Abstract
The Runx2 (Cbfa1/AML3) transcription factor and matrix metalloproteinase 9 (MMP9) are key regulators of growth plate maturation and bone formation. The genes for both proteins are characteristic markers of breast and prostate cancer cells that metastasize to bone. Here we experimentally addressed the compelling question of whether Runx2 and MMP are functionally linked. By cDNA expression array analysis, we identified MMP9 as a novel downstream target of Runx2. Like that of MMP13, MMP9 expression is nearly depleted in Runx2 mutant mice. Chromatin immunoprecipitation and electrophoretic mobility shift assays revealed the recruitment of Runx2 to the MMP9 promoter. We show by mutational analysis that the Runx2 site mediates transactivation of the MMP9 promoter in osteoblasts (MC3T3-E1) and nonosseous (HeLa) cells. The overexpression of Runx2 by adenovirus delivery in nonmetastatic (MCF-7) and metastatic breast (MDA-MB-231) and prostate (PC3) cancer cell lines significantly increases the endogenous levels of MMP9. The knockdown of Runx2 by RNA interference decreases MMP9 expression, as well as that of other Runx2 target genes, including the genes for MMP13 and vascular endothelial growth factor. Importantly, we have demonstrated using a cell invasion assay that Runx2-regulated MMP9 levels are functionally related to the invasion properties of cancer cells. These results are consistent with Runx2 control of multiple genes that contribute to the metastatic properties of cancer cells and their activity in the bone microenvironment.
Runx2 (Cbfa1/AML3) is a member of the runt family of transcription factors and is a key regulator of bone development that is requisite for the maturation of hypertrophic chondrocytes and osteoblasts (4, 18). Mutation of the Runx2 gene is associated with human cleidocranial dysplasia (41). Runx2 null mice die at birth with the absence of a mineralized skeleton (11, 29). While the requirement of Runx2 for osteoblast differentiation is well documented, Runx2 is also expressed at significant levels in nonosseous tissues, but its function there is unknown. These tissues include the testes (30, 37), mammary epithelium (25), thymus (49), endothelial cells (55), and transformed cells (9). Furthermore, elevated Runx2 expression has been reported for breast and prostate tumors and for cancer cell lines that metastasize to the bone environment (6, 62). Runx2 is associated with growth control, and Runx2-deficient cells show increased proliferation compared to wild-type cells (21, 44). The family of Runx proteins functions in both cell growth and differentiation. Runx1 is required for hematopoiesis (39, 61), Runx3 is required for nerve cell development (31), and mutations of Runx1 and Runx3 are associated with leukemia (39) and gastric cancer (32), respectively. Thus, the Runx genes, which encode essential transcription factors for organogenesis, are all associated with various types of cancer (8).
The identification of Runx2 target genes can provide insight into the mechanisms by which Runx2 functions in metastatic cancer cells as well as in normal cellular differentiation. Importantly, the expression of C-terminally deleted Runx2 in vivo disrupted normal skeletal development (11, 17), and in metastatic breast cancer lines, prevented tumor-mediated osteolysis in the bone environment (5). Studies have shown that metastatic breast cancer cell lines carrying mutations in the Runx2 protein that inhibit its normal functional activity in subnuclear domains result in blocking of the osteolytic lesions induced by the parental metastatic cell (5, 26). For this study, we have examined the mechanisms by which Runx2 may contribute to the metastatic properties of cancer cells by identifying novel target genes from a comparison of the expression profiles of genes present in bone tissue from wild-type mice and in tissue from Runx2ΔC/ΔC mice, whose Runx2 lacks C-terminal functions and results in embryonic lethality and the absence of a mineralized skeleton (11).
We have identified a novel Runx2 target gene, the gene for matrix metalloproteinase 9 (MMP9), which is well characterized for its activity during metastasis (13, 19). The MMPs are a family of extracellular matrix-degrading enzymes that share common functional domains and activation mechanisms. These enzymes are synthesized as secreted transmembrane proenzymes and processed to the active form by removal of an amino-terminal propeptide. The MMPs are endopeptidases that are regulators of cell growth, migration, and extracellular matrix remodeling and are expressed in both bone-forming osteoblasts and bone-resorbing osteoclasts (40). MMP13 is expressed in osteoblastic cells and has been identified as a Runx2 target gene (23, 43, 52). MMP9 is highly expressed in monocytes (osteoclast precursors) and in multinucleated osteoclasts that resorb bone (46, 59). Osteoclasts that resorb calcified cartilage at the growth plate are particularly enriched in MMP9 at the ossification front (58). Genetic models of MMP13 and MMP9 deficiency exhibit skeletal defects resulting from impaired enzyme activity (40). Stimulated endothelial cells also express MMP9 in response to cytokines (3) and shear stress (35) during development and after birth. Thus, both MMP9 and Runx2 are expressed in a variety of cell types.
MMPs have long been associated with cancer cell invasion and metastasis (19). For three decades, MMPs have been projected as potential targets for cancer therapy based on their up-regulation in virtually all human tumors and their ability to degrade all components of the extracellular matrix. The effects of MMP suppression in tumor models were so compelling in preclinical studies that synthetic metalloproteinase inhibitors of enzyme activity were rapidly developed and tested in human clinical trials. The results of these trials, however, have been disappointing (13). Thus, a better understanding of the regulatory mechanisms that control MMP transcription in cancer cells could provide new avenues for therapeutic intervention that are more specific and effective.
Here we report that MMP9 is a direct target of Runx2 in bone tissue and in metastatic breast and prostate cells. Several lines of evidence are presented to establish a Runx2 regulatory element in the proximal promoter and a definitive role for Runx2 in MMP9-induced expression in cancer cells. We further show that the modulation of Runx2 activity by either forced expression or RNA interference is directly correlated with MMP9 expression and the invasive properties of metastatic cancer cells. Thus, our studies suggest a regulatory linkage between Runx2, the expression of metastatic markers (MMP9 and MMP13), and the invasive behavior of cancer cells.
MATERIALS AND METHODS
Cell culture, transient transfection, and plasmids.
Mouse osteoblastic MC3T3-E1 cells and the nonmetastatic MCF-7 and metastatic MDA-MB-231 human breast cancer cell lines were cultured in alpha minimal essential medium (α-MEM) containing 10% fetal bovine serum (FBS; Invitrogen Inc., CA). Prostate cancer PC3 cells (a gift from L. Chung, University of Virginia, Charlottesville, Virginia) were maintained in T-medium supplemented with 5% FBS. Human cervical carcinoma (HeLa) cells were cultured and maintained in Dulbecco's modified Eagle's medium supplemented with 5% FBS. HeLa or MC3T3 cells were transfected at 50% confluence with 200 ng of promoter reporter and 0.2 to 1.6 μg of Runx2 expression plasmid using Superfect reagent (QIAGEN, Valencia, CA). Cells were harvested after 48 h of transfection or viral infection and assayed for luciferase reporter activity. Experiments were performed in triplicate at least three times. A cytomegalovirus promoter-driven Runx2 expression vector and an adenovirus expressing wild-type Runx2 were reported previously (2). Mouse MMP9 promoter fragments (from −1.3 kbp to +1 bp and −250 bp to +1 bp) were PCR amplified from MC3T3 genomic DNA using the forward primers 5′-TATCGGGGTACCGAGAGTTTTGTAGAGAGCGTA-3′ (1.3 kb) and 5′-TATCGGGGTACCGTCTGGGGGTCCTGCCTGACTTGG-3′ (−250 bp) and the common reverse primer 5′-ATACCGCTCGAGTGGCTAACGCTGCCTTTGCAGA-3′, which contains an XhoI site (in italics). PCR products were doubly digested with KpnI and XhoI and cloned upstream of the luciferase gene in the pGL2 vector. A 2-bp substitution mutation (boldface and lowercase) was introduced into the Runx binding element (boldface) using the following primers: 5′-TATCGGGGTACCTTTGCAGAAACTAAACCCTGAGTTCTGTacTTTCCTGTG-3′ for the fragment starting at −250 bp and 5′-ACAAAGTCTGCAGTTTGCAGAAACTAAACCCTGAGTTCTGTacTTTCCTGTG-3′ for the full-length MMP9 promoter. The incorporation of the mutation was confirmed by automated sequencing. Plasmid constructs containing the mouse MMP9 promoters fused to the luciferase coding sequence were transfected as described previously (27).
