Abstract
Utilizing the principle of competitive PCR, we developed two assays to enumerate Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and nitrite-oxidizing bacteria belonging to the genus Nitrospira. The specificities of two primer sets, which were designed for two target regions, the amoA gene and Nitrospira 16S ribosomal DNA (rDNA), were verified by DNA sequencing. Both assays were optimized and applied to full-scale, activated sludge wastewater treatment plant (WWTP) samples. If it was assumed that there was an average of 3.6 copies of 16S rDNA per cell in the total population and two copies of the amoA gene per ammonia-oxidizing bacterial cell, the ammonia oxidizers examined represented 0.0033% ± 0.0022% of the total bacterial population in a municipal WWTP. N. oligotropha-like ammonia-oxidizing bacteria were not detected in an industrial WWTP. If it was assumed that there was one copy of the 16S rDNA gene per nitrite-oxidizing bacterial cell, Nitrospira spp. represented 0.39% ± 0.28% of the biosludge population in the municipal WWTP and 0.37% ± 0.23% of the population in the industrial WWTP. The number of Nitrospira sp. cells in the municipal WWTP was more than 62 times greater than the number of N. oligotropha-like cells, based on a competitive PCR analysis. The results of this study extended our knowledge of the comparative compositions of nitrifying bacterial populations in wastewater treatment systems. Importantly, they also demonstrated that we were able to quantify these populations, which ultimately will be required for accurate prediction of process performance and stability for cost-effective design and operation of WWTPs.
Nitrification, the bio-oxidation of ammonia (NH3) to nitrate (NO3−), is a key process in the removal of ammonia from wastewater, which alleviates problems of aquatic toxicity, high oxygen demand in receiving waters, and nutrient contributions to eutrophication (3, 13). The number and physiological activity of nitrifying bacteria in wastewater treatment reactors are considered the rate-limiting parameters for the bioconversion of nitrogen in sewage (48). The slow growth rate of nitrifying bacteria and the sensitivity of these organisms to several environmental factors (e.g., pH, O2, temperature, substrate concentration, and the presence of toxic substances) influence the minimum sludge age during wastewater treatment required to establish stable nitrification (34). Consequently, early detection of a decline in the nitrifying population by rapid and reliable molecular methods may improve process control and prevent washout of these organisms from a wastewater treatment system.
Chemolithotrophic nitrification is a two-step biochemical process involving sequential transformation of NH3 to NO3− via NO2− (6). In wastewater treatment plants (WWTPs), the reactions are catalyzed by two phylogenetically distinct groups of bacteria, the ammonia-oxidizing bacteria (AOB) belonging to the β subclass of the class Proteobacteria and nitrite-oxidizing bacteria (NOB). In AOB, oxidation of NH3 to NO2 is carried out in two steps: NH3 is oxidized to hydroxylamine (NH2OH) by ammonia monooxygenase (30), and then NH2OH is oxidized to NO2− by hydroxylamine oxidoreductase (41). In NOB, the oxidation of NO2− is catalyzed by the enzyme nitrite oxidoreductase (18).
The use of quantitative molecular techniques with environmental samples is challenging, mainly due to small quantities of templates in complex biological backgrounds and the potential presence of inhibitory compounds that can copurify with the nucleic acids (9). The competitive PCR (cPCR) assay is based on competitive coamplification of a specific target sequence together with an internal standard whose concentration is known (50). This approach has excellent sensitivity and quantitative accuracy and has the added advantage, because of the competitive principle on which it is based, of stringent internal control (36). cPCR has been used to enumerate both culturable and nonculturable organisms in environmental samples, including AOB (16, 23, 31, 35, 45), although its use with this group of bacteria in activated sludge has been very limited (23). No cPCR assay has been developed previously for quantification of NOB in activated sludge.
The goal of this research was to design and evaluate cPCR assays for quantification of AOB and Nitrospira spp., the dominant NOB in activated sludge (17), in mixed-liquor suspended-solids (MLSS) samples from a municipal WWTP. The amoA gene was used to characterize the indigenous ammonia-oxidizing population, and the information obtained was used to design a specific cPCR assay for this gene. Both assays were also applied to samples retrieved from an industrial WWTP. All the data obtained were compared with total 16S ribosomal DNA (rDNA) data determined by dot blot hybridization with a universal oligonucleotide.
MATERIALS AND METHODS
Samples.
MLSS samples were collected from the aeration basins of a municipal WWTP in June 1999 and between February 2000 and January 2001. MLSS samples were collected from an industrial WWTP between June and September 2000. Both WWTPs employed a complete-mix suspended growth, aerobic activated sludge process (with biomass recycling). The bioreactors were operated in the neutral pH range and in the mesophilic temperature range. However, the chemical compositions of the influent wastewater, in addition to various operational and environmental parameters, differed considerably for the two facilities.
The municipal WWTP (27) biologically treats an average of 40 million gallons per day in a tank reactor with a 6-h hydraulic retention time. The wastewater consists primarily of sanitary sewage collected from a local municipality; however, industrial and hospital discharges contribute to the overall flow. Typical influent 5-day biochemical oxygen demand and ammonia nitrogen levels are 200 and 20 mg/liter, respectively. Organic carbon oxidation and nitrification occur in the same treatment reactor which operates with an average biological solids retention time of 10 days. The average level of ammonia removal in the plant for the 12 samples analyzed was 86.7% ± 7.8%.
The industrial WWTP (24, 26) treats approximately 27 million gallons of wastewater per day with a hydraulic retention time of 24 h. The waste flows result from the manufacture of fibers, plastics, and chemicals and consist mainly of acetic acid, propionic acid, n-butyric acid, ethylene glycol, ethanol, methanol, isopropanol, and acetone. No sanitary sewage is discharged into this WWTP. The carbonaceous influent is supplemented with both nitrogen (ammonia) and phosphorus (phosphoric acid). The average influent 5-day biological oxygen demand and ammonia nitrogen levels are 750 and 45 mg/liter, respectively. The WWTP is operated with a biological solids retention time of between 12 and 14 days. The average level of ammonia removal in the plant for the three samples analyzed was 98.4% ± 1.79%.
DNA extraction.
