Abstract
Stem cell factor (SCF) delays differentiation and enhances the expansion of erythroid progenitors. Previously, we performed expression-profiling experiments to link signaling pathways to target genes using polysome-bound mRNA. SCF-induced phosphoinositide-3-kinase (PI3K) appeared to control polysome recruitment of specific mRNAs associated with neoplastic transformation. To evaluate the role of mRNA translation in the regulation of expansion versus differentiation of erythroid progenitors, we examined the function of the eukaryote initiation factor 4E (eIF4E) in these cells. SCF induced a rapid and complete phosphorylation of eIF4E-binding protein (4E-BP). Overexpression of eIF4E did not induce factor-independent growth but specifically impaired differentiation into mature erythrocytes. Overexpression of eIF4E rendered polysome recruitment of mRNAs with structured 5′ untranslated regions largely independent of growth factor and resistant to the PI3K inhibitor LY294002. In addition, overexpression of eIF4E rendered progenitors insensitive to the differentiation-inducing effect of LY294002, indicating that control of mRNA translation is a major pathway downstream of PI3K in the regulation of progenitor expansion.
Leukemia may arise through mechanisms that enable the proliferation and survival of normal cells when expansion of the progenitor pool is required. The control of the balance between expansion and maturation by cytokines and growth factors is such a mechanism. While erythroid progenitors require erythropoietin (Epo) to mature into erythrocytes, the progenitor pool can be expanded both in vivo and in vitro in response to the cooperative action of Epo, stem cell factor (SCF), and glucocorticoids (6, 9, 16, 49). SCF cooperates with Epo to suppress differentiation and sustain renewal divisions of erythroid progenitors. Cooperation of SCF and cytokines has also been observed in other hematopoietic progenitors (10). A mutated SCF receptor (v-Kit) was initially identified as a viral oncogene in a feline leukemia virus (40). Mutations in c-Kit that induce constitutive activity are found specifically in t(8;21) and inv (16) leukemia, but autocrine loops activating c-Kit are also reported in other types of leukemia (7, 51). In an avian model system, SCF cooperates with oncogenic MLL fusion genes to transform lymphomyeloid multipotent progenitors (42). Furthermore, activating c-Kit mutations are common in gastrointestinal stromal tumors (29) and in bilateral testicular germ cell tumors (33). This suggests that the inhibition of erythroid differentiation by SCF may exemplify a more general role of SCF in the regulation of cell growth and differentiation.
The expansion of erythroid progenitors in the presence of SCF is abrogated by the phosphatidylinositol 3 kinase (PI3K) inhibitor LY294002 (49), resulting in terminal differentiation instead. An important effector of PI3K is protein kinase B (PKB), which controls cellular processes such as cell cycle progression, apoptosis, and mRNA translation through the phosphorylation of, e.g., Forkhead transcription factors, the proapoptotic protein Bad, and the mammalian target of rapamycin (mTOR) (1, 5, 13, 19, 25, 39). To identify critical pathways and targets downstream of SCF signaling, we performed profiling experiments using polysome-bound mRNA to detect those mRNAs that are expressed and translated into protein. The expression of the putative oncogene Ndpk-B (nucleoside diphosphate kinase B) (also known as Nm23-M2 or Nme2) appeared to be regulated by SCF. Regulation did not occur at the level of gene transcription but exclusively through the recruitment of its mRNA into polysomes, which was fully dependent on PI3K activity (28). As selective recruitment of mRNAs into polysomes appears to be an important regulatory mechanism in cell growth control and tumorigenesis (4, 41), we examined the role of mRNA translation in PI3K-dependent control of the expansion and differentiation of erythroid progenitors.
The mRNA cap-binding eukaryote initiation factor 4E (eIF4E) recruits the scaffolding protein eIF4G, which associates with, among others, the mRNA helicase eIF4A and the small subunit of the ribosome. This complex scans the 5′ untranslated region (5′UTR) of mRNA until an appropriate AUG codon is recognized. In binding to the limiting factor eIF4E, eIF4G has to compete with 4E-binding proteins (4E-BP). Unphosphorylated 4E-BP binds and inhibits eIF4E, but eIF4E is released upon the phosphorylation of 4E-BP by the mTOR kinase (23). The increased availability of eIF4E is associated with cell proliferation (19). Increased levels of eIF4E are detected in a number of solid tumors, especially in breast, colon, and head/neck tumors (15). Interference with translation initiation via the overexpression of an mTOR-insensitive 4E-BP1 results in the inhibition of the cell cycle progression (19). These observations suggest that the availability of eIF4E in translation initiation contributes to neoplastic transformation. In addition, it has also been suggested that eIF4E enhances nucleocytoplasmic transport of specific transcripts. The aberrant regulation of eIF4E-dependent mRNA transport contributed to leukemogenesis, as it impaired granulocytic and monocytic differentiation (46).
Although eIF4E and its associated factors are general translation factors, they bind and scan mRNAs with a short and simple 5′UTR much more efficiently than mRNAs with a long and structured 5′UTR. The 5′UTR of the nm23-M2 transcript, which is strictly dependent on Epo- and SCF-induced PI3K activity, begins with a terminal oligopyrimidine (TOP) sequence and contains an inverted repeat (28). The TOP sequence confers selective translation to a given mRNA as polysome recruitment becomes dependent on mTOR activation (27). TOP sequences are present in mRNAs encoding ribosomal proteins and elongation factors to render the energy-consuming process of ribosome biogenesis dependent on the presence of mitogenic factors and nutrients. Additional secondary structures within the 5′UTR may also control mRNA translation, such as the iron response element that renders translation of the transferrin receptor dependent on the availability of iron (45).
In this paper, we show that control of mRNA translation is an important PI3K-dependent pathway regulating progenitor expansion. We demonstrate that SCF induces rapid PI3K-dependent phosphorylation of 4E-BP and that overexpression of eIF4E in erythroid progenitors delays differentiation and enhances renewal divisions in the absence of SCF. We identified mRNAs upregulated by SCF-induced polysome recruitment. These mRNAs contain a long and structured 5′UTR (Y-box-binding protein 1), a short 5′UTR starting with a TOP sequence (the splicing factor U2-Snrpb"), or both a TOP sequence and an inverted repeat (Nm23-M2). Polysome recruitment of all three mRNAs was strongly enhanced by eIF4E overexpression in cells that were factor deprived, while eIF4E overexpression did not affect polysome association in the presence of SCF.
MATERIALS AND METHODS
Cells.
I/11 cells were cultivated in StemPro-34 medium (Life Technologies) as described previously (49). For expansion, the medium was supplemented with 0.5 U/ml Epo (kind gift from Ortho-Biotech, Tilburg, The Netherlands), 100 ng/ml SCF (supernatant of CHO producer cells), and 10−6M dexamethasone (Dex; Sigma-Aldrich). To induce differentiation, cells were cultivated in StemPro-34 medium supplemented with 5 U/ml Epo and 0.5 mg/ml iron-loaded transferrin (Intergene). Cell numbers and cell size distribution were determined using an electronic cell counter (CASY-1; Schärfe-System, Reutlingen, Germany). LY294002 and rapamycin were obtained from Alexis (Switzerland).
Hemoglobin content determination and cell morphology.
Small aliquots of the cultures were removed and analyzed for hemoglobin content by photometry as described earlier (5, 31). The values obtained were the averages of triplicate measurements after normalization for cell number and mean single cell volume. Cell morphology was analyzed in cytospins stained with histological dyes and neutral benzidine (8) using an Olympus BX40 microscope (40× lens objective; numerical aperture, 0.65), an Olympus DP50 charge-coupled-device camera, and Viewfinder Lite 1.0 acquisition software. Images were cropped using Adobe Photoshop 6.0.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting, immunoprecipitation, and antibodies.