Zymography and Western blot analysis.
MMP9 activity was analyzed with gelatin zymograms as described previously (45). Bone tissue extracts, cell lysates, and culture medium were diluted in 50 mM Tris-HCl, pH 7.4, without reducing agent and separated by electrophoresis in 7.5% sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE) containing 2 mg/ml gelatin. After electrophoresis, the gels were washed in 2.5% Triton X-100 for 30 min and then incubated for 16 h at 37°C in 50 mM Tris-HCl, pH 7.4, 200 mM NaCl, 10 mM CaCl2. Gels were then stained with Coomassie brilliant blue R-250 and destained with 40% methanol-10% acetic acid until clear bands appeared. Images were captured with an AlphaImager gel documentation system (Alpha Innotech, San Leandro, CA). For Western blot analysis, whole-cell lysates were mixed with Laemmli buffer under reducing conditions and separated by 10% SDS-PAGE as described previously (11). Proteins were transferred to a polyvinylidene difluoride membrane and incubated with either a 1:200 dilution of rabbit polyclonal MMP9 antibody (Abcam Inc., Cambridge, MA) or a 1:2,000 dilution of mouse monoclonal Runx2 antibody (2), followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz, CA). Immunoreactive proteins were detected using an enhanced chemiluminescence kit (Pierce, Rockford, IL).
Osteogenesis expression array.
The total RNA (5 μg) isolated from craniofacial skeletons of wild-type or Runx2ΔC/ΔC embryos (at 17.5 days postcoitum [dpc]) was subjected to a mouse osteogenesis cDNA expression array (SuperArray Bioscience Corporation, Frederick, MD) to examine the expression levels of osteogenic markers according to the manufacturer's protocol.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (22). Briefly, formaldehyde cross-linking was performed for 10 min in rat osteoblastic cells, and samples were sonicated to obtain DNA fragments with an average size of 0.3 kb. Protein-DNA complexes were immunoprecipitated using Runx2 antibody (M-70; Santa Cruz) or immunoglobulin G (IgG) as a control. DNAs were purified and subjected to PCR amplification. The MMP9 promoter fragment (−380 to −130 bp) containing the Runx element was amplified using the forward primer 5′-CTCAGAAGCCCAAGGAAGAGT-3′ and the reverse primer 5′-CACTACCCACTCCTTTATGCCC-3′. Each PCR was performed using 10% (3 μl) of the bound DNA fraction from the chromatin precipitate or 1% (1 μl) of the input fraction. The PCR products were separated in 2.0% agarose gels containing 0.5 μg/ml ethidium bromide. DNA bands were visualized using UV light and recorded with an AlphaImager gel documentation system. All experiments were repeated at least three times, and representative results are presented in the figures.
Immunostaining.
Endogenous MMP9 was detected in bone tissues from wild-type or Runx2ΔC/ΔC embryos at 17.5 dpc. Embryos were fixed in paraformaldehyde and embedded in paraffin as described previously (64). Heads were serially sectioned at 8 μm for immunolabeling with a 1:800 dilution of mouse monoclonal MMP9 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at room temperature, followed by incubation with a 1:800 dilution of fluorescein isothiocyanate-conjugated secondary antibody (Molecular Probes, Eugene, OR) for 30 min. Sections were further stained with DAPI (4′,6′-diamidino-2-phenylindole) to stain nuclei, and images were captured with an epifluorescence Axioplan 2 microscope (Zeiss, Inc., Thorwood, NY).
Electrophoretic mobility shift analysis.
Nuclear extracts were prepared from osteoblastic ROS 17/2.8 cells or primary rat osteoblasts cultured for 12 and 20 days according to the method of Dignam et al. (15). Aliquots of supernatant enriched with nuclear proteins were quick-frozen in a dry ice-ethanol bath and stored at −80°C until further use. The following complementary oligonucleotides representing the wild-type (WT) and mutant (MT) Runx binding elements of the rat MMP9 promoter were synthesized: WT, ACCCTGAGTTCTGTGGTTGTTTCCTGTGGGTCT; and MT, ACCCTGAGTTCTGTacTTTCCTGTGGGTCT (lowercase nucleotides were mutated). The plus strand (10 pmol) was labeled with [γ-32P]ATP for 1 h at 37°C with T4 polynucleotide kinase (New England Biolabs, Beverly, MA). The Runx binding cassette was generated by annealing with a threefold excess amount of the minus strand. The unincorporated nucleotides were removed using a quick-spin G-25 Sephadex column (Roche Molecular Biochemicals, Indianapolis, IN). DNA-protein binding reactions were carried out using 10 fmol of radiolabeled probe and 4 to 8 μg of nuclear extract at room temperature for 20 min. Protein-DNA complexes were separated in a 6.5% (40:0.5) nondenaturing polyacrylamide-Tris-borate-EDTA gel. For immunoshift analysis, 200 ng of Runx2 antibody was incubated with nuclear extract at 22°C for 30 min prior to the addition of probe. The samples were electrophoresed at 200 V for 3 h at 4°C. Gels were dried and subjected to autoradiography at −70°C.
siRNA treatment and adenoviral infection.
Breast and prostate cancer cells were transfected with small interfering RNA (siRNA) duplexes at 30 to 50% confluence using Oligofectamine (Invitrogen Life Technologies). siRNAs specific for human Runx2 [r(CUCUGCACCAAGUCCUUUU)d(TT), r(UGCCUCUGCUGUUAUGAAA)d(TT), r(UCUGUUUGGCGACCAUAUU)d(TT), andr(GGUUCAACGAUCUGAGAUU)d(TT)] were obtained from QIAGEN Inc. (Stanford, CA). Cells were transfected with either Runx2 or control siRNA duplexes specific for green fluorescent protein (GFP) at concentrations of 25 and 50 nM. For optimal transfection, a reduced serum medium (Opti-MEM) was used to dilute the siRNA duplexes and Oligofectamine per the manufacturer's instructions. After 4 h, the cells were supplemented with α-MEM containing 30% FBS for a final concentration of 10% in the medium. The siRNA treatment was carried out for 72 h, after which cells were harvested to isolate total protein and RNA. A knockdown of endogenous Runx2 and its effect on metastatic markers were examined by real-time quantitative PCR (QPCR). For Runx2 overexpression, cells were transduced with an adenovirus (multiplicity of infection [MOI], 10) expressing either Runx2 or a LacZ control for 1.5 h with gentle shaking in medium supplemented with 1% FBS as previously described (2). Cells were washed twice with serum-free medium after viral infection and cultured in regular medium for another 48 h before being harvested.
Real-time reverse transcription-PCR (RT-PCR) analysis.
The expression levels of MMP2, MMP9, MMP13, and vascular endothelial growth factor (VEGF) in PC3, MCF-7, and MDA-MB-231 cells were analyzed after Runx2 siRNA treatment or adenovirus transduction. Total RNA was isolated using Trizol reagent according to the manufacturer's specifications. RNA was primed with oligo(dT), and cDNAs were synthesized at 42°C with SuperScript II RNA polymerase (Invitrogen). The cDNA was then used for real-time PCR with TaqMan chemistry (Applied Biosystems, Inc., Foster City, CA). Amplicon quantities relative to a standard curve generated from serial dilutions of pooled cDNAs from all samples were determined using ABI Prism software (ABI7900 HT sequence detection system). For PCR amplification, the following primers were used: MMP2 forward, 5′-TAGGCCATAGCAGACG-3′; MMP2 reverse, 5′-TTAAGGCCCATATCAGG-3′; MMP9 forward, 5′-ATAGACTACTACAGGCT-3′; MMP9 reverse, 5′-TAGCACGGATAGACCA-3′; MMP13 forward, 5′-ATGAGCCAGAGTGTCGGTTC-3; MMP13 reverse, 5′-GTTAGTAGCGACGAGCAGGAC-3′; VEGF forward, 5′-AGTTAGTCACACT GGAGATTGAC-3′; and VEGF reverse, 5′-ATAGGATCGACAGTTGTAACC-3′.
