved line represents the exponential relationships between these two variables. Cluster A includes strains supposed to be of the same species; cluster B includes strains of different species. Reference lines at 97% 16S rDNA and 70% DNA-DNA similarity were the empirical criteria recommended to be used to define a candidatus species. The error bars represent the standard errors of the similarity estimates.
Colony formation on ZoBell versus AC medium.
Of the isolated strains, 15 were found to grow on both the rich medium (ZoBell) and the poorer solid medium (AC medium). Nine of these belonged to the genus Pseudomonas. Also, two of the numerically dominant bacteria in the total community, Flavobacterium sp. strain GOBB3-309 and Flavobacterium sp. strain GOBB3-C101-1, grew on both ZoBell and AC media. Pseudomonas sp. strain GOBB3-106 and Flavobacterium sp. strain GOBB3-109 (not dominating) only grew on the AC medium plates. The majority of the isolated taxa (25 species) were found only on ZoBell medium plates.
DISCUSSION
The main objective with this study was to define the culturable bacterial taxa introducing new energy and carbon from allochthonous RDOM to the northern Baltic and to show if RDOM catabolism is a general characteristic of estuarine bacteria or restricted to a limited group of species. Fresh RDOM was therefore used as an energy and carbon substrate for a natural estuarine bacterial community (Table 2). Since a small amount of estuarine DOM is unavoidably introduced together with the inoculum, we compared the enrichment cultures to controls with a similar amount of inoculum diluted with estuarine salt medium. Major salts, macronutrients, and the concentration of the estuarine DOM were thus similar in both treatments. Growth of a bacterial population exceeding the control must therefore be as a result of metabolism of at least one substrate in the allochthonous RDOM pool. The energy and carbon supply for this growth could come from any of the substrates or colloids passing the 0.2-μm (pore-size) filter used for preparation of the medium. Taxa increasing in numbers with RDOM present were therefore probably RDOM consumers in the natural environment. We cannot identify other chemical constituents in the experimental water (e.g., trace metals or organic inhibitors) that were likely to hamper or limit the growth of some RDOM-consuming populations, although we also cannot exclude this possibility. If limiting or inhibiting substances were present, some RDOM-consuming culturable populations would be overlooked in this study.
TABLE 2.
DOC utilization, initial DOC concentration (To), and the concentration at the end (Tend)a
| Season | Treatment | DOC concn (μM)
|
Utilized DOC (μg of C liter−1) | Bacterial production (μg of C liter−1) | |
|---|---|---|---|---|---|
| To | Tend | ||||
| March | +RDOM+I | 479 ± 0 | 459 ± 3.0 | 240 ± 36 | 14 ± 0.2 |
| −RDOM+I | 46 ± 0 | 44 ± 2.9 | 24 ± 36 | 3.8 ± 0.1 | |
| +RDOM−I | 479 ± 0 | 477 ± 0 | 24 ± 0 | ||
| May | +RDOM+I | 800 ± 0 | 743 ± 11.8 | 684 ± 142 | 57 ± 0.7 |
| −RDOM+I | 71 ± 17.9 | 72 ± 5.8 | −12 ± 216 | 14 ± 1.4 | |
| +RDOM−I | 800 ± 0 | 797 ± 41.3 | 36 ± 496 | ||
| November | +RDOM+I | 921 ± 0 | 863 ± 5.9 | 696 ± 71 | 31.2 ± 0.9 |
| −RDOM+I | 179 ± 35 | 171 ± 0 | 96 ± 420 | 11.6 ± 0.9 | |
| +RDOM−I | 921 ± 0 | 906 ± 23.6 | 180 ± 391 | ||
Utilized DOC represents the DOC which was degraded at the end of the experiment. Bacterial production was calculated from ΔTNB.
