Abstract
The Hoxb1 autoregulatory enhancer directs segmental expression in vertebrate hindbrain. Three conserved repeats (R1, R2, and R3) in the enhancer have been described as Pbx-Hoxb1 (PH) binding sites, and one Pbx-Meinox (PM) binding site has also been characterized. We have investigated the importance and relative roles of PH and PM binding sites with respect to protein interactions and in vivo regulatory activity. We have identified a new PM site (PM2) and found that it cooperates with the R3 PH site to form ternary Prep1-Pbx1-Hoxb1 complexes. In vivo, the combination of the R3 and PM2 sites is sufficient to mediate transgenic reporter activity in the developing chick hindbrain. In both chicken and mouse transgenic embryos, mutations of the PM1 and PM2 sites reveal that they cooperate to modulate in vivo regulatory activity of the Hoxb1 enhancer. Furthermore, we have shown that the R2 motif functions as a strong PM site, with a high binding affinity for Prep1-Pbx1 dimers, and renamed this site R2/PM3. In vitro R2/PM3, when combined with the PM1 and R3 motifs, inhibits ternary complex formation mediated by these elements and in vivo reduces and restricts reporter expression in transgenic embryos. These inhibitory effects appear to be a consequence of the high PM binding activity of the R2/PM3 site. Taken together, our results demonstrate that the activity of the Hoxb1 autoregulatory enhancer depends upon multiple Prep1-Pbx1 (PM1, PM2, and PM3) and Pbx1-Hoxb1 (R1 and R3) binding sites that cooperate to modulate and spatially restrict the expression of Hoxb1 in r4 rhombomere.
Hox proteins belong to a large family of transcription factors that control cell identity, differentiation, and patterning in embryonic development. The ordered expression of these genes along the body axis is a critical aspect of regional identity (24, 37). For example, the functional identity of segmental units, rhombomeres (r), in the vertebrate hindbrain depends on the combinatorial activity of Hox proteins along the anteroposterior axis (reviewed in references 22, 24, 27, and 38). Members of the Hox protein family participate in a cascade that involves regulation of their spatial and temporal expression by direct auto-, para-, and/or cross-regulatory mechanisms. Initial expression of Hox genes in the hindbrain is regulated by a variety of transient inputs, including retinoic acid (RA) and fibroblast growth factors (FGFs) (2, 21, 35, 36, 48, 51) along with several transcription factors, such as retinoic acid receptors, Kreisler, and Krox20 (33, 34, 44, 54). Following this initial phase, direct cross talk and feedback circuits between the Hox genes are one of the important mechanisms that serve to maintain restricted segmental expression once the early cues are no longer functioning (17, 29, 32, 40, 49, 50).
Some of the cofactors and protein players involved in regulation of Hox target sites are highly conserved from flies to mammals (1, 8, 26). One of the most conserved regulatory mechanisms involves the Hox/PBC protein complexes, which are essential for maintaining rhombomeric expression of a number of Hox genes in the vertebrate hindbrain (29, 30, 32, 38, 40, 57). Interactions between Hox proteins and the TALE (three amino acids loop extension) family member Pbx/Exd can confer or modulate the DNA binding specificity and selectivity of Hox proteins (11, 12, 15, 31, 53). While Hox proteins recognize the 5′-NNAT-3′ sequence, Hox-Pbx dimers recognize the bipartite composite 5′-TGATNNAT-3′-type sequences, which are termed, in this paper, Pbx-Hox binding (PH) sites (16, 31, 32). In the Pbx-Hox heterodimer, the Pbx protein binds the 5′ part of the bipartite sequence, while Hox protein contacts the 3′-NNAT sequence motif. The two base pairs (NN) predicted to contact the N-terminal arm of the Hox homeodomain seem to have a crucial role in selecting which Hox partner is preferred in dimerization (11, 12, 23, 32). This bipartite PH consensus sequence is found in several Hox enhancers, including Hoxb1, Hoxb2, Hoxa3, and Hoxb4 in vertebrates and labial in Drosophila melanogaster, and enhancer activities showed a dependence primarily on the PH sites (16, 17, 19, 32, 42). However, it is important that other studies have also found that additional cofactors can determine the specificity of Hox response elements, in a manner that is largely independent of the binding preferences of PH sites (25). For example, an Exd/labial binding site can alter its specificity to that of Deformed if moved into a different context, suggesting that, in addition to the specific DNA binding site motif, the enhancer context exerts an important role in determining PH expression specificity (25).
Additional Hox regulators have subsequently been identified in the Meinox subfamily of TALE proteins, comprising Prep and Meis genes (6, 7, 13, 20, 45, 47). These proteins form specific DNA-independent heterodimers with the Pbx proteins and bind to the hexameric sequence 5′-TGACAG-3′, which in this paper is referred to as a Pbx-Meinox binding (PM) site (4, 16). Several Hox enhancers (Hoxb1, Hoxb2, Hoxa3, and labial) contain a combination of PM and PH elements, which suggest that together they have an important role in modulating Hox expression (16, 19, 32, 42). Since Pbx employs different interaction surfaces to bind Meinox and Hox proteins, it is able to form ternary Meinox-Pbx-Hox complexes, which can bind the combined PM-PH sites, thus enhancing the DNA binding selectivity of the Hox component (16, 19, 32, 42).
Both Hoxb1 and Hoxb2 enhancers mediate similar patterns of r4-restricted expression in vivo (29, 40) but differ in the organization and number of their PH and PM sites (Fig. 1A). The Hoxb2 enhancer has single PH and PM sites, while the Hoxb1 enhancer has one previously characterized PM1 site and three putative PH sites (R1, R2, and R3) within a highly conserved 331-bp region (b1-ARE) (16, 19, 29, 40). In addition to the PH sites in the Hoxb1 enhancer required to mediate expression in the region of r4, flanking repressor elements are also essential to restrict the activity of the enhancer specifically to r4 (40, 51). The PM-PH motifs in the Hoxb1 (PM1-R3, Fig. 1) and Hoxb2 enhancers are separated by 17 and 8 bp, respectively, and form trimeric complexes in vitro (16, 19). The functional role of the PM sites has been tested in vivo using regulatory assays in transgenic mice. Mutation of the PM site in the Hoxb2 r4 enhancer abolished its ability to mediate reporter expression in r4, demonstrating that it was an essential component. In contrast, mutation of the PM1 site in the Hoxb1 r4 enhancer (b1-ARE) did not eliminate reporter activity (16, 19). This suggested that other uncharacterized elements or additional PM sites might function in the Hoxb1 enhancer to modulate activity.
FIG. 1.