Invasion assay.
Cells were analyzed for invasion/migration through Matrigel (BD Biosciences) according to the manufacturer's protocol. Briefly, cells were placed in Matrigel inserts or control inserts at 1 × 105 cells/ml in serum-free medium and were allowed to migrate for 20 h at 37°C. Nonmigrating cells were removed from the top of the filter by scrubbing with a cotton swab. Cells that migrated were fixed and stained with a Hema 3 kit (Fisher Chemicals). The numbers of cells that migrated to the bottom side of the insert were counted manually and are presented as percentages of invasion.
RESULTS
MMP9 expression is lost in bone tissue of Runx2-deficient mice.
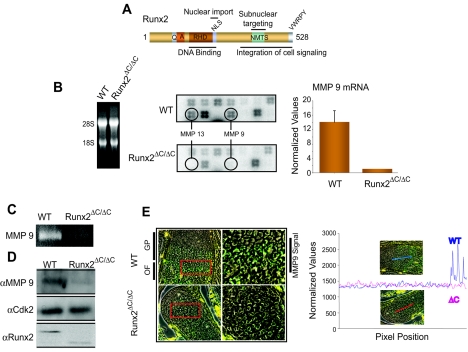
To characterize downstream targets and understand the pathways where Runx2 is involved, we performed a limited gene expression profiling analysis using an osteogenic cDNA array (see Materials and Methods) with total RNAs from bone tissue of WT and Runx2ΔC/ΔC mice. We identify MMP9 as a novel target of Runx2 (Fig. 1B). The absence of MMP13, a known target of Runx2 (52), from the expression profile of the Runx2ΔC/ΔC mouse validated the hypothesis that the differences observed between the WT and Runx2 mutant bone samples reflect biological consequences. These changes were observed in replicate samples and further confirmed by RT-PCR analysis (Fig. 1B). The expression of MMP9 was significantly downregulated in Runx2ΔC/ΔC mice compared to wild-type littermates. A zymographic analysis of gelatinase activity (Fig. 1C) in total protein extracts from the long bones of wild-type and Runx2ΔC/ΔC mice showed a significant decrease in pro-MMP9 activity. These results are consistent with the decreased MMP9 protein levels in Runx2ΔC/ΔC mice relative to wild-type littermates (Fig. 1D).
FIG. 1.
MMP9 (also called gelatinase B) is a novel downstream target of Runx2. (A) Schematic illustration of domain organization of Runx2. QA, polyglutamate and -alanine stretch; RHD, runt homology domain; NLS, nuclear localization signal; NMTS, nuclear matrix targeting signal. (B) Total RNA (5 μg) from wild-type (WT) and Runx2ΔC/ΔC mutant mice (at 17.5 dpc) was hybridized to an osteogenesis-related cDNA array (left panel). The signals of MMP13 and -9 on blots are indicated in circles (middle panel). The MMP9 mRNA levels (normalized to glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) in WT and Runx2ΔC/ΔC mutant mice were detected by RT-QPCR analysis (right panel). Protein extracts from the long bones of WT and Runx2ΔC/ΔC littermates were resolved by 8% SDS-PAGE with 2% gelatin and processed for zymography (C) and Western blotting (D) as described in Materials and Methods. (E) Immunolocalization of MMP9 in wild-type and Runx2ΔC/ΔC mutant embryos (at 17.5 dpc). Embryos were fixed in paraformaldehyde and embedded in paraffin, and the heads were serially sectioned at 8 μm. The left panels show MMP9 staining of the growth plate (GP) area of exoccipital bone. The ossification front (OF) is further magnified in the right panels showing the MMP9 signal. (The image was generated by overlaying the MMP9 image with an image that reflects sample autofluorescence. The MMP9 signal is shown in green, the background is shown in red, and autofluorescence is shown in yellow). Quantitation of the MMP9 signal in WT and Runx2 mutant tissue sections is shown by line scanning (the position is indicated in the micrograph) using Metamorph software, which measures the intensity of the fluorescent signal.
To examine the tissue distribution of MMP9 in bones of WT and Runx2 mutant animals, we performed in situ immunofluorescence analysis using an MMP9 antibody on sections of day 17.5 embryos (Fig. 1E). The developing exoccipital bone showed abundant MMP9 expression in the growth plate region, where MMP9 has been shown to be maximally expressed (58), for the WT mice. For the Runx2 mutant mice, MMP9 expression was significantly reduced as confirmed by line scan quantitation of the images. Taken together, these findings implicate MMP9 as a downstream target of Runx2 in bone tissues.
Runx2 mediates induction of MMP9 gene transcription by direct association with the proximal promoter.
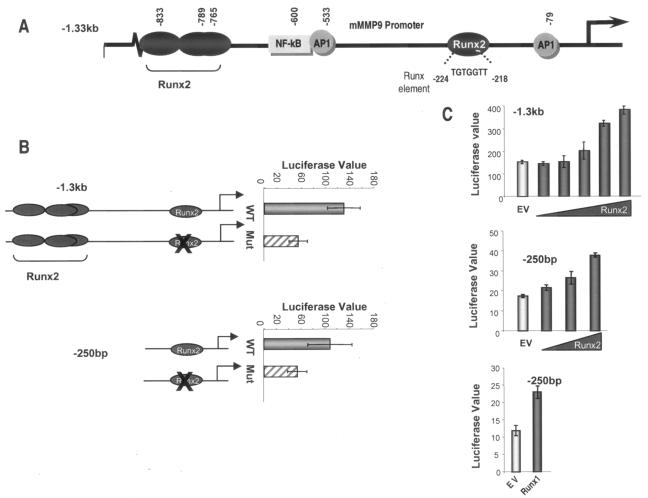
Four consensus Runx elements are located within 1.3 kb of the 5′ flanking sequences of the mouse MMP9 gene (Fig. 2A). Three closely spaced putative sites retain only the CCA motif, while the proximal site is a perfect match to the seven-nucleotide consensus TGTGGTT. We assessed which of these Runx consensus sites may contribute to MMP9 transcription by the transient transfection of osteoblastic MC3T3-E1 cell lines which express endogenous Runx2. Mutation of the proximal Runx binding element resulted in a twofold decrease in the basal activity of the −1.3-kb and −250-bp MMP9 promoter deletion fragments (Fig. 2B). To further show that Runx2 is an activator of MMP9 transcription, the −1.3-kb or −250-bp MMP9 promoter reporter construct was cotransfected into HeLa cells, which do not express Runx2, with increasing amounts of a Runx2 expression plasmid. Our results demonstrate a dose-dependent increase in the activity of both the −1.3-kb (Fig. 2C, upper panel) and −250-bp (middle panel) MMP9 promoter fragments. The Runx site in the MMP9 promoter is also responsive to Runx1, which increases the −250-bp MMP9 promoter activity to the same extent as Runx2 (Fig. 2C, lower panel). Therefore, Runx factors transactivate the MMP9 promoter for regulation of its expression in osseous and nonosseous cells.
FIG. 2.