Our results suggested that a limited group of species primarily exploited the pool of RDOM supplied to the Öre estuary. On the basis of DNA-DNA hybridization as few as six strains of the 42 different taxa (14%) showed a numeric response when RDOM comprised the major energy and carbon source, a finding independent of the season. The fact that the same set of species dominated independently of the season suggested that they were both present and competitive consumers of RDOM at various environmental and hydrographic conditions during the year. We infer from this that these strains had the catabolic competence to either directly or indirectly exploit substrates in the RDOM pool and produce biomass. The majority of the taxa in the group belonged to the CFB class; only one species was affiliated with the γ-Proteobacteria class. As shown in Table 1, all of the isolates in the CFB class belonged to the Flavobacteriaceae family, and four were in the Flavobacterium genus. None of the dominating species showed similarity (<97%) to bacterial sequences hitherto submitted to the BLAST database. Thus, all RDOM-catabolizing and -culturable isolates appeared to be new species.
The taxonomic affiliation of the RDOM-catabolizing marine bacteria to the CFB class was in accordance with current knowledge of the catabolic competence of this phylum (2). The CFB group is recognized to catabolize highly polymeric substances that are likely common in the RDOM pool in the form of polyaromatic substances and sugar polymers from higher plants. A recent combined phylogenic and functional study of bacteria also showed that bacteria in the CFB group respond to high-molecular-weight substrates (6).
During the productive season in the Öre estuary dominating bacterial taxa are distributed among all taxonomic groups, except during the spring flood, when bacteria in the CFB group are common (26). The latter finding was in good agreement with our results, since RDOM is expected to be the main source of carbon and energy during the spring flood. Pinhassi et al. (27) also found that bacteria in the CFB group are more common in the northern Baltic than in samples from the southern California Bight. Our study suggested that this is explained by the competence of bacterial species in the CFB phylum to catabolize complex substrates in the allochthonous DOM discharged by the numerous rivers, which is characteristic of the Baltic brackish water environment. Most of the taxa isolated in our study clearly represented a different set of taxa than that found in the estuarine in situ samples analyzed by Pinhassi et al. (27). This suggested that RDOM-catabolizing bacteria constituted a numerically small fraction of the bacterioplankton community in the estuary during the vegetation period and in the photic zone.
Our controls with estuarine DOM had a distribution of species between different phyla similar to that found in the in situ samples by Pinhassi et al. (26). Thus, the dilution of estuarine DOM did not appear to cause a dramatic change to the diversity expected in situ. It was therefore likely that the RDOM pool provided an entirely different group of compounds than is present in the estuarine water, constituting the basis for the clear shift in species abundance recorded. It was unlikely that similar compounds dominated in the two substrate pools, since a mere concentration effect would most likely have resulted in an increase in cell numbers of species already present in the control bottles. This was not observed.
According to a multivariate randomization test (22), the species composition was very close in duplicate bottles, especially when enriched with RDOM. Also, the same limited group of species dominated all of the investigated seasons (e.g., Fig. 3). This was remarkable given the potential seasonal variability in substrate diversity of the RDOM and the seasonally different diversity in the estuarine inoculum (26). Both factors could potentially contribute to create a seasonal variability in the group of RDOM consumers. This was, however, not observed and suggested a low diversity of culturable RDOM-catabolizing bacterioplankton in the estuary. Consequently, the isolated group of bacteria catabolizing RDOM appeared to be tolerant to the varying hydrographical and other environmental conditions during the year. They were also competitive relative to the other species exploiting RDOM as a substrate.
However, due to our approach to focus on culturable strains the applied strategy may have overlooked some bacterial species not growing sufficiently well on the solid media provided (i.e., “not-yet-cultured” bacteria). This most likely occurred in March, when the discrepancy between total species abundance and direct counts was greatest (cf. DNA devoid cells and cross-hybridization results below). Consequently, this also means that our estimate of species richness in the group of RDOM consumers was a minimum estimate of the true number.
Two observations indicated that the culturable species isolated constituted the majority of the bacteria in the investigated communities. First, the culturable dominating species accounted for a significant portion of the total bacterial numbers in two of the seasons investigated. However, cells lacking DNA may explain some of the discrepancy between total species abundance and observed direct count. On the other hand DNA-DNA cross-hybridization between different species could have contributed to some overestimation of the species abundance (see below). Second, the total species richness found in our study was no lower than that found in studies using culture-independent techniques and was similar to that found in other studies. The 48 different taxa observed by Pinhassi et al. (27) during their 7-month survey in the same study area was close to the 42 taxa of cultured bacteria found in our dilution cultures. The species richness observed in the northern Baltic was also comparable to that found in studies with the culture-independent molecular approach in various aquatic environments (7, 10, 28). However, only four strains similar to those found in the field study at the same site by Pinhassi et al. (27) were isolated in our study. This suggested that species richness based on the isolation of bacteria from in situ samples could underestimate the true number due to the fact that species with a low abundance in the samples with specific substrate requirements are disregarded in the isolation procedure. Enrichment with an alternate carbon source prior to cultivation provided a markedly different bacterial community despite the same analytical strategy and sampling site.
