Abstract
Combinations of 10 Cryptosporidium parvum oocysts, with various ratios of genotype I to genotype II, were isolated and subjected to PCR-restriction fragment length polymorphism analysis. Amplification of both genotypes in these samples ranged from 31 to 74% and yielded no information about the genotype proportions. In addition, since both genotypes were not always detected, amplification of a single genotype is not conclusive evidence that the sample contains only a single genotype.
Two genotypes of Cryptosporidium parvum are considered to be responsible for most of the burden of cryptosporidiosis in humans—Type I, reported to infect predominantly humans, and Type II, which infects both animals and humans (8, 10, 14). As many as 20 other Cryptosporidium species have been reported to infect other mammals and birds and reptiles (19) but were thought to be host specific and not believed to cause disease in humans. Recently, several studies have reported one or more of these other species to be the cause of human infections (4, 6, 17).
The diversity of Cryptosporidium species that can infect humans has important implications for epidemiological studies and environmental screening. Studies on the molecular epidemiology have found that multiple genotypes can circulate within a geographic region (5, 9, 17, 18). Oocysts can survive for more than 3 months in the environment under appropriate conditions and are resistant to normal water treatment disinfection practices (13). This persistence in the environment allows the environmental samples to be contaminated with more than one genotype. Multiple genotypes have been detected within single outbreaks (9), indicating that single sources of exposure can contain mixed genotypes. Coinfection of humans with multiple genotypes is also possible, as multiple genotypes have been isolated from a single patient (1, 5, 9).
Cryptosporidiosis is generally self-limiting in immunocompetent patients (2). However, there remains no effective chemotherapy for the disease (15). This lack of drug treatment is of concern for the immunocompromised patients, for whom the parasite’s self-perpetuating life cycle can result in long-term diarrhea and major fluid loss (2). Without treatment, control becomes the best method to reduce the burden of disease. Environmental screening has been undertaken to ensure the safety of water supplies and identify sources of contamination if necessary (12). Studies of molecular epidemiology have also been done to determine infection and transmission patterns (17, 18).
Accurate and sensitive methods of detection and genotyping are needed to adequately address these issues. In both epidemiological and environmental studies, PCR has become an efficient means to detect and genotype the organism (7). However, there have been no attempts to evaluate its ability to characterize a mixed genotype population of oocysts. With the increased use of PCR, there remains a need to fully evaluate this technique. This report evaluates the sensitivity of PCR-restriction fragment length polymorphism (RFLP) analysis applied to mixed ratios of C. parvum genotypes.
For this study, the ability of PCR to detect multiple genotypes in a single source was evaluated with mixed ratios of C. parvum genotypes I and II. Using a method previously described (16), oocysts of each genotype were isolated via micromanipulation and transferred into 10 μl of 1× PCR buffer in a thin-walled PCR tube. Using nested PCR, detection of a single oocyst is possible, and 100% detection can be achieved at the level of 10 oocysts (16).
For each reaction, a set of 10 oocysts was isolated. The sets differed in their ratios of genotype I to genotype II oocysts. The five ratios evaluated were (genotype I:genotype II) 1:9, 3:7, 5:5, 7:3, and 9:1. DNA was liberated and PCR was performed following a procedure described previously (16). The nested PCR results in a final 593-bp amplicon for genotype I and a 590-bp amplicon for genotype II (indistinguishable within the agarose gel).
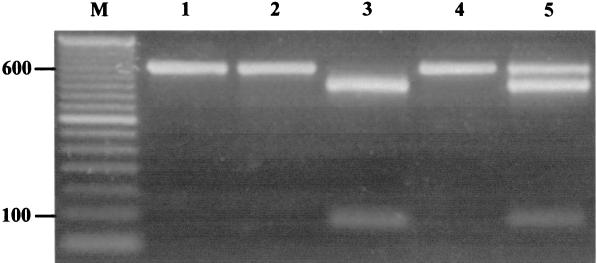
Following PCR, 15 μl of the nested amplicon was digested with 3 U of VspI (Promega, Madison, Wis.) in the supplied buffer for 8 h at 37°C. Digested products were visualized on 2% agarose gels stained with ethidium bromide (0.5 μg/μl) followed by UV transillumination. The VspI digest allows differentiation between genotypes I and II, as only the genotype I amplicon contains a restriction site—producing 503- and 90-bp fragments (Fig. 1). To demonstrate that genotype I amplicons were completely digested, approximately 200 ng of PCR amplicon from both genotypes were digested singly or in the presence of the other genotype (Fig. 1).
