Abstract
A high-rate fluidized-bed bioreactor has been treating polychlorophenol-contaminated groundwater in southern Finland at 5 to 8°C for over 6 years. We examined the microbial diversity of the bioreactor using three 16S ribosomal DNA (rDNA)-based methods: denaturing gradient gel electrophoresis, length heterogeneity-PCR analysis, and restriction fragment length polymorphism analysis. The molecular study revealed that the process was dependent on a stable bacterial community with low species diversity. The dominant organism, Novosphingobium sp. strain MT1, was isolated and characterized. Novosphingobium sp. strain MT1 degraded the main contaminants of the groundwater, 2,4,6-trichlorophenol, 2,3,4,6-tetrachlorophenol, and pentachlorophenol, at 8°C. The strain carried a homolog of the pcpB gene, coding for the pentachlorophenol-4-monooxygenase in Sphingobium chlorophenolicum. Spontaneous deletion of the pcpB gene homolog resulted in the loss of degradation ability. Phenotypic dimorphism (planktonic and sessile phenotypes), low growth rate (0.14 to 0.15 h−1), and low-copy-number 16S rDNA genes (single copy) were characteristic of strain MT1 and other MT1-like organisms isolated from the bioreactor.
Pentachlorophenol (PCP) and various chlorophenol mixtures have been extensively used in wood treatment against blue staining and soft rot fungi all over the world. Several factors, such as temperature, pH, oxygen concentration, and the composition of the microbial community, influence the degradation of organic compounds. The bioremediation potential of polychlorophenols at low temperatures has been considered limited, since it has mainly been studied using mesophilic bacteria (for a review, see reference 34). Long-term experience with fluidized-bed processes at cold temperatures (33, 35) together with the description of the boreal chlorophenol-contaminated aquifer microbiology (24), however, has conclusively shown the potential for low-temperature biodegradation and bioremediation of polychlorophenols.
Chlorophenol contamination of soil and groundwater has occurred in Finland in several sawmill locations (16, 18). Large-scale contamination by polychlorophenols was discovered in 1987 at Kärkölä groundwater aquifer in southern Finland (18, 24). Early attempts to treat the contaminated water included a batch process with immobilized Mycobacterium chlorophenolicum PCP-1 culture (44), an activated-sludge process in a municipal treatment plant (9), and activated carbon filtration (12). A fluidized-bed polychlorophenol bioremediation process at ambient groundwater temperature was developed (13, 14, 22, 25, 26). The process was extended to full-scale treatment in 1995 and continues to operate (33, 34). Disruptions in the degradation efficiency in the Kärkölä fluidized-bed reactor (FBR) have occurred only as a consequence of failures in aeration. In 1999, the period for recovery of stable operation lasted several months, suggesting that the process depends on slow-growing bacteria. PCP degradation recovery times ranging from 10 to 40 days have also been shown in an aerobic chlorophenol treatment process, in which interruption of the oxygen supply for several hours resulted in accumulation of PCP but not 2,4,6-trichlorophenol (TCP) or 2,3,4,6-tetrachlorophenol (TeCP) (22).
Only a few studies have used modern molecular tools to address the impact of polychlorophenols on bacterial diversity. Beaulieu et al. (2) reported that adding PCP to soil slurry resulted in a selective enrichment of the genus Sphingomonas. The taxonomy of the genus Sphingomonas has recently been revised by Takeuchi et al. (43). Based on the results of phylogenetic and chemotaxonomic analyses, several species of the genus Sphingomonas have been transferred to three new genera, Sphingobium, Novosphingobium, and Sphingopyxis. In this study, members of this cluster and their close relatives are referred as sphingomonads. Sphingobium chlorophenolicum and Novosphingobium subarcticum are sphingomonads that are well known for the capability to degrade polychlorinated phenols (5, 15, 29, 30). The first chlorine elimination from the benzene ring in S. chlorophenolicum is oxygenolytic through the action of PCP-4-monooxygenase, encoded by the gene pcpB (20, 31).
In this research, molecular and cultivation studies were applied in parallel to analyze the structure of the polychlorophenol-degrading bacterial community and to isolate an ecologically relevant psychrotolerant organism from the Kärkölä fluidized-bed process. The diversity and stability of the polychlorophenol-degrading bacterial community was investigated using universal PCR amplification of eubacterial 16S rRNA genes. Subsequent analysis of the PCR products was performed by denaturing gradient gel electrophoresis (DGGE [27]), length heterogeneity analysis of amplified PCR products (LH-PCR [3, 37, 42]), restriction fragment length polymorphism (RFLP) analysis, and sequencing. The study was further focused on characterization of Novosphingobium sp. strain MT1, isolated from the bioreactor.
MATERIALS AND METHODS
Description of bioreactor.