Overexpression of Runx2 results in transcriptional activation of MMP9 promoter. (A) Promoter region (−1.3 kb) of mouse MMP9 showing Runx binding elements at positions −224 (TGTGGTT), −765 (TGAGGTC), −789 (ACCCCAG), and −833 (ACCCCAA) and NF-κB and two AP1 binding sites. (B) Basal activities of wild-type (WT) and Runx binding mutant (Mut) full-length (−1.3 kb, upper panel) and proximal (−250 bp, lower panel) MMP9 promoters in mouse osteoblastic MC3T3-E1 cells. Cells were transfected with either the WT or mutant MMP9 promoter (200 ng/well in six-well plates) using the Fugene 6 reagent. (C) Effect of Runx2 expression on −1.3-kb MMP9 promoter (upper panel) and of Runx1 or Runx2 on −250-bp MMP9 promoter (middle and lower panels) in HeLa cells. Samples were analyzed for luciferase activity after 36 h of transient transfection with increasing amounts of Runx2 (200 ng to 1.6 μg) or Runx1 (1.6 μg) expression plasmid. EV, empty vector control for Runx2 expression construct. The promoter activity (luciferase value × 1,000) was normalized by cotransfection with Renilla luciferase.
To examine the direct association of Runx2 with the MMP9 promoter, we performed in vitro (electrophoretic mobility shift assays) and in vivo (chromatin immunoprecipitation) binding assays (Fig. 3). Nuclear extracts from rat osteosarcoma (ROS17/2.8) or primary calvarial osteoblasts, a rich source of Runx2 protein, showed a specific protein-DNA interaction (open arrow, Fig. 3A) at the Runx motif in the proximal MMP9 promoter. The presence of Runx2 in this complex was confirmed by a supershift using a polyclonal antibody against Runx2 (filled arrow, Fig. 3A). The specificity of binding to the putative Runx sites was established by competition analyses with the cold wild-type probe, which completely abolished binding of the complex, while the mutant Runx oligonucleotide failed to compete (Fig. 3B). A ChIP assay was then performed to examine the in vivo association of Runx2 with the MMP9 gene (Fig. 3C). Using primary rat osteoblasts, we observed recruitment of Runx2 to the proximal MMP9 promoter. Together, these analyses demonstrate that Runx2 activates transcription of the MMP9 gene by directly binding to a functional Runx2 regulatory element.
FIG. 3.
Runx2 occupancy of MMP9 promoter. (A) Nuclear extracts were prepared from ROS 17/2.8 and primary rat osteoblasts (ROB) (day 12 [matrix maturation stage] and day 20 [mineralization]). An oligonucleotide containing the Runx binding element from the mouse MMP9 promoter (−212 bp to −246 bp) was incubated with 4 μg (lanes 3, 5, and 8) or 8 μg (lanes 6 and 9) of nuclear extract. The open arrow represents the specific DNA-protein complex, and the filled arrow shows a supershift with 1 μl of Runx2 polyclonal antibody (lanes 4, 7, and 10). (B) Competition assay performed with increasing amounts (25-, 50-, and 100-fold molar excess) of either cold wild-type (WT) or Runx mutant (mt) oligonucleotide showing the specificity of the Runx2-DNA complex. (C) In vivo occupancy of Runx2 protein at the proximal MMP9 promoter, as shown by ChIP of nuclei from day 12 ROB cells. The primers used to amplify the key regulatory elements present in the proximal MMP9 promoter fragment in the ChIP assays are indicated. The arrows indicate the positions of the forward (−380) and reverse (−130) primers. The agarose gel presented is representative of three experiments and shows selective amplification of the MMP9 proximal promoter in the input and Runx2 immunoprecipitate lanes, while IgG did not show any product.
Runx2 regulates activity of MMP9 in bone metastatic cancer cells.
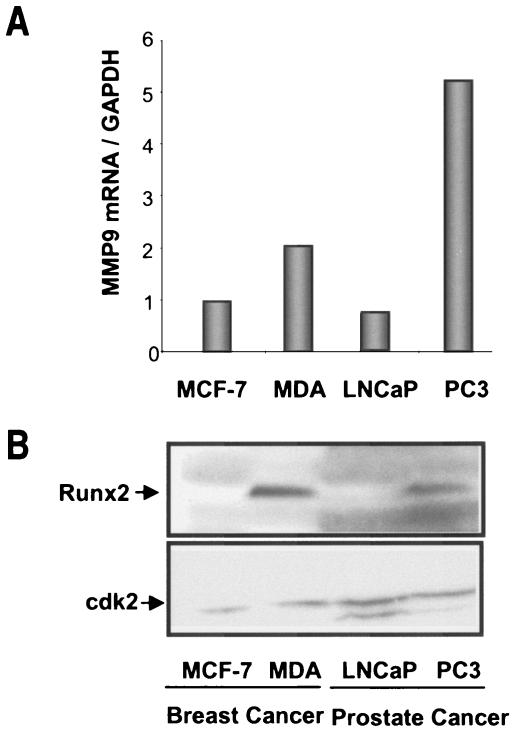
Matrix metalloproteinases are upregulated in metastatic cancer cells, and MMP9 is a key marker of these cells. We therefore examined whether Runx2 expression is correlated with MMP9 levels in cancer cells. We found MMP9 mRNAs to be highly expressed in breast (MDA-MB-231) and prostate (PC3) cells that metastasize to bone, in contrast to nonmetastatic (MCF-7 and LNCaP) tumor cells (Fig. 4A). Interestingly, the Runx2 protein was only detected in bone metastatic breast and prostate tumor cells (Fig. 4B).
FIG. 4.
Basal expression of MMP9 and Runx2 in nonmetastatic and bone metastatic cancer cells. (A) Total RNA from bone metastatic breast cancer cells (MDA-MB-231) and prostate cells (PC3), as well as from nonmetastatic breast (MCF-7) and prostate (LNCaP) tumor cells, was utilized to examine endogenous MMP9 mRNA levels by RT-QPCR analysis. Data were normalized to the GAPDH signal. (B) Whole-cell lysates from cancer cells were utilized to detect Runx2 protein with a monoclonal antibody by Western blotting. cdk2 protein levels are shown as an internal loading control.
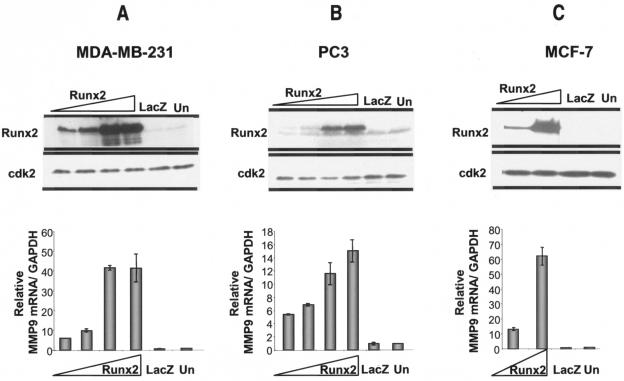
These observations provided an opportunity to address the transcriptional regulation of MMP9 by Runx2 in bone metastatic cancer cells. Breast (MDA-MB-231) and prostate (PC3) cancer cells were infected with an adenovirus expressing Runx2 at increasing MOIs and harvested after 48 h of infection (Fig. 5). The overexpression of Runx2 was confirmed by using a Runx2 monoclonal antibody in Western blot analyses (Fig. 5A and B, upper panels). We found a dose-dependent increase in endogenous MMP9 mRNA levels as a consequence of Runx2 expression in MDA-MB-231 (Fig. 5A) and PC3 (Fig. 5B) cells. To further assess whether Runx2 is sufficient to upregulate MMP9 in primary tumor cells also, we infected nonmetastatic breast cancer cells (MCF-7) with increasing MOIs of the Runx2-expressing adenovirus. Figure 5C shows a dose-dependent increase in MMP9 mRNA levels with increasing Runx2 expression levels compared to the case with the control virus. These results demonstrate that Runx2 upregulates endogenous MMP9 expression in bone metastatic cancer cells (Fig. 5A and B) as well as having the capacity to induce MMP9 transcription in nonmetastatic tumor cells (Fig. 5C). Therefore, Runx2 is a potent activator of the MMP9 gene in cancer cells.