Genomic DNA was extracted in triplicate from 2 ml of an MLSS sample by using a FastDNA kit (Bio 101, Vista, Calif.), with the following minor modifications: the binding matrix-DNA complex was washed twice with 80% (vol/vol) ethanol after the recommended salt-ethanol wash step, and the DNA was eluted in 100 μl of 10 mM Tris-HCl buffer (pH 8.0). The three extracts were combined before the DNA was analyzed. The integrity of DNA samples was analyzed by electrophoresis by using 0.8% (wt/vol) agarose (Fisher Scientific, Pittsburgh, Pa.), 1× TBE (40), and 1× GelStar nucleic acid gel stain (FMC Corporation, Rockland, Maine).
Total 16S rDNA enumeration by dot blot hybridization.
Genomic DNA samples were denatured in a 0.4 N NaOH solution and incubated in a boiling water bath for 10 min. Nucleic acids (0.4 to 0.6 μg) were transferred to BIOTRANS nylon membranes (ICN Pharmaceuticals, Costa Mesa, Calif.) by using a dot blot apparatus (Bio-Rad, Hercules, Calif.). The membranes were baked for 30 min at 80°C, washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (40), and then baked for 60 min at 80°C. The filters were prehybridized at 37°C in a solution containing 0.5 M NaH2PO4, 1 mM EDTA, and 7% (wt/vol) sodium dodecyl sulfate. Hybridizations were performed overnight at 37°C by adding 32P-labeled oligonucleotide 1392r (25) (1.5 × 107 to 3.0 × 107 cpm) to the prehybridization solution. After hybridization, the membranes were washed twice for 15 min each time at the same temperature in a solution containing 10 mM NaCl, 20 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 0.5% (wt/vol) sodium dodecyl sulfate. The membranes were exposed to a phosphor screen, and the 16S rDNA was quantified with a Molecular Dynamics Storm 840 phosphor imager (Molecular Dynamics, Sunnyvale, Calif.) equipped with ImageQuant 5.0 analysis software. The total number of 16S rDNA molecules was determined as previously described (2).
amoA gene libraries.
The gene encoding the enzyme ammonia monooxygenase (amoA) was amplified by PCR from genomic DNA extracted from MLSS from the municipal WWTP collected in June 1999 by using primers amoA1-F and amoA2-R (38). PCR amplification was performed in 25-μl (total volume) mixtures by using Ready-To-Go PCR beads (Amersham Pharmacia, Piscataway, N.J.). The program used for the amplification was as follow: 5 min at 94°C, followed by 50 cycles consisting of 30 s at 94°C, 30 s at 62°C, and 30 s at 72°C and a final cycle consisting of 7 min at 72°C. The PCR product was cloned into the pCR 2.1 vector according to the instructions of the manufacturer (TA cloning kit; Invitrogen, Carlsbad, Calif.). Clones were screened by colony hybridization as previously described (40), using the amoA gene from Nitrosomonas europaea as the probe (30). Plasmid DNA from randomly selected clones were isolated with an RMP AFS kit (Bio 101). DNA were sequenced at the Molecular Biology Resource Facility of the University of Tennessee (Knoxville) by using an ABI PRISM dye terminator cycle sequencing kit with AmpliTaq DNA polymerase (protocol P/N 402078, revision A), an Applied Biosystems 373 DNA sequencer (Perkin-Elmer, Foster City, Calif.), and M13f or M13r located on the pCR 2.1 plasmid as the primer.
amoA cPCR.
A 100-bp oligonucleotide corresponding to the target DNA (121 bp of the M-20 clone from the amoA gene library, with a 21-bp internal deletion; 5′ GAA TAT GTT CGC CTG ATT GAG CAA GGC TCA CTG CGT ACC TTT GGT GGA CAC TTC GCA GCG TTC GTA TCC ATG TTG ATG TTC TGC GTA TGG TGG TAC TTT G 3′) was synthesized with an Oligo 1000 DNA synthesizer (Beckman, Palo Alto, Calif.). PCR amplification of the competitor was performed in a 25-μl (total volume) mixture by using Ready-To-Go PCR beads (Amersham Pharmacia), 10 pmol of primer amo598f (5′ GAATATGTTCGCCTGATTG 3′; corresponding to positions 598 to 616 of the N. europaea amoA gene [30]), 10 pmol of primer amo718r (5′ CAAAGTACCACCATACGCAG 3′; corresponding to positions 699 to 718 of the N. europaea amoA gene), and 100 pmol of the 100-bp competitor as the template. The program used for amplification of the competitor was as follow: 5 min at 94°C, followed by 40 cycles consisting of 15 s at 94°C, 15 s at 60°C, and 15 s at 72°C and a final cycle consisting of 7 min at 72°C. The PCR product was cloned in the pCR 2.1-topo vector (TA cloning kit; Invitrogen) and sequenced to confirm its identity. To prepare a stock solution of the competitor, PCR amplification was conducted in a 100-μl (total volume) reaction mixture containing 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 0.001% [wt/vol] gelatin), 50 nmol of each deoxynucleoside triphosphate, 40 pmol of each primer, 1 ng of plasmid DNA, and 2.5 U of Taq DNA polymerase (Gibco BRL, Gaithersburg, Md.) by using the same program. The amplified fragment was cleaned with a QIAquick PCR purification kit (QIAGEN Inc., Valencia, Calif.). The DNA was quantified by using a DyNA Quant200 fluorometer (Hoefer Pharmacia Biotech, San Francisco, Calif.) and agarose gels with a mass ladder (GenSura Laboratories, San Diego, Calif.).