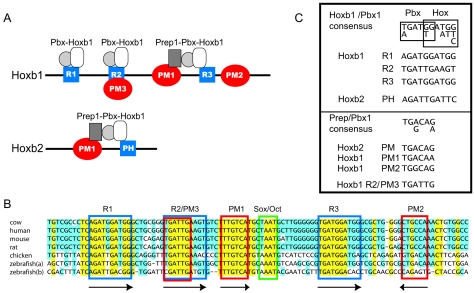
Hoxb1 enhancer. (A) Schematic representation of r4-Hoxb1 and r4-Hoxb2 enhancers including the PM and PH sites binding to the various Prep-Pbx complexes. The blue squares indicate the PH sites (R1, R2/PM3, and R3 in Hoxb1 and PH in Hoxb2), and the red circles indicate the PM sites (PM1 and PM2). The sequence of the Hoxb2 PM-PH site is included at the bottom of the Hoxb2 enhancer scheme. Notice that the R2 site is also called R2/PM3. (B) Sequence conservation between mammal, chicken, and zebra fish r4-Hoxb1 regulatory regions. The conserved PH (blue), the PM (red), and the Oct1 (green) sites are boxed. Arrows below the sites indicate site orientation. (C) Consensus sequences of the PH and PM sites based upon the mouse r4-Hoxb1 and r4-Hoxb2 enhancers.
In this study, we investigate the basis for the different in vivo requirement of PM sites between the Hoxb1 and Hoxb2 regulatory regions. We have found that Hoxb1 has a second PM site (PM2), 9 bp downstream of R3. To investigate whether interplay between the multiple PH and/or PM sites in Hoxb1 might be important for regulatory activity, we have performed in vitro and in vivo experiments to evaluate the function of the novel PM2 site and of the R2 and R1 motifs. Our results demonstrate that PM2 in combination with PM1 has an important role in Hoxb1 regulation. Furthermore, R2 can function as a PM site, which we now call R2/PM3. Interactions between various combinations of PM and PH sites play a role in determining whether the enhancer can successfully form a ternary complex in vitro or retain regulatory activity in vivo. Our findings have important implications in helping to define critical cis-regulatory components, protein interactions, and constraints that govern Hox response elements in downstream target genes of the Hox cascade.
MATERIALS AND METHODS
Cell extracts.
P19 cells were grown in Dulbecco modified Eagle medium supplemented with 10% newborn calf serum (Gibco-BRL), 100 U/ml penicillin, and 100 μg/ml of streptomycin.
RA induction of P19 cells and nuclear extracts.
To induce differentiation, P19 cells were treated with trans-retinoic acid to a final concentration of 10−5 M. To obtain nuclear extract, the cells were collected after 12 h of incubation, washed with phosphate-buffered saline, scraped, and recovered by centrifugation. Nuclear extracts were prepared as described previously (3).
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed with in vitro-translated proteins as previously described (5) using 2 μl of reticulocyte lysate containing the desired combination of proteins mixed with 18 μl of PPH binding buffer [10 mM Tris-Cl (pH 7.5), 75 mM NaCl, 1 mM EDTA, 6% glycerol, 3 mM spermidine, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg poly(dI-dC), 30,000 cpm 32P-labeled oligonucleotide] to a total volume of 20 μl. After 30 min of incubation on ice the reaction mixtures were separated by 5% polyacrylamide gel electrophoresis in 0.5× Tris-buffered EDTA. For the competition assays, a 50- or 100-fold molar excess of unlabeled competitor oligonucleotide was added to the binding reaction mixture 10 min before the labeled probe.
Oligonucleotides.
Oligonucleotides used in EMSA were obtained commercially from Biosense (Belgium) or Roche (Italy). Oligonucleotides were resuspended and diluted in Tris-EDTA. Double-stranded oligonucleotides were prepared by mixing equal molar ratios of single-stranded oligonucleotides. The mixture was heated at 90°C for 5 min and slowly cooled at room temperature. Purification of double-stranded oligonucleotides was performed on preparative 15% acrylamide electrophoresis gels in 0.5× Tris-buffered EDTA. The oligonucleotides were eluted from the gel in 500 μl TEN (10 mM Tris, 1 μM EDTA, 300 mM NaCl, pH 8), shaken at 37°C overnight, and recovered by ethanol precipitation. The pellet was washed, dried, and resuspended in 50 μl Tris-EDTA. The sequences of oligonucleotides used for EMSA are shown in Table 1.
TABLE 1.
List of the oligonucleotides employed for EMSA and electroporation analyses in this studya
| Probe | Sequence |
|---|---|
| b2-PM-PH | 5′-GGAGCTGTCAGGGGGCTAAGATTGATCGCCTCA-3′ |
| PM1-R3 | 5′-TCTTTGTCATGCTAATGATTGGGGGGTGATGGATGGGCGCTG-3′ |
| R2-PM1-R3 | 5′-TCAGAGTGATTGAAGTGTCTTTGTCATGCTAATGATTGGGGGGTGATGGATGGGCG-3′ |
| m-R2-pm1-R3 | 5′-TCAGAGTGATTGAAGTGTCTTTcTtATGCTAATGATTGGGGGGTGATGGATGGGCG-3′ |
| m-r2-PM1-R3 | 5′-TCAGAGTcgTTcgAGTGTCTTTGTCATGCTAATGATTGGGGGGTGATGGATGGGCG-3′ |
| m-R2-PM1-R3 | 5′-TCAGAGTGATTGAAGTGTCTTTGTCATGCTAATGATTGGGGGGTcgTGcgTGGGCG-3′ |
| R2-PM1 | 5′-TCAGAGTGATTGAAGTGTCTTTGTCATGCTA-3′ |
| PM1-R2 | 5′-TCTTTGTCATGCTAATGATTGGGGGGTGATTGAAGGGCGCTG-3′ |
| R3-PM1 | 5′-TCAGAGTGATGGATGTGTCTTTGTCATGCTA-3′ |
| R3-PM1-R2 | 5′-TCAGAGTGATGGATGTGTCTTTGTCATGCTAATGATTGGGGGGTGATTGAAGGGCG-3′ |
| m-r3-PM1-R2 | 5′-TCAGAGTcgTGcgTGTGTCTTTGTCATGCTAATGATTGGGGGGTGATTGAAGGGCG-3′ |
| R3-PM2 | 5′-GGGGTGATGGATGGGCGCTGGGACTGCCAAACT-3′ |
| m-R3-pm2a | 5′-GGGGTGATGGATGGGCGCTGGGctTGCCAAACT-3′ |
| m-R3-pm2b | 5′-GGGGTGATGGATGGGCGCTGGGACTtCgAAACT-3′ |
Oligonucleotide probes are named based on the combination and position of the individual PM or PH elements in the Hoxb1 and Hoxb2 r4 enhancer. Probes in which the wild-type sequence has been mutated are indicated with an “m” preceding the name and by lowercase letters for the mutated element. Letters in boldface indicate the sequence of the specific sites in each probe (i.e., R3 or PM1). The noncapitalized letters indicate the specific sequence changes used to mutate the sites.
Constructs for in vivo analysis in chicken and mouse embryos.