FIG. 5.
Overexpression of Runx2 results in transcriptional activation of endogenous MMP9. Bone metastatic (MDA-MB-231 and PC3) and nonmetastatic (MCF-7) cancer cells were transduced with a LacZ- or Runx2-expressing adenovirus at increasing MOIs (for MDA-MB-231 and PC3 cells, 5, 10, 20, and 40 MOI; for MCF-7 cells, 10 and 40 MOI) for 48 h. Whole-cell lysates were utilized to detect the Runx2 protein with a mouse monoclonal antibody, and protein levels of cdk2 were measured as an internal loading control. Total RNA isolated from cells infected with the Runx2- or LacZ-expressing adenovirus (control) or from untreated (Un) cells was utilized to examine the MMP9 mRNA, as detected by RT-QPCR analysis. The amount of MMP9 mRNA was normalized to that of GAPDH.
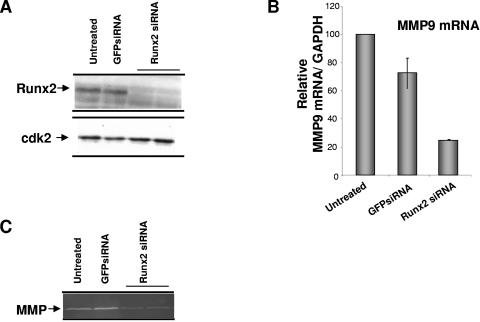
To further document the role of Runx2 in MMP9 transcriptional regulation, we depleted Runx2 in bone metastatic breast cancer cells by using RNA interference and measured endogenous levels of MMP9 mRNA and its functional activity by zymography (Fig. 6). With increasing amounts (25 nM and 50 nM) of siRNA oligonucleotides for Runx2, >80% of the Runx2 protein was knocked down, as detected by Western blot analysis (Fig. 6A). siRNA treatment did not affect cdk2 levels, and the control GFP oligonucleotide did not change the endogenous Runx2 levels. Thus, the RNA interference for Runx2 was specific. The decrease in Runx2 resulted in a more-than-threefold decrease in MMP9 mRNA, as detected by real-time RT-PCR analysis (Fig. 6B). We also observe a significant decrease in MMP9 activity in Runx2 siRNA-treated samples compared to GFP-treated or untreated controls (Fig. 6C). Thus, the inhibition of Runx2 expression results in a downregulation of MMP expression. These results further confirm that MMP9 is significantly regulated by Runx2.
FIG. 6.
Runx2 knockdown results in decreased endogenous MMP9 levels in metastatic breast cancer cells. (A) MDA-MB-231 breast cancer cells were treated with siRNAs for Runx2 (25 nM) and for GFP as a control with Oligofectamine for 72 h. Whole-cell lysates were analyzed for Runx2 protein and cdk protein as an internal loading control by Western blotting. (B) mRNA levels of endogenous MMP9 were detected by real-time RT-PCR of total RNA isolated from cells treated with siRNA for Runx2 or GFP or from untreated cells. (C) MMP9 protein activity was detected using gelatin zymography.
Runx2 influences migration of metastatic cancer cells.
MMP9 expression has been implicated in tumor invasion and metastasis. Our results show that Runx2 knockdown decreases MMP9 expression in bone metastatic breast cancer cells (Fig. 6). Thus, to examine the consequences of Runx2 depletion in metastatic cancer cells on invasive properties, we performed a Matrigel invasion assay with MDA-MB-231 breast cancer cells (Fig. 7). Cells treated with Runx2 siRNA showed significantly less (16%) invasion than untreated (62%) or GFP control oligonucleotide (47%)-treated cells (Fig. 7A). In a complementary experiment, we examined the invasion properties of MCF-7 (nonmetastatic) cells overexpressing Runx2 by adenovirus infection (Fig. 7B). The parental MCF-7 tumorigenic cells expressed MMP9 at 50% the levels found in metastatic MDA-MB-231 cells. The in vitro invasive capability of the MCF-7 cells was far lower (62% in MDA-MB-231 cells versus 7% in MCF-7 cells) (Fig. 7A and B, controls). The overexpression of Runx2 in MCF-7 cells resulted in a threefold increase in the invasion of MCF-7 cells (Fig. 7B) and was consistent with the increase in MMP9 levels (Fig. 5C). Thus, these findings indicate that Runx2 can modulate the invasive properties of nonmetastatic cancer cells, in part by increasing MMP9 activity. Because our findings also suggest that Runx2 is linked to the invasive properties of bone metastatic cancer cells, we considered the extent to which the expression of other known Runx2 target genes related to tumorigenic properties is altered in cancer cells.
FIG. 7.
Runx2 influences invasion of metastatic and nonmetastatic cancer cells. (A) Untreated breast cancer (MDA-MB-231) cells (control) or cells treated with either Runx2 or GFP siRNA (25 nM) for 48 h (2 × 105 cells) were plated on Matrigel or control inserts for an invasion assay. Cells were allowed to migrate for 20 h at 37°C. Cells that migrated were counted in 10 random fields, and the data are presented as percentages of invasion relative to control inserts. (B) MCF cells (4 × 105 cells) infected with Runx2 or LacZ (control) adenovirus were plated on a Matrigel-coated filter (12-μm pore size) and allowed to migrate for 24 h at 37°C.
Several markers of metastasis were examined in MDA-MB-231 breast cancer cells treated with Runx2 siRNA by real-time PCR analysis (Fig. 8A). We found significant reductions in the mRNA levels of MMP2, MMP13, and VEGF with Runx2 knockdown in comparison to the GFP control oligonucleotide treatment (Fig. 8A). The expression of β-actin did not change with Runx2 siRNA treatment (data not shown). Taken together, these findings suggest that Runx2 plays an important role in cell invasion by affecting the expression of the family of matrix metalloproteinases, as well as that of vascular endothelial growth factor, which was recently characterized as a target of Runx2 in hypertrophic chondrocytes (65). Figure 8B schematically illustrates the induction of Runx2 target genes expressed by metastatic cells with known functions in the bone environment that may contribute to the osteolysis associated with breast cancer.
FIG. 8.
Runx2 knockdown decreases mRNA levels of metastatic markers (MMP2, MMP13, and VEGF) in MDA-MB-231 breast cancer cells. (A) Real-time RT-PCR analysis of total cellular RNA (5 μg) prepared from cells treated with Runx2 or GFP siRNA for 48 h. GAPDH mRNA was used as a control for normalization. (B) Transcriptional regulation of metastatic markers and Runx2 target genes known to be expressed in cancer cells. Breast cancer cells from primary tumors express metastatic markers (VEGF and MMPs) that are directly regulated by Runx2. It is not clear when during the metastatic process the Runx2 protein is first detected. The presence of Runx2 in cancer cells from bone metastatic tumors further activates the expression of tumor-related target genes.
DISCUSSION
The family of MMPs, Runx1, and Runx2 are expressed in osteoprogenitors, chondrocytes, and osteoblasts. Our identification of MMP9 as a novel target gene of both Runx1 and Runx2 in osteoblasts supports the concept that Runx factors are important transcriptional regulators of MMP9 and other MMPs for mediating multiple roles of these enzymes during bone formation.
MMP9 is a key inducer of events associated with pathological conditions that include rheumatoid arthritis, tumor invasion, and metastasis. In the present study, we demonstrate by chromatin immunoprecipitation that Runx2 is recruited to the MMP9 promoter and show that Runx2 activates endogenous MMP9 gene expression in normal skeletal, breast, and prostate cancer cells that metastasize to bone. Importantly, Runx2 has been linked to the activities of metastatic cancer cells in the bone microenvironment which result in osteolytic and osteoblastic lesions induced by secreted factors (5, 26, 36, 38, 60). Our studies show that Runx2 regulates the family of MMPs and VEGF in metastatic cell lines. Importantly, the depletion of Runx2 expression in metastatic MDA-MB-231 cells by RNA interference reduces the invasion properties of these cancer cells. Significantly, the forced expression of Runx2 in nonmetastatic MCF-7 cells induces MMP9 and invasive properties threefold. Thus, Runx2 is a positive regulator of genes associated with metastasis and osteolytic disease.
