Abstract
The lantibiotic mersacidin is an antimicrobial peptide of 20 amino acids which inhibits bacterial cell wall biosynthesis by binding to the precursor molecule lipid II and which is produced by Bacillus sp. strain HIL Y-85,54728. The structural gene of mersacidin as well as accessory genes is organized in a biosynthetic gene cluster which is located on the chromosome and contains three open reading frames with similarities to regulatory proteins: mrsR2 and mrsK2 encode two proteins with homology to bacterial two-component systems, and mrsR1 shows similarity to a response regulator. Both mrsR2/K2 and mrsR1 were inactivated by insertion of an antibiotic resistance marker. Disruption of mrsR1 resulted in loss of mersacidin production; however, producer self-protection was not impaired. In contrast, inactivation of mrsR2/K2 led to an increased susceptibility to mersacidin whereas biosynthesis of the lantibiotic remained unaffected. Binding of mersacidin to intact cells was significantly enhanced in the mrsR2/K2 knockout mutant. Reverse transcription-PCR analysis from total RNA preparations showed that in contrast to the wild-type strain, the structural genes of the ABC transporter MrsFGE were not transcribed in the knockout mutant. It was therefore concluded that producer self-protection against mersacidin is conferred by the ABC transporter MrsFGE and that the transcription of mrsFGE is regulated by MrsR2/K2, whereas production of the antibacterial peptide is solely activated by MrsR1.
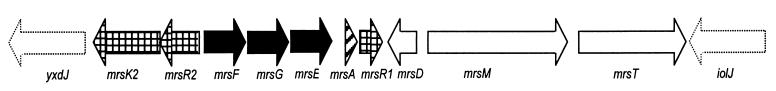
Mersacidin, an antimicrobial peptide of 20 amino acids, is produced by Bacillus sp. HIL Y-85,54728 (7) and belongs to a subgroup of bacteriocins called lantibiotics (reviewed in references 41 and 29). After translation, lantibiotic prepeptides undergo extensive posttranslational modification reactions which result in biosynthesis of several nonproteinogenic amino acids, such as lanthionine and 3-methyllanthionine. These rare amino acids form intrachain thioether bridges and represent the unique structural feature of the lantibiotics (43). Apart from the structural gene, which encodes the prepeptide, the genes involved in modification, proteolytic processing of the prepeptide, and producer self-protection and genes encoding proteins with putative regulatory functions are organized in biosynthetic gene clusters (41). Recently, we reported the complete nucleotide sequence of the 12.3-kb biosynthetic gene cluster of mersacidin which contains, in contrast to all other lantibiotic gene clusters, three putative regulatory genes (1). Two open reading frames at the 5′ end of the cluster, mrsR2 and mrsK2, encode a typical two-component regulatory system (Fig. 1). MrsK2 is a 477-amino-acid protein which shows homology to a histidine kinase, and MrsR2 is an OmpR-like response regulator protein of 240 amino acids (1). Such systems have been found to mediate signal transduction in several bacteriocin gene clusters (20). For example, NisR/NisK and SpaR/SpaK regulate biosynthesis of the lantibiotics nisin and subtilin, respectively, as well as producer self-protection against the antimicrobial peptides, which is called “immunity” (22, 23). A further open reading frame, mrsR1, which is located downstream of the mersacidin structural gene mrsA, encodes another putative single regulatory protein of 213 amino acids with lower similarities to the OmpR family. However, a gene for a corresponding histidine kinase is not present (Fig. 1) (1).
FIG. 1.
Organization of the biosynthetic gene cluster of mersacidin, which is located between the genes yxdJ and iolJ (broken lines) on the chromosome of the producer strain: structural gene (striped arrow), genes necessary for modification and export of mersacidin (white arrows), genes involved in regulation (checkered arrows), and genes for producer self-protection (black arrows).
In contrast to the elongated pore-forming peptides, mersacidin is a globular molecule and acts by binding to the ultimate cell wall precursor lipid II, thereby inhibiting cell wall biosynthesis (4). The mrsFGE operon (Fig. 1), located in the reverse orientation to mrsR2 and mrsK2, encodes three proteins forming a type B ABC (ATP-binding cassette) transporter. Homologous transporters in other lantibiotic producers mediate protective functions (21, 35, 40, 44). In the case of epidermin, the type B ABC transporter EpiFEG confers immunity of the producing organism, most probably by extruding bound peptide from the bacterial membrane (33).
Here we present evidence that MrsR2/K2 exclusively regulate transcription of the mrsFGE operon without affecting the biosynthesis of mersacidin. Moreover, it is shown that transcription of the above operon is necessary for self-protection of the producer towards mersacidin and that binding of mersacidin to lipid II is inhibited in the presence of MrsFGE. The single regulatory protein MrsR1, however, is essential for biosynthesis of mersacidin, but not for the expression of the immunity proteins.
MATERIALS AND METHODS
Bacterial cultures and isolation of DNA.
All strains were maintained as glycerol cultures at −70°C and grown on blood agar or tryptic soy agar. Appropriate antibiotics were added to media used for culturing and selecting mutants. For detection of mersacidin production, the wild-type producer strain Bacillus sp. strain HIL Y-85,54728 and the respective mutants were grown in a synthetic medium, as previously described (2). Antibacterial activities in the supernatant were determined by agar diffusion assays using Micrococcus luteus ATCC 4698 as an indicator organism. Quantitation of mersacidin by reversed-phase high-performance liquid chromatography (RP-HPLC) was performed as previously described (2).
DNA was prepared using QIAgen (Hilden, Germany) genomic tips according to the recommendations of the supplier. General protocols were followed for cloning strategies and enzymatic DNA modifications (42). Digested DNA fragments were eluted from agarose gels employing the QIAquick gel extraction kit (QIAgen).
Inactivation of mrsR2/K2 and mrsR1 by allelic exchange.