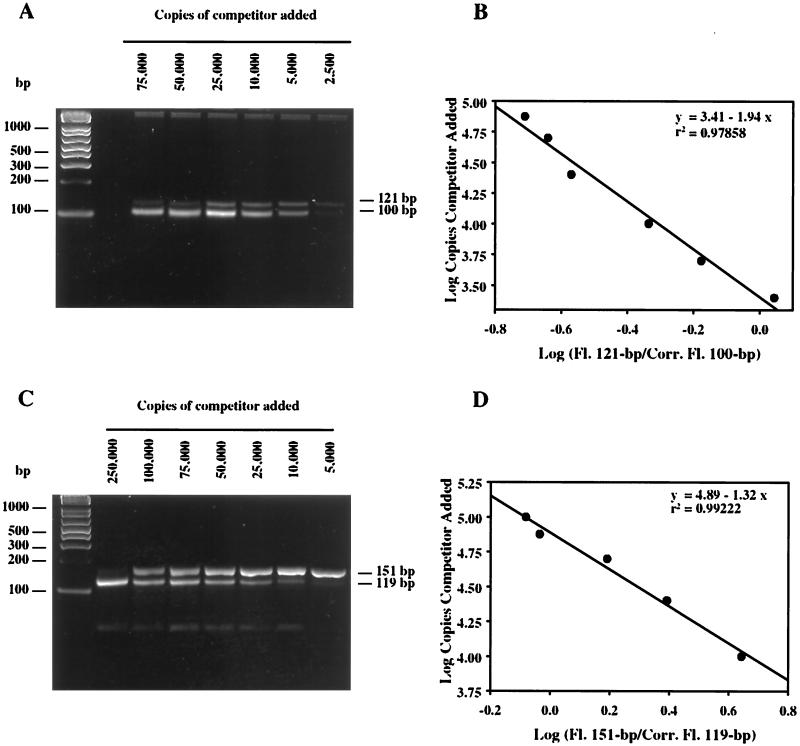
Amplification of the cPCR product was performed in 25-μl (total volume) mixtures containing Ready-To-Go PCR beads (Amersham Pharmacia), 10 pmol of primer amo598f, 10 pmol of primer amo718r, dilutions of the competitor (2,500 to 100,000 copies), and 15 to 30 ng of MLSS genomic DNA. Amplification was performed by using the program described above, shortened to 30 cycles. PCR products were separated by electrophoresis by using 3% (wt/vol) NuSieve 3:1 agarose (BMA, Rockland, Maine), 1× TBE (40), and 1× GelStar nucleic acid gel stain (FMC Corporation, Rockland, Maine). The intensities of the individual bands were quantified with an AlphaImager 1220 documentation and analysis system (Alpha Innotech Corporation, San Leandro, Calif.). The fluorescence of the competitor product was corrected by a factor of 121/100 so that the corrected fluorescence intensity of the 100-bp band could be directly compared with the measured fluorescence intensity of the 121-bp band. The competition equivalence point was determined as previously described (9).
Primer design and amplification of Nitrospira sp. 16S rDNA from suspended-solids genomic DNA.
Nitrospira 16S rDNA primers NSR1113f (5′ CCTGCTTTCAGTTGCTACCG 3′) and NSR1264r (5′ GTTTGCAGCGCTTTGTACCG 3′) were deduced from 16S rDNA sequences aligned with the CLUSTAL W program (47). The alignment contained two Nitrospira partial 16S rDNA sequences obtained from industrial MLSS (K. C. Cook, A. C. Layton, H. M. Dionisi, and G. S. Sayler, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. N-35, 2000) and Nitrospira 16S rDNA sequences available from GenBank (National Center for Biotechnology Information). The Nitrospira clones from the industrial MLSS, 931 (AF420301) and 9335, were 99% similar to each other and 98% similar to other Nitrospira 16S rDNA sequences, including Y14639, AF155154, and AJ224038.
PCR amplification of the Nitrospira sp. 16S rDNA was performed in 25-μl mixtures containing Ready-To-Go PCR beads (Amersham Pharmacia), 10 pmol of primer NSR1113f, 10 pmol of primer NSR1264r, and 2 ng of genomic DNA from MLSS samples as the template. The program used for amplification was 5 min at 94°C, followed by 40 cycles consisting of 30 s at 94°C, 30 s at 65°C, and 30 s at 72°C and a final cycle consisting of 15 min at 72°C. The PCR product was cloned and analyzed as described above.
Nitrospira 16S rDNA cPCR.
A 119-bp oligonucleotide, corresponding to the target DNA (151 bp of Nitrospira sp. 16S rDNA positions 1113 to 1264 [Escherichia coli numbering] with 32-bp internal deletion; 5′ CCT GCT TTC AGT TGC TAC CGG GTC ATG CCG AGC ACT CTG AAA GGA CTG CCC AGG AGA ACG GGG AGG AAA TGC CTG GGG CCA CAC ACG TGC TAC AAT GGC CGG TAC AAA GCG CTG CAA AC 3′) was synthesized and amplified by using primers NSR1113f and NSR1264r as described above. The PCR product was cloned and sequenced. The stock solution of the competitor was prepared as described above for the amoA competitor.
Amplification of the cPCR product was performed in 25-μl (total volume) mixtures containing Ready-To-Go PCR beads (Amersham Pharmacia), 10 pmol of primer NSR1113f, 10 pmol of primer NSR1264r, dilutions of the competitor (2,500 to 100,000 copies), and 0.5 to 3 ng of MLSS genomic DNA. The program used for amplification was 5 min at 94°C, followed by 35 cycles consisting of 15 s at 94°C, 15 s at 65°C, and 15 s at 72°C and a final cycle consisting of 7 min at 72°C. PCR products were separated by electrophoresis and analyzed as described above for the amoA cPCR products, except that a correction factor of 151/119 was used.
Analysis of sequence data.
Partial Nitrospira 16S rDNA and amoA sequences were compared with sequences in publicly accessible databases by using the program Basic Local Alignment Search Tool (BLAST) (1). Sequences were aligned by using ClustalW. Trees were inferred from distance matrix analyses by using FITCH with global rearrangements and randomized input order. Phylogenetic analysis of amoA sequences was carried out by using the Phylogeny Inference Package (PHYLIP, version 3.57c).
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank database under accession numbers AF420290 to AF420300.
RESULTS
Analysis of AOB from the municipal WWTP.