Oligonucleotides or subcloned fragments of the regulatory regions of the mouse Hoxb1 and Hoxb2 genes were cloned into the BGZ40 vector (29). A 622-bp genomic fragment which contains the 331-bp StuI-HindIII fragment of Hoxb1 spanning the R1, R2, and R3 PH sites and the PM1 and PM2 PM sites and functions as an r4 autoregulatory enhancer (40) was used as a control (wild-type [WT]) for comparison with variants in which PM1 and/or PM2 was mutated. The 622-bp enhancer was isolated by PCR from mouse genomic DNA using 5′-CG CGG CTA GTC ATC CTT TTG TCC CAA GA-3′ as the forward primer and 5′-CCG CGG TCT TGC CCT ACA ACC TTT CG-3′ as the reverse primer. The fragment was cloned into pGEM-T (Promega) and then transferred as a SacII fragment into the BGZ40 vector (29). The PM1, PM2, and PM1+PM2 sites were mutated (nucleotide substitution in PM1 is underlined, and deletion in PM2 is boldfaced; compare with the sequence in Fig. 1B) using the Quick Change kit (Stratagene) with the following oligonucleotides: PM1, 5′-GGG CTC AGA GTG ATT GAA GTG TCT TGC TGT AGC TAA TGA TTG GGG GGT GAT GGA TGG-3′; PM2, 5′-GGG GGG TGA TGG ATG GGC GCT GG DG G AAA CTC TGG CCC GCT TAG CCC ATT GGC C-3′; PM1+PM2, 5′-GGC TCA GAG TGA TTG AAG TGT CTT GCT GTA GCT AAT GAT TGG GGG GTG ATG GAT GGG CGC TGG DG GAA ACT CTG GCC CGC TTA GCC CAT TGG CCT GGG-3′. All constructs were sequenced to verify the mutations. The 622-bp WT and variant PM1 and/or PM2 constructs were assayed in chicken and mouse embryos. In mouse experiments, purified insert sequences were isolated from the vector backbone by digestion with ScaI and XhoI and gel electrophoresis; the entire plasmid was used for chick electroporation.
Generation of transgenic chicken and mouse embryos.
Transgenic mouse embryos were generated by pronuclear injection of purified DNA into fertilized mouse eggs from an intercross of F1 hybrids (CBA × C57BL/6) and stained for lacZ reporter activity as described previously (56). Constructs were assayed in founder (F0) transgenic embryos at 9.5 days postcoitum (dpc). Generation of transgenic chicken embryos was performed by in ovo electroporation, as previously described (18, 39, 43). Chicken embryos at Hamburger-Hamilton stages 8 to 11 (5 to 12 somites) were coinjected in the neural tube with pBGZ40 lacZ reporter construct derivatives (0.5 to 1.0 mg/ml), Fast Green, and a cytomegalovirus (CMV) green fluorescent protein (GFP) electroporation control vector. Unilateral in ovo electroporation into the left side of the neural tube was performed (18, 39, 43), and the embryos were permitted to develop overnight before being scored for reporter activities. Chicken embryos at the appropriate stage of development were dissected from maternal tissues, and only embryos showing strong GFP expression throughout the neural tube (indicative of efficient uptake and expression of electroporated DNAs) were analyzed further for lacZ expression. Embryos were then washed in phosphate buffer, pH 7.4 (phosphate-buffered saline), and immediately fixed for 5 to 20 min, depending on development stage, at room temperature in 2% paraformaldehyde, 0.2% glutaraldehyde, 2 mM MgCl2, 5 mM EGTA, pH 7.3, in phosphate buffer. The embryos were washed three times for 15 min at room temperature (0.01% sodium deoxycholate, 0.02% Nonidet P-40, 2 mM MgCl2, in phosphate-buffered saline) and stained for 1 hour to overnight at 30°C or 37°C depending on the level of lacZ activity (50 mg X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside], 0.106 g potassium ferrocyanide, and 0.082 g dissolved in wash buffer). The efficiency of reporter expression in chick electroporation experiments was calculated as the percentage of the total number of embryos with strong or robust lacZ expression in r4 to the total number of embryos strongly expressing the GFP control reporter for each construct. Embryos with very weak or patchy or no staining were counted as negative.
RESULTS
Differential protein binding properties of distinct sites in the Hoxb1 r4 enhancer.
A highly conserved 331-bp Hoxb1 r4 enhancer (b1-ARE, autoregulatory element) contains three conserved and functionally important sites, known as R1, R2, and R3 (40), which, because of their sequence identity, have been designated PH sites (Fig. 1). Single and multiple mutations in these sites, in transgenic embryos, revealed that they all contribute to regulatory activity but that site R3 was the most important in vivo (40). There is a PM site, PM1, positioned between R2 and R3 (Fig. 1A and B), and it was previously shown that this motif is able to form ternary Prep-Pbx-Hoxb1 complexes in association with R3 but is not essential for r4-restricted activity of this enhancer (16). Therefore, the Hoxb1 enhancer might not depend upon functional Pbx-Meinox sites, or a role for the PM1 site might be obscured by the presence of other elements that contribute to enhancer activity.
To address this question, we aligned the sequence of this region from six vertebrate species using Vector NTI's integrated ClustalW (52) global alignment program (Fig. 1B). In addition to the known components, we identified a new potential PM site (PM2) downstream of R3. The PM2 motif is conserved in all cases, with the exception of the zebra fish Hoxb1b gene (Fig. 1B). This is interesting because the zebra fish Hoxb1a gene is expressed in r4 of the developing hindbrain, in a manner similar to the Hoxb1 gene of other vertebrates, while the duplicated paralogous Hoxb1b gene is not expressed in r4 (41). The lack of Hoxb1b expression in r4 of the zebra fish hindbrain was intriguing because repeats R1 to R3 and PM1 are all present and conserved, implying that changes in other motifs must contribute to the absence of segmental expression of this duplicated gene. Together these observations led us to investigate the potential role of this new PM2 site in the in vitro and in vivo properties of the Hoxb1 enhancer.
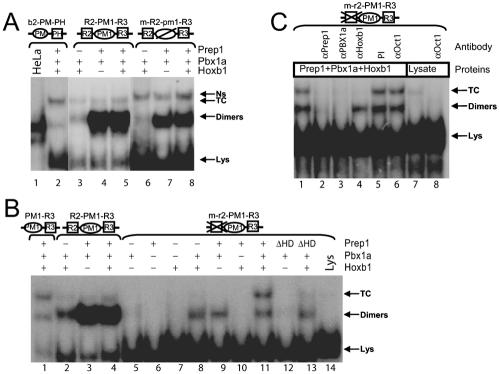
The multiple PH and PM sites in Hoxb1 contrast with the single PH and PM sites in the Hoxb2 r4 enhancer (Fig. 1A), which have both been shown to be essential for activity (16, 19). In order to evaluate interactions between the PH and PM sites in Hoxb1, we performed EMSA using in vitro-translated proteins with a set of wild-type and mutated double-stranded oligonucleotides derived from the Hoxb1 enhancer (Table 1). In the figures, putative PH sites (previously assigned in references 16 and 19) are indicated as squares and PM sites are indicated as spheres. Oligonucleotide probes are named based on the combination and position of the individual PM or PH elements. Probes in which the wild-type sequence has been mutated are indicated with an “m-” preceding the name and by lowercase letters for the specifically mutated element (e.g., R2-PM1-R3 versus m-R2-pm1-R3). As a positive control to monitor formation of both dimeric and trimeric complexes, we routinely used a previously characterized (16) probe containing the combined PM-PH sites from the Hoxb2 enhancer (b2-PM-PH) (Fig. 1A and Table 1). The b2-PM-PH control probe binds Prep1-Pbx1 dimers, if incubated with HeLa cell nuclear extracts (Fig. 2A, lane 1), and forms a ternary Prep1-Pbx1-Hoxb1 complex (Fig. 2A, lane 2) in the presence of in vitro-translated proteins, in agreement with our previous results (16).