Inactivation of putative regulatory genes of the mersacidin gene cluster was performed employing the pTV1ts-derived temperature-sensitive vector pTV0 (1), which carries a chloramphenicol resistance gene. The multiple cloning site of pUC18 was amplified using the primers MCS-5′ and MCS-3′ (Table 1) to introduce blunt-end EcoRV restriction sites on both ends. The EcoRV-digested PCR product was ligated into the PvuII site of pTV0, yielding pTV0[MCS]. Bacillus sp. and Staphylococcus carnosus TM300 were transformed by protoplast transformation (15, 16); Escherichia coli strains were transformed by electroporation.
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| RK1 | 5′-CCTCTGAATTCCCGATCAAATAAACATTCGACCG-3′ |
| RK2 | 5′-ATCTTGGATCCAGAATTTGTCTAAAGAAAAATATGAAG-3′ |
| RK3 | 5′-ATATTCTGCAGTTCCTTGATCGATTTTCAGAAGGG-3′ |
| RK4 | 5′-TTTGCAAGCTTAGATAACCTTACAGTTTCCAATACT-3′ |
| MCS-5′ | 5′-TCCCTGATATCCAGTGAATTCGAGCTCGG-3′ |
| MCS-3′ | 5′-CATCGGATATCGCCAAGCTTGCATGCC-3′ |
| ptv0-2 | 5′-ACCCCTCTTTCCATGTATTCACT-3′ |
| Reko-4.2 | 5′-TTATCTGGAACATCTGTGG-3′ |
| Reko-5 | 5′-CCACGTATTCAGATCGATAAGTAAG-3′ |
| Reko-8 | 5′-ATCCATGGACACAGCCTATGTTAG-3′ |
| RT1 | 5′-AAGTCGGTGTTAGTGACGTAATCG-3′ |
| RT2 | 5′-ATTCCCACTACGATAGGCAATACAA-3′ |
| RT4 | 5′-TGAGTCAAGAAGCTATCATTCGTTC-3′ |
| RT5 | 5′-ATTAACAAATACATTCAGAAGTTAGAGTAC-3′ |
For inactivation of the mrsR2/K2 operon, a resistance cassette was constructed using a 723-bp PCR fragment corresponding to the central part of mrsK2 (primers RK1 and RK2), the erythromycin resistance gene of pUC19E (24), and a 780-bp PCR fragment covering the 5′ end of mrsR2, the intergenic region, and the 5′ end of mrsF (primers RK3 and RK4). Both PCR fragments were cloned downstream and upstream of the erythromycin resistance gene into pUC19E, yielding pUC19RKIEryRKII. In the final step, the whole cassette was cut out with EcoRI-HindIII, ligated into the EcoRI/HindIII-digested vector pTV0[MCS], and transformed into S. carnosus TM300, yielding pRK-In.
For inactivation of mrsR1, a cassette was constructed from the 0.6-kb EcoRV fragment harboring the complete mrsA gene as well as the first 202 nucleotides of the mrsR1 gene, the erythromycin resistance gene of pUC19E, and a 0.9-kb KpnI-EcoRI fragment covering the last 65 nucleotides of the mrsR1 gene, mrsD, and the first 26 nucleotides of mrsM. The KpnI-EcoRI fragment was cloned downstream of the erythromycin cassette into pUC19E, yielding pEry1, and the EcoRV fragment covering mrsA was cloned into the HindIII site (made blunt) of the E. coli-Staphylococcus shuttle vector pBT2 (6), yielding pCS3. Then, the fragment harboring the KpnI-EcoRI fragment and erythromycin resistance gene was cut out from pEry1 with EcoRI/PstI and inserted into pCS3. In the final step, the whole cassette was cut out with EcoRI/EcoRV, cloned into the EcoRI/SmaI-digested pTV0[MCS], and transformed into S. carnosus TM300, yielding pR1-In.
pR1-In and pRK-In were purified in appropriate amounts and transformed into Bacillus sp. strain HIL Y-85,54728 at 30°C. Colonies in which the plasmid had integrated into the chromosome were selected by a temperature shift to 45°C in the presence of erythromycin (25 μg/ml) and chloramphenicol (20 μg/ml). Integration by single crossover was confirmed by PCR using the specific primers Reko-8 (annealing to mrsE) or Reko-5 (annealing to mrsG), respectively, and ptv0-2, which is specific for the vector sequence of pTV0[MCS] (Table 1). Fragments of the predicted length could be amplified for both clones. For the selection of clones that had performed a double crossover, the single-crossover mutants were grown for about 100 generations in tryptic soy broth in the presence of erythromycin (25 μg/ml). Aliquots of the cultures were plated on tryptic soy agar. Single colonies were transferred to agar plates with erythromycin (25 μg/ml) and chloramphenicol (20 μg/ml) as well as to agar plates containing only erythromycin (25 μg/ml). Possible double-crossover mutants, which had lost their capability to grow in the presence of chloramphenicol but still were resistant to erythromycin, were confirmed by PCR with one primer specific for the sequence of the erythromycin gene (Reko-4.2) and the other primer specific for a gene of the mersacidin biosynthetic gene cluster (Reko-5 or Reko-8) as well as by Southern blots with Reko-4.2 as a probe. The strains were designated Bacillus sp. strain TTΔmrsR2/K2 and Bacillus sp. strain TTΔmrsR1, respectively.
Mersacidin immunity assay and MIC determinations.
For determination of strain-specific immunity, the wild-type producer strain Bacillus sp. strain HIL Y-85,54728 and the respective double-crossover mutants were grown to an optical density at 600 nm (A600) of 0.5 in 10 ml of half-concentrated Mueller-Hinton broth. Mersacidin was added in concentrations ranging from 7.5 to 25 μg/ml.
Determination of the MIC of mersacidin was performed by a microtiter plate assay with half-concentrated Mueller-Hinton broth.