FIG. 1.
Two percent ethidium bromide-stained agarose gel illustrating nested-PCR amplification of an 18S rRNA segment within C. parvum and genotype determination with VspI RFLP analysis. Approximately 200 ng of nested-PCR amplicon from singly or mixed genotypes was subjected to VspI digestion for 8 h at 37°C. Lane 1, genotype I amplicon; lane 2, genotype II amplicon; lane 3, digested genotype I amplicon illustrating 503- and 90-bp fragments; lane 4, digested genotype II; lane 5, digestion of both genotypes within a single reaction tube; lane M, 50-bp molecular weight marker (Gibco BRL, Grand Island, N.Y.).
In total, at least 50 replicates for each ratio were subjected to PCR-RFLP analysis. Results are summarized in Table 1. Both genotypes were detected in various proportions at all ratios, and thus, mere amplification during PCR yielded no information about the proportion of genotypes in a single sample. In addition and more significantly, both genotypes were not always detected, even at a 5:5 ratio, which consistently provided the greatest sensitivity. Genotype I was singly detected at all ratios, while genotype II was singly detected at all ratios but one. Because no ratio amplified both genotypes universally, a PCR amplification of a single genotype is not a conclusive indication that the sample contains only a single genotype. Thus, when reporting the presence of one C. parvum genotype in a sample, it should be mentioned that the other genotype was not detected. This result raises the issue of false-negative samples with epidemiological implications. It has been reported that oocyst concentration in environmental samples is likely to be low (range, <0.007 to 484 oocysts/liter; n = 66; geometric mean, 2.7 oocysts/liter) (3), and thus the presence of a given genotype may go undetected.
TABLE 1.
Detection of C. parvum genotypes I and II within a single-source environment of mixed genotype ratiosa
| Type I:type II genotype ratio | Positive detection/no. of replicates (%)
|
||
|---|---|---|---|
| Genotype I only | Genotype II only | Both genotypes | |
| 1:9 | 1/53 (2) | 25/53 (47) | 27/53 (51) |
| 3:7 | 9/50 (18) | 7/50 (14) | 34/50 (68) |
| 5:5 | 10/50 (20) | 3/50 (6) | 37/50 (74) |
| 7:3 | 27/52 (52) | 3/52 (6) | 22/52 (42) |
| 9:1 | 35/51 (69) | 0/51 (0) | 16/51 (31) |
Mixed C. parvum genotypes I and II ratios were established by micromanipulation, and the presence of a particular genotype was determined by nested-PCR amplification followed by RFLP analysis with VspI.
Some variation in the detection of genotypes I and II at the different ratios may be due to oocyst age. By the end of the study, the sample of genotype I oocysts used was 8 months old while the sample of genotype II oocysts used was 5 months old. However, oocysts can remain infective 6 to 9 months in the environment (13) and environmental samples can easily contain oocysts of various ages.
Although the overall detection percentages may vary slightly with oocyst age and population size, the implications are clear. Both genotypes were detected, even at a fairly disparate oocyst ratio (1:9). Conversely, both genotypes were not always detected, even at an even ratio. This supports the need for analysis at the single-oocyst level for molecular epidemiology studies.
We acknowledge the current limitations of PCR amplification and genotyping at the single-oocyst level due to the fact that PCR sensitivity is reduced with low oocyst numbers (16). With environmental samples containing low numbers of oocysts (3, 11), the low sensitivity of PCR may make the process less effective (16). For fecal samples and other samples in which the quantity of oocysts may be large, amplifying and typing single manipulated oocysts may provide more accurate information about genotype distribution. Therefore, with the current sensitivity of PCR to single-source mixed populations, additional techniques, i.e., micromanipulation, RFLP analysis, cloning, and/or sequencing, should be performed to determine the true distribution of genotypes in a sample.