For acute stimulation with growth factors, proliferating I/11 cells were washed twice with phosphate-buffered saline (PBS) and seeded at 4 × 106 cells/ml in plain Iscove's modified Dulbecco's medium (Life Technologies). After 4 h of factor deprivation, cells were stimulated at 37°C with SCF (100 ng/ml) or Epo (5 U/ml). Cells were harvested after the indicated time points by the addition of ice-cold PBS. Cell lysates, SDS-PAGE, immunoprecipitation, and Western blotting were performed as described previously (48). To analyze eIF4E and 4E-BP, 10 μl of protein extract (≈1 × 106 cells) was loaded onto a 15% polyacrylamide gel. The antibodies used were the following: anti-eIF4E, anti-4E-BP1, anti-phospho-4E-BP1(Thr37/46), anti-phospho-4E-BP1(Ser65) (Cell Signaling Technology, Inc.), anti-ERK1(K-23), and anti-Myc (gE10, Santa Cruz Biotechnology, Inc.). For immunoprecipitation, Myc-eIF4E was immunoprecipitated from the lysate of 15 × 106 cells by an overnight incubation at 4°C with monoclonal anti-Myc antibodies (1 μg antibody/15 × 106 cells), followed by an hour of incubation at 4°C with 15 μl of a 50% solution of protein G-Sepharose beads (Pharmacia LKB).
m7GTP-Sepharose affinity chromatography.
For the isolation of eIF4E and associated proteins, 15 × 106 cells were lysed in buffer C (50 mM MOPS [morpholinepropanesulfonic acid]-KOH [potassium hydroxide] [pH 7.2], 0.5 mM EDTA, 0.5 mM EGTA, 100 mM KCl, 14 mM 2-mercaptoethanol, 50 mM NaF, 100 μM GTP, and protease inhibitor cocktail) and subjected to m7GTP-Sepharose chromatography as described previously (35). Briefly, the lysed cells were incubated at 4°C with 25 μl of equilibrated Sepharose resin (7-methyl GTP-Sepharose 4B; Amersham Biosciences) for 1 h. The resin was washed three times with buffer C, and recovered proteins were eluted directly into sample buffer for SDS-PAGE analysis.
Generation of myc-eIF4E-expressing I/11 clones.
The myc-eIF4E cDNA (NCBI accession number M61731) containing six myc tags at the 5′ start of the coding sequence was isolated from the pCS3MT vector and inserted into the eukaryotic retroviral expression vector pBabe using EcoRI and BamHI restriction sites. Retroviral transduction was performed as described previously (5). Briefly, 0.5 × 106 ecotropic Phoenix cells were transfected with 12 μg plasmid DNA (myc-eIF4E-pBabe) using a calcium-phosphate coprecipitation assay. After 40 h, cells were treated with 10 μg/ml mitomycin C (Kyowa Hakko Kogyo, Tokyo, Japan) for 1 h and washed three times with PBS. A total of 2 × 106 I/11 cells was added in 4 ml StemPro-34 medium supplemented with Epo, SCF, and Dex and cocultured for 24 h. Subsequently, I/11 cells were removed from the Phoenix cells and cultured in semisolid medium (methocel-containing StemPro-34, supplemented with factors) containing 2 μg/ml puromycin (Sigma). After 7 days, well-separated colonies were picked, expanded, and analyzed for myc-eIF4E expression.
Immunofluorescence microscopy.
Cells (eIF4E-overexpressing and empty vector clones) were spun onto a microscope slide. The cells were fixed in 4% paraformaldehyde and permeabilized for 30 min with 0.2% Triton X-100. After being blocked for 1 h in PBS containing 1% bovine serum albumin and 0.05% Tween 20, the fixed cells were incubated for 1 h at room temperature with anti-eIF4E antibody and with anti-Myc antibodies for the eIF4E-overexpressing cells. The slides were washed and incubated for 1 h at room temperature with fluorescein isothiocyanate (FITC) anti-rabbit or tetramethyl rhodamine isothiocyanate (TRITC) anti-mouse secondary antibody (DakoCytomation), respectively. Coverslips were mounted with a drop vector shield (Vector Laboratories, Inc.), including DAPI (4′,6′-diamidino-2-phenylindole) (0.3 ng/μl). Imaging of the cells was done with 543-nm, 488-nm, and 405-nm excitation provided by an argon laser and a 63 × 1.4-numerical-aperture Apochromat objective lens (Carl Zeiss MicroImaging) for FITC, TRITC, and DAPI, respectively. Zeiss AIM software, version 3.2, was used for merging the images.
Flow cytometry.
To distinguish between live and dead cells, the DNA content was determined. A total of 0.5 × 106 to 1.0 × 106 cells was fixed and permeabilized with ice-cold methanol (0.5 ml; 30 min), washed two times with PBS, and incubated for 30 min with 0.5% (wt/vol) RNase A in PBS under constant shaking. DNA was stained with propidium iodide (PI) (50 mg/ml in PBS), and fluorescence was measured by flow cytometry.
RNA isolation and cDNA synthesis.
Total RNA was isolated using the Trizol reagent (Life Technologies) as recommended in the manufacturer's protocol. Isolation of polysomal RNA by sucrose gradient fractionation was performed as described previously (28, 36). Cell extracts were layered on a 4-ml linear sucrose gradient (15 to 40% [wt/vol] sucrose), and eight fractions were collected. Northern blotting indicated that fractions 1 to 4 contain nonpolysomal and subpolysomal mRNA, and that fractions 5 to 8 consisted of polysome-bound RNA. These fractions were pooled to generate subpolysomal and polysomal mRNA for each sample. RNA was quantified by UV absorbance. Poly(A)+ mRNA was purified, and cDNA was generated as described previously (28).
Real-time PCR.
The real-time PCR assay involved TaqMan technology (PE Applied Biosystems; model 7700 sequence detector) using the double-stranded-DNA-specific fluorescence dye SYBR green I to detect PCR product as previously described (30). The amplification program consisted of 1 cycle of 50°C with a 2-min hold (AmpErase UNG incubation) and 1 cycle of 95°C with a 10-min hold (AmpliTaq Gold activation), followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 62°C for 30 s, and extension at 72°C for 30 s. All the different primer pairs had similar optimal PCR annealing temperatures. Acquisition of the fluorescence signal from the samples was carried out at the end of the elongation step. To confirm amplification specificity, the PCR products from each primer pair were subjected to agarose gel electrophoresis, and the dissociation curve was checked at the end of each run. Gene-specific primers corresponding to Ndpk-B (X68193), eukaryotic translation elongation factor eEF-1β2 (MGC:6763), eIF4E (M61731), Fli-1 (X59421), ornithine decarboxylase (ODC; M12330), mammalian RNase inhibitor (IMAGE:1366946), U2 splicing factor Snrpb" (AA146248), and Y-box-binding protein 1 (YB-1; X57621) were obtained from Invitrogen Life Technologies or Sigma-Genosys, Ltd. The sequences of the primers used for the amplification are listed in Table 1.
TABLE 1.
Primer sequences used for real-time PCR amplification
| Primer name | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| Ndpk-B | 5′ATG GGA TTC GGA GAC CTG AA3′ | 5′TCA GCA GGT GGT GGA CCA GA3′ |
| eEF-1β2 | 5′ATG GGA TTC GGA GAC CTG AA3′ | 5′TCA GCA GGT GGT GGA CCA GA3′ |
| eIF4E | 5′TCT AAT CAG GAG GTT GCT AAC3′ | 5′TAG ACA ACT GGA TAT GGT TGTA3′ |
| Fli-1 | 5′TGC AGC CAC ATC CAA CAG AG3′ | 5′TGA AGG CAC GTG GGT GTT AG3′ |
| ODC | 5′TG ACG TCA TTG GTG TGA GC3′ | 5′TAT CAA GCA GAT GCA TGC TGT3′ |
| RI | 5′TCC AGT GTG AGC AGC TGA G3′ | 5′TGC AGG CAC TGA AGC ACC A3′ |
| U2 Snrpb" | 5′TCA GTT TGG ACA CGT GGT AG3′ | 5′TCC TTG TCA GCG AAA GTA CCA3′ |
| YB-1 | 5′TGC AGG AGA GCA AGG TAG AC3′ | 5′TGG TGG ATC GGC TGC TTT TG3′ |
RESULTS
Epo and SCF control eIF4E levels available for mRNA translation.