Labeling of mersacidin and purification of radiolabeled mersacidin.
Mersacidin was labeled in vitro by a reductive alkylation method as described in reference 11. Briefly, 0.01 mmol of mersacidin was dissolved in 1 ml of methanol and mixed with 9 ml of 50 mM MOPS-100 mM K2SO4, pH 6.5, and 0.015 mmol of [14C]formaldehyde (ICN Pharmaceuticals, Frankfurt, Germany) (58 Ci/mol). NaCNBH3 was added to a final concentration of 10 mM. After 30 min of incubation at room temperature, fresh NaCNBH3 was added to a final concentration of 20 mM, and this was followed by a further incubation for 30 min. The radiolabeled mersacidin was purified by RP chromatography on POROS R2 10 (Applied Biosystems, Weiterstadt, Germany) (2), and the methylation was confirmed by mass spectrometry.
Binding studies employing [14C]dimethyl-mersacidin.
Bacillus sp. strain HIL Y-85,54728 and Bacillus sp. strain TTΔmrsR2/K2 were grown at 30°C in half-concentrated Mueller-Hinton broth. [14C]dimethyl-mersacidin was diluted 1:100 with methylated, unlabeled mersacidin. At an A600 of 0.5, [14C]dimethyl-mersacidin was added to final concentrations of 7.5, 10, 20, and 25 μg/ml. Culture aliquots of 1 ml were taken at 60-min intervals. In order to determine the number of bound mersacidin molecules per cell the aliquots were filtered employing hydrophilic Durapore filters with pore diameters of 0.2 μm (Millipore, Eschborn, Germany). The filters were dried and counted in Quickzint 100 (Zinsser, Frankfurt, Germany) in a model 1900 CA Tri-Carb liquid scintillation counter (Packard, Zurich, Switzerland). For calculation purposes, an A600 of 1.0 was assumed to correspond to 2 × 109 CFU.
Transcription analysis of mrsFGE by RT-PCR.
Isolation of total RNA from Bacillus sp. strain HIL Y-85,54728 or Bacillus sp. strain TTΔmrsR2/K2 was performed with the QIAgen RNeasy total RNA kit. Cultures of both the producer and the knockout mutant were harvested after 8 and 16 h, which correspond to mid-exponential phase and mid-stationary phase, respectively. The cells were lysed by incubation with lysozyme in (20 mg/ml) Tris-EDTA buffer containing RNase Block RNase inhibitor (20 U/ml; Stratagene, La Jolla, Calif.) for 30 min at 37°C. The further procedure was performed as described by the manufacturer. RNA eluates were quantified by determining the E260/E280 quotient in RNase-free water. Before reverse transcription-PCR (RT-PCR) analysis, RNA samples were freed from contaminating DNA by extensive incubation with DNase during RNA purification employing the QIAgen RNase-free DNase set. If contaminating DNA was still present, 0.5 μg of the eluted RNA was digested in 17.55 μl of RDD buffer, which contained 0.2 U of RNase-free DNase I and 0.25 μl of RNase Block RNase inhibitor in a total volume of 20 μl. After 30 min of incubation at 37°C the reaction was halted by addition of 2 μl of 50 mM EDTA and a temperature shift to 65°C for 5 min.
DNase I-pretreated RNA (50 μg) was subjected to RT-PCR with the primers RT-1 and RT-2 amplifying a central 1,217-bp part of the mrsFGE transcript. In order to rule out any inhibitory effect on the RT-PCR, coamplification of a 206-bp fragment of the mrsA transcript was performed employing the primers RT-4 and RT-5. The RT-PCR was carried out in a PCRexpress Thermal Cycler (Hybaid, Heidelberg, Germany) using the QIAgen OneStep RT-PCR kit. After RT for 30 min at 50°C, the samples were heated to 95°C for 15 min in order to inactivate the reverse transcriptase enzyme. Simultaneously, negative controls were performed by introducing parallel samples into the thermal cycler during the 95°C step, thereby ensuring immediate denaturation of the reverse transcriptase enzyme. Subsequently, 30 PCR cycles were performed with 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C. The program was terminated by a 10-min step at 72°C. The samples were analyzed on a 0.8% agarose gel.
DNA sequence determination and analysis.
Double-stranded plasmid DNA and PCR products were sequenced by Sequiserve (Vaterstetten, Germany). Processing of the nucleotide sequence data was performed by using the Blitz program at the EMBL web site (http://www2.ebi.ac.uk/bic) and the Omiga 2.0 software package (Oxford Molecular, Oxford, United Kingdom).
Nucleotide sequence accession number.
The nucleotide sequence of the complete gene cluster of mersacidin is available under accession number AJ250862 at the EMBL database.
RESULTS
Inactivation of mrsR2/K2 and mrsR1.
The two-component system MrsR2/K2 and the single regulatory protein MrsR1 were inactivated by insertion of an erythromycin resistance cassette into the respective genes (Fig. 1), yielding Bacillus sp. strain TTΔmrsR2/K2 and Bacillus sp. strain TTΔmrsR1. The correct insertion of the resistance marker into the genome of Bacillus sp. strain HIL Y-85, 54728 was confirmed by PCR, which revealed amplification products of the expected size (data not shown). Southern blotting of genomic DNA preparations of Bacillus sp. strain TTΔmrsR2/K2 and Bacillus sp. strain TTΔmrsR1, respectively, employing the digoxigenin-labeled oligonucleotide Reko-4.2 as a probe, indicated that single insertion events of the resistance cassette had taken place (data not shown).
Production of mersacidin.