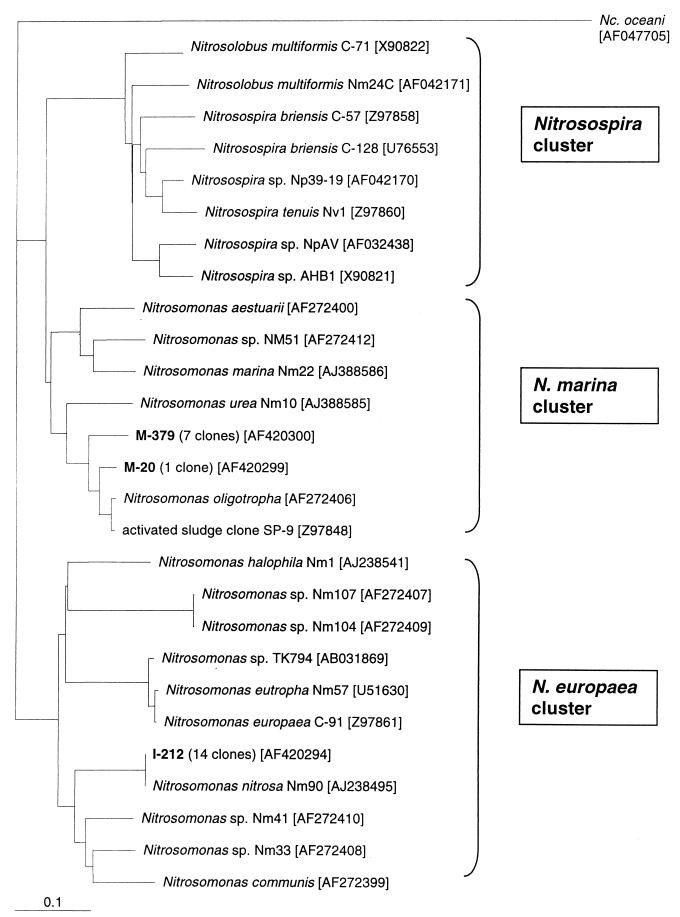
The amoA gene was amplified by using genomic DNA isolated from MLSS from a municipal WWTP as the template and primers amoA1-F and amoA2-R (38). The PCR products were used for generation of an amoA library. The sequences of randomly selected clones indicated that non-amoA sequences that were approximately the expected size (491 bp) were coamplified with amoA sequences. Such nonspecific amplification occurred with this sample even after the annealing temperature was increased to 62°C. The resulting sequences were not similar to sequences available in the GenBank database. Hybridization with the amoA gene from N. europaea was employed to identify amoA sequences in the library. Colony hybridization indicated that 1 of every 10 clones hybridized with N. europaea amoA. Sequences from these clones were affiliated with amoA sequences from AOB belonging to the β subclass (Fig. 1). All amoA sequences retrieved from the municipal WWTP plant belonged to the Nitrosomonas marina cluster and were between 92 and 95% identical to the amoA gene from Nitrosomonas oligotropha. The closest environmental clone was SP-9 (92 to 95% identical), which was retrieved from activated sludge from a sewage treatment plant in Germany (Fig. 1) (38).
FIG. 1.
Phylogenetic Fitch-Margoliash amoA dendrogram (PHYLIP, version 3.57c, global rearrangements and randomized input [three jumbles]), showing the relationship of cloned amoA sequences from an industrial WWTP (I-212) and a municipal WWTP (M-379 and M-20) with cultured ammonia oxidizers (37). The root was determined by using the gamma-subclass AOB sequence from Nitrosococcus oceani C-107 as an outgroup. The numbers in brackets are accession numbers for the amoA sequences. Clusters were labeled by using the assignments of Purkhold et al. (37).
Validation of the amoA cPCR assay.
A new set of primers was designed for cPCR analysis of the amoA gene due to the lack of specificity of the amoA1-F-amoA2-R primer set with the municipal WWTP sample. Amplification of genomic DNA from MLSS with primers amo598f and amo718r produced an approximately 120-bp fragment. The PCR products were cloned, and 10 clones were randomly selected and sequenced. All sequences were identified as the amoA gene, and no false-positive results were obtained.
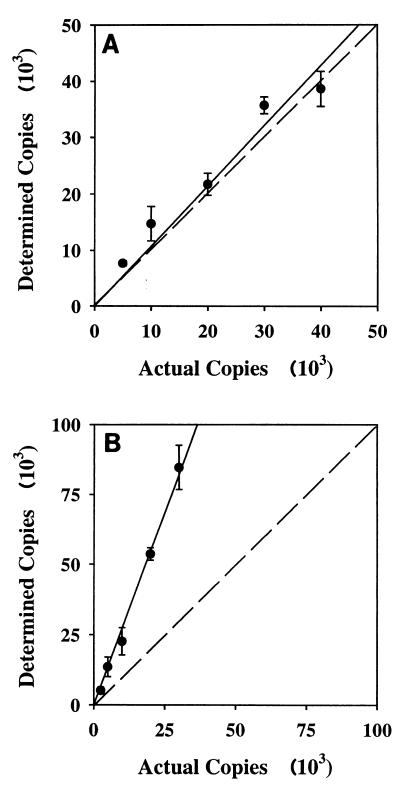
The 121-bp fragment amplified from clone M-20 was used to test the cPCR assay. Good agreement between the expected copy number and the copy number determined was observed for the amoA gene (Fig. 2A). The efficiencies of amplification of the competitor and target DNA were the same for between 10 and 50 cycles of the PCR (data not shown). When the amoA cPCR technique was applied to total genomic DNA from an MLSS sample, bands were detected in the gel at approximately 121 and 100 bp, which corresponded to the target and competitor sizes (Fig. 3A). Addition of 15 to 30 ng of MLSS genomic DNA to the PCR mixture resulted in reproducible quantitative PCR analysis. However, when more than 50 ng of DNA was added, amplification of both competitor and target DNA was inhibited. These results indicate that this cPCR assay is suitable for use with a complex DNA sample and is capable of detecting and quantifying the N. oligotropha-like populations of AOB in activated sludge samples.
FIG. 2.
Quantification of copies of the amoA gene (A) and the Nitrospira 16S rDNA gene (B). Numbers of determined copies were calculated by using cPCR as described in Materials and Methods with a PCR product of clone M-20 (A) or a plasmid containing the 16S rDNA gene of Nitrospira (B). Error bars indicate the standard deviations of the means (n = 3 or 4). The dashed lines indicate a ratio of actual number of copies to determined number of copies of 1:1.
FIG. 3.
cPCR for amoA gene and Nitrospira sp. 16S rDNA. (A) Agarose gel containing cPCR products of the amoA gene from a mixed-liquor sample from a municipal WWTP (sample M-09-07-00), obtained by procedures described in Materials and Methods. (B) Determination of the competition equivalence point for the data shown in panel A. (C) Agarose gel containing cPCR products of the Nitrospira 16S rDNA gene from a mixed-liquor sample from a municipal WWTP (sample M-04-01-00). (D) Determination of the competition equivalence point for the data shown in panel C. Fl., fluorescence; Corr. Fluor., corrected fluorescence. The equations and fits of the lines are shown.