Since 4E-BP sequester eIF4E in the absence of growth factors, we examined 4E-BP phosphorylation in erythroid progenitors in response to Epo and SCF. Upon factor deprivation, only unphosphorylated 4E-BP are detected. Epo induced partial phosphorylation of 4E-BP, while only SCF induced complete phosphorylation of 4E-BP (Fig. 1A). A comparison of phosphospecific and total 4E-BP staining showed that the migration of 4E-BP is directly related to its phosphorylation state. An unphosphorylated α-isoform, a partially phosphorylated β-isoform, and a fully phosphorylated γ-isoform can be discerned in agreement with the proposed two-step phosphorylation mechanism (23). Importantly, SCF but not Epo is able to induce phosphorylation of Ser64, the last step in a cascade involving the sequential phosphorylation of T36/T45, T69, and S64 to release eIF4E (Fig. 1B) (25). Epo/SCF-induced phosphorylation of 4E-BP was abrogated in the presence of the PI3K inhibitor LY294002 and the mTOR inhibitor rapamycin (Fig. 1B). The MEK1 inhibitor U0126 partially inhibited the hyperphosphorylation of 4E-BP (Fig. 1C), which may indicate that part of the tuberous sclerosis complex/mTOR pathway is controlled through MEK1 (37, 44). However, the major effect of LY294002 and the minor effect of U0126 are in accordance with the observation that the inhibition of PI3K but not MEK1 abolished polysome association of Ndpk-B (28).
FIG. 1.
Phosphorylation of 4E-BP is controlled by SCF and Epo in erythroid progenitors. (A and B) I/11 cells were factor deprived for 4 h, stimulated with Epo (lane E; 5 U/ml), SCF (lane S; 100 ng/ml), or Epo plus SCF (lane ES) for 10 min, or left untreated (lane NF). (C) I/11 cells were factor deprived in the presence of 10 μM LY294002 (lane LY), 10 nM rapamycin (lane R), or 20 μM U0126 (lane U) and stimulated for 10 min with Epo and SCF (ES) in the presence of the inhibitors. Minus lanes indicate no inhibitor added. Western blots with total cell lysates (A and C) or with 4E-BP immunoprecipitates (B) were stained with antibodies recognizing T36/T45-phosphorylated 4E-BP (α-P[T36/T45]-4E-BP), S64-phosphorylated 4E-BP (α-P[S64]-4E-BP), or total 4E-BP (α-4E-BP). ERK 1/2 staining was used as a sample loading control (α-ERK1/2). The unphosphorylated α-isoform, a partially phosphorylated β-isoform, and a fully phosphorylated γ-isoform of 4E-BP are indicated in panel A. (D) Samples were taken at various time points (0, 10, 30, 60, 120, and 180 lanes) following the addition of Epo or SCF (lane ss, steady state; cells not subjected to factor deprivation). Western blots were stained with antibodies recognizing 4E-BP. (E) I/11 cells were factor deprived (lane NF) or restimulated (lanes E and S) as indicated in panels A and B, and eIF4E was precipitated with m7GTP-Sepharose. Precipitates were stained for eIF4G, 4E-BP, and eIF4E on Western blots.
Since only SCF induced full phosphorylation of 4E-BP during a 10-min stimulation, we examined the kinetics of 4E-BP phosphorylation to exclude the possibility that the difference was due to different kinetics of induction and feedback modulation. Even after prolonged stimulation of erythroid progenitors with Epo, phosphorylation of 4E-BP was partial and only small amounts of the fully phosphorylated γ-isoform were detected. In contrast, SCF rapidly induced complete phosphorylation of 4E-BP, which was not dephosphorylated in the continuous presence of the factor (Fig. 1D). Subsequently, we tested whether the phosphorylation of 4E-BP actually resulted in reduced 4E-BP binding to eIF4E and increased association of eIF4E with the scaffolding protein eIF4G. m7GTP-Sepharose was used to precipitate eIF4E from an extract of 15 × 106 cells that were factor deprived or restimulated with Epo and/or SCF. Surprisingly, the amount of 4E-BP pulled down from restimulated cells increased, concomitant with increased precipitation of eIF4E. However, eIF4G binding to eIF4E also increased (Fig. 1E). This indicates that 4E-BP is in excess but that stimulation of erythroid progenitors with SCF does induce eIF4F complexes.
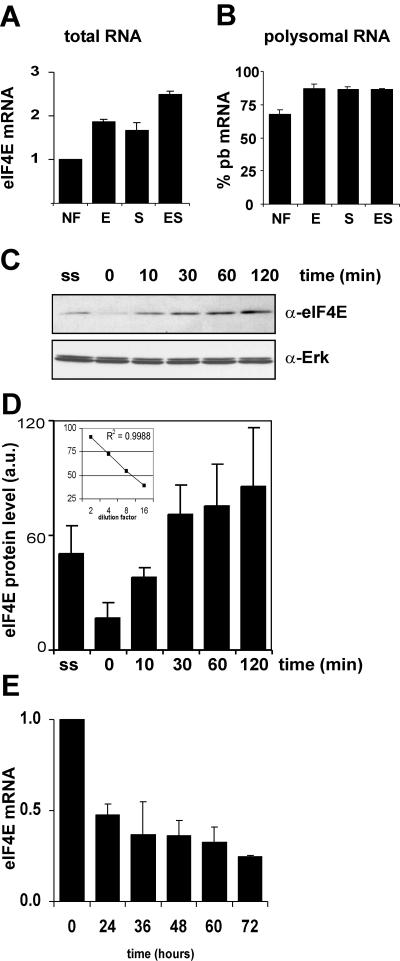
The level of eIF4E available for mRNA translation initiation is regulated by 4E-BP phosphorylation, but we also observed factor-induced upregulation of eIF4E expression in mRNA profiling experiments (30). Real-time quantitative PCR showed that, following a 2-h Epo, SCF, or Epo/SCF exposure, eIF4E mRNA increased approximately twofold in I/11 erythroblasts compared to that in factor-deprived I/11 cells (Fig. 2A). This upregulation was due to an increase in total transcript because the relative distributions between subpolysome- and polysome-associated mRNA did not change in response to factor deprivation and restimulation (Fig. 2B). Increased mRNA expression results in increased protein expression (Fig. 2C and D), which is in accordance with the data shown in Fig. 1E. During terminal erythroid differentiation, eIF4E mRNA expression was downmodulated approximately fourfold in 72 h (Fig. 2E).
FIG. 2.
eIF4E expression is upregulated upon growth factor stimulation in I/11 cells. (A and B) I/11 cells were factor deprived for 4 h and stimulated with Epo (E), SCF (S), or Epo plus SCF (ES) or left untreated (NF) for 2 h. Total (A) and free and polysome-associated mRNA (B) was isolated, and eIF4E mRNA was quantified using real-time PCR. (A) eIF4E expression is given as a change (n-fold) ratio of that in factor-stimulated cells to that in untreated cells. (B) The percentage of eIF4E mRNA present in the polysome-bound (pb) fraction is calculated. (C) I/11 cells were factor deprived and restimulated with Epo plus SCF for various time intervals (0, 10, 30, 60, and 120 lanes) as indicated. Protein samples were analyzed on Western blots for eIF4E expression. The Western blots were stained with α-actin to control for equal loadings. ss, steady state. (D) Quantification of eIF4E intensities. Bars indicate the averages from three independent experiments, a representative of which is shown in 2C. Error bars indicate standard deviations. ss, steady state. The insert demonstrates the linearity of the eIF4E antibody used (R2 = 0.9988). Twofold dilutions of total cell lysate were tested for eIF4E expression. Expression is given in arbitrary units (a.u.). (E) I/11 cells were induced to differentiate, and samples were harvested for mRNA isolation at the indicated time points. Expression of eIF-4E was examined by real-time RT-PCR and is given as a change (n-fold) ratio of eIF-4E expression to expression in expanding I/11 cells (time zero).