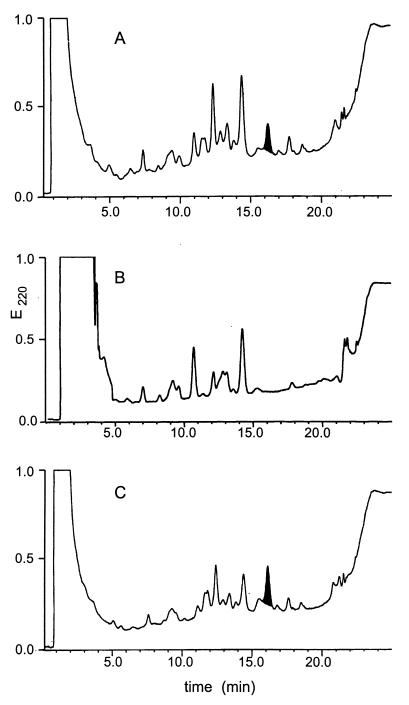
In order to analyze the potential effects of the gene inactivations on mersacidin biosynthesis, supernatants from cultures of Bacillus sp. strain TTΔmrsR2/K2, Bacillus sp. strain TTΔmrsR1, and the wild-type producer strain Bacillus sp. strain HIL Y-85,54728 were analyzed by an agar diffusion assay and RP-HPLC. Surprisingly, inactivation did not affect mersacidin biosynthesis in Bacillus sp. strain TTΔmrsR2/K2, since comparable amounts of the mature lantibiotic could be detected by the activity assay and HPLC in the supernatants of the MrsR2/K2 knockout mutant strain and the wild-type producer strain (Fig. 2). In the case of Bacillus sp. strain TTΔmrsR1, however, neither antibacterial activity exceeding that of the mrsA knockout mutant Bacillus sp. strain HIL Y-85,54728 Rec1 (1) nor a peak containing mersacidin could be detected in the culture supernatant (Fig. 2). These results clearly demonstrate that the two-component system consisting of MrsR2 and MrsK2 is involved neither in the regulation of transcription of the mersacidin structural gene nor of genes involved in posttranslational modification of the mersacidin prepeptide. In contrast, MrsR1 obviously has an essential regulatory function in the biosynthesis of the mature lantibiotic.
FIG. 2.
RP-HPLC of culture supernatants from the wild-type producer strain (A), Bacillus sp. strain TTΔmrsR1 (B), and Bacillus sp. strain TTΔmrsR2/K2 (C). The peak that displayed antibacterial activity and coeluted with mersacidin is marked in black.
Producer self-protection or immunity.
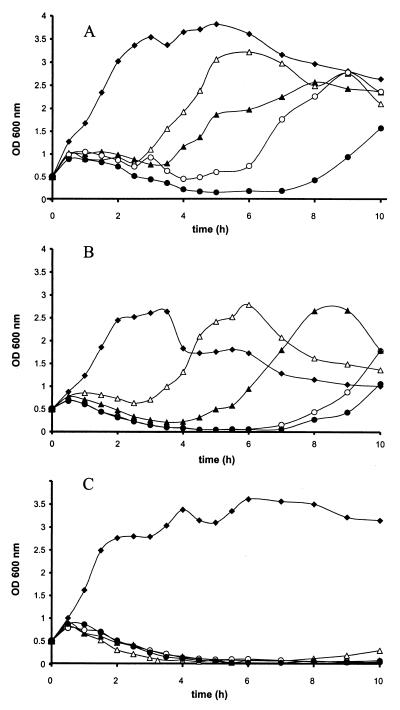
In order to test the susceptibility of the strains to exogenous mersacidin, the wild-type producer strain and the knockout mutants were cultured in half-concentrated Mueller-Hinton broth. When an optical density of 0.5 had been reached, mersacidin was added, and significant differences in susceptibility between the strains could be observed. The wild-type producer strain and Bacillus sp. strain TTΔmrsR1 showed more or less similar growth curves. After a dose-dependent lag time, both cultures were able to resume exponential growth, although the final optical density reached by Bacillus sp. strain TTΔmrsR1 was lower than that of the wild-type strain (Fig. 3A and B). Under the same conditions, almost no growth of Bacillus sp. strain TTΔmrsR2/K2 could be observed (Fig. 3C). Only with the lowest concentration of mersacidin (7.5 μg/ml) was a slight increase of optical density noticed after 8 h of incubation. MIC determinations in half-concentrated Mueller-Hinton broth corroborated the results gained by the immunity assays. While Bacillus sp. strain TTΔmrsR1 and the wild-type producer strain displayed identical MICs (12.5 μg/ml), the respective values resulting from tests with Bacillus sp. strain TTΔmrsR2/K2 were significantly lower (4.6 μg/ml). Therefore, it was concluded that the two-component regulatory system MrsR2/K2 is involved in conferring immunity to the producing cell. Since disruption of the genes did not influence synthesis of mature mersacidin, a separate regulation circuit has to be assumed for transcription of the immunity genes. MrsR1 exclusively takes part in the regulation of peptide biosynthesis and does not affect transcription of the immunity genes.
FIG. 3.
Growth of the wild-type producer strain (A), Bacillus sp. strain TTΔmrsR1 (B), and Bacillus sp. strain TTΔmrsR2/K2 (C) in the presence of mersacidin at concentrations of 7.5 (▵), 10 (▴), 20 (○), and 25 (•) μg/ml; ⧫, control.
Binding studies employing [14C]dimethyl-mersacidin.