The strategy to isolate all morphotypes developing on the plates and also to examine an appreciable number of replicates within those morphotypes results in a higher probability of isolating species with a low abundance on the solid media. Of 23 colony morphotypes, 42 different taxa were found by using the present species definition of the 16S rDNA technique. The reason that species richness exceeded the number of observed morphotypes was a taxonomic difference within colony morphotype, also previously reported by others (see, for example, reference 21). As we limited our screening procedure to three replicates of each morphotype, the species richness of culturable bacteria in our dilution cultures might have been even greater. Consequently, studies not performing extensive screening for species within colony morphotypes may markedly underestimate the true bacterial diversity in the culturable population (10). That microbial cultivation techniques have not yet been exhaustively employed for determining the taxonomic diversity has previously been emphasized (34).
The power of the quantitative DNA-DNA hybridization or chromosomal painting method is its direct association with one of the present species definitions for bacteria (20, 27, 35) and the fact that the detection limit of this method is low (∼5,000 cells ml−1). In comparing sequence conservation within whole genomes versus 16S rDNA genes, it was found that the latter approach is clearly less powerful for phylogenetic resolution, especially at the species level (32). Regarding the confidence in the phylogenetic determinations, it is also important to note that, according to others (see, for example, reference 32) and our own DNA-DNA similarity data, 16S rDNA information alone can possibly underestimate the true species richness. At least two of our 16S rDNA sequences that were 99% similar had only a 4.4% genomic DNA similarity (Fig. 4). This meant that species appearing to be similar at the rDNA level could be different species according to DNA-DNA homology criteria.
A limitation of quantitative DNA-DNA hybridization is the risk of cross-hybridization within one genus. An abundance of strains can be overestimated if there are other strains of the same genus present in the sample because DNA-DNA similarity at the genus level can be as high as 20% (31). This finding is in good agreement with an analysis based on the relationship between 16S rDNA and DNA-DNA hybridization, wherein changes of 16S RNA similarity from 97 to 96% markedly decreased the genomic DNA similarity from ∼70 to 30% (Fig. 4). About 30% cross-hybridization should be a maximum estimate based on this analysis. However, Gonzales et al. (12) showed that DNA-DNA hybridization and quantitative 16S rDNA probes of natural communities give similar results. Since short oligomers are expected to have a higher stringency and not to suffer from the cross-hybridization bias, this suggests that cross-hybridization is not a severe problem in analyzing natural communities with whole DNA hybridization.
Pinhassi et al. (27; A. Hagström, unpublished results) also state that overestimation of single strain abundance due to cross-hybridization is not a severe problem and estimate 2.5% ± 9.8% cross-hybridization. However, the bacteria included in their comparison (n = 120) also included clearly distant species, which would be expected to have a low level of cross-hybridization. If dominance of species within a genus is common, this would underestimate the influence of cross-hybridization. According to the more closely related isolates selected for our comparison (16S rDNA similarity of 93 to 96%, n = 11), there was a fivefold-higher risk of overestimating closely related strains (on average, 12.2% ± 2.2%). Clearly, cross-hybridization can overestimate both the abundance of single species and the number of total culturable bacteria if several closely related species are abundant. However, the present precision is sufficiently good to provide new knowledge of bacterial population ecology in both cultures and the field, given that species abundance may vary by orders of magnitude (26).
A rough idea of the potential overestimation of the total abundance of species by DNA hybridization in our study can be provided by a simple hypothetical example that disregards the not-yet-cultured bacteria. We assume that the measured abundance is the true abundance in the experiments to obtain realistic ratios between species. This is plausible given the observed cross-hybridization values. By using the specific cross-hybridization values between species determined and the true abundance of all closely related species, the relative measurement error for a species can be calculated. The relative overestimation for each species probed in the samples was calculated by adding all cross-hybridization values. The overestimation of total abundance of all quantified species was determined by this calculation to be 0.7 to 9.7% as a function of season and treatment.