FIG. 2.
The R2 site inhibits ternary complex formation mediated by PM1-R3 sites. EMSA were performed using combinations of in vitro-translated Prep1, Pbx1a, and Hoxb1 proteins, as indicated. The labeled DNA targets are shown on the top. A square represents a PH site; a circle represents a PM site. The sequence of oligonucleotides and the nature of mutations are shown in Table 1. Mutant oligonucleotides are preceded by “m-,” and the mutated site is in lowercase. Note that the reticulocyte lysate contains nonspecific factors forming a slower-migrating complex marked Ns and a faster-migrating one marked Lys. (A) The control b2-PM-PH oligonucleotide, but not the R2-PM1-R3 and m-R2-pm1-R3 oligonucleotides, forms a ternary Prep1-Pbx1a-Hoxb1 ternary complex. (B) Mutations in R2 restore the ability of PM1 and R3 to form the ternary complex. In vitro-translated Prep1, Pbx1a, and Hoxb1 form a ternary complex on the mutated m-r2-PM-R3 oligonucleotide but not on the wild-type R2-PM-R3. Note the different abilities of the two oligonucleotides to bind Prep1-Pbx1a dimers. ΔHD lanes replace Prep1 with a deleted homeodomain form. (C) The Prep1-Pbx-Hoxb1 ternary complex and the Oct1 transcription factor bind the mutated m-r2-PM-R3 oligonucleotide, producing two closely migrating but distinct bands. Addition of anti-Pbxa, anti-Prep1, and anti-Hoxb1 antibodies inhibits the DNA binding activity of the ternary complex (lanes 2 to 4). Anti-Oct1 antibody inhibits the endogenous reticulocyte band (lanes 7 and 8).
To test if a functional PM1 site is required for ternary complex formation in the presence of R2 and R3, we analyzed a wild-type R2-PM1-R3 probe and a variant (m-R2-pm1-R3) probe in which the PM1 site is mutated (Table 1). The two Hoxb1 probes strongly bind a Prep1-Pbx1a heterodimer (Fig. 2A, lanes 4 and 7) and very weakly bind a Pbx1a-Hoxb1 heterodimer (lane 3). Surprisingly, no ternary complex was detected with either probe (Fig. 2A, lanes 5 and 8). Since probes spanning PM1 and R3 have been shown to be successful at forming ternary complexes (16), this result indicates that the presence of the R2 site interferes with, rather than enhances, the formation of a ternary complex. The mutation in the PM1 site increased the binding activity of an endogenous, more slowly migrating factor, present in the reticulocyte lysate (Ns) (Fig. 2A, lanes 6 to 8), which, in a subsequent section, we show contains Oct proteins. It is surprising that deletion of the known PM1 site did not decrease the Prep1/Pbx1a heterodimer binding (Fig. 2A, lanes 7 and 8), suggesting that there are additional unexpected PM binding sites in this region of the oligomer.
To address this issue, we analyzed the binding properties of the R2 site in conjunction with PM1 (R2-PM1). R2-PM1 binds the Prep1-Pbx1a heterodimer but does not bind Hoxb1-Pbx1a dimers or form a ternary complex (data not shown, but see below). Also, an increase in the physical distance between R2 and PM1 by 2 bp (R2-GG-PM1) or 4 bp (R2-GGGG-PM1) did not change the binding properties compared with normal spacing (data not shown). Thus, the R2 site is unable to efficiently bind Pbx1a-Hoxb1 dimers and to cooperate with the PM1 site to form a ternary complex. However, R2 does function as a high-affinity binding site for Prep1-Pbx dimers. These results highlight distinct differences in the abilities of R2 and R3 to cooperate with PM1 and suggest that R2 may be more accurately defined as a PM site, which we now rename R2/PM3 (Fig. 1), although it is unable to synergize with R3 to form a ternary complex.
R2/PM3 inhibits ternary complex formation by PM1-R3.
To further explore differences in the properties of R2/PM3 versus R3, we investigated the possibility that R2/PM3 had an inhibitory effect on ternary complex formation in probes containing the combined R2-PM1-R3 sites. In this context, mutation of the R2/PM3 site (m-r2-PM1-R3) has no effect on binding of Pbx1a-Hoxb1 and Prep1-Pbx1a heterodimers but restores the ability of PM1-R3 to form a slower-migrating ternary complex. However, the mutation appeared to decrease the overall affinity of the probe for Prep1-Pbx1 (Fig. 2B, compare lanes 3 and 9). Ternary complex formation required an intact Prep1 protein, as no complex was formed with a variant lacking the homeodomain (ΔHDPrep1) (Fig. 2B, lane 13). The presence of all three proteins in the ternary complex was verified using specific antibodies against Prep1, Pbx1a, and Hoxb1 (Fig. 2C). This result shows that mutation of the R2/PM3 site rescues the ability of PM1 and R3 to form ternary complexes and suggests that R2/PM3 directly interferes with this process. This reveals a cross talk between R2/PM3, PM1, and R3 elements.
Previous studies have shown that other factors, besides Pbx, Hoxb1, and Prep1, can bind the region between R2 and R3 (14). One of these factors is Oct1, which binds the octamer-like sequence 5′-ATGCTAAT-3′ located next to R3 in the PM1-R3 sequence (Fig. 1B) (14). Oct1 is very abundant in the reticulocyte lysate (data not shown), and we found that the endogenous binding activity of the reticulocyte lysate, which comigrates with the ternary complex, does correspond to Oct1. Indeed, this slow-migrating band in the reticulocyte lysate was inhibited by an anti-Oct1 antibody, which did not affect the formation of the ternary Prep1-Pbx-Hoxb1 complex (Fig. 2C, lanes 6 to 8). Therefore, probes spanning the R3 and PM1 sites produce two distinct comigrating bands that include Oct1 and the Prep1-Pbx1a-Hoxb1 ternary complex.
The ability of R2/PM3 to inhibit ternary complex formation by PM1 and R3 is context dependent.