Together, the data show that the level of eIF4E available for mRNA translation is controlled by SCF on at least two levels: phosphorylation of 4E-BP allowing increased eIF4G binding and increased expression.
eIF4E overexpression in I/11 erythroid progenitor cells impairs differentiation and enhances cell renewal.
If regulation of eIF4E is a major effector of SCF-induced PI3K, overexpression of eIF4E is expected to affect the PI3K-dependent balance between the expansion and differentiation of erythroid progenitors (49). Using retroviral expression vectors, we established multiple clones expressing myc-tagged eIF4E (eIF4E-myc) (Fig. 3A). The eIF4E-myc could efficiently bind 4E-BP, which enabled the exogenously expressed eIF4E-myc to titrate the inhibitory function of 4E-BP (Fig. 3B). Using m7GTP-Sepharose, we pulled down eIF4E from control and eIF4E-overexpressing cells that were factor deprived and restimulated. In contrast to control cells, factor-deprived eIF4E-overexpressing cells contained eIF4E-eIF4G complexes (Fig. 3C). More eIF4G was recruited by eIF4E in the eIF4E-overexpressing erythroid progenitors under all conditions (Fig. 3C and D). Since all unphosphorylated 4E-BP was bound by the excess of eIF4E, induction of 4E-BP phosphorylation upon stimulation of the cells with SCF resulted in a more pronounced decrease in 4E-BP binding than that observed in control cells (compare Fig. 1E and 3D). In conclusion, although the level of eIF4E-myc overexpression detected in Fig. 3A is modest compared to that of endogenous eIF4E, the increase in eIF4G association indicates that the expression of eIF4E-myc significantly enhances the level of free eIF4E (not bound to 4E-BP).
FIG. 3.
Ectopic eIF4E-myc protein is able to interact and titrate out endogenous 4E-BP and is localized mainly in the cytoplasm of I/11 cells. (A) Lysates of an empty-vector-transduced clone (eV lane) and various eIF4E-myc-transduced clones (eIF4E overexpressor lanes) were tested for eIF4E expression on Western blots using antibodies recognizing eIF4E (top panel) or the myc-tagged protein (bottom panel). The eIF4E-myc and endogenous eIF4E proteins have different mobilities and are indicated by arrows. (B) myc-eIF4E-expressing I/11 cells expanding in the presence of Epo, SCF, and Dex (steady state [ss] lanes) or factor deprived for 4 h (NF lanes) were lysed, and eIF4E-myc was immunoprecipitated with anti-Myc antibody (α-myc, + lanes). Mock immunoprecipitations using Sepharose beads without antibody (− lanes) served as controls. Western blots were stained for 4E-BP (top) or the myc-tagged protein (bottom). Unphosphorylated and hypophosphorylated 4E-BP (α- and β-forms) were coimmunoprecipitated with eIF4E-myc. The myc-tagged eIF4E could efficiently bind to endogenous 4E-BP. (C and D) eIF4E was precipitated by m7GTP-Sepharose and tested for eIF4G and 4E-BP association on Western blots. Endogenous eIF4E was detected by specific antibody, and eIF4E-myc was detected by anti-Myc antibody. Lysates were prepared from eIF4E-myc-expressing and control I/11 cells that were steady-state expanding (lanes ss) or factor deprived (lanes NF) (C) or factor deprived and restimulated with Epo (lanes E; 5 U/ml, 60 min), SCF (lanes S; 100 ng/ml; 60 min), or Epo plus SCF (lanes ES) (D). (E) Cytospins of empty vector (upper left panel) and eIF4E-myc (middle and lower left panels) expressing I/11 cells are fixed and stained for eIF4E (upper and middle left panels) or the myc-tagged protein (lower left panel) using FITC- and TRITC-labeled second antibodies, respectively. In addition, nuclei of all cells are stained with DAPI (middle panels). The right panels represent the overlay of eIF4E or myc-tagged protein with DAPI. A small fraction of endogenous eIF4E appeared in nuclear bodies, whereas eIF4E-myc seemed to be exclusively cytoplasmic.
Using immunofluorescence, we analyzed the subcellular localization of endogenous eIF4E and of eIF4E-myc. Previously, it has been shown that eIF4E may accumulate in nuclear bodies to prevent cytoplasmic translocation of specific mRNAs (46). A small fraction of endogenous eIF4E appeared in nuclear bodies, but eIF4E-myc appeared to be exclusively cytoplasmic (Fig. 3E). Thus, it is not likely that eIF4E-myc in erythroid progenitors functions through a nontranslational mechanism such as retaining specific mRNAs in the nucleus. Finally, we also expressed an eIF4E mutant in which tryptophan-73 was mutated to alanine (W73A mutant) in I/11 cells. The W73A mutant is unable to bind the scaffolding protein eIF4G or 4E-BP. A low level of eIF4E(W73A) expression was detected in mass cultures within a week after transduction, but expression of this mutant was rapidly lost and we were unable to establish single-cell-derived clones expressing the W73A mutant (data not shown). This suggests that positive factors can also be competed for by eIF4E-myc overexpression.
To examine the effect of eIF4E overexpression on the balance between the expansion and differentiation of erythroid progenitors, we exposed vector control cells and eIF4E-myc-expressing clones to conditions inducing expansion (Epo, SCF, and Dex), delayed differentiation (Epo and SCF), differentiation (Epo), or apoptosis (no factor). Both control and eIF4E-myc-expressing cells failed to proliferate or mature in the absence of growth factors (Fig. 4A and E). Under differentiation conditions, i.e., in the presence of Epo only, control cells undergo prompt differentiation, characterized by transient proliferation, decreased cell size, and the accumulation of hemoglobin (Fig. 4A to C). Notably, this differentiation is markedly delayed in eIF4E-myc-expressing cells, which is indicated by prolonged expansion, maintenance of a blast-like cell size, and lack of hemoglobin accumulation (Fig. 4E to G). In the presence of Epo plus SCF, control cells were delayed in differentiation, but cell size reduction started almost as rapidly as in the presence of Epo only. Under these conditions, differentiation of eIF4E-myc-expressing cells was essentially absent. The delay of differentiation in eIF4E-myc-expressing cells is also evident from the cell morphology of the distinct cultures after 4 days of exposure to Epo (Fig. 4D and H). The effect of eIF4E-myc expression on Epo-dependent differentiation was analyzed for multiple independent clones, which showed consistent inhibition of differentiation (Fig. 5).
FIG. 4.
Overexpression of eIF4E impairs differentiation of I/11 erythroid progenitors. I/11 cells transduced with an empty control vector (A to D) or with an eIF4E expression vector (E to H) were seeded in medium without factor (−) or supplemented with Epo (E; 2 U/ml), Epo plus SCF (ES; 100 ng/ml SCF), or Epo, SCF, and dexamethasone (ESD; 10−6M dexamethasone). (A and E) Cumulative cell number, (B and F) cell size, and (C and G) hemoglobin content per cell volume (arbitrary units [a.u.]) were analyzed at regular intervals for 7 days. At day 4, cells seeded in Epo were harvested for cytospins and stained for hemoglobin (brown) and histological dyes (D and H). Hemoglobinized and enucleated erythrocytes were present in control cells, while eIF4E-expressing cells contain mainly blasts.
FIG. 5.