Acknowledgments
We thank B. Helen Jost and Marilyn M. Marshall for their assistance and helpful discussions.
REFERENCES
- 1.Carraway, M., S. Tzipori, and G. Widmer. 1997. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect. Immun. 65:3958–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark, D. P. 1999. New insights into human cryptosporidiosis. Clin. Microbiol. Rev. 12:554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeChevallier, M., W. Norton, and R. Lee. 1991. Occurrence of Giardia and Cryptosporidium spp. in surface water supplies. Appl. Environ. Microbiol. 57:2610–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLauchlin, J., C. Amar, S. Pedraza-Diaz, and G. L. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLauchlin, J., S. Pedraza-Diaz, C. Amar-Hoetzeneder, and G. L. Nichols. 1999. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J. Clin. Microbiol. 37:3153–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan, U., R. Weber, L. Xiao, I. Sulaiman, R. C. Thompson, W. Ndiritu, A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan, U. M., and R. C. Thompson. 1998. Molecular detection of parasitic protozoa. Parasitology 117:S73–S85. [DOI] [PubMed] [Google Scholar]
- 8.Morgan, U. M., L. Xiao, B. D. Hill, P. O’Donoghue, J. Limor, A. A. Lal, and R. C. Thompson. 1998. Detection of the Cryptosporidium parvum “human” genotype in a Dugong (Dugong dugon). J. Parasitol. 86:1352–1354. [DOI] [PubMed] [Google Scholar]
- 9.Patel, S., S. Pedraza-Diaz, J. McLauchlin, and D. P. Casemore. 1998. Molecular characterisation of Cryptosporidium parvum from two large suspected waterborne outbreaks. Outbreak Control Team South and West Devon 1995, Incident Management Team and Further Epidemiological and Microbiological Studies Subgroup North Thames 1997. Commun. Dis. Public Health. 1:231–233. [PubMed] [Google Scholar]
- 10.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. Mac Kenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose, J., J. Lisle, and M. LeChevallier. 1997. Waterborne cryptosporidiosis: incidence, outbreaks, and treatment strategies, p.93–109. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Inc., New York, N.Y.
- 12.Sinclair, J. L. 2000. Enumeration of Cryptosporidium spp. in water with US EPA method 1622, USA. J. AOAC Int. 83:1108–1114. [PubMed] [Google Scholar]
- 13.Smith, H. V., and J. B. Rose. 1998. Waterborne cryptosporidiosis: current status. Parasitol. Today 14:14–22. [DOI] [PubMed] [Google Scholar]
- 14.Spano, F., L. Putignani, A. Crisanti, P. Sallicandro, U. M. Morgan, S. M. Le Blancq, L. Tchack, S. Tzipori, and G. Widmer. 1998. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J. Clin. Microbiol. 36:3255–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sterling, C. R. 2000. Cryptosporidiosis: the treatment dilemma. J. Med. Microbiol. 49:207–208. [DOI] [PubMed] [Google Scholar]
- 16.Sturbaum, G. D., C. Reed, P. J. Hoover, B. H. Jost, M. M. Marshall, and C. R. Sterling. 2001. Species-specific, nested PCR-restriction fragment length polymorphism detection of single Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 67:2665–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492–497. [DOI] [PubMed] [Google Scholar]
- 18.Xiao, L., J. Limor, C. Bern, and A. A. Lal. 2001. Tracking Cryptosporidium parvum by sequence analysis of small double-stranded RNA. Emerg. Infect. Dis. 7:141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao, L., U. M. Morgan, R. Fayer, R. C. Thompson, and A. A. Lal. 2000. Cryptosporidium systematics and implications for public health. Parasitol. Today 16:287–292. [DOI] [PubMed] [Google Scholar]