Amplification of Nitrospira 16S rDNA.
Primers NSR1113f and NSR1264r were designed to amplify 16S rDNA sequences specific for Nitrospira spp. These primers encompass the clade II group as described by Burrell et al. (8). The amplification specificity of primers NSR1113f and NSR1264r was tested by using genomic DNA extracted from municipal MLSS as the template. A library was generated by cloning the PCR product, and 18 clones were randomly selected and sequenced. All sequences were identified as Nitrospira sequences, and they exhibited more than 98% similarity to one another. No false-positive results were obtained, indicating the specificity of this primer set.
Validation of cPCR for Nitrospira 16S rDNA.
A cPCR assay was developed to quantify Nitrospira 16S rDNA in MLSS. Although cPCR assumes that the amplification efficiencies of target and competitor are equal, control experiments suggested that the target sequence (151 bp) was amplified about threefold more efficiently than the competitor sequence (119 bp) (data not shown), independent of the number of cycles used (10 to 50 cycles). Amplification of the competitor and amplification of the target reached saturation at the same number of cycles (40 cycles) (data not shown), demonstrating that the assay can be used to quantify Nitrospira 16S rDNA if a correction factor is used. When the expected copy number based on the concentration of the cloned Nitrospira 16S rDNA gene was plotted versus the copy number calculated by the assay, the copy number was overestimated, but the relationship was linear with a slope of 2.74 (Fig. 2B). Therefore, this value was utilized as a correction factor for the cPCR assay. Application of this assay to environmental samples showed that two bands of the expected sizes (119 and 151 bp) were amplified (Fig. 3C). Between 0.5 and 3 ng of suspended solids genomic DNA in the PCR mixture produced reproducible results.
Assessment of MLSS samples from full-scale WWTPs.
Genomic DNA were extracted from 12 MLSS samples obtained from the municipal WWTP between February 2000 and January 2001. The total number of 16S rDNA copies was determined for each sample by using dot blot hybridization with oligonucleotide 1392r as the probe (25). The total 16S rDNA values were not significantly different for different samples and values ranged from 1.64 × 1013 to 3.76 × 1013 copies per liter of mixed liquor (Table 1). Similar total 16S rDNA values (2.91 × 1013 to 3.00 × 1013 copies per liter) were obtained for suspended solids collected over a 3-month period from an industrial WWTP (Table 1). The traditional method used to quantify biomass in WWTPs, the mixed-liquor volatile suspended-solids (MLVSS) method, was also used to assess total organism concentrations in both systems (Table 1). The MLVSS values ranged from 1,631 to 3,264 mg/liter.
TABLE 1.
Numbers of total 16S rDNA, amoA gene, and Nitrospira sp. 16S rDNA copies per liter and MLVSS concentrations in samples from a municipal WWTP and an industrial WWTP
| Samplea | No. of 16S rDNA copies per literb | No. of amoA copies per literc | No. of Nitrospira 16S rDNA copies per literc | MLVSS concn (mg/liter) |
|---|---|---|---|---|
| M-02-02-00 | (2.31 ± 0.88) × 1013 | (4.79 ± 1.64) × 108 | (1.49 ± 0.39) × 1010 | 1,960 |
| M-03-13-00 | (1.65 ± 0.26) × 1013 | (2.80 ± 0.62) × 108 | (1.18 ± 0.43) × 1010 | 1,736 |
| M-04-01-00 | (1.78 ± 0.39) × 1013 | (5.81 ± 3.14) × 108 | (1.88 ± 0.37) × 1010 | 2,226 |
| M-05-01-00 | (2.51 ± 1.01) × 1013 | (8.10 ± 0.61) × 108 | (2.53 ± 0.51) × 1010 | 2,051 |
| M-06-02-00 | (2.29 ± 0.78) × 1013 | (4.50 ± 0.48) × 108 | (3.04 ± 0.35) × 1010 | 1,631 |
| M-07-14-00 | (1.93 ± 0.46) × 1013 | (1.17 ± 0.27) × 108 | (2.71 ± 0.19) × 1010 | 2,191 |
| M-08-02-00 | (1.71 ± 0.39) × 1013 | (8.67 ± 1.10) × 107 | (1.40 ± 0.24) × 1010 | 1,939 |
| M-09-07-00 | (3.76 ± 0.27) × 1013 | (1.38 ± 0.48) × 108 | (2.44 ± 0.37) × 1010 | 2,184 |
| M-10-05-00 | (1.64 ± 0.04) × 1013 | (5.02 ± 0.77) × 108 | (5.47 ± 0.20) × 1010 | 1,974 |
| M-11-17-00 | (1.94 ± 0.28) × 1013 | (1.12 ± 0.44) × 108 | (5.82 ± 0.98) × 109 | 1,890 |
| M-12-01-00 | (3.59 ± 0.88) × 1013 | (2.58 ± 1.60) × 108 | (2.95 ± 0.24) × 1010 | 1,974 |
| M-01-04-01 | (2.67 ± 0.03) × 1013 | (2.16 ± 0.66) × 108 | (2.56 ± 0.52) × 1010 | 1,890 |
| I-06-26-00 | (2.97 ± 0.35) × 1013 | ND | (8.14 ± 1.20) × 109 | 2,635 |
| I-08-07-00 | (2.91 ± 0.04) × 1013 | ND | (4.29 ± 1.32) × 1010 | 2,516 |
| I-09-04-00 | (3.00 ± 0.07) × 1013 | ND | (3.90 ± 0.14) × 1010 | 3,264 |
Samples whose designations begin with M were obtained from the municipal WWTP, and samples whose designations begin with I were obtained from the industrial WWTP. Sample numbers correspond to the month-day-year of sampling.
The data were generated by dot blotting and hybridization with the universal 1392r probe of equal amounts of template DNA (genomic DNA extracted from suspended solids). Means ± standard deviations were calculated from data for three or four replicate dots for each of the samples which was hybridized with the probe, quantified by phosphor imaging, and compared with known amounts of 16S rDNA applied to the membrane.
The numbers of amoA gene and Nitrospira 16S rDNA copies per liter of mixed liquor were estimated by cPCR as indicated in Materials and Methods. The results are means ± standard deviations based on three to five determinations. ND, not detected.