The mechanism of Runx2-mediated transcriptional regulation of MMP9 should be considered in the context of other factors. Mutation of the Runx2 sites in the MMP9 promoter results in a two- to threefold decrease in transcription. However, other factors (e.g., tumor necrosis factor alpha) stimulate MMP9 transcription through binding to NF-κB, AP1, and Sp-1 sites (20, 56, 57, 63). Among MMP family members, MMP13 activation by parathyroid hormone has been shown to be regulated by interactions of Runx2 with c-Fos and c-Jun at AP-1 sites (14, 23, 43, 52, 53). Bertrand-Philippe et al. (7) showed that JunD and Runx factors assemble at the adjacent SRE and UTE-1 sites in the TIMP1 promoter and form functional interactions that stimulate transcription. These two examples of genes in cancer cells represent indirect mechanisms of Runx2-mediated activation. Thus, there exists the possibility that Runx2 binding to the MMP9 promoter may lead to a cooperative interaction with other factors, including Ets (16). Thus, the Runx2 transcriptional effect on MMP9 and other target genes in cancer cells could be amplified through its interaction with other transcription factors in coregulatory complexes.
During osteogenesis, Runx2 functions as a “platform protein” that interacts with a spectrum of coregulatory proteins, including chromatin remodeling factors, to provide combinatorial mechanisms for either inducing or repressing gene transcription. The recruitment of several coregulatory proteins to the MMP9 promoter has been shown to coordinate cell signaling (extracellular signal-regulated kinase and NF-κB). Chromatin remodeling by Brg-1, histone modifications, and the sequential recruitment of transcriptional regulators are also critical to the regulation of MMP9 gene expression (34). We show that the Runx2 binding element in the proximal promoter (−250), which is necessary for MMP9 gene transcription, is located in a region with neighboring AP-1, NF-κB, and Sp-1 sites required for synergistic cooperation on the MMP9 promoter (50). This complexity of gene transcription involving Runx2 occurs on other gene promoters, including that of the bone-specific osteocalcin gene. Runx2 sites flank the vitamin D response element and are required for chromatin remodeling (28, 42). Thus, a key feature of Runx regulation is its ability to function in the combinatorial control of gene expression by forming coregulatory interactions at multiple elements and organizing transcriptional complexes in subnuclear domains (33, 54). Significantly, disruption of the subnuclear organization of Runx2 regulatory complexes blocks the Runx2-mediated transcription of target genes in metastatic breast cancer cells in vivo (26).
MMP9 has been linked to the metastasis of primary tumors. Hiratsuka et al. (24) demonstrated that MMP9 is specifically induced in premetastatic lung endothelial cells and macrophages by distant primary tumors via VEGFR-1/Flt-1 tyrosine kinase (TK) and that it promotes lung metastasis. Furthermore, they showed using a genetic approach that the suppression of MMP9 induction by the deletion of either VEGFR-1TK or MMP9 markedly reduced lung metastasis. Cancer cells that metastasize to bone have higher Runx2 levels than primary tumors and also exhibit elevated MMP9 expression and activity. The blocking of MMP9 activity has also been related to a decreased invasion of cancer cells through Matrigel (1). The knockdown of MMP9 in cells derived from invasive and metastatic Ewing's sarcoma inhibits migration toward fibronectin (48), suggesting that depleting key markers for metastasis decreases invasion. Here we show that a Runx2 knockdown by RNA interference in bone metastatic cancer cells downregulates MMP9 and reduces cell migration.
The upregulation of Runx2 expression in breast (6, 51) and prostate (10) cancer cells suggests an important functional contribution of Runx2 in tumorigenic cells. Our siRNA studies also indicate a role for Runx2 in regulating the invasion of bone metastatic cancer cells (MDA-MB-231 cells), in part through the transcriptional regulation of MMP9, but also through other Runx target genes, such as VEGF. Notably, VEGF is a primary component of tumor formation and metastasis (12). The VEGF gene is a characterized target of Runx2, and it has been shown that Runx2 is a necessary component of a tissue-specific genetic program that regulates VEGF for the progression of endochondral bone formation (65). Runx2 controls osteoblast differentiation from mesodermal cell populations by regulating bone-specific genes that include osteocalcin, bone sialoprotein (BSP), VEGF, MMP13, and TIMP (illustrated in Fig. 8B). These genes are associated with various tumors; for example, human malignant melanoma cells express BSP in relation to Runx2 in vivo as a function of the extent of local invasion (47). Because Runx2 regulates the expression of key markers implicated in tumor invasion and metastasis (VEGF, BSP, and matrix metalloproteinases [collagenase 3, membrane type 1 matrix metalloproteinase, and MMP9]), the presence of Runx2 appears to be an earlier upstream event in the further activation of tumor-related genes in the bone microenvironment (Fig. 8B). The perturbation of Runx2 regulatory function in breast cancer cells has been shown to abolish the cellular activity that causes osteolytic lesions in vivo in the bone environment (5, 26). The present study provides insight into mechanisms that include the direct control of cell invasion and the expression of metastasis-related factors by Runx2. Other studies are required to definitively establish that Runx2 mediates the metastasis of cells from a primary tumor to secondary sites.
Acknowledgments
We are appreciative of the editorial assistance of Elizabeth Bronstein and Judy Rask. We thank members of the laboratory for stimulating discussions and support throughout the course of these studies.
This work was supported by NIH grants AR39588, P01 PO1 AR48818, PO1 CA82834, and P30 DK32520.