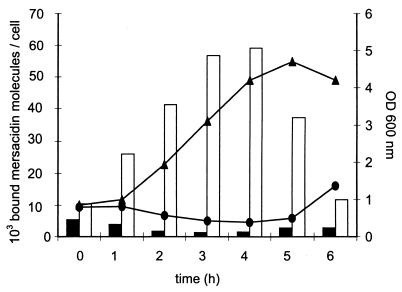
In order to examine the nature of the immunity mechanism involved, the experiment shown in Fig. 3 was repeated with Bacillus sp. strain TTΔmrsR2/K2 and the wild-type producer strain, employing 10 μg of radiolabeled [14C]dimethyl-mersacidin per ml. Again, the mutant strain displayed a higher susceptibility towards mersacidin than the wild-type producer strain (Fig. 4). Enhanced susceptibility was correlated with increased binding of mersacidin to the cells of the MrsR2/K2-knockout mutant. After 3 h of incubation the amount of mersacidin bound by Bacillus sp. strain TTΔmrsR2/K2 was 45-fold higher than the amount bound by the wild-type strain (Fig. 4). It should be noted that the [14C]dimethyl-mersacidin utilized for binding studies was about six times less active than the unmodified mersacidin. Consequently, in this experiment, the mutant cells were able to resume growth at a higher concentration than in presence of the unmodified mersacidin (Fig. 3). Similar results were obtained with 7.5 and 20 μg of [14C]dimethyl-mersacidin per ml (data not shown).
FIG. 4.
Binding of [14C]dimethyl-mersacidin (10 μg/ml) to cells of Bacillus sp. strain TTΔmrsR2/K2 (white bars) and the wild-type strain (black bars) and growth of Bacillus sp. strain TTΔmrsR2/K2 (•) and the wild-type strain (▴) in the presence of [14C]dimethyl-mersacidin (10 μg/ml).
This experiment indicated that the self-protection against mersacidin depends either on an active extrusion of the bound peptide from the bacterial membrane or a shielding mechanism that inhibits the binding of mersacidin to the cells. In Staphylococcus epidermidis Tü 3298, the producer strain of the lantibiotic epidermin, the immunity mechanism is mediated by the type B ABC transporter EpiFEG (33). The ABC transporter MrsFGE of the mersacidin gene cluster displays homology to the epidermin transporter proteins, which makes MrsFGE a likely candidate for conferring immunity to the mersacidin producing cell. Consequently, transcription analyses of mrsFGE were performed in the wild-type producer strain and the mrsR2/K2 knockout mutant.
RT-PCR analysis of the mrsFGE operon.
Previous work had shown that the three genes mrsF, mrsG, and mrsE are transcribed as one operon (1). Therefore, primers were chosen to flank a 1,217-bp RNA region covering the 3′ end of the mrsF gene, the complete mrsG, and the 5′ end of the mrsE gene.
RT-PCRs of total RNA from cultures of Bacillus sp. strain TT ΔmrsR2/K2 and the wild-type strain, harvested after 8 and 16 h, were performed. The amplification of a 206-bp fragment from the mrsA transcript served as a positive control. Furthermore, each RT-PCR sample was prepared with a parallel negative control lacking the RT step to ensure that none of the PCR products was amplified from contaminating genomic DNA. Figure 5 shows that, in contrast to the wild-type strain, no amplification product of the mrsFGE transcript could be detected in Bacillus sp. strain TT ΔmrsR2/K2. Only when the number of PCR cycles had been increased from 30 to 40 rounds of amplification could a very weak signal in the knockout mutant be observed. In contrast, amplification of the 206-bp product of the mrsA transcript serving as a control was observed in both the wild type and Bacillus sp. strain TT ΔmrsR2/K2. These results indicate that transcription of mrsFGE depends on the presence of the two-component system MrsK2/MrsR2 and that producer self-protection towards mersacidin is indeed mediated by the action of MrsF, MrsG, and MrsE, most probably by inhibiting binding of the peptide to lipid II or actively removing bound peptide.
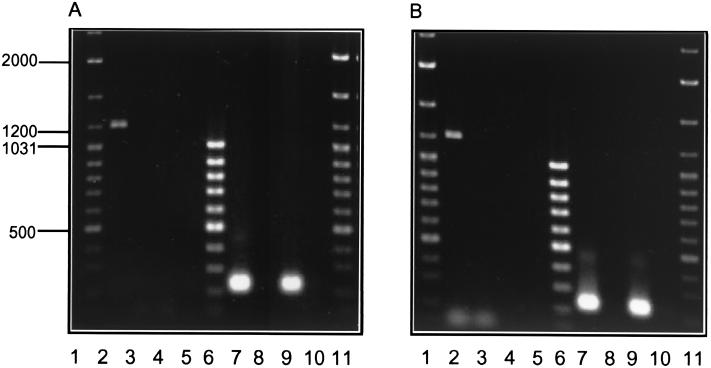
FIG. 5.
RT-PCR of total RNA of Bacillus sp. strain TTΔmrsR2/K2 and the wild-type strain. Cells were harvested after 8 (A) or 16 (B) h of incubation. Slots 3, 5, 8, and 10 contain control reactions for the samples presented in slots 2, 4, 7, and 9, respectively. These controls were performed to rule out contamination with chromosomal DNA by introducing parallel samples into the thermal cycler during the 95°C step, thereby ensuring immediate denaturation of the reverse transcriptase enzyme. Slots 1 and 11, GeneRuler DNA ladder mix; slot 2, RT-PCR of the mrsEFG transcript in the wild-type Bacillus sp. strain HIL Y-85,54728 with RT1/RT2; slot 3, control; slot 4, RT-PCR of the mrsEFG-transcript in Bacillus sp. strain TTΔmrsR2/K2; slot 5, control; 6, GeneRuler 100-bp DNA ladder mix (1.031, 0.9, 0.8, 0.7, 0.6, 0.5, 0.4, 0.3, and 0.2 kb); slot 7, RT-PCR of the mrsA transcript in Bacillus sp. strain HIL Y-85,54728 using RT4/RT5; slot 8, control; slot 9, RT-PCR of the mrsA-transcript in Bacillus sp. strain TTΔmrsR2/K2; slot 10, control.