The amoA cPCR assay was used for the same set of municipal and industrial samples. The number of amoA gene copies per liter of mixed liquor varied between 8.67 × 107 and 8.10 × 108 for municipal WWTP samples, and there were not apparent seasonal differences (Table 1). In contrast, no amplification of N. oligotropha-like amoA was observed with the samples from the industrial WWTP. This result suggests that this type of AOB was not present in the industrial activated sludge samples.
The values for Nitrospira 16S rDNA content determined by cPCR were considerably higher than the values for amoA gene content, ranging from 5.82 × 109 to 5.47 × 1010 copies per liter of mixed liquor from the municipal WWTP (Table 1). Similar values were obtained for samples from the industrial WWTP, in which the Nitrospira 16S rDNA content ranged from 8.14 × 109 to 4.29 × 1010 copies per liter (Table 1). These values for Nitrospira 16S rDNA content represent 0.113% ± 0.076% and 0.101% ± 0.064% of the total 16S rDNA found in 12 samples from the municipal WWTP and three samples from the industrial WWTP, respectively.
DISCUSSION
Many questions regarding the quantitative dynamics of nitrifying bacteria in wastewater treatment remain unanswered due to the lack of reliable quantification techniques. To our knowledge, this is the first description of quantification of NOB belonging to the genus Nitrospira with cPCR and the Nitrospira spp. in a WWTP activated sludge process. A specific cPCR assay for quantification of N. oligotropha-like AOB was also designed, and the data obtained was compared with total 16S rDNA values.
The amoA gene, encoding the ammonia monooxygenase active site, has been used increasingly as a phylogenetic biomarker for molecular diversity studies of AOB in environmental samples (12, 14, 16, 17, 23, 31, 37, 38, 39, 45), as 16S rDNA-based approaches may result in coamplification of sequences belonging to nonnitrifying members of the β subclass of the Proteobacteria (38). Three clusters can be identified by amoA analysis: the Nitrosospira cluster, the N. marina cluster, and the N. europaea-Nitrosococcus mobilis cluster (37). Using the amoA1-F-amoA2-R primer set (38), we studied the AOB population in a municipal WWTP. Only amoA sequences related to N. oligotropha belonging to the N. marina cluster were detected (Fig. 1). Members of the N. oligotropha lineage were recently detected in different WWTPs (12, 37, 38, 46), as well as in freshwater and soil (22, 44), indicating the ecological versatility in this group of AOB.
cPCR assays have been developed previously for enumeration of the amoA gene (16, 31, 45). However, the primer set designed by Rotthauwe and coworkers (38) could not be used for quantification with our samples because of nonspecific amplification. Therefore, we developed a cPCR assay specific for the amoA gene found in the municipal WWTP examined. The technique allowed us to quantify the N. oligotropha-like amoA gene content in all 12 samples analyzed (Table 1). Use of this technique with activated sludge samples from an industrial WWTP, by contrast, resulted in no amplification (Table 1). An amoA library from this plant suggested that sequences similar to Nitrosomonas nitrosa (99.8 to 100% identity) (Fig. 1) were present. These sequences had two mismatches with the amo598f primer and eight mismatches with the amo718r primer. Interestingly, N. nitrosa was originally isolated from activated sludge from a chemical-processing facility (22) but has not been detected previously in WWTP samples by amoA analysis (37). The N. nitrosa-like amoA gene expands the list of AOB species found in activated sludge with this technique.
The wide diversity of the amoA sequences retrieved from different WWTPs (37, 38; this study) indicates that it may be difficult to develop a quantitative assay able to detect all clusters simultaneously. As multiple populations can coexist, several primer sets may be needed to perform ecological studies of different AOB subpopulations activated sludge. Quantification of the amoA gene, like quantification of any other target gene, can be biased easily by incorrect specificity of the primer set used. Overestimation of the AOB population could result from the use of a broad set of primers due to nonspecific amplification. On the other hand, although the intolerance of PCR to base pair mismatches enhances assay specificity, it can reduce the sensitivity of a PCR for detecting genes that exhibit high levels of sequence degeneracy, such as the amoA gene.
cPCR relies on measurement of PCR products at the end point, after gel electrophoresis. An internal standard minimizes quantification problems caused by impurities in the samples, which can affect PCR amplification. However, the basic assumption that the internal standard (competitor) and the target product are amplified equally needs to be verified. In this study, the target and the competitor were amplified at the same rate in the amoA cPCR assay. In contrast, the Nitrospira sp. 16S rDNA target product was amplified more efficiently than the competitor molecule constructed. Consequently, an extra correction factor accounting for the differences was incorporated into calculations of the Nitrospira 16S rDNA copy number. Preferential amplification of the target gene over a competitive template or vice versa can occur due to differential denaturation or as a result of different target lengths (9), although only a 1°C difference in melting temperature and 32 bp differentiate the two amplicons. Similar differences in amplification efficiencies (3.1-fold) have been reported for another cPCR system (28).
Cultivated NOB have been assigned to the genera Nitrobacter, Nitrospira, Nitrococcus, and Nitrospina (5). Recent findings obtained by 16S rDNA sequence analysis and fluorescence in situ hybridization (FISH) demonstrated that Nitrospira spp. are the dominant nitrite-oxidizing microorganisms in freshwater aquaria, nitrifying fluidized bed reactors, phosphate-removing biofilms, and activated sludge (8, 12, 15, 17, 42, 43, 49). Although the AOB population varies within and between different WWTP systems, the levels of NOB (i.e., Nitrospira 16S rDNA) were remarkably similar in the municipal activated sludge and the industrial activated sludge (5.82 × 109 to 5.47 × 1010 and 8.14 × 109 to 4.29 × 1010 copies per liter, respectively) (Table 1). In addition, Nitrospira sp. 16S rDNA sequences from the two libraries were 98% similar (data not shown), suggesting that NOB in activated sludge are less diverse and phylogenetically distinct than AOB. Because of the similarity of Nitrospira 16S rDNA this cPCR assay could be applied to different activated sludge systems without further optimization.
The number of rDNA operons per bacterial genome varies from 1 to as many as 15, and it has been suggested that the copy number affects the ability of bacteria to adapt quickly to changing environmental conditions (10, 19). If it was assumed that there was an average of 3.6 copies of 16S rDNA per cell (20), the total cell numbers in samples from both plants calculated by the dot blot hybridization method ranged from 4.55 × 1012 to 1.04 × 1013 cells per liter of suspended solids. When the MLVSS values calculated for the test samples and a conversion value of 2.8 × 10−13 g/cell (29) were used, total cell concentrations between 5.8 × 1012 and 1.2 × 1013 per liter were obtained. Thus, the cell concentrations obtained with the two methods were comparable.