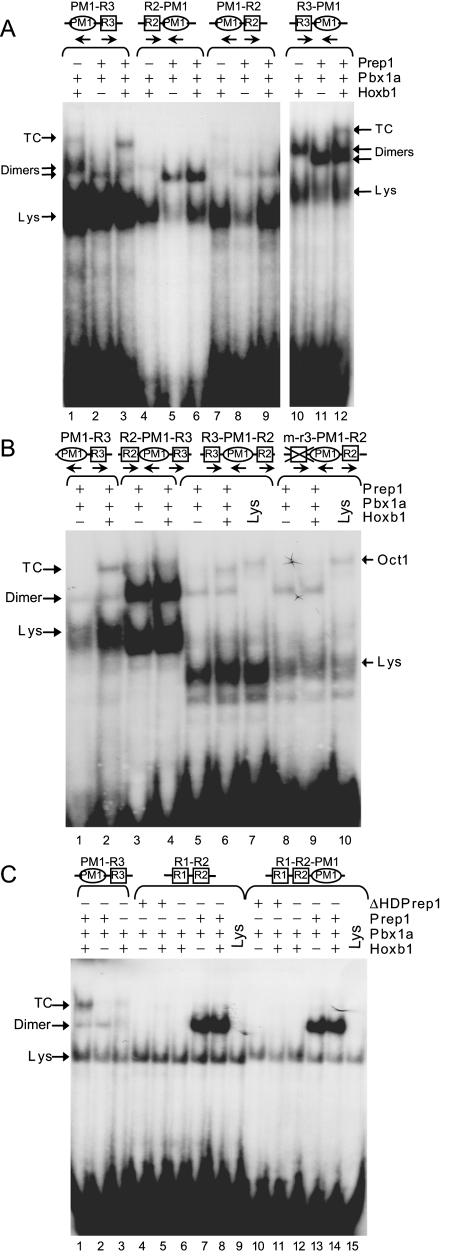
We further analyzed the influence of the spacing and relative orientation of R2/PM3, PM1, and R3 sites on ternary complex formation in vitro. First we tested whether R2/PM3 would be able to mediate ternary complex formation if located 3′ of PM1 (PM1-R2) in a position with the same orientation and distance as R3. In this context, the R2/PM3 site still did not cooperate with PM1 to form a ternary complex (Fig. 3A, lanes 3, 6, and 9). Conversely, transposing R3 upstream of PM1 (R3-PM1) did not diminish its ability to cooperate with PM1 and form a ternary complex (Fig. 3A, lane 12). These results imply that the inhibitory activity of R2/PM3 is dependent on the sequence itself.
FIG. 3.
Characterization of the interactions of the various PH sites of the Hoxb1 enhancer with the PM1 site. EMSA were performed using combinations of in vitro-translated Prep1, Pbx1a, Hoxb1, and ΔHDPrep1 proteins, as indicated. The labeled DNA targets are shown on the top. The activities present in reticulocyte lysate are indicated by Lys. (A) Ternary complex (TC) formation is independent of the orientation of PM1 with respect to R2 and R3. R2 is unable to cooperate with PM1 even when located downstream (PM1-R2), while R3 is able to mediate the ternary complex formation also in the opposite orientation (R3-PM1). (B) The position of R2/R3 relative to PM1 is important for the ternary complex formation. Switching of R2 and R3 (R3-PM1-R2) decreases the Prep1-Pbx1a binding and restores ternary complex (TC) formation; the R3 site plays a fundamental role in the ternary complex formation (compare R3-PM1-R2 and m-r3-PM1-R2). The endogenous binding activities present in reticulocyte lysate are indicated by Lys for the nonspecific factor and by Oct1 for the characterized factor. (C) Both R1-R2 and R1-R2-PM1 oligonucleotides bind with high-affinity Prep1-Pbx dimers but are unable to form a ternary complex.
We also investigated whether R2/PM3 would still inhibit R3 when the two sites were transposed (R3-PM1-R2). Surprisingly, this swap restored the ability of the probe to form ternary complexes and decreased the binding affinity for Prep1-Pbx1a heterodimers (Fig. 3B, lanes 2 to 6). In this transposed context, R3 is essential for the ternary complex, since mutation of R3 (m-r3-PM1-R2) prevented its formation but had no effect on the Oct1-dependent complex (Fig. 3B, lanes 7 to 10). These experiments show that, while spacing and orientation between PM1 and R3 have no influence on ternary complex formation in vitro, the relative position of R2/PM3 and R3 is important in determining the inhibitory activity. When R2/PM3 lies 5′ of PM1 and R3, it effectively inhibits ternary complex formation and forms a high-affinity DNA-bound Prep1-Pbx complex. Therefore, the R2/PM3 motif is not simply an independent Prep1-Pbx1 binding site or dominant inhibitor but also serves to differentially modulate activity of the other elements in the Hoxb1 r4 enhancer in a context-dependent manner.
We also analyzed whether the presence of the R1 site could alter the properties of R2/PM3 and its interaction with PM1 in an analogous manner. A probe containing the R1 and R2/PM3 sites alone (R1-R2) or in combination with PM1 (R1-R2-PM1) was assayed for activity. In both cases, probes bound the Prep1-Pbx1a heterodimers with high affinity, but there was no evidence for Hoxb1-Pbx1a binding or formation of a ternary complex (Fig. 3C). Since we have previously shown that R1 binds Hoxb1/Exd heterodimers (40), this implies that R2/PM3 may also alter the binding properties of R1. In conclusion, R1 does not influence the ability of the R2/PM3 site to cooperate with PM1 and form a ternary complex. This illustrates that the binding and cooperative properties of R2/PM3 differ from those of R1 and R3.
The PM2 site in the Hoxb1 enhancer cooperates with R3 in forming a Prep1-Pbx1-Hoxb1 ternary complex.