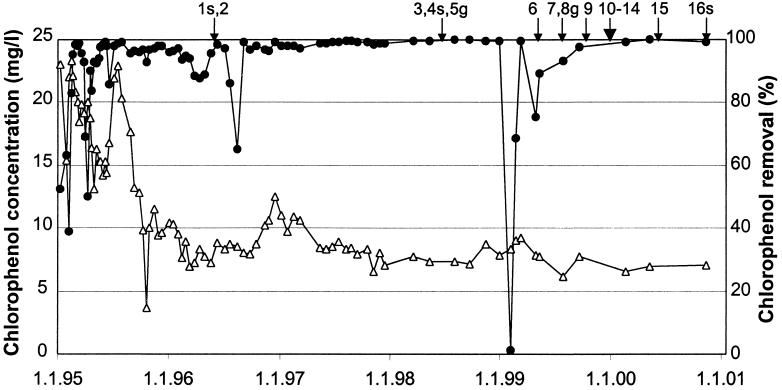
The operation of the full-scale fluidized-bed process (34) began in January 1995. At start-up, the FBR was inoculated with unacclimated sludge from a municipal wastewater plant. The reactor has a volume of 5 m3, coarse sand as the biomass carrier, and a feed rate of 45 to 50 m3/day. The groundwater has a low concentration of dissolved oxygen (typically <1 mg/liter) and a high concentration of ferrous iron (1 to 10 mg/liter). The chlorophenols of the contaminant are TeCP (2,3,4,6-TeCP; 70 to 75%), TCP (2,4,6-TCP; 15 to 20%), PCP (5%), and minor amounts of other chlorophenols (2,4,5-TCP, 3,4-dichlorophenol, and 2,4-dichlorophenol). Contaminated groundwater is aerated and supplemented with (NH4)2HPO4 (Prayon Rupel, Engis, Belgium). The efficiency of polychlorophenol degradation has been over 99% during stable operation (Fig. 1). Failures in oxygen supply occurred in July 1996 and January 1999. During January 1999, the breakdown of the air compressor resulted in temporary loss of chlorophenol degradation. Recovery from the upsets was facilitated by significantly decreasing the groundwater feed rate, followed by gradual increase to the normal rate.
FIG. 1.
Bioreactor function and time points of sample collection (arrows). Feed (▵) is the total concentration of 2,4,6-TCP, 2,3,4,6-TeCP, and PCP in the influent water. Chlorophenol removal (•) is shown as the removal percentage. Bioreactor sand samples (s) and influent groundwater samples (g) are marked. Dates are given below the graph as day.month.year. (Data reproduced with permission from Häme Regional Environment Centre, Lahti, Finland.)
Sample collection.
Water and carrier sand samples (samples 1 to 16) were collected from 1996 to 2001 (Fig. 1). Samples were collected weekly over January and February 2000 (samples 10 to 14). Water samples from the bioreactor (50 ml) and the influent groundwater (500 ml) were filtered through a Filtropur S 0.2-μm-pore-size acetylacetate filter unit (Sarstedt, Nümbrecht, Germany). Sample 2, however, was collected through centrifugation of 500 ml of water, and sample 9 was filtered through a PS 0.2-μm-pore-size glass fiber filter unit (Whatman International Ltd., Maidstone, England). The samples were stored at −20°C until DNA extraction.
Preparation of DNA.
Pieces of the membrane from the filter unit or approximately 0.1 g of the carrier sand was placed in a tube that was filled with 0.4 ml of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The samples were then incubated with lysozyme (5 mg/ml) at 37°C for 1 h following the addition of sodium dodecyl sulfate and proteinase K to final concentrations of 1% and 0.2 mg/ml, respectively. A phenol-chloroform-isoamyl alcohol mixture (25:24:1; 0.4 ml) was added to dissolve the filter and to extract proteins from nucleic acids. Cell disruption was ensured by bead milling the cells in a cell homogenizer (Mikro-U Dismembrator; B. Braun Biotech International, Melsungen, Germany) with 0.6 g of glass beads (diameter, 0.1 mm) and by homogenizing them at 1,600 rpm for 3 min. The samples were centrifuged at 13,000 × g for 10 min, and the upper phase was reextracted one to three times with chloroform-isoamyl alcohol (24:1). The DNA was finally precipitated with ethanol and dissolved in 100 μl of molecular biology grade water (5 Prime-3 Prime, Boulder, Colo.). The preparation of DNA from bacterial isolates and S. chlorophenolicum ATCC 39723 (a kind gift from L. Nohynek, University of Helsinki, Finland) was performed without bead milling.
LH-PCR analysis.
A set of universal primers, eubacterial forward primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′; IRD700 labeled) (19) and bacterial reverse primer PRUN518 (5′-ATTACCGCGGCTGCTGG-3′) (27), was used to amplify a fragment of 16S ribosomal DNA (rDNA) approximately 500 bp in length, covering variable regions V1 to V3. To study the effect of the degenerate nucleotides of primer 27F, parallel PCRs were performed using primer fD1 (45). In this primer, the degenerate nucleotide M (C or A) was replaced with C. In the PCRs, 1 μl of purified DNA was used as a template in a 50-μl PCR mixture containing 0.2 mM deoxynucleoside triphosphates dNTPs, 0.3 μM (each) primer, 1× DynaZyme reaction buffer, and 2 U of DynaZyme F501-KL polymerase (Finnzymes, Espoo, Finland). The PCR procedure included an initial denaturation step at 95°C for 5 min and 30 cycles of amplification (94°C for 30s, 55°C for 1 min, and 72°C for 3 min). The LH-PCR patterns were analyzed by gel electrophoresis with an automated model 4200 sequencer (LI-COR BioTech, Lincoln, Nebr.) using 6% Long Range polyacrylamide gel (FMC Bioproducts, Rockland, Maine). Band detection and calculation were performed using the program QuantityOne (Bio-Rad Laboratories, Hercules, Calif.). The LH-PCR sizes were compared against the simulated LH-PCR lengths of 1,200 bacterial sequences of 10 major phylogenetic bacterial groups (M. Tiirola, J. Suvilampi, M. Kulomaa, and J. Rintala, submitted for publication) obtained from the EMBL database.
DGGE analysis.
In the DGGE analysis, the DNA was amplified as described above, except that the forward primer, 27F, included a 40-bp GC clamp (5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG-3′) at the 5′ end. Polyacrylamide gel electrophoresis was done on 6% (wt/vol) gel containing a gradient of 30 to 60% denaturant according to the protocol of Muyzer et. al (27). DGGE bands were excised from the gel and eluted overnight in 200 μl of TE buffer at 37°C. The supernatant was used as a template for reamplification and sequencing.