If it was assumed that there were 3.6 copies of 16S rDNA per cell in the total population and two copies of the amoA gene per AOB cell (the number of copies per chromosome in N. europaea [30]), N. oligotropha-like AOB represented 0.0033% ± 0.0022% of the total bacterial population in the municipal MLSS. Unfortunately, the number of 16S rDNA copies per cell of the most closely related Nitrospira species isolated so far, Nitrospira moscoviensis (11), has not been determined. However, only one copy of the rDNA operon is present in the chromosome of a Nitrobacter species that is similar physiologically but not phylogenetically (33). Therefore, if it was assumed that there was one copy of the 16S rDNA gene per NOB cell, Nitrospira spp. represented 0.39% ± 0.28% of the total bacterial population in the municipal WWTP and 0.37% ± 0.23% of the population in the industrial WWTP.
The average ratio of the Nitrospira spp. population to the N. oligotropha-like population was 194.97 ± 133.4 during the 12 months studied for the municipal WWTP (if it was assumed that there were one copy of 16S rDNA per Nitrospira cell and two copies of the amoA gene per AOB cell). Other studies, in which FISH was used to quantify nitrifying populations in a phosphate-removing biofilm and in a nitrifying fluidized bed reactor, indicated that the NOB population was more than 1 order of magnitude larger than the AOB population and could be as much as 30 times larger (12, 42). Another study of the nitrifying bacteria present in a water treatment lagoon also showed that nitrite-oxidizing autotrophs outnumbered the ammonia oxidizers (32). Similarly, in natural aerobic environments, such as water-saturated grassland soils with low nitrite concentrations, the number of lithotrophic NOB was 2 orders of magnitude greater than the number of lithotrophic AOB (7).
Due to the low percentages of the bacterial populations belonging to the AOB and NOB groups in MLSS samples, use of dot blot rRNA or DNA hybridization analysis to study these populations resulted in signals below the detection limit (unpublished data). On the other hand, although quantification of the nitrifying community by FISH analysis has been used successfully for organisms growing in biofilms (42), it is difficult to use this method with activated sludge aggregates. PCR-based quantification, such as cPCR analysis, due to its high analytical sensitivity and precision, is a more appropriate technique for quantifying organisms present at very low levels in activated sludge.
Engineering design, efficient operation, and assessment of alternative process control strategies for nitrification require information about the quantities and activities of AOB and NOB in activated sludge treatment systems. It is important to note that engineering biokinetic parameters for WWTP design models estimated that nitrifying populations accounted for 5 to 8% of the total MLSS (4). Such an estimate, while highly conservative relative to WWTP performance design, can substantially increase capital and operation costs due to overcapacity. In contrast, a more recent model estimated that the autotrophic biomass was negligible, accounting for only 2 to 3% of the total activated sludge mass in municipal wastewater (21). Our values suggest that the amount of autotrophic biomass in municipal WWTP may be even smaller. Given these discrepancies, additional data for nitrifying populations collected by cPCR under a wide range of operational conditions are needed.
Acknowledgments
This work was funded by a research grant from the Water Environment Research Foundation (WERF project 98-CTS-2) and in part by the Waste Management Research and Education Institute of the University of Tennessee. H.M.D. is a recipient of a postdoctoral fellowship from CONICET.
We thank Neil Quigley of the Molecular Biology Resource Facility (University of Tennessee, Knoxville) for DNA sequencing and Arthur Meyers of Eastman Chemical Company (Kingsport, Tenn.) for his thorough review of the manuscript, technical advice, and collection of samples. We also thank Shawn Hawkins for technical assistance, the Knoxville Utility Board for access to the WWTP, and Jack Parker and Brent Wood for collecting samples.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 2.Applegate, B. M., U. Matubutham, J. Sanseverino, and G. S. Sayler. 1995. Biodegradation genes as marker genes in microbial ecosystems, p.1–14. In Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Arthur, J. W., C. W. West, K. N. Allen, and S. F. Hedke. 1987. Seasonal toxicity of ammonia to five fish and nine invertebrate species. Bull. Environ. Contam. Toxicol. 38:324–331. [DOI] [PubMed] [Google Scholar]
- 4.Benefield, L. D., and C. W. Randall. 1985. Biological process design for wastewater treatment. Prentice Hall Inc., Charlottesville, Va.
- 5.Bock, E., and H.-P. Koops. 1992. The genus Nitrobacter and related genera, p.2302–2309. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 6.Bock, E., H.-P. Koops, B. Ahlers, and H. Harms. 1992. Oxidation of inorganic nitrogen compounds as energy source, p.414–430. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 7.Both, G. J. 1990. The ecology of nitrite oxidizing bacteria in grassland soils. Ph.D. thesis. Institute for Ecological Research, Heteren, The Netherlands.
- 8.Burrell, P. C., J. Keller, and L. L. Blackall. 1998. Microbiology of a nitrite-oxidizing bioreactor. Appl. Environ. Microbiol. 64:1878–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler, D. P. 1998. Redefining relativity: quantitative PCR at low template concentrations for industrial and environmental microbiology. J. Ind. Microbiol. Biotechnol. 21:128–140. [Google Scholar]
- 10.Condon, C., D. Liveris, C. Squires, I. Schwartz, and C. L. Squires. 1995. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177:4152–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrich, S., D. Behrens, E. Lebedeva, W. Ludwig, and E. Bock. 1995. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov., and its phylogenetic relationship. Arch. Microbiol. 164:16–23. [DOI] [PubMed] [Google Scholar]
- 12.Gieseke, A., U. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall, G. H. 1986. Nitrification in lakes, p.127–156. In J. I. Prosser (ed.), Nitrification, vol. 20. IRL Press, Oxford, United Kingdom.