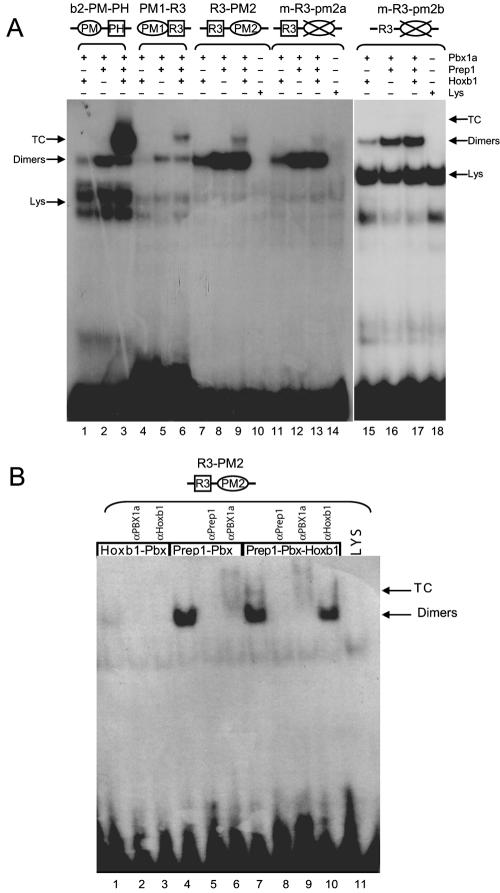
The identification of a new conserved PM2 site (PM2; 5′-TGGCAG-3′), located, on the minus strand, in the reverse orientation, 9 bp downstream of the R3 site, opened the possibility that it might also contribute to Hoxb1 enhancer activity (Fig. 1B and C). To test the binding ability of this putative PM2 site, we generated probes for R3-PM2 and performed EMSA. As shown in Fig. 4A (lane 9), R3-PM2 forms a ternary complex with Hoxb1, Prep1, and Pbx1a. Furthermore, mutations of PM2 (m-R3-pm2a or m-R3-pm2b, Table 1) strongly decrease the binding of the trimer but not of the Prep1-Pbx1a or of the Pbx1a-Hoxb1 dimer (Fig. 4A, lanes 13, 15, and 17). The trimeric and dimeric complexes are inhibited by antibodies against each of the three proteins but not by anti-Oct1 antibodies (Fig. 4B). In addition, competition assays show that the wild-type R3-PM1 probe competes with the R3-PM2 site for binding to the heterodimer and the ternary complex (data not shown). These data confirm that R3 represents the major binding site for the dimeric Hoxb1-Pbx1a complexes and that R3 and PM2 sites can synergize to promote ternary complex formation, in a manner similar to R3 and PM1. This suggests that both PM2 and PM1 may contribute to ternary complex formation and regulatory activity in vivo. Taking into account the newly defined PM2 and PM3 sites from Hoxb1, a revised PM consensus sequence appears to be 5′-TG(A/C)(C/T)(A/T)(G/A)-3′ (Fig. 1C). However, it is important that our experiments (unpublished) have also revealed that flanking sequences can dramatically influence relative binding affinities of PM sites for Prep1-Pbx1a heterodimers.
FIG. 4.
The PM2 site cooperates with R3 to form a ternary complex. EMSA were performed with in vitro-translated proteins, as indicated above each lane. (A) The R3-PM2 oligonucleotide forms a ternary complex (TC) with Prep1, Pbx1a, and Hoxb1 proteins in a manner similar to that seen with PM1-R3 and b2-PM-PH probes. Mutation of PM2 prevents ternary complex formation in combination with R3. (B) The binding specificity of proteins in the ternary complex was tested using specific antibodies as indicated above each lane.
R2 inhibits the formation of ternary complexes also in P19 cell extracts.
To complement the EMSA with in vitro-translated proteins, we characterized the binding properties of the probes described above in a cellular context, by using nuclear extracts from control and RA-induced P19 cells which express Hoxb1 (or other Hox proteins only after treatment with retinoic acid) (46). We found that a ternary complex was formed with extracts of retinoic acid-treated cells only with PM1-R3 but not with R2-PM1-R3 (not shown). Again, mutation of the R2 site (m-r2-PM1-R3) restored ternary complex formation (data not shown). Thus, the R2/PM3 site also inhibits the Prep1-Pbx-Hoxb1 ternary complex formation in P19 nuclear extracts, demonstrating a modulatory cross talk between R2/PM3 and the PM1-R3 binding motifs in the Hoxb1 enhancer. R2/PM3 inhibits the cooperation between PM1 and R3 which is necessary for Prep1-Pbx-Hoxb1 ternary complex formation.
In vivo analysis of PH-PM sites by electroporation in chicken embryos.
In light of these findings on the new PM2 site and of the inhibitory activity of the R2/PM3 site, we investigated the in vivo roles of these motifs in Hoxb1 enhancer activity. Minimal Hoxb1 (b1-PM1-R3) and Hoxb2 (b2-PM-PH) sequences required to bind the ternary complex in vitro were linked to a lacZ reporter gene under the control of a minimal human beta-globin promoter (pBGZ40), which requires added enhancer activity to mediate expression in vivo (29). Constructs were assayed for regulatory activity by in ovo electroporation unilaterally into the left side of a developing neural tube in chicken embryos (Fig. 5). To control for uptake and expression of DNAs, a CMV enhancer/promoter linked to a GFP reporter was coelectroporated with test constructs. Only embryos displaying strong GFP expression, indicative of efficient electroporation, were stained for β-galactosidase and scored for activity. Table 2 summarizes the results of the electroporation experiments for the different constructs.
FIG. 5.
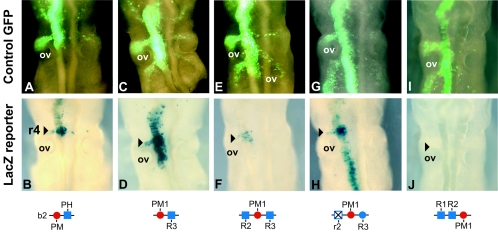
In vivo analysis of enhancer activity by in ovo electroporation in chicken embryos. The top panels (A, C, E, G, and I) represent dorsal views of the expression patterns of a CMV-GFP vector used as a control to monitor the efficiency of unilateral electroporation into the left side of the neural tube of a developing chicken embryo. Below each of the top panels (B, D, F, H, and J) is the pattern of transgene expression in the same embryo coelectroporated with Hoxb2 (B) and Hoxb1 (D, F, H, and J) lacZ reporter constructs. The respective constructs carrying wild-type and mutated variants of the PH and PM sites are noted below each panel. In each case, the left side of chick neural tube was electroporated at HH (Hamburger and Hamilton) stages 6 to 7 and assayed at HH17 for lacZ and GFP reporter expression. Note that in panel D expression mediated by the minimal PM1-R3 element is broader than r4 because this region lacks repressor sequences shown to be important for restricting regulatory activity to r4 (51). The arrowhead marks the position of r4 as defined by its position relative to the otic vesicle (ov).
TABLE 2.
Summary of chicken electroporation experiments indicating the structure of the oligonucleotides electroporated (construct), the number of injected embryos, the level of reporter gene activity, and the ternary complex formation in vitro
| Construct | No. of embryos | Strong r4 expression (%) | Weak, patchy, or no expression in r4 (%) | In vitro ternary complex formation |
|---|---|---|---|---|
| R1-R2-PM1-R3-PM2 | 165 | 79 | 21 | + |
| R1-R2 | 31 | 0 | 100 | − |
| R1-R2-PM1 | 34 | 0 | 100 | − |
| PM1-R3 | 52 | 56 | 44 | + |
| R2-PM1-R3 | 47 | 15 | 85 | − |
| m-r2-PM1-R3 | 27 | 55 | 45 | + |
| R3-PM2 | 33 | 64 | 36 | + |
| m-R1-R2-pm1-R3-PM2 | 18 | 39 | 61 | NAa |
| m-R1-R2-PM1-R3-pm2 | 18 | 11 | 89 | NA |
| m-R1-R2-pm1-R3-pm2 | 17 | 0 | 100 | NA |
NA, not available.