DISCUSSION
The mersacidin biosynthesis gene cluster harbors the two-component regulatory system MrsR2/K2 as well as a single regulator protein, MrsR1. Only the bacteriocin production of L. plantarum is regulated by a similar complex system which comprises three regulatory proteins, one histidine kinase (PlnB) and two response regulators that function as activator (PlnC) and repressor (PlnD) of the promoters of the entire bacteriocin regulon (10, 10a). Single two-component regulatory systems are frequently involved in biosynthesis of unmodified and modified bacteriocins in gram-positive bacteria (20). They often form part of a regulatory autoinduction circuit which enables bacteria to sense cell-to-cell signals excreted in a cell density-dependent manner, a mechanism commonly referred to as quorum sensing (14, 17). In gram-positive bacteria the signal molecule usually represents a processed and sometimes modified peptide. It is secreted by an ABC transporter and is recognized by the sensor domain of the histidine kinase. The corresponding response regulator activates transcription of genes encoding the regulatory proteins as well as the precursor of the signal peptide, thereby forming an autoregulation loop. This model has been shown to apply to, e.g., the biosynthesis of unmodified antimicrobial peptides like sakacin P (12) as well as to lantibiotics like nisin, subtilin, and salivaricin (23, 45; M. Kleerebezem, W. M. de Vos, and O. P. Kuipers, Abstr. 3rd Int. Workshop Lantibiot. Related Modified Antibiot. Pept., p. 33, 1998). In the case of the lantibiotics mentioned above, the mature antimicrobial peptides function as signal molecules and induce both transcription of genes involved in their biosynthesis and transcription of genes mediating the immunity of the producer strain. In this study, we have shown that MrsR2 exclusively activates transcription of the mrsFGE immunity operon. Since traces of the amplification product of mrsFGE could also be detected in the mutant after increasing the number of PCR cycles, inactivation of the two-component system downregulates rather than shuts off synthesis of mrsFGE RNA.
The production of active mersacidin depended on the presence of an intact MrsR1. It is not yet clear whether MrsR1 has to be phosphorylated since a dedicated kinase is missing. However, the experiments performed above rule out phosphorylation by MrsK2, since production of mersacidin was observed in the MrsR2/K2 knockout mutant. Single regulatory proteins without corresponding histidine kinase have been described for several lantibiotic gene clusters and include EpiQ (34), MutR (37), LasX (M. Skaugen, V. Hoel Christie, and I. F. Nes, Abstr. 3rd Int. Workshop Lantibiot. Related Modified Antibiot. Pept., p. 36, 1998), and LtnR (28). Again, EpiQ and MutR stimulate both biosynthesis of the lantibiotic and transcription of the immunity transporters (35, 37). Although a linkage between transcription of the immunity genes and the biosynthesis genes of mersacidin seems to be meaningful to prevent transcription of the immunity genes in the absence of mersacidin, our experiments suggest two independent regulatory systems, MrsR2/K2 and MrsR1. One other lantibiotic immunity system with a dedicated regulatory protein has been described. In the lacticin 3147 system, LtnR is a transcriptional repressor of the ltnRIFE operon, and by regulating its own synthesis, it keeps up a steady-state level of immunity within the cell (28).
Research on the phospho-relay system regulating initiation of sporulation in Bacillus subtilis led to the discovery of a highly sophisticated cellular network which controls most of the stationary-phase responses such as sporulation, synthesis of degradative enzymes, competence, and biosynthesis of antibiotics (31). In this network most, if not all, regulatory pathways involving two-component systems are interconnected, thereby enabling the cell to exhibit a response based on integration of multiple environmental signals. Analysis of the complete sequence data available for B. subtilis 168 (25) revealed the presence of 36 putative histidine kinases and 34 putative response regulators (13). Mersacidin is produced from the beginning of stationary phase onwards; however, the link between the mersacidin regulatory system and the cellular regulation network of B. subtilis remains to be discovered. The production of classical peptide antibiotics, e.g., tyrocidin and surfactin, is well integrated into the network and controlled by AbrB and ComA/ComS, respectively (26, 32). For the expression of subtilin in B. subtilis 6633, the presence of sigma H, whose activity strongly increases during early stationary phase, seems to be essential (P. Kiesau, S. Borchert, S. Kloess, T. Stein, and K.-D. Entian, Abstr. 3rd Int. Workshop Lantibiot. Related Modified Antibiot. Pept., p. 57, 1998).
The type B ABC transporter encoded by mrsF, mrsG, and mrsE conferred immunity towards exogenous mersacidin and inhibited binding of mersacidin to whole Bacillus cells. Homologous type B ABC transporters are part of the biosynthetic gene clusters of the lantibiotics nisin (44), subtilin (22), epidermin (35), lacticin 3147 (27), streptococcin A FF-22 (30), mutacin II (8), and lacticin 481 (40). For other lantibiotics, single proteins are sufficient to achieve immunity, e.g., CylI, PepI, and EpiI (9, 18, 39). In MIC determinations with mersacidin, Bacillus sp. strain TT ΔmrsR2/K2, and the wild-type producer, the level of immunity in the MrsR2/K2 knockout mutant was decreased to 37% of the wild type. The viability of the producing cells was not affected, nor was there any decrease in mersacidin yields. Thus, the effect of the inactivation of MrsR2/K2, which downregulates expression of mrsFGE, does not seem to be very dramatic in comparison to other lantibiotic systems. For example, after inactivation of NisI in L. lactis, the resulting clone showed only 30% of the wild-type production and 20% of the wild-type immunity (38). The limited effect observed in the mersacidin system may be explained by the very low level transcription of mrsFGE that still took place in the absence of the regulatory system. Additionally, the physiology of production and the mode of action of mersacidin have to be considered. Production of mersacidin occurs during stationary phase, and the yields in our synthetic medium are not very high. The lantibiotic reaches its maximum concentration (25 μg/ml) when the cells are no longer growing, possess relatively little lipid II (4) and, therefore, are less susceptible than during exponential phase when cell wall biosynthesis runs at maximum speed. In consequence, addition of mersacidin to cultures during exponential phase demonstrated the increased susceptibility of the MrsR2/K2 knockout mutant. The lantibiotic producer strains of cationic elongated pore-forming peptides, which possess the LanEFG transporter (for lantibiotics, the collective locus designation lan is used when homologous genes of different gene clusters are referred to), harbor additional membrane-associated immunity proteins such as NisI (24), SpaI (21), and EpiH (19), and in the case of nisin only the coexpression of nisFEG, nisI, and nisABTC confers full immunity (38). Similar results have been reported for epidermin, for which the highest level of immunity was conferred by coexpression of epiFEG, epiH, and epiT (19). From these data, it seems that the pore-forming peptides, which often are produced during exponential growth and which possess a more efficient mode of action, need a more complex immunity system than mersacidin, which inhibits cell wall biosynthesis. In the case of nisin this system most probably is organized in a complex with the biosynthetic system (38).