16S RFLP analysis.
The PCR product was generated using primer 27F and either primer rD1 (5′-AAGGAGGTGATCCAGCC-3′) (45) or primer Com2-Ph (5′-CCGTCAATTCCTTTGAGTTT-3′) (39). The primer pair 27F-rD1 yielded fragments approximately 1,500 bp in size and was used for selecting bacterial isolates in the primary analysis. Later comparisons were done using the primer pair 27F–Com2-Ph, which amplified a shorter (900-bp) fragment of the ribosomal gene and facilitated production of stronger PCR products from the bioreactor samples. The PCR products were digested with the restriction enzymes MspI and HaeIII/HinfI (double digestion) (Promega, Madison, Wis.). The digested PCR products were analyzed by electrophoresis in 12% native polyacrylamide gel followed by ethidium bromide staining and were documented with the GelDoc2000 digital imaging system (Bio-Rad Laboratories).
Bacterial isolation and cultivation.
Bacterial samples were plated on PYG agar (41) and DSM726 agar (the composition is available at http://www.dsmz.de/media/) immediately after sample collection and incubated at room temperature (22 ± 1°C). Fifty randomly selected colonies were transferred to PYG medium. After incubation at room temperature for 3 to 21 days, the bacterial DNAs were extracted and analyzed by LH-PCR and 16S RFLP. Selected isolates were characterized by sequencing the 5′ region (about one-third of the whole length) of the 16S rRNA gene. Later isolations of MT1-like organisms and cultivation of the isolates were performed using primarily R2A medium (36). For measurement of the growth rate, bacterial culture in the late exponential growth phase was diluted 1:10 in R2A medium and cultured in a rotary shaker (125 rpm) at 22°C. The optical density at 595 nm of the triplicate samples was measured periodically.
Degradation of polychlorophenols.
Degradation of 2,4,6-TCP, 2,3,4,6-TeCP, and PCP at 8 ± 1°C was studied in 120-ml glass serum bottles in a gyratory shaker as described previously (24). Cells were cultured in PYG medium, centrifuged, and suspended (approximately 107/ml) in autoclaved water and sand from the bioreactor. Controls with bacteria killed with formaldehyde (2%) were used in the experiments. Duplicate samples were spiked four times with a chlorophenol mixture containing 2,4,6-TCP, 2,3,4,6-TeCP, and PCP at approximately the same ratio as in the influent groundwater of the Kärkölä FBR. Chlorophenols were derivatized with acetic anhydride, extracted with hexane, and analyzed by gas chromatography according to the method of Männistö et al. (24).
Southern blot analyses.
Genomic DNA was digested with restriction enzymes (ClaI, EcoRI, EcoRV, HindIII, PstI, and SalI) and analyzed by electrophoresis on a 1% agarose gel. The DNA was transferred onto a nylon membrane (Hybond N+; Amersham Pharmacia Biotech, Little Chalfont, England) by an alkaline blotting procedure (6). Preparation of the probes, hybridization, and chemiluminescent CDP-Star detection of the signal were performed with the DIG DNA labeling and detection kit (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer’s instructions. Probe pcpB-5 specific for the PCP-4-monooxygenase gene was generated with the primers that amplified the region corresponding to nucleotides 444 to 616 from the beginning of the coding sequence of the pcpB gene (accession no. M98557). R. Crawford (University of Idaho, Moscow, Idaho) kindly provided the template, plasmid pCCL3. To determine the 16S rDNA copy number, a mixture of the DNAs of S. chlorophenolicum ATCC 39723 and strain MT1 was amplified using the universal primers that amplified the 16S rDNA region 338 to 534 (according to Escherichia coli numbering [4]). Overnight hybridization was performed at 61.5°C for pcpB-5 and 63.5°C for the 16S-338 probe.
Cloning and sequencing.
Reamplified DGGE fragments were analyzed by dye terminator cycle sequencing with model 377 sequencer (Applied Biosystems, Foster City, Calif.) using 27F and PRUN518 as sequencing primers. For sequencing of the pcpB gene homolog, the degenerate primers pcpB-G (5′-GGSTTCACSTTCAAYTTCGA-3′) and pcpB-D2 (5′-TCCTGCATSCCSACRTTCAT-3′) (2) were used. The PCR product was cloned into the pGEM-T vector (Promega), and the clones were analyzed by bidirectional cycle sequencing with the IRD-labeled universal primers T7 and SP6 with a SequiTherm Excel II kit (Epicentre Technologies, Madison, Wis.), using the LI-COR 4200 sequencer. Sequences were compiled with the program DNAman (Lynnon BioSoft, Vaudreuil, Quebec, Canada) and compared to sequences in gene banks by using the program BLAST (1).
Nucleotide sequence accession numbers.
The sequences were deposited in the EMBL database under accession number AJ319678 (MT1 pcpB gene homolog, partial sequence) and the numbers shown in Table 1. The nearly complete (>95%) sequence of the 16S rDNA gene from strain MT1 was previously deposited under accession number AJ303009 (23).
TABLE 1.