The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Aalinkeel, R., M. P. Nair, G. Sufrin, S. D. Mahajan, K. C. Chadha, R. P. Chawda, and S. A. Schwartz. 2004. Gene expression of angiogenic factors correlates with metastatic potential of prostate cancer cells. Cancer Res. 64:5311-5321. [DOI] [PubMed] [Google Scholar]
- 2.Afzal, F., J. Pratap, K. Ito, Y. Ito, J. L. Stein, A. J. van Wijnen, G. S. Stein, J. B. Lian, and A. Javed. 2005. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J. Cell. Physiol. 204:63-72. [DOI] [PubMed] [Google Scholar]
- 3.Amorino, G. P., and R. L. Hoover. 1998. Interactions of monocytic cells with human endothelial cells stimulate monocytic metalloproteinase production. Am. J. Pathol. 152:199-207. [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee, C., L. R. McCabe, J.-Y. Choi, S. W. Hiebert, J. L. Stein, G. S. Stein, and J. B. Lian. 1997. Runt homology domain proteins in osteoblast differentiation: AML-3/CBFA1 is a major component of a bone specific complex. J. Cell. Biochem. 66:1-8. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, G. L., K. E. Hebert, M. Kamal, A. Javed, T. A. Einhorn, J. B. Lian, G. S. Stein, and L. C. Gerstenfeld. 2004. Fidelity of Runx2 activity in breast cancer cells is required for the generation of metastases associated osteolytic disease. Cancer Res. 64:4506-4513. [DOI] [PubMed] [Google Scholar]
- 6.Barnes, G. L., A. Javed, S. M. Waller, M. H. Kamal, K. E. Hebert, M. Q. Hassan, A. Bellahcene, A. J. van Wijnen, M. F. Young, J. B. Lian, G. S. Stein, and L. C. Gerstenfeld. 2003. Osteoblast-related transcription factors Runx2 (Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res. 63:2631-2637. [PubMed] [Google Scholar]
- 7.Bertrand-Philippe, M., R. G. Ruddell, M. J. Arthur, J. Thomas, N. Mungalsingh, and D. A. Mann. 2004. Regulation of tissue inhibitor of metalloproteinase 1 gene transcription by RUNX1 and RUNX2. J. Biol. Chem. 279:24530-24539. [DOI] [PubMed] [Google Scholar]
- 8.Blyth, K., E. R. Cameron, and J. C. Neil. 2005. The runx genes: gain or loss of function in cancer. Nat. Rev. Cancer 5:376-387. [DOI] [PubMed] [Google Scholar]
- 9.Blyth, K., A. Terry, N. Mackay, F. Vaillant, M. Bell, E. R. Cameron, J. C. Neil, and M. Stewart. 2001. Runx2: a novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene 20:295-302. [DOI] [PubMed] [Google Scholar]
- 10.Brubaker, K. D., R. L. Vessella, L. G. Brown, and E. Corey. 2003. Prostate cancer expression of runt-domain transcription factor Runx2, a key regulator of osteoblast differentiation and function. Prostate 56:13-22. [DOI] [PubMed] [Google Scholar]
- 11.Choi, J.-Y., J. Pratap, A. Javed, S. K. Zaidi, L. Xing, E. Balint, S. Dalamangas, B. Boyce, A. J. van Wijnen, J. B. Lian, J. L. Stein, S. N. Jones, and G. S. Stein. 2001. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc. Natl. Acad. Sci. USA 98:8650-8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claffey, K. P., and G. S. Robinson. 1996. Regulation of VEGF/VPF expression in tumor cells: consequences for tumor growth and metastasis. Cancer Metastasis Rev. 15:165-176. [DOI] [PubMed] [Google Scholar]
- 13.Coussens, L. M., B. Fingleton, and L. M. Matrisian. 2002. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 295:2387-2392. [DOI] [PubMed] [Google Scholar]
- 14.D'Alonzo, R. C., N. Selvamurugan, G. Karsenty, and N. C. Partridge. 2002. Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J. Biol. Chem. 277:816-822. [DOI] [PubMed] [Google Scholar]
- 15.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dittmer, J. 2003. The biology of the Ets1 proto-oncogene. Mol. Cancer 2:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducy, P., M. Starbuck, M. Priemel, J. Shen, G. Pinero, V. Geoffroy, M. Amling, and G. Karsenty. 1999. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 13:1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ducy, P., R. Zhang, V. Geoffroy, A. L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754. [DOI] [PubMed] [Google Scholar]
- 19.Egeblad, M., and Z. Werb. 2002. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2:161-174. [DOI] [PubMed] [Google Scholar]
- 20.Fini, M. E., J. D. Bartlett, M. Matsubara, W. B. Rinehart, M. K. Mody, M. T. Girard, and M. Rainville. 1994. The rabbit gene for 92-kDa matrix metalloproteinase. Role of AP1 and AP2 in cell type-specific transcription. J. Biol. Chem. 269:28620-28628. [PubMed] [Google Scholar]
- 21.Galindo, M., J. Pratap, D. W. Young, H. Hovhannisyan, H. J. Im, J. Y. Choi, J. B. Lian, J. L. Stein, G. S. Stein, and A. J. van Wijnen. 2005. The bone-specific expression of RUNX2 oscillates during the cell cycle to support a G1 related anti-proliferative function in osteoblasts. J. Biol. Chem. 280:20274-20285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez, S., J. Liu, A. Javed, M. Montecino, G. S. Stein, J. B. Lian, and J. L. Stein. 2004. The vitamin D response element in the distal osteocalcin promoter contributes to chromatin organization of the proximal regulatory domain. J. Biol. Chem. 279:43581-43588. [DOI] [PubMed] [Google Scholar]
- 23.Hess, J., D. Porte, C. Munz, and P. Angel. 2001. AP-1 and Cbfa/Runt physically interact and regulate PTH-dependent MMP13 expression in osteoblasts through a new OSE2/AP-1 composite element. J. Biol. Chem. 276:20029-20038. [DOI] [PubMed] [Google Scholar]
- 24.Hiratsuka, S., K. Nakamura, S. Iwai, M. Murakami, T. Itoh, H. Kijima, J. M. Shipley, R. M. Senior, and M. Shibuya. 2002. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell 2:289-300. [DOI] [PubMed] [Google Scholar]
- 25.Inman, C. K., and P. Shore. 2003. The osteoblast transcription factor Runx2 is expressed in mammary epithelial cells and mediates osteopontin expression. J. Biol. Chem. 278:48684-48689. [DOI] [PubMed] [Google Scholar]
- 26.Javed, A., G. L. Barnes, J. Pratap, T. Antkowiak, L. C. Gerstenfeld, A. J. van Wijnen, J. L. Stein, J. B. Lian, and G. S. Stein. 2005. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc. Natl. Acad. Sci. USA 102:1454-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Javed, A., S. Gutierrez, M. Montecino, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 1999. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D-responsive transcription and contribute to chromatin organization. Mol. Cell. Biol. 19:7491-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javed, A., M. van Rees, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 1999. C/EBP family members are expressed in osteoblasts and are involved in regulation of osteocalcin gene transcription. J. Bone Miner. Res. 14:S469. [Google Scholar]
- 29.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y.-H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755-764. [DOI] [PubMed] [Google Scholar]
- 30.Lengner, C. J., H. Drissi, J.-Y. Choi, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 2002. Activation of the bone related Runx2/Cbfa1 promoter in mesenchymal condensations and developing chondrocytes of the axial skeleton. Mech. Dev. 114:167-170. [DOI] [PubMed] [Google Scholar]
- 31.Levanon, D., D. Bettoun, C. Harris-Cerruti, E. Woolf, V. Negreanu, R. Eilam, Y. Bernstein, D. Goldenberg, C. Xiao, M. Fliegauf, E. Kremer, F. Otto, O. Brenner, A. Lev-Tov, and Y. Groner. 2002. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 21:3454-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Q. L., K. Ito, C. Sakakura, H. Fukamachi, K. Inoue, X. Z. Chi, K. Y. Lee, S. Nomura, C. W. Lee, S. B. Han, H. M. Kim, W. J. Kim, H. Yamamoto, N. Yamashita, T. Yano, T. Ikeda, S. Itohara, J. Inazawa, T. Abe, A. Hagiwara, H. Yamagishi, A. Ooe, A. Kaneda, T. Sugimura, T. Ushijima, S. C. Bae, and Y. Ito. 2002. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109:113-124. [DOI] [PubMed] [Google Scholar]
- 33.Lian, J. B., A. Javed, S. K. Zaidi, C. Lengner, M. Montecino, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 2004. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit. Rev. Eukaryot. Gene Expr. 14:1-41. [PubMed] [Google Scholar]