Overexpression of eIF4E consistently impairs differentiation of I/11 erythroid progenitors. (A and B) Vector and eIF4E-myc-transduced clones were subjected to differentiation as described for Fig. 4. The average cell volume (A) and hemoglobin content (B), as measured 3 and 4 days following induction of differentiation, are shown for six clones. Error bars represent standard deviations.
Measuring the DNA content by flow cytometry at day 3 following reseeding in differentiation medium confirmed that control cells arrest in the G1 phase of the cell cycle, while eIF4E-myc cells can be detected in all phases of the cell cycle (Fig. 6A; the G1 peak of control cells is shifted to the left because the highly condensed DNA of late erythroblasts binds less propidium iodine; no significant number of apoptotic cells is detected by annexin staining or the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling assay). However, eIF4E-myc-expressing cells are not transformed and fail to undergo prolonged renewal in the presence of Epo. Seven days following reseeding in differentiation conditions, the cultures of eIF4E-myc-expressing cells accumulate cells with a sub-G1 DNA content and annexin-positive staining (Fig. 6B and data not shown), suggesting increased apoptosis. In the control cultures, all cells enucleated. These results were confirmed in four control clones and six eIF4E-myc-expressing clones. The failure to undergo differentiation could be due to the failure to downregulate eIF4E (Fig. 2C). Indeed, expression of eIF4E protein decreased during the differentiation of control cells, followed by decreased expression of 4E-BP. However, expression of endogenous eIF4E remained high in eIF4E-myc progenitors (Fig. 6C).
FIG. 6.
eIF4E overexpression in I/11 cells impairs differentiation but fails to prolong renewal in the presence of Epo. (A and B) Empty vector (EV) control clones and eIF4E-myc-expressing cells (eIF4E) were fixed and permeabilized, stained with PI, and analyzed by flow cytometry. (A) Three days following reseeding of the cells in differentiation medium (supplemented with Epo only), the EV control cells were predominantly arrested in the G1 phase of the cell cycle, and the G2 peak was absent. The level of PI staining is relatively low because the nuclei are condensed and bind less PI. A prominent peak representing the G2 phase of the cell cycle is present in eIF4E-overexpressing cells. (B) At day 6 and day 7 following seeding in differentiation medium, the eIF4E-overexpressing cells were still in cycle as demonstrated by the G2 peak, but in time, the number of cells with a sub-G1 DNA content (indicating dead cells) accumulates. (C) Protein lysates of empty vector (EV lanes) control clones and eIF4E-myc-expressing cells (eIF4E lanes) were harvested at increasing intervals following the reseeding of the cells in differentiation medium. Western blots were stained with antibodies recognizing endogenous eIF4E, 4E-BP, or ERK 1/2 as indicated. Erk served as a loading control.
PI3K inhibition does not abrogate cell expansion of eIF4E-overexpressing I/11 cells.
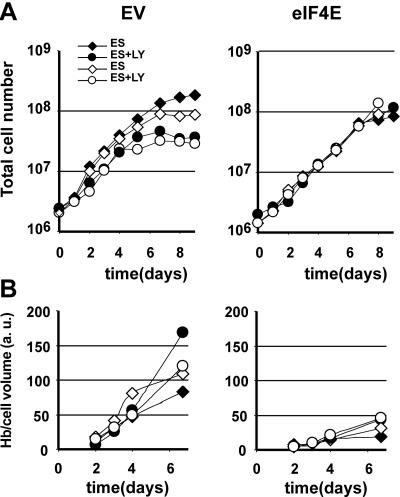
We showed that PI3K activity rather than MEK1/extracellular signal-regulated kinase (ERK) activation is essential to maintain cell renewal divisions in erythroid progenitors (49). Since the phosphorylation of 4E-BP and the release of eIF4E is downstream of PI3K, we analyzed whether constitutive expression of eIF4E maintains cell renewal in the presence of the PI3K-inhibitor LY294002. The addition of LY294002 to renewal conditions decreased the expansion of control cultures and accelerated cell differentiation evidenced by increased hemoglobinization. In contrast, eIF4E-myc-expressing cells did not alter cellular renewal or differentiation in response to LY294002 (Fig. 7). This indicates that control of translation initiation is a major target of PI3K in erythroid development.
FIG. 7.
Overexpression of eIF4E renders cells insensitive to the PI3K inhibitor LY294002. Two empty-vector control clones and two eIF4E-overexpressing clones (black and white symbols indicate separate clones) were seeded in medium supplemented with Epo and SCF (ES) in the absence (diamonds) or presence (circles) of LY294002 (LY; 10 μM). (A) Total cell number and (B) hemoglobin (Hb) content per cell volume (in arbitrary units [a.u.]) were monitored daily.
Polysome association of Ndpk-B, U2-Snrpb", and YB-1 transcripts depends on PI3K activation or eIF4E expression.
Recently we screened for Epo- and SCF-induced genes in a profiling assay using polysome-bound mRNA. The expression of Ndpk-B appeared to be controlled specifically at the level of polysome recruitment by a mechanism involving SCF-activated PI3K activity (28). Since other targets upregulated by SCF may be regulated by the same mechanism, the expression of Epo- and SCF-upregulated genes was examined by quantitative reverse transcription-PCR in total, free, and polysome-associated mRNA fractions (data not shown). In addition to Ndpk-B, we found expression of the splicing factor U2-Snrpb" and the Y-box-binding protein 1 (YB-1) to be regulated by polysome association (data not shown and Fig. 8). In the absence of growth factor, less than 10, 5, and 25% of Ndpk-B, YB-1, and U2-Snrpb" mRNAs, respectively, were detected in the polysomal fractions. Upon stimulation with Epo and SCF, 75% of the Ndpk-B, 80% of the U2-Snrpb", and 50% of the YB-1 messengers were found in the polysomal fractions, and polysome association was dependent on PI3K activation (Fig. 8A to C).
FIG. 8.
eIF4E overexpression increases polysome association of structured mRNAs in the absence of growth factors or in the presence of growth factors and the PI3K inhibitor LY294002. I/11 control cells (filled bars) and eIF4E-overexpressing cells (open bars) were factor deprived in the absence or presence of LY294002 (LY; 10 μM) or rapamycin (R; 10 nM) and restimulated for 2 h with Epo (E; 5 U/ml), SCF (S; 100 ng/ml), or Epo plus SCF (ES). In addition, cells were left untreated (NF) or were harvested from steady-state proliferating cultures (ss). Free and polysome-bound mRNA was isolated and assayed for the expression of (A) NdpkB, (B) YB-1, (C) U2-Snrpb", (D) eIF1β2, (E) ODC, and (F) Fli-1. The percentage of mRNA associated with polysomes (pb mRNA) was calculated for the different genes under the different conditions.
Whereas the 5′UTR of Ndpk-B starts with a TOP tract and contains an inverted repeat, the 5′UTR of U2-Snrpb" is short and contains a TOP sequence (AA146248), and the 5′UTR of YB-1 is long and highly structured and lacks a TOP sequence (21) (X57621). Thus, these three genes represent different types of mRNAs and may be representative panels to study the pathways involved in gene-specific mRNA recruitment to polysomes. We examined the effect of eIF4E overexpression on factor-dependent polysome association of Ndpk-B, U2-Snrpb", and YB-1 mRNA. Polysome recruitment of Fli-1 mRNA was analyzed as a negative control to assess the general effect of LY294002 and rapamycin on polysome formation. Polysome association of eukaryote elongation factor 1β (eEF1β) and ODC was examined as a positive control. They contain a TOP sequence and a highly structured 5′UTR, respectively. Enhanced eIF4E expression increased polysome association of Ndpk-B, YB-1, and U2-Snrpb" mRNA in factor-deprived cells, and it rendered factor-induced polysome association resistant to the PI3K inhibitor LY294002 (Fig. 8). The polysome association of the mRNAs in Epo- or SCF-stimulated cells was the same for control and eIF4E-overexpressing cells.