Both the b2-PM-PH and the b1-PM1-R3 motifs direct expression of the lacZ transgene reporter at high levels in the hindbrain, in particular in the region of r4 (Fig. 5A to D). Thus, even a single copy of the minimal b1-PM1-R3 or b2-PM-PH motif of the Hoxb1 and Hoxb2 enhancers is sufficient to mediate hindbrain expression of the transgene. Note that the expression of the PM1-R3 lacZ transgene is not restricted to r4, as weaker or patchy transgene expression is detected rostral and caudal to r4 and in r4 neural crest (Fig. 5D). However, r4 corresponds to the region of most intense staining. This more widespread hindbrain expression by the minimal PM1-R3 region is consistent with our previous observations indicating that short-range repressor elements (missing in this minimal probe) are necessary to restrict the activity of the PM and PH sites in the Hoxb1 enhancer to r4 (51). Nonetheless, these results indicate that the minimal PM1-R3 region is sufficient to activate reporter staining in the general region of r4.
In contrast, expanding the Hoxb1 minimal PM1-R3 region to include the R2/PM3 motif (R2-PM1-R3) leads to strongly reduced and often undetectable reporter expression in the hindbrain (Fig. 5E and F; Table 2). The residual expression in some embryos appears to be r4 specific, which may be a consequence of the overall decrease in activity and/or reflect a role for the additional R2/PM3 sequences in helping to restrict expression to r4. Conversely, mutation of the R2/PM3 site in this same context (m-r2-PM1-R3) restores the ability of the PM1 and R3 elements to stimulate reporter expression mostly in r4 (Fig. 5G and H). These results are consistent with the in vitro EMSA and show that, in vivo, the presence of R2/PM3 inhibits regulatory activity and transgene expression in the hindbrain, mediated by PM1 and R3. R2/PM3 may also contribute to spatially restricting activity to r4. We also tested the regulatory activities of the tandem R1-R2 sites alone or in combination with the PM1 site (R1-R2-PM1). In both cases, these motifs were unable to mediate the expression of the transgene in r4 of the chicken hindbrain (Fig. 5I and J; Table 2 and data not shown). This correlates with the inability of these motifs to form ternary complexes with Hoxb1-Pbx-Prep1 and highlights the absolute requirement for R3 for robust enhancer function.
As PM2 forms ternary complexes with R3, it is possible that, like PM1, it might also potentiate the activity of R3 in vivo. Therefore, we assayed the regulatory activity of the R3-PM2 motif in the chick hindbrain and found strong lacZ transgene expression in the hindbrain (Fig. 6A). The R3-PM2 minimal region, like that of R2-PM1, mediates broad hindbrain expression in the chick, with the highest levels in the region of r4. The ability of both PM1 and PM2 to form ternary complexes with R3 and mediate rhombomeric expression opens the possibility of synergy or functional compensation between these motifs in the Hoxb1 enhancer, which may have been masked in previous studies examining the role of PM1 alone (16, 19). Therefore, we investigated the relative roles of the PM1 and/or PM2 site in the context of a larger Hoxb1 622-bp fragment containing R1 toR3, as well as PM1 and PM2 sites and sequences that serve to restrict expression to r4 (Fig. 6B). This wild-type fragment functions efficiently in vivo as an r4 enhancer, as 79% of chicken embryos electroporated with this construct display strong staining in r4 (Fig. 6B and Table 2). Mutation of the PM1 site leads to a reduction in efficiency (39% versus 79%), and many of these embryos display patchy reporter staining further suggestive of a decrease in activity (Fig. 6C; Table 2). Mutation of the PM2 site in this context has a more pronounced effect on the regulatory activity, as this variant is expressed in only 11% of the embryos, again with a weak, patchy expression only in r4 (Fig. 6D; Table 2). Combining mutations in both PM1 and PM2 sites completely eliminates efficient reporter staining in r4 (Fig. 6E; Table 2). These results show that the PM1 and the PM2 sites are required for expression of the reporter in the chick hindbrain in terms of number of positive embryos and relative level of expression in r4.
FIG. 6.
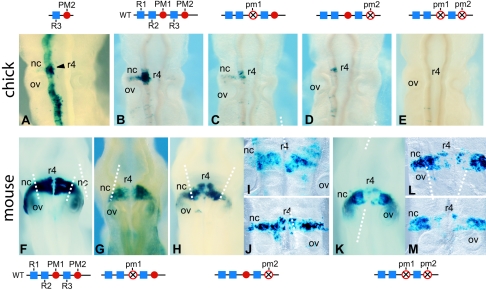
Transgenic analysis of the Hoxb1 enhancer in chicken and mouse embryos. (A to E) Dorsal views of transgene expression in electroporated chicken hindbrains. (A) The PM2 site in combination with R3 is able to mediate hindbrain expression with the highest level in r4 correlating with its ability to form a ternary complex. Note that the expression mediated by this minimal PM2-R3 element is broader than r4 because this region lacks repressor sequences shown to be important for restricting regulatory activity to r4 (51). (B to E) Expression patterns mediated by a wild-type 622-bp Hoxb1 fragment (B) and variants carrying mutations in the PM1 (C), PM2 (D), or PM1+PM2 (E) site. The constructs used are noted above each panel. (F to M) Transgene expression patterns in 9.5-dpc mouse embryos carrying the same wild-type (F) and mutant PM1 (G), PM2 (H to J), or PM1+PM2 (K to M) sites in the Hoxb1 constructs. (I, J, L, and M) Flat-mount preparations of embryos to clearly indicate the patchy and reduced level of expression in r4. The dashed vertical white lines indicate the border between the neural crest cells and the rhombomere or neural tube. Note in panels K to M that expression is greatly reduced or nearly absent in the r4 territory but is unaffected in the more lateral migrating cranial neural crest cells. This indicates a different requirement for PM sites in r4 than in neural crest. The arrowhead denotes rhombomere 4. nc, neural crest cells; ov, otic vesicle.
To further evaluate the in vivo regulatory activity of PM1 and PM2, we have tested these same 622-bp constructs in transgenic mouse assays, which have been previously employed to evaluate the role of PM1 and R3 in the Hoxb1 and Hoxb2 enhancers (16, 19). In these experiments, the DNA is stably integrated into the chromosome rather than working as an episome as in the chick electroporation experiments. The wild-type fragment from Hoxb1, containing all the PH and PM motifs, mediates strong reporter staining in the hindbrain of 9.5-dpc mouse embryos (Fig. 6F), similar to that observed in the chick electroporation experiments (Fig. 6B). Reporter staining is slightly reduced in the constructs carrying a mutation in the PM1 (Fig. 6G) or PM2 (Fig. 6H) sites, but the expression remains restricted to r4. Flat-mount preparations of the hindbrains make it easier to see the patchy and reduced expression in r4 of embryos carrying mutations in PM2 (Fig. 6I and J). Combined mutation of both the PM1 and PM2 sites results in a further decrease in expression in the mouse hindbrain (Fig. 6K). Again flat mounts of hindbrains with the double mutation illustrate that, in some cases, expression is abolished in all but a few cells in r4, in particular in the basal plate (Fig. 6L and M). It is interesting that, while there is a loss of expression in r4 and the basal plate in the PM1 and PM1+PM2 mutants, reporter staining is maintained laterally in neural crest cells migrating from r4 (Fig. 6H to M). This demonstrates the specificity of the PM site mutations and indicates that expression in migrating cranial neural crest cells derived from r4 has different requirements for PM sites than that of cells in r4 or the neural tube. This is consistent with our previous studies indicating that separate enhancers direct the expression of Hoxa2 in r4 and r4-derived neural crest cells (28).