- 34.Ma, Z., R. C. Shah, M. J. Chang, and E. N. Benveniste. 2004. Coordination of cell signaling, chromatin remodeling, histone modifications, and regulator recruitment in human matrix metalloproteinase 9 gene transcription. Mol. Cell. Biol. 24:5496-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magid, R., T. J. Murphy, and Z. S. Galis. 2003. Expression of matrix metalloproteinase-9 in endothelial cells is differentially regulated by shear stress. Role of c-Myc. J. Biol. Chem. 278:32994-32999. [DOI] [PubMed] [Google Scholar]
- 36.Mundy, G. R. 2002. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat. Rev. Cancer 2:584-593. [DOI] [PubMed] [Google Scholar]
- 37.Ogawa, S., H. Harada, M. Fujiwara, S. Tagashira, T. Katsumata, and H. Takada. 2000. Cbfa1, an essential transcription factor for bone formation, is expressed in testis from the same promoter used in bone. DNA Res. 7:181-185. [DOI] [PubMed] [Google Scholar]
- 38.O'Keefe, R. J., and T. A. Guise. 2003. Molecular mechanisms of bone metastasis and therapeutic implications. Clin. Orthop. Relat. Res. 2003(Suppl.):S100-S104. [DOI] [PubMed] [Google Scholar]
- 39.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 40.Ortega, N., D. J. Behonick, and Z. Werb. 2004. Matrix remodeling during endochondral ossification. Trends Cell Biol. 14:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otto, F., H. Kanegane, and S. Mundlos. 2002. Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum. Mutat. 19:209-216. [DOI] [PubMed] [Google Scholar]
- 42.Paredes, R., G. Arriagada, F. Cruzat, J. Olate, A. van Wijnen, J. Lian, G. Stein, J. Stein, and M. Montecino. 2004. The Runx2 transcription factor plays a key role in the 1α,25-dihydroxy vitamin D3-dependent upregulation of the rat osteocalcin (OC) gene expression in osteoblastic cells. J. Steroid Biochem. Mol. Biol. 89-90:269-271. [DOI] [PubMed] [Google Scholar]
- 43.Porte, D., J. Tuckermann, M. Becker, B. Baumann, S. Teurich, T. Higgins, M. J. Owen, M. Schorpp-Kistner, and P. Angel. 1999. Both AP-1 and Cbfa1-like factors are required for the induction of interstitial collagenase by parathyroid hormone. Oncogene 18:667-678. [DOI] [PubMed] [Google Scholar]
- 44.Pratap, J., M. Galindo, S. K. Zaidi, D. Vradii, B. M. Bhat, J. A. Robinson, J.-Y. Choi, T. Komori, J. L. Stein, J. B. Lian, G. S. Stein, and A. J. van Wijnen. 2003. Cell growth regulatory role of Runx2 during proliferative expansion of pre-osteoblasts. Cancer Res. 63:5357-5362. [PubMed] [Google Scholar]
- 45.Quesada, A. R., M. M. Barbacid, E. Mira, P. Fernandez-Resa, G. Marquez, and M. Aracil. 1997. Evaluation of fluorometric and zymographic methods as activity assays for stromelysins and gelatinases. Clin. Exp. Metastasis 15:26-32. [DOI] [PubMed] [Google Scholar]
- 46.Reponen, P., C. Sahlberg, C. Munaut, I. Thesleff, and K. Tryggvason. 1994. High expression of 92-kD type IV collagenase (gelatinase B) in the osteoclast lineage during mouse development. J. Cell Biol. 124:1091-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riminucci, M., A. Corsi, K. Peris, L. W. Fisher, S. Chimenti, and P. Bianco. 2003. Coexpression of bone sialoprotein (BSP) and the pivotal transcriptional regulator of osteogenesis, Cbfa1/Runx2, in malignant melanoma. Calcif. Tissue Int. 73:281-289. [DOI] [PubMed] [Google Scholar]
- 48.Sanceau, J., S. Truchet, and B. Bauvois. 2003. Matrix metalloproteinase-9 silencing by RNA interference triggers the migratory-adhesive switch in Ewing's sarcoma cells. J. Biol. Chem. 278:36537-36546. [DOI] [PubMed] [Google Scholar]
- 49.Satake, M., S. Nomura, Y. Yamaguchi-Iwai, Y. Takahama, Y. Hashimoto, M. Niki, Y. Kitamura, and Y. Ito. 1995. Expression of the Runt domain-encoding PEBP2 alpha genes in T cells during thymic development. Mol. Cell. Biol. 15:1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato, H., and M. Seiki. 1993. Regulatory mechanism of 92 kDa type IV collagenase gene expression which is associated with invasiveness of tumor cells. Oncogene 8:395-405. [PubMed] [Google Scholar]
- 51.Selvamurugan, N., S. Kwok, and N. C. Partridge. 2004. Smad3 interacts with JunB and Cbfa1/Runx2 for transforming growth factor-beta1-stimulated collagenase-3 expression in human breast cancer cells. J. Biol. Chem. 279:27764-27773. [DOI] [PubMed] [Google Scholar]
- 52.Selvamurugan, N., and N. C. Partridge. 2000. Constitutive expression and regulation of collagenase-3 in human breast cancer cells. Mol. Cell Biol. Res. Commun. 3:218-223. [DOI] [PubMed] [Google Scholar]
- 53.Selvamurugan, N., M. R. Pulumati, D. R. Tyson, and N. C. Partridge. 2000. Parathyroid hormone regulation of the rat collagenase-3 promoter by protein kinase A-dependent transactivation of core binding factor alpha1. J. Biol. Chem. 275:5037-5042. [DOI] [PubMed] [Google Scholar]
- 54.Stein, G. S., S. K. Zaidi, C. D. Braastad, M. Montecino, A. J. van Wijnen, J.-Y. Choi, J. L. Stein, J. B. Lian, and A. Javed. 2003. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol. 13:584-592. [DOI] [PubMed] [Google Scholar]
- 55.Sun, L., M. Vitolo, and A. Passaniti. 2001. Runt-related gene 2 in endothelial cells: inducible expression and specific regulation of cell migration and invasion. Cancer Res. 61:4994-5001. [PubMed] [Google Scholar]
- 56.Takahra, T., D. E. Smart, F. Oakley, and D. A. Mann. 2004. Induction of myofibroblast MMP-9 transcription in three-dimensional collagen I gel cultures: regulation by NF-kappaB, AP-1 and Sp1. Int. J. Biochem. Cell Biol. 36:353-363. [DOI] [PubMed] [Google Scholar]
- 57.Van den Steen, P. E., B. Dubois, I. Nelissen, P. M. Rudd, R. A. Dwek, and G. Opdenakker. 2002. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9). Crit. Rev. Biochem. Mol. Biol. 37:375-536. [DOI] [PubMed] [Google Scholar]
- 58.Vu, T. H., J. M. Shipley, G. Bergers, J. E. Berger, J. A. Helms, D. Hanahan, S. D. Shapiro, R. M. Senior, and Z. Werb. 1998. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell 93:411-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wucherpfennig, A. L., Y. P. Li, W. G. Stetler-Stevenson, A. E. Rosenberg, and P. Stashenko. 1994. Expression of 92 kD type IV collagenase/gelatinase B in human osteoclasts. J. Bone Miner. Res. 9:549-556. [DOI] [PubMed] [Google Scholar]
- 60.Yang, J., K. Fizazi, S. Peleg, C. R. Sikes, A. K. Raymond, N. Jamal, M. Hu, M. Olive, L. A. Martinez, C. G. Wood, C. J. Logothetis, G. Karsenty, and N. M. Navone. 2001. Prostate cancer cells induce osteoblast differentiation through a Cbfa1-dependent pathway. Cancer Res. 61:5652-5659. [PubMed] [Google Scholar]
- 61.Yergeau, D. A., C. J. Hetherington, Q. Wang, P. Zhang, A. H. Sharpe, M. Binder, M. Marin-Padilla, D. G. Tenen, N. A. Speck, and D. E. Zhang. 1997. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat. Genet. 15:303-306. [DOI] [PubMed] [Google Scholar]
- 62.Yeung, F., W. K. Law, C. H. Yeh, J. J. Westendorf, Y. Zhang, R. Wang, C. Kao, and L. W. Chung. 2002. Regulation of human osteocalcin promoter in hormone-independent human prostate cancer cells. J. Biol. Chem. 277:2468-2476. [DOI] [PubMed] [Google Scholar]
- 63.Yoshizaki, T., H. Sato, M. Furukawa, and J. S. Pagano. 1998. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc. Natl. Acad. Sci. USA 95:3621-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young, D. W., S. K. Zaidi, P. S. Furcinitti, A. Javed, A. J. van Wijnen, J. L. Stein, J. B. Lian, and G. S. Stein. 2004. Quantitative signature for architectural organization of regulatory factors using intranuclear informatics. J. Cell Sci. 117:4889-4896. [DOI] [PubMed] [Google Scholar]
- 65.Zelzer, E., D. J. Glotzer, C. Hartmann, D. Thomas, N. Fukai, S. Soker, and B. R. Olsen. 2001. Tissue specific regulation of VEGF expression during bone development requires Cbfa1/Runx2. Mech. Dev. 106:97-106. [DOI] [PubMed] [Google Scholar]