Environmental 16S rDNA sequences from the Kärkölä FBR and groundwater
| Sample origin | Sequence | Accession no. | LH-PCR sizea (bp) | Phylogenetic affiliation | Closest relative | % Identityb |
|---|---|---|---|---|---|---|
| FBR | DGGE band A | AJ132327 | 470 | α-Proteobacteria | Sphingomonas suberifaciens IFO15211T | 97.1 |
| FBR | DGGE band B | AJ132328 | 500 | Gram-positive; high G + C | Nocardioides sp. strain NCFB3005 | 96.3 |
| FBR | DGGE band C | AJ132329 | 504 | Gram-positive; high G + C | Corynebacterium-like QSSCO-20 | 97.2 |
| GWc | DGGE band D | AJ132330 | 522 | β-Proteobacteria | Janthinobacterium lividum DSM1522T | 98.4 |
| FBR | Clone E | AJ319679 | ND | β-Proteobacteria | Rhodoferax fermentas ATCC49787T | 95.8 |
Size measured in LH-PCR analysis with primers 27F and PRUN518. ND, not determined.
Identity percentage is based on alignments of about 430 bp.
GW, groundwater.
RESULTS AND DISCUSSION
Microbial diversity in the fluidized-bed bioreactor.
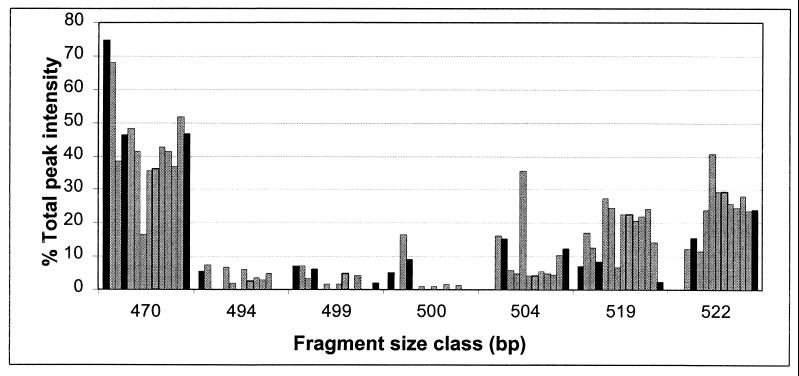
The LH-PCR patterns of the eubacterial 16S rRNA genes amplified from bioreactor samples consisted of few (four to nine) peaks, suggesting low phylogenetic diversity in the bacterial community. The most prominent peak was at a length of 470 bp (Fig. 2). Simulation of the EMBL database (Tiirola et al., submitted) showed that this size would indicate cyanobacteria or α-proteobacteria; however, cyanobacteria can be excluded because of the dark reactor conditions. Thus, the LH-PCR analysis suggested strong enrichment of α-proteobacteria in the bioreactor. Other notable size classes accounting for >6% of the total peak area in at least one of the samples are shown in Fig. 2. The choice between a degenerate (27F) and a nondegenerate (fD1) forward primer had a slight influence on the LH-PCR patterns: the dominance of the 470-bp band was equally obvious with both primers, but the nondegenerate primer, fD1, did not yield a band of 522 bp but bands of 521 and 523 bp instead. This was surprising, since the degenerate primer was expected to amplify more diverse rDNA sequences and thus yield more bands. LH-PCR amplification of the groundwater samples (5g and 8g) showed that the vast majority (61 and 77%) of the amplified PCR products were 522 bp, whereas only a minor proportion (6 and <1%) of the total fluorescence was observed at 470 bp, showing a striking difference between the bacterial communities in the bioreactor and groundwater.
FIG. 2.
Proportions of the main fragment size classes in LH-PCR analysis of the bioreactor samples with primer pair 27F-PRUN518. The bars represent relative band densities in each size class in samples 1s (solid), 2, 3, 4s (solid), 6, 7, 9, 10, 11, 12, 13, 14, 15, and 16s (solid) from left to right.
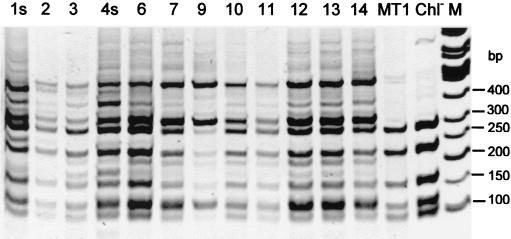
The 16S RFLP band patterns showed fairly constant bacterial profiles in all of the samples over the study period (MspI patterns are shown in Fig. 3), even at the times when the bioreactor was not working optimally (samples 6 and 7). An exception was sample 9, which was collected from the reactor water with different filter material. The different diversity profile of sample 9 may have been caused by a methodological bias. The filter material did not dissolve in the organic solvents used, and therefore, the bead-milling step in the DNA extraction protocol was not efficient. The 27F–Com2-Ph PCR product of sample 9 was cloned, and the 16S rDNA clone that represented the dominant 16S RFLP banding patterns of the sample with different enzymes was sequenced (clone E). The closest match for the sequence was Rhodoferax sp. (Table 1). The effects of the choice between degenerate primer 27F and nondegenerate primer fD1 were also compared with some of the bioreactor samples, but there was no noticeable effect on the RFLP patterns (data not shown).
FIG. 3.
16S RFLP patterns of MspI digestions from bioreactor samples 1s to 14 compared to the patterns obtained from strain MT1 and the MT1 Chl− mutant. The molecular size marker bands (lane M) are indicated on the right.