- 14.Horz, H., J. Rotthauwe, T. Lukow, and W. Liesack. 2000. Identification of major subgroups of ammonia-oxidizing bacteria in environmental samples by T-RFLP analysis of amoA PCR products. J. Microbiol. Methods 39:197–204. [DOI] [PubMed] [Google Scholar]
- 15.Hovanec, T. A., L. T. Taylor, A. Blakis, and E. F. Delong. 1998. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl. Environ. Microbiol. 64:258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanova I. A., J. R. Stephen, Y.-I. Chang, J. Brüggemann, P. E. Long, J. P. McKinley, G. A. Kowalchuk, D. C. White, and S. J. Macnaughton. 2000. A survey of 16S rRNA and amoA genes related to autotrophic ammonia-oxidizing bacteria of the β-subdivision of the class Proteobacteria in contaminated groundwater. Can. J. Microbiol. 46:1012–1020. [DOI] [PubMed] [Google Scholar]
- 17.Juretschko, S., G. Timmermann, M. Schmid, K.-H. Schleifer, A. Pommerening-Röser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirstein, K., and E. Bock. 1993. Close genetic relationship between Nitrobacter hamburgensis nitrite oxidoreductase and Escherichia coli nitrate reductases. Arch. Microbiol. 160:447–453. [DOI] [PubMed] [Google Scholar]
- 19.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch, G., M. Kühni, and H. Siegrist. 2001. Calibration and validation of an ASM3-based steady-state model for activated sludge systems. Part I. Prediction of nitrogen removal and sludge production. Water Res. 35:2235–2245. [DOI] [PubMed] [Google Scholar]
- 22.Koops, H.-P., B. Böttcher, U. C. Möller, A. Pommerening-Röser, and G. Stehr. 1991. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J. Gen. Microbiol. 137:1689–1699. [Google Scholar]
- 23.Kowalchuk, G. A., Z. S. Naoumenko, P. J. Derikx, A. Felske, J. R. Stephen, and I. A. Arkipchenko. 1999. Molecular analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in compost and composted material. Appl. Environ. Microbiol. 65:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lajoie, C. A., A. C. Layton, I. R. Gregory, G. S. Sayler, D. E. Taylor, and A. J. Meyers. 2000. Zoogleal clusters and sludge dewatering potential in an industrial activated-sludge wastewater treatment plant. Water Environ. Res. 72:56–64. [Google Scholar]
- 25.Lane, D. J. 1991. 16S/23S rRNA sequencing, p.115–175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, England.
- 26.Layton, A. C., P. N. Karanth, C. A. Lajoie, A. J. Meyers, I. R. Gregory, R. D. Stapleton, D. E. Taylor, and G. S. Sayler. 2000. Quantification of Hyphomicrobium populations in activated sludge from an industrial wastewater treatment system as determined by 16S rRNA analysis. Appl. Environ. Microbiol. 66:1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Layton, A. C., B. W. Gregory, J. R. Seward, T. W. Schultz, and G. S. Sayler. 2000. Mineralization of steroid hormones by biosolids in wastewater treatment systems in Tennessee, USA. Environ. Sci. Technol. 34:3925–3931. [Google Scholar]
- 28.Lee, S.-Y., J. Bollinger, D. Bezdicek, and A. Ogram. 1996. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62:3787–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madigan, M. T., J. M. Martinko, and J. Parker. 1997. Brock biology of microorganisms, 8th ed. Prentice-Hall, New York, N.Y.
- 30.McTavish, H., J. A. Fuchs, and A. B. Hooper. 1993. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J. Bacteriol. 175:2436–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendum, T. A., R. E. Sockett, and P. R. Hirsch. 1999. Use of molecular and isotopic techniques to monitor the response of autotrophic ammonia-oxidizing populations of the β subdivision of the class Proteobacteria in arable soils to nitrogen fertilizer. Appl. Environ. Microbiol. 65:4155–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitsuhashi, K., Y. Yoshida, and H. Kadota. 1983. Distribution and activities of nitrifying and denitrifying bacteria in the lagoon with intermittent aeration. Bull. Jpn. Soc. Sci. Fish. 49:1871–1874. [Google Scholar]
- 33.Navarro, E., M. P. Fernandez, F. Grimont, A. Clays-Josserand, and R. Bardin. 1992. Genomic heterogeneity of the genus Nitrobacter. Int. J. Syst. Bacteriol. 42:554–560. [Google Scholar]
- 34.Okabe, S., H. Satoh, and Y. Watanabe. 1999. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65:3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips, C. J., E. A. Paul, and J. I. Prosser. 2000. Quantitative analysis of ammonia-oxidizing bacteria using competitive PCR. FEMS Microbiol. Ecol. 32:167–175. [DOI] [PubMed] [Google Scholar]
- 36.Piatak, M.-C., Jr., K.-C. Luk, B. Williams, and J. D. Lifson. 1993. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. Bio/Techniques 14:70–80. [PubMed] [Google Scholar]
- 37.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rotthauwe, J.-H., K.-P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakano, Y., and L. Kerkhof. 1998. Assessment of changes in microbial community structure during operation of an ammonia biofilter with molecular tools. Appl. Environ. Microbiol. 64:4877–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sayavedra-Soto, L. A., N. G Hommes, and D. J. Arp. 1994. Characterization of the gene encoding hydroxylamine oxidoreductase in Nitrosomonas europaea. J. Bacteriol. 176:504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schramm, A., D. de Beer, J. C. van den Heuvel, S. Ottengraf, and R. Amann. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl. Environ. Microbiol. 65:3690–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schramm, A., D. de Beer, M. Wagner, and R. Amann. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stehr, G., B. Böttcher, P. Dittberner, G. Rath, and H.-P. Koops. 1995. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol. Ecol. 17:177–186. [Google Scholar]
- 45.Stephen, J. R., Y.-J. Chang, S. J. Macnaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suwa, Y., T. Sumino, and K. Noto. 1997. Phylogenetic relationships of activated sludge isolates of ammonia oxidizers with different sensitivities to ammonium sulfate. J. Gen. Appl. Microbiol. 43:373–379. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgens, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 222:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner, M., G. Rath, R. Amann., H.-P. Koops, and K.-H. Schleifer. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251–264. [Google Scholar]
- 49.Wagner, M., G. Rath, H.-P. Koops, J. Flood, and R. Amann. 1996. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci. Technol. 34:237–244. [Google Scholar]
- 50.Zimmermann, K., and J. W. Mannhalter. 1996. Technical aspects of quantitative competitive PCR. Bio/Techniques 21:268–279. [DOI] [PubMed] [Google Scholar]