Both control mRNAs, eEF1β and ODC, known to be regulated by polysome association, showed the same levels of regulation as Ndpk-B, YB-1, and U2-Snrpb". In contrast, at least half of all Fli-1 mRNA remained associated with polysomes independently of PI3K activity. Upon factor deprivation and inhibition of PI3K, 20 to 25% of the Fli-1 mRNA was lost from polysomes. This can be considered the general effect of growth factor stimulation on mRNA translation initiation, which is much lower than the effect observed for structured mRNAs.
DISCUSSION
Control of the expansion versus maturation of hematopoietic progenitors requires a tight regulation of the gene expression program. It becomes increasingly evident that the expression of genes critical for progenitor renewal and tumorigenesis is regulated not only at the level of gene transcription, but also by control of mRNA translation (14). The expansion of erythroid progenitors is critically dependent on SCF-induced PI3K activity. SCF-induced PI3K/PKB activity controls the level of eIF4E available for translation initiation, and we showed that overexpression of eIF4E in erythroid progenitors impairs their differentiation and enhances renewal divisions in the absence of SCF. Notably, the inhibition of PI3K did not lead to the differentiation of eIF4E-overexpressing erythroid progenitors as it does in control cells, indicating that control of eIF4E is a pathway of major importance downstream of PI3K in expanding erythroid progenitors. We observed that either SCF-induced signal transduction or eIF4E overexpression resulted in a significant increase in the polysome recruitment of a specific set of mRNAs with a structured 5′UTR, such as Ndpk-B, YB-1, and U2-Snrpb". This supports the notion that the expression of not only ribosomal proteins and translation factors but also other proteins with an important role in cellular metabolism and proliferation is controlled at the level of polysome recruitment. Therefore, we suggest that gene-specific recruitment of mRNAs into polysomes by SCF-induced PI3K/PKB activity may contribute to the control of expansion and maturation of erythroid progenitors.
Release of eIF4E is a major pathway downstream of PI3K in erythroid progenitors.
Overexpression of eIF4E specifically inhibits differentiation. Notably, the induction of erythroid maturation does not require the inhibition of cell cycle progression but involves three to four cell divisions with loss of cell size control, resulting in 8 to 16 erythrocytes (4 μm, in mice) from a single proerythroblast (12 μm) (16, 49). In consequence, maintenance of cell size is crucial in renewal divisions, which requires a proper balance between protein synthesis and G1 progression through the restriction point in the cell cycle (17, 50). This indicates an important role for protein synthesis and for a limiting factor in mRNA translation, such as eIF4E in particular, in the control of the expansion and differentiation of erythroid progenitors.
We previously showed that inhibition of PI3K abrogates the ability of erythroid progenitors to maintain a renewal program (49). Because overexpression of eIF4E rendered the expansion of erythroid progenitors insensitive to the PI3K inhibitor LY294002, selective protein synthesis appears to be a major pathway downstream of SCF-induced progenitor expansion. However, we also demonstrated that SCF-induced activation of the PI3K/PKB pathway results in the phosphorylation of Foxo3, which results in its cytoplasmic retention and inhibition of its transcriptional activity (5). Among the Foxo3 targets are p27, p130Rb2, Btg1, and cyclin G2, which all inhibit cell cycle progression (5; W. J. Bakker, submitted for publication). These genes are upregulated when PI3K activity is inhibited in control cells. Yet, erythroid progenitors overexpressing eIF4E undergo normal renewal divisions in the absence of PI3K activity. This seeming contradiction may have two explanations. First, the expression of Foxo family members is low in erythroid progenitors, and Foxo3 expression is markedly increased during differentiation, resulting in Foxo3 activity 48 h after induction of differentiation when cells arrest in G1. The low levels of Foxo3 protein in erythroid progenitors may result in insufficient expression of the cell cycle inhibitors to arrest cells when all other signals are activated. Second, preliminary analysis of translationally controlled genes suggests that increased eIF4E expression results in the enhanced expression of proteins that inhibit the cell cycle inhibitors such as kinase interacting with stathmin (KIS) (unpublished data). Thus, control of eIF4E availability is a major pathway downstream of PI3K to maintain renewal divisions of erythroid progenitors. Upon downregulation of PI3K, other pathways, such as Foxo3 activation, are activated to execute the differentiation program.
Constitutive eIF4E expression does not render erythroid progenitors independent of growth factors.
It has been shown that the PI3K/PKB/mTOR pathway is required for the tumorigenic phenotype of various tumors and sufficient for oncogenic transformation of chicken embryo fibroblasts (2, 3, 15). In breast tumor-derived cell lines and eIF4E-transformed fibroblasts, increased expression of eIF4E and phosphorylation of 4E-BP prevent apoptosis upon factor deprivation. Erythroid progenitors overexpressing eIF4E are not factor independent for either renewal or differentiation and are not prevented from apoptosis in the absence of growth factors. Moreover, eIF4E expression impairs differentiation but is not able to sustain long-term renewal. Upon moderate overexpression, the cells undergo delayed differentiation, but high levels of eIF4E eventually induce erythroid progenitors to die under differentiation conditions. Whereas eIF4E availability may be the crucial pathway downstream of PI3K signaling, it is not sufficient for sustained expansion or differentiation of erythroid progenitors. The distinct effects of eIF4E overexpression in different cell types, i.e., control of apoptosis versus control of differentiation, is in accordance with the different functions of PI3K and Foxo transcription factors in these cells. Whereas PI3K is known mainly as a survival factor in fibroblastoid cells (20), it controls the balance between the expansion and differentiation of erythroid progenitors (5, 47). The tyrosine kinase receptor RON/Stk is a downstream target of the EpoR (47). Direct activation of RON, using a NGF-inducible TrkA-RON fusion, induced phosphorylation of Gab1 and Gab2 and strong PI3K activation, but it was not able to induce renewal divisions in the absence of SCF or differentiation in the absence of Epo. Thus, expansion requires additional signaling pathways initiated by the SCF receptor cKit, and differentiation requires Epo-induced Stat5 activation (18, 47). Whether the biological effect of eIF4E is the rescue from apoptosis or impaired differentiation, in either case, it contributes to enhanced expansion of progenitors and to neoplastic transformation.
Increased eIF4E expression recruits specific RNA transcripts to polysomes.
Although eIF4E is predominantly a translation initiation factor, overexpression of eIF4E in myeloid progenitors was suggested to contribute to leukemogenesis by increased nuclear export of cyclin D1 (46). The proline-rich homeodomain protein, PRH/Hex, disrupts eIF4E nuclear bodies and reduces export of D-cyclins (46). We did observe nuclear bodies in control cells using antibodies against endogenous eIF4E, but upon overexpression of eIF4E, these nuclear bodies disappeared. Interestingly, we found that PRH/Hex is upregulated in response to SCF. This involved both transcriptional and translational control, as the 28-bp-long 5′UTR of the PRH transcript contains a TOP sequence (G. Grech, unpublished results). Possibly, the increased expression of PRH/Hex suppressed the nuclear localization of eIF4E.