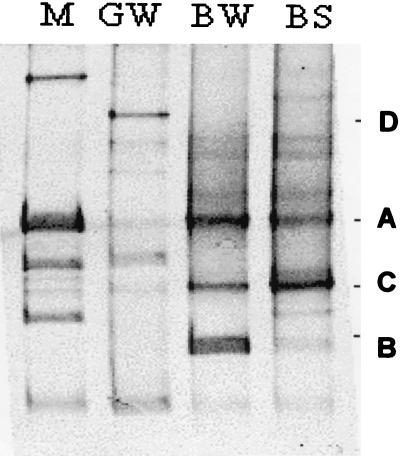
A typical DGGE pattern of the 16S rDNA genes from the bioreactor samples is shown in Fig. 4. The main bands A, B, and C from the bioreactor water sample could be sequenced directly after reamplification. Sequencing of the bands gave clear nucleotide sequences, showing that these bands were largely homogeneous, i.e., each band represented mainly one type of PCR product. Reamplification confirmed that the size of band A was 470 bp (Table 1). This size matched that of the major band in LH-PCR, as well as the predicted LH-PCR size of many of the sphingomonads in the EMBL sequence database. The sequence of the fragment was closest to that of Rhizomonas suberifaciens 16S rDNA. The other sequences from the bioreactor were gram-positive bacteria with high G + C contents. Only one sequence was obtained from the Kärkölä groundwater (band D), and the LH-PCR size of this band matched that of the major band in the LH-PCR analysis. When this Janthinobacterium-like sequence was compared to the 16S rDNA sequences of previously isolated bacteria from the same groundwater, the closest sequence identity was only 94% (strain K1 [24]; accession number AJ001384). Without further experiments, however, it is difficult to evaluate the relevance of Janthinobacterium sp. in the chlorophenol-contaminated groundwater.
FIG. 4.
DGGE separation patterns of amplified DNA fragments coding for eubacterial 16S rDNA. Lane M, reference fragments (the G + C contents of these fragments from top to bottom were 51.7, 53.7, 54.2, 55.9, and 57.7%); lane GW, influent groundwater sample 5 g; lane BW, bioreactor water sample 3; lane BS, carrier sand sample 4s. Band D from lane GW and bands A, B, and C from lane BW were reamplified and sequenced (Table 1).
Isolation of Novosphingobium sp. strain MT1.
Incubation of bioreactor water (sample 7) at room temperature yielded 2 × 106 CFU/ml on DSM726 agar and 3 × 106 CFU/ml on PYG agar (sample 7). Three of the 50 randomly selected colonies possessed patterns that matched the dominant LH-PCR and 16S RFLP (MspI and HaeIII/HinfI) patterns obtained from bioreactor samples. One of these isolates was further subcultivated and designated strain MT1. The partial 16S rDNA sequence of MT1 was identical with the sequence of the DGGE band A. However, the nearly full-length 16S rDNA sequence (AJ303009) clustered with Novosphingobium sequences, with 96.6% sequence identity with Novosphingobium aromaticivorum and Novosphingobium subterraneae type species (AB0251012 and AB025014). The 16S RFLP pattern of MT1 together with the pattern of clone E accounted for the most intense peaks of the 16S RFLP patterns of the bioreactor samples. A year later, three additional MT1-like organisms (MT101, MT103, and MT104) were isolated (sample 15), showing identical morphological characteristics and 16S RFLP patterns and the partial 16S rDNA sequence. These strains belong to sphingomonads that are, interestingly, well known for the capability to degrade various xenobiotic substances and their potential to produce exopolysaccharides (46, 47).
Two morphotypes of Novosphingobium sp. strain MT1 and MT1-like strains.
After sequential transfers in liquid PYG and DSM726 medium and agar, isolate MT1 was found to carry two phenotypically different forms with identical 16S rDNA sequences. Phenotypic dimorphism (sessile and planktonic forms) was also seen during subcultivation of isolates MT101, MT103, and MT104 on R2A agar. The sessile phenotype was characterized by dry, elevated colonies on agar that were difficult to culture in DSM726 medium and a tendency to grow in large rafts of thousands of cells or in rosette-like formations in PYG and R2A media. The planktonic phenotype was characterized by flat, semitranslucent colonies on agar and the capability to grow in DSM726 medium without cell aggregation. Cell aggregation is a property that requires synthesis of exopolysaccharides. Polysaccharide production may thus be essential for the bacteria in the high-rate FBR to aid attachment to the surface of the biomass carrier. Therefore, the planktonic phenotype may not reflect the natural state of the bacterium. Both phenotypes maintained their characteristic properties after several subculturing steps on R2A agar. However, after short-term storage in liquid nitrogen, the sessile form generated two colony types on agar, demonstrating that the phenotype was unstable. The tendency to form two different morphotypes has also been documented for other sphingomonads (32). The colony’s appearance on agar and growth style in the liquid medium exactly matched those described for Sphingomonas sp. strain S88 and its sphingan-polysaccharide-negative (Sps−) mutants (32, 47). The gene cluster (sps cluster) that appears to regulate the synthesis of sphingan polysaccharides has been identified in strain S88, although the signal that regulates the synthesis is not yet known (32). The ability of sphingomonads to produce exopolysaccharides has been suggested to improve sludge floc formation and proper settling in activated sludge samples (28).
Chlorophenol degradation by Novosphingobium sp. strain MT1.