It is therefore plausible that the dominating species found in this study are RDOM-degrading bacteria and that they constitute a limited set of the total bacterial community. However, the percentage of not-yet-cultured species might be larger than suggested by our estimates, and thus the species richness of RDOM-catabolizing bacteria was probably underestimated. This also supports the application of culture-independent techniques to provide complementary information for a more complete identification of RDOM-degrading estuarine bacteria.
Acknowledgments
We are grateful for valuable advice from J. Pinhassi and K. Winka during the course of this project and for linguistic corrections from Mark Dopson. Technical assistance was provided by Heta Rompainen and Magnus Lind.
Financial support was provided by the Foundation for Strategic Environmental Research (MISTRA, Dnr. 97238) and the Swedish National Research Council (B-AA/BU 08583-319) to Johan Wikner.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 2.Bergey, D. 1984. Bergey’s manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore, Md.
- 3.Blackburn, N., A. Hagström, J. Wikner, R. Cuadros-Hansson, and P. K. Bjornsen. 1998. Rapid determination of bacterial abundance, biovolume, morphology, and growth by neural network-based image analysis. Appl. Environ. Microbiol. 64:3246–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böckelmann, U., W. Manz, T. R. Neu, and U. Szewzyk. 2000. Characterization of the microbial community of lotic organic aggregates (‘river snow’) in the Elbe River of Germany by cultivation and molecular methods. FEMS Microbiol. Ecol. 33:157–170. [DOI] [PubMed] [Google Scholar]
- 5.Carlsson, P., and E. Granéli. 1993. Availability of humic bound nitrogen for coastal plankton. Estuarine Coast Shelf Sci. 36:433–447. [Google Scholar]
- 6.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devereux, R., S. H. He, C. L. Doyle, S. Orkland, D. A. Stahl, J. Legall, and W. B. Whitman. 1990. Diversity and origin of Desulfovibrio species: phylogenetic definition of a family. J. Bacteriol. 172:3609–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobson, S. J., T. A. McMeekin, and P. D. Franzmann. 1993. Phylogenetic relationships between some members of the genera Deleya, Halomonas, and Halovibrio. Int. J. Syst. Bacteriol. 43:665–673. [DOI] [PubMed] [Google Scholar]
- 10.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Findlay, S., M. Pace, D. Lints, J. Cole, N. Caraco, and B. Peierls. 1991. Weak coupling of bacterial and algal production in a heterotrophic ecosystem: the Hudson river estuary. Limnol. Oceanogr. 36:268–278. [Google Scholar]
- 12.Gonzales, J., W. Whitman, R. Hodson, and M. Moran. 1996. Identifying numerically abundant culturable bacteria from complex communities: an example from lignin enrichment culture. Appl. Environ. Microbiol. 62:4433–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagström, A., J. Pinhassi, and U. L. Zweifel. 2000. Biogeographical diversity among marine bacterioplankton. Aquat. Microb. Ecol. 21:231–244. [Google Scholar]
- 14.Hessen, D. 1992. Dissolved organic carbon in a humic lake: effects on bacterial production and respiration. Hydrobiology 229:115–123. [Google Scholar]
- 15.Hobbie, I. E., R. J. Daley, and S. Jasper. 1977. Use of nucleopore filters for counting by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoch, M. P., and D. L. Kirchman. 1993. Seasonal and inter-annual variability in bacterial production and biomass in a temperate estuary. Mar. Ecol. Prog. Ser. 98:283–295. [Google Scholar]
- 17.Kenzaka, T., N. Yamaguchi, K. Tani, and M. Nasu. 1998. rRNA-targeted fluorescent in situ hybridization analysis of bacterial community structure in river water. Microbiology 144:2085–2093. [DOI] [PubMed] [Google Scholar]
- 18.Kuparinen, J., L. Leonardsson, J. Mattila, and J. Wikner. 1996. Food web structure and function in the Gulf of Bothnia, the Baltic Sea. Amb. Spec. Rep. 8:12–20. [Google Scholar]
- 19.Lane, D. 1991. 16S/23S rRNA sequencing, p.115–175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 20.Lanoil, B. D., and S. J. Giovannoni. 1997. Identification of bacterial cells by chromosomal painting. Appl. Environ. Microbiol. 63:1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebaron, P., J. F. Ghiglione, C. Fajon, N. Batailler, and P. Normand. 1998. Phenotypic and genetic diversity within a colony morphotype. FEMS Microbiol. Lett. 160:137–143. [DOI] [PubMed] [Google Scholar]
- 22.Manly, B. 1997. Multivariate data, p.260–283. In B. F. J. Manly (ed.), Randomization, bootstrap and Monte Carlo methods in biology. Chapman & Hall, London, England.