Otto et al. (33) showed that cells which express the epidermin transporter EpiFGE are able to prevent binding of gallidermin to their membrane and, by actively extruding the peptide, maintain a concentration of gallidermin in the culture supernatant that is four times higher than that of the control. In this context it should be noted that pore formation by epidermin-gallidermin and nisin is mediated via binding to the docking molecule lipid II (5). Only in the presence of lipid II, can efflux from vesicles be seen with nanomolar nisin concentrations, i.e., in the range of the MIC (5). However, the interaction with lipid II is not essential. When micromolar concentrations are employed, release of carboxyfluorescein has been observed with nisin in the absence of lipid II in vitro (3). Thus, mersacidin, epidermin, gallidermin, and nisin possess LanEFG immunity systems and make use of the identical target molecule, although nisin and epidermin use lipid II primarily for pore formation, whereas mersacidin acts only by inhibition of cell wall biosynthesis. In contrast, the lantibiotic Pep5 forms pores but does not utilize lipid II as a docking molecule (5) and possesses a single immunity protein that does not show any homology to transport proteins (39). This leads to the question whether the LanEFG systems act by detaching lantibiotics from lipid II or by extrusion from the outer leaflet of the membrane. Our experiments revealed that binding of mersacidin was significantly decreased in the presence of the MrsFGE transporter system. Expression of an MrsFGE transporter with a truncated nonfunctional MrsE did not increase the immunity of the respective B. subtilis W23 clone, indicating that a functional transporter is necessary (1). Since labeled mersacidin does not bind to phosphatidylcholine liposomes in the absence of lipid II (4), an interaction of MrsFGE with mersacidin bound to lipid II is more likely. Homology (45.8% identical amino acids) of MrsF to BcrA of the bacitracin immunity transporter BcrABC, for which an active detaching of bacitracin from its target molecule, lipid I, was reported, further supports this hypothesis (36). In the case of epidermin-gallidermin, the protective effect mediated by EpiFGE was most pronounced when concentrations about 20 times the MIC were employed (33), a concentration level that would probably allow pore formation in the absence of lipid II. Therefore, it is not yet clear whether the substrate of EpiFGE was the membrane-bound gallidermin, gallidermin bound to lipid II, or both. In conclusion, further research into lantibiotic mode of action and immunity will show whether the immunity transporter LanEFG is specific for peptides that bind to lipid II and elucidate its mechanism of action.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Bi 504/1–3) and the BONFOR program of the Medizinische Einrichtungen, University of Bonn.
We gratefully acknowledge R. Jack for mass spectrometry of methylated mersacidin, K. Altena for providing pUC19RKIEryRKII, and C. Szekat for expert technical assistance and thank Aventis Pharma AG, Frankfurt am Main, Germany, for making the mersacidin producer available.
REFERENCES
- 1.Altena, K., A. Guder, C. Cramer, and G. Bierbaum. 2000. Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl. Environ. Microbiol. 66:2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bierbaum, G., H. Brötz, K. P. Koller, and H. G. Sahl. 1995. Cloning, sequencing and production of the lantibiotic mersacidin. FEMS Microbiol. Lett. 127:121–126. [DOI] [PubMed] [Google Scholar]
- 3.Breukink, E., C. van Kraaij, R. A. Demel, R. J. Siezen, O. P. Kuipers, and B. de Kruijff. 1997. The C-terminal region of nisin is responsible for the initial interaction of nisin with the target membrane. Biochemistry 36:6968–6976. [DOI] [PubMed] [Google Scholar]
- 4.Brötz, H., G. Bierbaum, K. Leopold, P. E. Reynolds, and H. G. Sahl. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brötz, H., M. Josten, I. Wiedemann, U. Schneider, F. Götz, G. Bierbaum, and H. G. Sahl. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317–327. [DOI] [PubMed] [Google Scholar]
- 6.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1–8. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee, S., S. J. Lad, M. S. Phansalkar, R. H. Rupp, B. N. Ganguli, H. W. Fehlhaber, and H. Kogler. 1992. Mersacidin, a new antibiotic from Bacillus. Fermentation, isolation, purification and chemical characterization. J. Antibiot. (Tokyo) 45:832–838. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P., F. Qi, J. Novak, and P. W. Caufield. 1999. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl. Environ. Microbiol. 65:1356–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coburn, P. S., L. E. Hancock, M. C. Booth, and M. S. Gilmore. 1999. A novel means of self-protection, unrelated to toxin activation, confers immunity to the bactericidal effects of the Enterococcus faecalis cytolysin. Infect. Immun. 67:3339–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diep, D. B., L. S. Havarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Diep, D. B., O. Johnsborg, P. A. Risøen, and I. F. Nes.2001. Evidence for dual functionality of the operon plnABCD in the regulation of bacteriocin production in Lactobacillus plantarum. Mol. Microbiol. 41:633–644. [DOI] [PubMed] [Google Scholar]
- 11.Dottavio-Martin, D., and J. M. Ravel. 1978. Radiolabeling of proteins by reductive alkylation with [14C]formaldehyde and sodium cyanoborohydride. Anal. Biochem. 87:562–565. [DOI] [PubMed] [Google Scholar]
- 12.Eijsink, V. G., M. B. Brurberg, P. H. Middelhoven, and I. F. Nes. 1996. Induction of bacteriocin production in Lactobacillus sake by a secreted peptide. J. Bacteriol. 178:2232–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Götz, F., and B. Schumacher. 1987. Improvement of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40:285–288. [Google Scholar]
- 16.Grosch, J. C., and K. L. Wollweber. 1982. Transformation of Bacillus licheniformis and Bacillus amyloliquefaciens protoplasts by plasmid DNA, p.97–105. In U. N. Streips, H. S. Goodgal, W. R. Guild, and G. A. Wilson (ed.), Genetic exchange. Marcel Dekker, New York, N.Y.