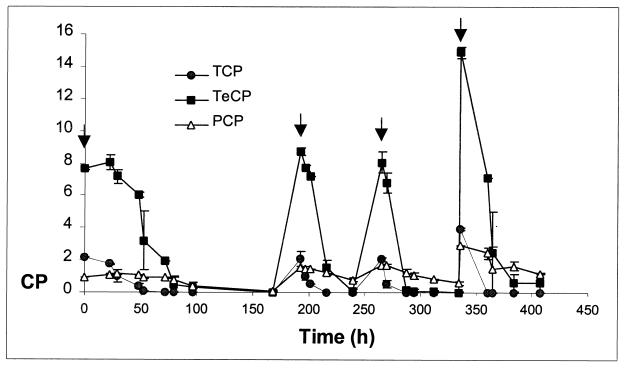
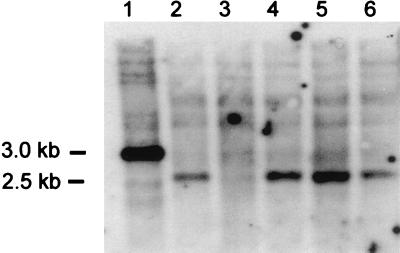
Chlorophenol degradation was tested in shake flasks at 8°C by spiking them with a synthetic chlorophenol mixture (Fig. 5). All three polychlorophenols were degraded by MT1. During characterization of the different morphotypes of strain MT1, a mutant that did not degrade chlorophenols was isolated, and it was designated MT1 Chl−. While S. chlorophenolicum carried the pcpB gene in a 3.0-kb EcoRI locus, strain MT1 and the three MT1-like strains carried pcpB gene homologs in a 2.5-kb locus in the DNA hybridization analysis (Fig. 6). However, MT1 Chl− lacked the gene. The results were confirmed by PCR amplification with the pcpB-specific primers pcpB-G and pcpB-D2, which yielded PCR products of the right size from S. chlorophenolicum, strain MT1, and the three MT1-like strains but did not yield any product from MT1 Chl−. The cloned fragment (661 bp in length) of the MT1 pcpB gene homolog had 99 to 100% identity with the environmental clones of pcpB homologs (AF270967, AF27974, and AF27966) obtained from PCP-enriched soil in Canada (2). The identity with the pcpB gene of S. chlorophenolicum ATCC 39723 (M98557) was 75%, and the homology between the translated amino acid sequences was 79%. The observation that spontaneous deletion of the pcpB gene homolog produced the nondegrading mutant (MT1 Chl−) suggests that Novosphingobium sp. strain MT1 initiates the degradation of PCP and other polychlorophenols by PCP-4-monooxygenase-like protein. The pcpB gene has not previously been reported to exist in sphingomonads other than different S. chlorophenolicum strains (7, 21), although the sequence has been unexpectedly found in the nondegrading isolates B2 and B4 identified as Pseudomonas and Ralstonia by 16S rDNA sequencing (38).
FIG. 5.
Degradation of the mixture of 2,4,6-TCP, 2,3,4,6-TeCP, and PCP by Novosphingobium sp. strain MT1 at 8°C. Chlorophenols (CP) were spiked four times (indicated with arrows). The error bars indicate standard deviations.
FIG. 6.
Southern blot analysis of genomic DNAs hybridized with a pcpB gene-specific probe. Each lane was loaded with 1 μg of DNA digested with EcoRI: lane 1, S. chlorophenolicum ATCC 39723; lane 2, strain MT1; lane 3, Chl− mutant of strain MT1; lane 4, strain MT101; lane 5, strain MT103; lane 6, strain MT104.
Growth characteristics isolated strains.
The multiplication times of Novosphingobium sp. strain MT1 and the MT1-like strains MT101 and MT104 in exponential growth phase were 7.4, 6.9, and 6.7 h, respectively. The growth rates correspond to 0.14 to 0.15 doublings per h and are about three times longer than the hydraulic retention time of the Kärkölä high-flow-rate FBR. Furthermore, bacterial growth rates in cold processes with chlorophenols as the sole carbon source are even lower. A net biomass yield of 0.03 mg of volatile suspended solids per mg of chlorophenols removed and a biomass decay rate of 0.06 to 0.08 day−1 have been reported for another fluidized-bed system (22). This indicates a very long mean cell residence time, thus explaining the maintainability of biodegradation processes with dilute chlorophenol solutions as the substrate. The 16S rDNA-specific hybridization analysis of the genomic DNA digested with six restriction enzymes showed that MT1 and MT1-like organisms had only one ribosomal operon, which is consistent with the relatively low growth rate of the organisms. The number of rRNA genes reflects the ecological strategy of bacteria, since it correlates with the rate at which bacteria can respond to varying resources (17). Slow-growing bacteria typically have one or two rRNA operons (10, 17), probably because rRNA overproduction is a metabolic expense under oligotrophic conditions in the long term.
Chlorophenols serve as the primary carbon and energy source in the Kärkölä FBR, since the humic substances in the Kärkölä groundwater (2.5 to 12.1 mg/liter, measured as dissolved organic carbon excluding chlorophenols) are mostly recalcitrant (40). The chlorophenol concentration in the high recycle flow bioreactor remains close to that of the effluent water (0.002 to 0.1 mg/liter) (33), and therefore bacteria have to possess high affinity for the substrate. According to our recent study (23), the half saturation constant, Ks, for 2,3,4,6-TeCP of Novosphingobium sp. strain MT1 is 2.1 mg/liter at +8°C, which is 10 times more efficient than that of S. chlorophenolicum strains (34). In the same study, microaerophilic growth and enhanced chlorophenol degradation at +8°C compared to that at room temperature were detected for strain MT1. Microaerophily may reflect the probable history of the strain in the low-oxygen-concentration groundwater in Kärkölä.
Conclusion.