In consequence, the delayed differentiation of erythroid progenitors upon overexpression of eIF4E must be due to the effects of eIF4E on mRNA translation. Increased availability of eIF4E allows more eIF4F, harboring the eIF4A helicase, to bind and scan structured mRNAs, i.e., to recruit the structured mRNAs into polysomes (24). Until recently, PI3K/PKB/mTOR-dependent polysome recruitment was described mainly for mRNAs containing a TOP sequence and encoding ribosomal proteins and translation factors (34, 43). Few other structured mRNAs were shown to be regulated at the level of translation initiation like c-Myc and ODC (14). In this paper, we describe PI3K- and eIF4E-dependent polysome recruitment of Ndpk-B, YB-1, and U2-Snrpb". Notably, both Ndpk-B and YB-1 are suggested to be involved in tumor progression (26, 38). The Ndpk-B 5′UTR contains a TOP tract and an inverted repeat predicted to form a stem-loop structure (28). Removal of the Ndpk-B 5′UTR caused the loss of Ndpk-B translational control. YB-1 mRNA contains a highly structured 5′UTR and lacks a TOP tract, whereas U2-Snrpb" has a short 5′UTR starting with a TOP sequence. All three mRNAs as well as the controls EF1β (TOP mRNA) and ODC (structured mRNA) were hardly or not associated with polysomes in the absence of growth factors. Of unstructured mRNAs, such as Fli-1 mRNA, at least 50% of the mRNA remains associated with polysomes in the absence of growth factors. Overexpression of eIF4E increased polysome association of Ndpk-B, YB-1, and U2-Snrpb" in the absence of growth factors but did not completely rescue polysome association relative to SCF-induced polysome association. This suggests that additional signaling still plays a role in mRNA translation, possibly by the phosphorylation of proteins that stabilize structures in the mRNAs.
Besides increased recruitment of structured mRNAs to polysomes, increased expression of eIF4E may act via other mechanisms as well, and such additional mechanisms may contribute to the observed phenotype of eIF4E-overexpressing erythroid progenitors. In conjunction with other initiation factors (eIF2 and eIF3), eIF4E affects the selection of the ATG initiation codon. Under conditions of suboptimal growth factors, protein synthesis will start at the first ATG start codon that is embedded in an optimal Kozak sequence (32). When translation initiation factors are abundantly available, less-optimal ATG codons can be selected to start protein synthesis, and eIF4F will continue mRNA scanning beyond a first open reading frame (ORF) to reinitiate at the next ATG. Short ORFs in the 5′UTR can serve to attenuate translation of the functional ORF (e.g., thrombopoietin) (22), but upstream ORFs can also affect the choice of the start codon for the functional protein and thereby the translation of antagonistic proteins from the same mRNA (e.g., CEBPβ and SCL) (11, 12).
Acknowledgments
We thank Jorie Vermissen for her help with the RNA helicase screening, I. Touw and R. Delwel for many fruitful discussions and critical reading of the manuscript, and Gert Scheper for his kind gift of myc-eIF4E-pCS3MT plasmids.
This work was supported by grants from the European Union (HPRN-CT-2000-00083) and The Netherlands Organization for Scientific Research (050-10-051).
REFERENCES
- 1.Alvarez, B., E. Garrido, J. A. Garcia-Sanz, and A. C. Carrera. 2003. Phosphoinositide 3-kinase activation regulates cell division time by coordinated control of cell mass and cell cycle progression rate. J. Biol. Chem. 278:26466-26473. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, M., E. Blazek, and P. K. Vogt. 2001. A role of the kinase mTOR in cellular transformation induced by the oncoproteins P3k and Akt. Proc. Natl. Acad. Sci. USA 98:136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avdulov, S., S. Li, V. Michalek, D. Burrichter, M. Peterson, D. M. Perlman, J. C. Manivel, N. Sonenberg, D. Yee, P. B. Bitterman, and V. A. Polunovsky. 2004. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cells 5:553-563. [DOI] [PubMed] [Google Scholar]
- 4.Bader, A. G., and P. K. Vogt. 2004. An essential role for protein synthesis in oncogenic cellular transformation. Oncogene 23:3145-3150. [DOI] [PubMed] [Google Scholar]
- 5.Bakker, W. J., M. Blazquez-Domingo, A. Kolbus, J. Besooyen, P. Steinlein, H. Beug, P. J. Coffer, B. Lowenberg, M. von Lindern, and T. B. van Dijk. 2004. FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1. J. Cell Biol. 164:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer, A., F. Tronche, O. Wessely, C. Kellendonk, H. M. Reichardt, P. Steinlein, G. Schutz, and H. Beug. 1999. The glucocorticoid receptor is required for stress erythropoiesis. Genes Dev. 13:2996-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beghini, A., P. Peterlongo, C. B. Ripamonti, L. Larizza, R. Cairoli, E. Morra, and C. Mecucci. 2000. C-kit mutations in core binding factor leukemias. Blood 95:726-727. [PubMed] [Google Scholar]
- 8.Beug, H., S. Palmieri, C. Freudenstein, H. Zentgraf, and T. Graf. 1982. Hormone-dependent terminal differentiation in vitro of chicken erythroleukemia cells transformed by ts mutants of avian erythroblastosis virus. Cell 28:907-919. [DOI] [PubMed] [Google Scholar]
- 9.Broudy, V. C., N. L. Lin, G. V. Priestley, K. Nocka, and N. S. Wolf. 1996. Interaction of stem cell factor and its receptor c-kit mediates lodgment and acute expansion of hematopoietic cells in the murine spleen. Blood 88:75-81. [PubMed] [Google Scholar]
- 10.Broxmeyer, H. E., S. Cooper, L. Lu, G. Hangoc, D. Anderson, D. Cosman, S. D. Lyman, and D. E. Williams. 1991. Effect of murine mast cell growth factor (c-kit proto-oncogene ligand) on colony formation by human marrow hematopoietic progenitor cells. Blood 77:2142-2149. [PubMed] [Google Scholar]
- 11.Calkhoven, C. F., C. Muller, and A. Leutz. 2000. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 14:1920-1932. [PMC free article] [PubMed] [Google Scholar]
- 12.Calkhoven, C. F., C. Muller, R. Martin, G. Krosl, H. Pietsch, T. Hoang, and A. Leutz. 2003. Translational control of SCL-isoform expression in hematopoietic lineage choice. Genes Dev. 17:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta, S. R., A. Brunet, and M. E. Greenberg. 1999. Cellular survival: a play in three Akts. Genes Dev. 13:2905-2927. [DOI] [PubMed] [Google Scholar]
- 14.De Benedetti, A., and J. R. Graff. 2004. eIF-4E expression and its role in malignancies and metastases. Oncogene 23:3189-3199. [DOI] [PubMed] [Google Scholar]
- 15.De Benedetti, A., and A. L. Harris. 1999. eIF4E expression in tumors: its possible role in progression of malignancies. Int. J. Biochem. Cell Biol. 31:59-72. [DOI] [PubMed] [Google Scholar]
- 16.Dolznig, H., F. Boulme, K. Stagl, E. M. Deiner, W. Mikulits, H. Beug, and E. W. Mullner. 2001. Establishment of normal, terminally differentiating mouse erythroid progenitors: molecular characterization by cDNA arrays. FASEB J. 15:1442-1444. [DOI] [PubMed] [Google Scholar]
- 17.Dolznig, H., F. Grebien, T. Sauer, H. Beug, and E. W. Mullner. 2004. Evidence for a size-sensing mechanism in animal cells. Nat. Cell Biol. 6:899-905. [DOI] [PubMed] [Google Scholar]
- 18.Dolznig, H., B. Habermann, K. Stangl, E. M. Deiner, R. Moriggl, H. Beug, and E. W. Mullner. 2002. Apoptosis protection by the Epo target Bcl-X(L) allows factor-independent differentiation of primary erythroblasts. Curr. Biol. 12:1076-1085. [DOI] [PubMed] [Google Scholar]