- 17.Hastings, J. W., and E. P. Greenberg. 1999. Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J. Bacteriol. 181:2667–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidrich, C., U. Pag, M. Josten, J. Metzger, R. W. Jack, G. Bierbaum, G. Jung, and H. G. Sahl. 1998. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl. Environ. Microbiol. 64:3140–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hille, M., S. Kies, F. Götz, and A. Peschel. 2001. Dual role of gdmh in producer immunity and secretion of the staphylococcal lantibiotics gallidermin and epidermin. Appl. Environ. Microbiol. 67:1380–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleerebezem, M., L. E. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895–904. [DOI] [PubMed] [Google Scholar]
- 21.Klein, C., and K. D. Entian. 1994. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 60:2793–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein, C., C. Kaletta, and K. D. Entian. 1993. Biosynthesis of the lantibiotic subtilin is regulated by a histidine kinase/response regulator system. Appl. Environ. Microbiol. 59:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuipers, O. P., M. M. Beerthuyzen, P. G. de Ruyter, E. J. Luesink, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299–27304. [DOI] [PubMed] [Google Scholar]
- 24.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281–291. [DOI] [PubMed] [Google Scholar]
- 25.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256. [DOI] [PubMed] [Google Scholar]
- 26.Marahiel, M. A., P. Zuber, G. Czekay, and R. Losick. 1987. Identification of the promoter for a peptide antibiotic biosynthesis gene from Bacillus brevis and its regulation in Bacillus subtilis. J. Bacteriol. 169:2215–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAuliffe, O., C. Hill, and R. P. Ross. 2000. Identification and overexpression of ltnI, a novel gene which confers immunity to the two-component lantibiotic lacticin 3147. Microbiology 146:129–138. [DOI] [PubMed] [Google Scholar]
- 28.McAuliffe, O., T. O’Keeffe, C. Hill, and R. P. Ross. 2001. Regulation of immunity to the two-component lantibiotic, lacticin 3147, by the transcriptional repressor LtnR. Mol. Microbiol. 39:982–993. [DOI] [PubMed] [Google Scholar]
- 29.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285–308. [DOI] [PubMed] [Google Scholar]
- 30.McLaughlin, R. E., J. J. Ferretti, and W. L. Hynes. 1999. Nucleotide sequence of the streptococcin A-FF22 lantibiotic regulon: model for production of the lantibiotic SA-FF22 by strains of Streptococcus pyogenes. FEMS Microbiol. Lett. 175:171–177. [DOI] [PubMed] [Google Scholar]
- 31.Msadek, T. 1999. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 7:201–207. [DOI] [PubMed] [Google Scholar]
- 32.Msadek, T., F. Kunst, and G. Rapoport. 1995. A signal transduction network in Bacillus subtilis includes the DegS/DegU and ComP/ComA two-component systems, p.447–471. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 33.Otto, M., A. Peschel, and F. Götz. 1998. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tü3298. FEMS Microbiol. Lett. 166:203–211. [DOI] [PubMed] [Google Scholar]
- 34.Peschel, A., J. Augustin, T. Kupke, S. Stevanoviç, and F. Götz. 1993. Regulation of epidermin biosynthetic genes by EpiQ. Mol. Microbiol. 9:31–39. [DOI] [PubMed] [Google Scholar]
- 35.Peschel, A., and F. Götz. 1996. Analysis of the Staphylococcus epidermidis genes epiF, -E, and -G involved in epidermin immunity. J. Bacteriol. 178:531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Podlesek, Z., A. Comino, B. Herzog-Velikonja, D. Zgur-Bertok, R. Komel, and M. Grabnar. 1995. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 16:969–976. [DOI] [PubMed] [Google Scholar]
- 37.Qi, F., P. Chen, and P. W. Caufield. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ra, R., M. M. Beerthuyzen, W. M. de Vos, P. E. Saris, and O. P. Kuipers. 1999. Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145:1227–1233. [DOI] [PubMed] [Google Scholar]
- 39.Reis, M., M. Eschbach-Bludau, M. I. Iglesias-Wind, T. Kupke, and H. G. Sahl. 1994. Producer immunity towards the lantibiotic Pep5: identification of the immunity gene pepI and localization and functional analysis of its gene product. Appl. Environ. Microbiol. 60:2876–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rincé, A., A. Dufour, P. Uguen, J. P. Le Pennec, and D. Haras. 1997. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 63:4252–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahl, H. G., and G. Bierbaum. 1998. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu. Rev. Microbiol. 52:41–79. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schnell, N., G. Engelke, J. Augustin, R. Rosenstein, V. Ungermann, F. Götz, and K. D. Entian. 1992. Analysis of genes involved in the biosynthesis of lantibiotic epidermin. Eur. J. Biochem. 204:57–68. [DOI] [PubMed] [Google Scholar]
- 44.Siegers, K., and K. D. Entian. 1995. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61:1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Upton, M., J. R. Tagg, P. Wescombe, and H. F. Jenkinson. 2001. Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J. Bacteriol. 183:3931–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]