Molecular analysis of the 16S rDNA gene sequences of the bacterial community suggest that the long-term operation of the Kärkölä bioreactor has resulted in a stable bacterial community with a low species diversity. No considerable shift in LH-PCR or RFLP patterns was observed even after the process was interrupted and degradation efficiency was fluctuating in 1999. Strain MT1 was among the dominant organisms in the bioreactor but not in the influent groundwater, as judged by the molecular analyses. The strain clustered with Novosphingobium species and had a tendency to form two different morphotypes. The growth characteristics (low growth rate, optimized number of rRNA operons, and ability to aggregate) observed for strain MT1 and MT1-like isolates may be essential for the bacteria that maintain the bioremediation process in the oligotrophic fluidized-bed process but may also explain the relatively long periods for recovery of stable operation after process disturbances. The ability to survive in nutrient-poor marine (8) and subsurface (11) environments has also been detected for some other sphingomonads and suggests that this group may be ubiquitous in oligotrophic environments as well. The pcpB gene homolog was essential for chlorophenol degradation. Novosphingobium sp. strain MT1 with high-rate polychlorophenol degradation at 8°C is the first isolated psychrotrophic bacterium that has been shown with molecular tools to have ecological relevance in groundwater bioremediation processes.
Acknowledgments
This study was supported by the Graduate School for Environmental Ecology, Ecotoxicology and Engineering (University of Jyväskylä), the Academy of Finland, and the Jenny and Antti Wihuri Foundation.
We thank Irene Helkala for technical assistance, Mervi Ahlroth for reviewing the manuscript, and Olli Valo and Juhani Alitalo (Häme Regional Environment Centre) for the bioreactor performance data and assistance in sample collection. We also thank Nordic Envicon Oy for the chance to study the microbiology of the bioreactor.
REFERENCES
- 1.Altschul, S. F., T. J. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaulieu, M., V. Becaert, L. Deschenes, and R. Villemur. 2000. Evolution of bacterial diversity during enrichment of PCP-degrading activated soils. Microb. Ecol. 40:345–355. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosius, J., M. L. Palmer, J. P. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of the ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassidy, M. B., H. Lee, J. T. Trevors, and R. B. Zablotowicz. 1999. Chlorophenol and nitrophenol metabolism by Sphingomonas sp. UG30. J. Ind. Microbiol. Biotechnol. 23:232–241. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski, P. 1992. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 201:134–139. [DOI] [PubMed] [Google Scholar]
- 7.Ederer, M. M., R. L. Crawford, R. P. Herwig, and C. S. Orser. 1997. PCP degradation is mediated by closely related strains of the genus Sphingomonas. Mol. Ecol. 6:39–49. [DOI] [PubMed] [Google Scholar]
- 8.Eguchi, M., T. Nishikawa, K. MacDonald, R. Cavicchioli, J. C. Gottschal, and S. Kjelleberg. 1996. Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 62:1287– 1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ettala, M., J. Koskela, and A. Kiesilä. 1992. Removal of chlorophenols in a municipal sewage treatment plant using activated sludge. Water Res. 26:797–804. [Google Scholar]
- 10.Fegatella, F., J. Lim, S. Kjelleberg, and R. Cavicchioli. 1998. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64:4433–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredrickson, J. K., D. L. Balkwill, M. F. Romine, and T. Shi. 1999. Ecology, physiology, and phylogeny of deep subsurface Sphingomonas sp. J. Ind. Microbiol. Biotechnol. 23:273–283. [DOI] [PubMed] [Google Scholar]
- 12.Järvinen, K. 1996. Active carbon filtration in bioremediation of Kärkölä contaminated groundwater. Uusimaa Environmental Research Centre publication no. 6. Uusimaa Environmental Research Centre, Helsinki, Finland. (In Finnish.)
- 13.Järvinen, K. T., and J. A. Puhakka. 1994. Bioremediation of chlorophenol contaminated ground water. Environ. Technol. 15:823–832. [DOI] [PubMed] [Google Scholar]
- 14.Järvinen, K. T., E. S. Melin, and J. A. Puhakka. 1994. High-rate bioremediation of contaminated groundwater at low temperatures. Environ. Sci. Technol. 28:2387–2392. [DOI] [PubMed] [Google Scholar]
- 15.Karlson, U., F. Rohjo, J. D. van Elsas, and E. Moore. 1995. Genetic and serological evidence for the recognition of four pentachlorophenol-degrading bacterial strains as a species of the genus Sphingomonas. Syst. Appl. Microbiol. 18:539–548. [Google Scholar]
- 16.Kitunen, V. H., R. Valo, and M. Salkinoja-Salonen. 1987. Contamination of soil around wood-preserving facilities by polychlorinated aromatic compounds. Environ. Sci. Technol. 21:96–101. [Google Scholar]
- 17.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lampi, P., T. Vartiainen, and J. Tuomisto. 1990. Population exposure to chlorophenols, dibenzo-p-dioxins and dibenzofurans after a prolonged ground water pollution by chlorophenols. Chemosphere 20:625–634. [Google Scholar]
- 19.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115–175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley & Sons, Chichester, United Kingdom.