- 23.Moran, M., and R. Hodson. 1994. Support of bacterioplankton production by dissolved humic substances from three marine environments. Mar. Ecol. Prog. Ser. 110:241–247. [Google Scholar]
- 24.Norrman, B. 1993. Filtration of water samples for DOC studies. Mar. Chem. 41:239–242. [Google Scholar]
- 25.Peltzer, E., B. Fry, P. Doering, J. McMenna, B. Norrman, and U.-L. Zweifel. 1996. A comparison of methods for the measurement of dissolved organic carbon in natural waters. Mar. Chem. 44:85–96. [Google Scholar]
- 26.Pinhassi, J., and A. Hagström. 2000. Seasonal succession in marine bacterioplankton. Aquat. Microb. Ecol. 21:245–256. [Google Scholar]
- 27.Pinhassi, J., U. L. Zweifel, and A. Hagström. 1997. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl. Environ. Microbiol. 63:3359–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappe, M. S., P. F. Kemp, and S. J. Giovannoni. 1997. Phylogenetic diversity of marine coastal picoplankton 16S rRNA gene cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol. Oceanogr. 42:811–826. [Google Scholar]
- 29.Rolff, C., and R. Elmgren. 2000. Use of riverine organic matter in plankton food webs of the Baltic Sea. Mar. Ecol. Prog. Ser. 197:81–101. [Google Scholar]
- 30.Smith, S., and F. Mackenzie. 1987. The ocean as a net heterotrophic system: implications from the carbon biogeochemical cycle. Global Biogeochem. Cycles 1:187–198. [Google Scholar]
- 31.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849. [Google Scholar]
- 32.Stackebrandt, E., and R. Pukall. 1999. Deriving taxonomic decisions from 16S rDNA: a case study. Mar. Biol. 133:159–161. (Response to Althoff et al., Mar. Biol. 130:529–536, 1996.)
- 33.Stigebrandt, A. 1991. Computations of oxygen fluxes through the sea-surface and the net production of organic-matter with application to the Baltic and adjacent seas. Limnol. Oceanogr. 36:444–454. [Google Scholar]
- 34.Suzuki, M., M. Rappe, Z. Haimberger, H. Winfield, N. Adair, J. Ströbel, and S. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voordouw, G., Y. Shen, C. S. Harrington, A. J. Telang, T. R. Jack, and D. W. S. Westlake. 1993. Quantitative reverse sample genome probing of microbial communities and its application to oil-field production waters. Appl. Environ. Microbiol. 59:4101–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Truper. 1987. Report of the ad-hoc-committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463–464. [Google Scholar]
- 37.Wikner, J., R. Cuadros, and M. Jansson. 1999. Differences in consumption of allochthonous DOC between a lake and an estuary in a temperate watershed. Aquat. Microb. Ecol. 17:289–299. [Google Scholar]
- 38.Wikner, J., and Å. Hagström. 1999. Bacterioplankton intra-annual variability: Importance of hydrography and competition. Aquat. Microb. Ecol. 20:245–260. [Google Scholar]
- 39.Wilmotte, A., G. Vanderauwera, and R. Dewachter. 1993. Structure of the 16S ribosomal-RNA of the thermophilic cyanobacterium chlorogloeopsis Htf (Mastigocladus-Laminosus Htf) strain PCC7518, and phylogenetic analysis. FEBS Lett. 317:96–100. [DOI] [PubMed] [Google Scholar]
- 40.Zweifel, U. L., and A. Hagström. 1995. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts). Appl. Environ. Microbiol. 61:2180–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]