- 19.Fingar, D. C., C. J. Richardson, A. R. Tee, L. Cheatham, C. Tsou, and J. Blenis. 2004. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 24:200-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franke, T. F., D. R. Kaplan, and L. C. Cantley. 1997. PI3K: downstream AKTion blocks apoptosis. Cell 88:435-437. [DOI] [PubMed] [Google Scholar]
- 21.Fukuda, T., M. Ashizuka, T. Nakamura, K. Shibahara, K. Maeda, H. Izumi, K. Kohno, M. Kuwano, and T. Uchiumi. 2004. Characterization of the 5′-untranslated region of YB-1 mRNA and autoregulation of translation by YB-1 protein. Nucleic Acids Res. 32:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghilardi, N., A. Wiestner, and R. C. Skoda. 1998. Thrombopoietin production is inhibited by a translational mechanism. Blood 92:4023-4030. [PubMed] [Google Scholar]
- 23.Gingras, A. C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 25.Gingras, A.-C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807-826. [DOI] [PubMed] [Google Scholar]
- 26.Janz, M., N. Harbeck, P. Dettmar, U. Berger, A. Schmidt, K. Jurchott, M. Schmitt, and H. D. Royer. 2002. Y-box factor YB-1 predicts drug resistance and patient outcome in breast cancer independent of clinically relevant tumor biologic factors HER2, uPA and PAI-1. Int. J. Cancer 97:278-282. [DOI] [PubMed] [Google Scholar]
- 27.Jefferies, H. B. J., S. Fumagalli, P. B. Dennis, C. Reinhard, R. B. Pearson, and G. Thomas. 1997. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 16:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joosten, M., M. Blazquez-Domingo, F. Lindeboom, F. Boulme, A. Van Hoven-Beijen, B. Habermann, B. Lowenberg, H. Beug, E. W. Mullner, R. Delwel, and M. Von Lindern. 2004. Translational control of putative protooncogene Nm23-M2 by cytokines via phosphoinositide 3-kinase signaling. J. Biol. Chem. 279:38169-38176. Epub 32004 Jul 38107. [DOI] [PubMed] [Google Scholar]
- 29.Kim, T. W., H. Lee, Y. K. Kang, M. S. Choe, M. H. Ryu, H. M. Chang, J. S. Kim, J. H. Yook, B. S. Kim, and J. S. Lee. 2004. Prognostic significance of c-kit mutation in localized gastrointestinal stromal tumors. Clin. Cancer Res. 10:3076-3081. [DOI] [PubMed] [Google Scholar]
- 30.Kolbus, A., M. Blazquez-Domingo, S. Carotta, W. Bakker, S. Luedemann, M. von Lindern, P. Steinlein, and H. Beug. 2003. Cooperative signaling between cytokine receptors and the glucocorticoid receptor in the expansion of erythroid progenitors: molecular analysis by expression profiling. Blood 102:3136-3146. [DOI] [PubMed] [Google Scholar]
- 31.Kowenz, E., A. Leutz, G. Doderlein, T. Graf, and H. Beug. 1987. ts-oncogene-transformed erythroleukemic cells: a novel test system for purifying and characterizing avian erythroid growth factors. Mod. Trends Hum. Leuk. 31:199-209. [DOI] [PubMed] [Google Scholar]
- 32.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Looijenga, L. H., H. de Leeuw, M. van Oorschot, R. J. van Gurp, H. Stoop, A. J. Gillis, C. A. de Gouveia Brazao, R. F. Weber, W. J. Kirkels, T. van Dijk, M. von Lindern, P. Valk, G. Lajos, E. Olah, J. M. Nesland, S. D. Fossa, and J. W. Oosterhuis. 2003. Stem cell factor receptor (c-KIT) codon 816 mutations predict development of bilateral testicular germ-cell tumors. Cancer Res. 63:7674-7678. [PubMed] [Google Scholar]
- 34.Meyuhas, O. 2000. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 267:6321-6330. [DOI] [PubMed] [Google Scholar]
- 35.Morley, S. J., and V. M. Pain. 1995. Hormone-induced meiotic maturation in Xenopus oocytes occurs independently of p70s6k activation and is associated with enhanced initiation factor (eIF)-4F phosphorylation and complex formation. J. Cell Sci. 108:1751-1760. [DOI] [PubMed] [Google Scholar]
- 36.Mullner, E. W., and J. A. Garcia-Sanz. 1997. Polysome gradients, p. 457-462. In I. Lefkovits (ed.), Manual of immunological methods, vol. 1. Academic Press, London, England. [Google Scholar]
- 37.Naegele, S., and S. J. Morley. 2004. Molecular cross-talk between MEK1/2 and mTOR signaling during recovery of 293 cells from hypertonic stress. J. Biol. Chem. 279:46023-46034. [DOI] [PubMed] [Google Scholar]
- 38.Postel, E. H. 1998. NM23-NDP kinase. Int. J. Biochem. Cell Biol. 30:1291-1295. [DOI] [PubMed] [Google Scholar]
- 39.Proud, C. G. 2002. Control of the translational machinery in mammalian cells. Eur. J. Biochem. 269:5337. [DOI] [PubMed] [Google Scholar]
- 40.Qiu, F. H., P. Ray, K. Brown, P. E. Barker, S. Jhanwar, F. H. Ruddle, and P. Besmer. 1988. Primary structure of c-kit: relationship with the CSF-1/PDGF receptor kinase family—oncogenic activation of v-kit involves deletion of extracellular domain and C terminus. EMBO J. 7:1003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajasekhar, V. K., A. Viale, N. D. Socci, M. Wiedmann, X. Hu, and E. C. Holland. 2003. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell 12:889-901. [DOI] [PubMed] [Google Scholar]
- 42.Schulte, C. E., M. von Lindern, P. Steinlein, H. Beug, and L. M. Wiedemann. 2002. MLL-ENL cooperates with SCF to transform primary avian multipotent cells. EMBO J. 21:4297-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stolovich, M., H. Tang, E. Hornstein, G. Levy, R. Cohen, S. S. Bae, M. J. Birnbaum, and O. Meyuhas. 2002. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol. Cell. Biol. 22:8101-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tee, A. R., R. Anjum, and J. Blenis. 2003. Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. J. Biol. Chem. 278:37288-37296. [DOI] [PubMed] [Google Scholar]
- 45.Thomson, A. M., J. T. Rogers, and P. J. Leedman. 1999. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int. J. Biochem. Cell Biol. 31:1139-1152. [DOI] [PubMed] [Google Scholar]
- 46.Topisirovic, I., M. L. Guzman, M. J. McConnell, J. D. Licht, B. Culjkovic, S. J. Neering, C. T. Jordan, and K. L. Borden. 2003. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol. Cell. Biol. 23:8992-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Akker, E., T. van Dijk, M. Parren-van Amelsvoort, K. S. Grossmann, U. Schaeper, K. Toney-Earley, S. E. Waltz, B. Lowenberg, and M. von Lindern. 2004. Tyrosine kinase receptor RON functions downstream of the erythropoietin receptor to induce expansion of erythroid progenitors. Blood 103:4457-4465. [DOI] [PubMed] [Google Scholar]
- 48.van Dijk, T. B., E. van Den Akker, M. P. Amelsvoort, H. Mano, B. Lowenberg, and M. von Lindern. 2000. Stem cell factor induces phosphatidylinositol 3′-kinase-dependent Lyn/Tec/Dok-1 complex formation in hematopoietic cells. Blood 96:3406-3413. [PubMed] [Google Scholar]
- 49.von Lindern, M., E. M. Deiner, H. Dolznig, M. Parren-Van Amelsvoort, M. J. Hayman, E. W. Mullner, and H. Beug. 2001. Leukemic transformation of normal murine erythroid progenitors: v- and c-ErbB act through signaling pathways activated by the EpoR and c-Kit in stress erythropoiesis. Oncogene 20:3651-3664. [DOI] [PubMed] [Google Scholar]
- 50.Zetterberg, A., O. Larsson, and K. G. Wiman. 1995. What is the restriction point? Curr. Opin. Cell Biol. 7:835-842. [DOI] [PubMed] [Google Scholar]
- 51.Zheng, R., K. Klang, N. C. Gorin, and D. Small. 2004. Lack of KIT or FMS internal tandem duplications but coexpression with ligands in AML. Leuk. Res. 28:121-126. [DOI] [PubMed] [Google Scholar]