- 20.Lange, C. C., B. J. Schneider, and C. S. Orser. 1996. Verification of the role of PCP 4-monooxygenase in chlorine elimination from pentachlorophenol by Flavobacterium sp. strain ATCC 39723. Biochem. Biophys. Res. Commun. 219:146–149. [DOI] [PubMed] [Google Scholar]
- 21.Leung, K. T., S. Campbell, Y. Gan, D. C. White, H. Lee, and J. T. Trevors. 1999. The role of the Sphingomonas species UG30 pentachlorophenol-4-monooxygenase in p-nitrophenol degradation. FEMS Microbiol. Lett. 173:247–253. [DOI] [PubMed] [Google Scholar]
- 22.Mäkinen, P. M., T. J. Theno, J. F. Ferguson, J. E. Ongarth, and J. A. Puhakka. 1993. Chlorophenol toxicity removal and monitoring in aerobic treatment: recovery from process upsets. Environ. Sci. Technol. 27:1434–1439. [Google Scholar]
- 23.Männistö, M. K., M. A. Tiirola, and J. A. Puhakka. Degradation of 2,3,4,6-tetrachlorophenol at low temperature and low dioxygen concentrations by phylogenetically different groundwater and bioreactor bacteria. Biodegradation, in press. [DOI] [PubMed]
- 24.Männistö, M. K., M. A. Tiirola, M. Salkinoja-Salonen, M. S. Kulomaa, and J. A. Puhakka. 1999. Diversity of chlorophenol degrading bacteria isolated from contaminated boreal groundwater. Arch. Microbiol. 171:189–197. [DOI] [PubMed] [Google Scholar]
- 25.Melin, E. S., K. T. Järvinen, and J. A. Puhakka. 1998. Effects of temperature on chlorophenol biodegradation kinetics in fluidized-bed reactors with different biomass carriers. Water Res. 32:81–90. [Google Scholar]
- 26.Melin, E. S., J. A. Puhakka, and J. F. Ferguson. 1998. Enrichment and operation strategies for polychlorophenol degrading microbial cultures in an aerobic fluidized-bed reactor. Water Environ. Res. 70:171–180. [Google Scholar]
- 27.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 62:2156–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neef, A., R. Witzenberger, and P. Kämpfer. 1999. Detection of sphingomonads and in situ identification in activated sludge using 16S rRNA-targeted oligonucleotide probes. J. Ind. Microbiol. Biotechnol. 23:261–267. [DOI] [PubMed] [Google Scholar]
- 29.Nohynek, L. J., E. L. Nurmiaho-Lassila, E. L. Suhonen, H. J. Busse, M. Mohammadi, J. Hantula, F. Rainey, and M. S. Salkinoja-Salonen. 1996. Description of chlorophenol-degrading Pseudomonas sp. strains KF1T, KF3, and NKF1 as a new species of the genus Sphingomonas, Sphingomonas subarctica sp. nov. Int. J. Syst. Bacteriol. 46:1042–1055. [DOI] [PubMed] [Google Scholar]
- 30.Nohynek, L. J., E. L. Suhonen, E. L. Nurmiaho-Lassila, J. Hantula, and M. Salkinoja-Salonen. 1995. Description of four pentachlorophenol-degrading bacterial strains as Sphingomonas chlorophenolica sp. nov. Syst. Appl. Microbiol. 18:527–538. [Google Scholar]
- 31.Orser, C. S., C. C. Lange, L. Xun, T. C. Zahrt, and B. J. Schneider. 1993. Cloning, sequence analysis, and expression of the Flavobacterium pentachlorophenol-4-monooxygenase gene in Escherichia coli. J. Bacteriol. 175:411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollock, T. J., and R. W. Armentrout. 1999. Planktonic/sessile dimorphism of polysaccharide-encapsulated sphingomonads. J. Ind. Microbiol. Biotechnol. 23:436–441. [DOI] [PubMed] [Google Scholar]
- 33.Puhakka, J. A., K. T. Järvinen, J. H. Langwaldt, E. S. Melin, M. K. Männistö, J. M. Salminen, and M. T. Sjölund. 2000. On-site and in situ bioremediation of wood-preservative contaminated groundwater. Water Sci. Technol. 31:371–376. [Google Scholar]
- 34.Puhakka, J. A., and E. S. Melin. 1998. Chlorophenol-contaminated groundwater bioremediation at low temperatures, p.1111–1120. In R. A. Meyer (ed.), Encyclopedia of environmental analysis and remediation. John Wiley and Sons, Inc., New York, N.Y.
- 35.Puhakka, J. A., E. S. Melin, K. T. Järvinen, P. M. Koro, J. A. Rintala, P. Hartikainen, W. K. Shieh, and J. F. Ferguson. 1995. Fluidized-bed biofilms for chlorophenol mineralization. Water Sci. Technol. 42:227–235. [Google Scholar]
- 36.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saboo, V. M., and M. A. Gealt. 1998. Gene sequences of the pcpB gene of pentachlorophenol-degrading Sphingomonas chlorophenolica found in nondegrading bacteria. Can. J. Microbiol. 44:667–675. [PubMed] [Google Scholar]
- 39.Schwieger F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sjölund, M. 1999. The effect of humic substances on biological remediation of chlorophenol contaminated aquifer. M.S. thesis. Institute of Water and Environmental Engineering, Tampere University of Technology, Tampere, Finland. (14 Finnish.)
- 41.Staley, J. T. 1968. Prosthecomicrobium and Ancalomicrobium: new prosthecate freshwater bacteria. J. Bacteriol. 95:1921–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki, M., M. S. Rappé, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi, M., K. Hamana, and A. Hiraishi. 2001. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. Bacteriol. 51:1405–1417. [DOI] [PubMed] [Google Scholar]
- 44.Valo, R. J., M. M. Häggblom, and M. S. Salkinoja-Salonen. 1990. Bioremediation of chlorophenol contaminated simulated groundwater by immobilized bacteria. Water Res. 24:253–258. [Google Scholar]
- 45.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White, D. C., S. D. Sutton, and D. B. Ringelberg. 1996. The genus Sphingomonas: physiology and ecology. Curr. Opin. Biotechnol. 7:301–306. [DOI] [PubMed] [Google Scholar]
- 47.Yamazaki, M., I. Thorne, M. Mikolajczak, R. W. Armentrout, and T. J. Pollock. 1996. Linkage of genes essential for synthesis of a polysaccharide capsule in Sphingomonas strain S88. J. Bacteriol. 178:2676–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]