Together these in vivo transgenic assays in mouse and chicken embryos indicate that the rhombomere-restricted activity of the Hoxb1 enhancer is dependent upon contributions from both the PM1 and PM2 sites. They also suggest that R2/PM3 may participate in serving to restrict expression to r4 in conjunction with other regulatory elements.
DISCUSSION
Hoxb1 plays an important role in directly regulating its own expression in r4, as well as that of Hoxb2. The affinity of Hoxb1 for DNA is specifically increased by dimerization with Pbx1, which facilitates binding to the autoregulatory element. This element was thought to contain three PH sites (R1, R2, and R3), of which the R3 site was most important for in vivo activity (16, 40). We and others have previously reported that in vitro the combination of PM-PH sites in both Hoxb1 and Hoxb2 allows a Prep1-Pbx-Hoxb1 ternary complex to bind DNA (16, 40). In vivo, the requirement for the PM sequence appeared to be different in different enhancer contexts, since the single PM site in Hoxb2, but not the PM1 site of Hoxb1, was essential for regulatory activity (16, 40). This suggested that additional components might contribute to the activity of the Hoxb1enhancer. In this study, we have discovered and characterized a second PM site (PM2) in the Hoxb1 enhancer, showing that it is functionally important. These results explain why the mutation of the PM1 site alone did not abolish Hoxb1 enhancer activity in transgenic embryos and confirms the important role of the cooperativity between PH and PM sites in Hoxb1 target sites. Moreover we have shown that R2 can function as a PM site (R2/PM3) binding the Prep1-Pbx1 but not the Pbx1-Hoxb1 heterodimer. This property of R2/PM3 results in inhibition of ternary complex formation mediated by flanking PM1 and R3 sites.
PM2 cooperates with R3 to form a trimeric Prep-Pbx-Hox complex in vitro and stimulate in vivo expression.
An additional Meinox-Pbx site (PM2) is present downstream of R3 (Fig. 1A and C). The PM2 site forms a trimeric Prep-Pbx-Hoxb1 complex in combination with R3 in vitro (Fig. 4) and stimulates robust reporter staining in the chicken hindbrain when linked to a lacZ reporter gene (Fig. 6A). The use of a 622-bp regulatory region that contains all PH and PM sites necessary for robust r4-restricted regulatory activity shows that both PM1 and PM2 are required for enhancer activity in transgenic mouse and chicken embryos (Fig. 6; Table 2). Our results indicate that mutation of PM2 alone may have a stronger influence on reporter expression and regulatory activity than alterations in PM1 alone, but when both sites are altered the activity is reduced even further or disappears completely in chicken embryos, indicating that the three remaining sites (R1, R2/PM3, and R3) are not sufficient to compensate for alterations in PM1 and PM2. The data on in vitro binding and in vivo transgene expression suggest that PM1 and PM2 have similar capabilities in cooperating with the R3 motif to form a ternary complex and stimulate reporter expression. In the mouse, the role of PM1 and PM2 is similar to that scored in chicken embryos. Furthermore, mutation of the PM sites specifically affected transgene expression in r4 cells and not in the r4-derived migrating neural crest cells. This demonstrates the distinct regulatory requirements for neural crest and rhombomeric cell populations. The synergy between the three PM sites and the R3 PH site is a key component in positive activation of the Hoxb1 enhancer in rhombomere 4.
R2/PM3 inhibits ternary complex formation.
We have found that the R2/PM3 site negatively interferes with the formation of the Prep1-Pbx1-Hoxb1 ternary complex in combination with the PM1-R3 sites and that mutations in R2/PM3 restore the ability of the PM1-R3 motif to assemble a ternary complex (Fig. 2). Furthermore, in vivo, the presence of R2/PM3 exerts a negative effect on PM1-R3 activity and inhibits reporter gene expression (Fig. 5) consistent with its ability to inhibit ternary complex formation in vitro. In addition, via this inhibitory activity R2/PM3 appears to contribute to the restriction of the Hoxb1 expression to r4. R2/PM3 represents a high-affinity binding site for the Prep1-Pbx1a dimer, and the same mutation that restores ternary complex formation also decreases the affinity for Prep1-Pbx1 dimers. High-affinity Prep1-Pbx1a binding to the Hoxb1 enhancer is due mainly to R2/PM3, suggesting that a strong binding site for Prep1-Pbx1 has an inhibitory effect on ternary complex formation.
Further proof that R2/PM3 is a PM site comes from the demonstration that R2/PM3 cannot form a ternary complex in combination with PM1. While the relative position of R2/PM3 and PM1 did not affect its inhibitory activity, transposition of R2/PM3 and R3 altered its ability to prevent ternary complex formation (Fig. 3B). These data are in agreement with previous results showing that the orientation and spacing between PM and PH sites can be very flexible (42). R2/PM3 also appears to limit the ability of the R1 motif to bind Hoxb1-Pbx1a heterodimers. How can we account for the unique properties of the R2/PM3 site in comparison to other PM sites? We favor the idea that differences in sequence context (flanking sequences) around the PM sites are important for modulating binding affinity, because we have found that flanking sequences adjacent to perfect core consensus PM sequences can dramatically alter binding properties (unpublished data).
Overall our data indicate that the high-affinity binding of Prep1-Pbx1a heterodimers to R2/PM3 is the only measured parameter that correlates with its ability to inhibit ternary complex formation. The high affinity of Prep1-Pbx1 for R2/PM3 may create a steric hindrance that prevents ternary complex formation at R3, since this is rescued by mutations in R2/PM3 that decrease the binding affinity for the dimer. Stable binding to R2/PM3 may also decrease the occupancy of the PM1-R3 site by Prep1-Pbx dimers, which is necessary to form the ternary complex with Hoxb1. In contrast, the PM1 site is a very weak site for Prep1-Pbx1a dimers, even though it displays specificity for the Prep1-Pbx complex and is essential for the ternary complex (16). It may therefore be better in luring and releasing Prep-Pbx dimers for interaction with Hoxb1-Pbx1a dimers in the generation of ternary complexes.
The Hoxb1 enhancer.
Our results show that highly homologous PM sequences display distinctly different biochemical (i.e., Prep-Pbx binding affinity) and functional (i.e., repressing or activating) properties. This is in agreement with previous studies of Drosophila labial and mouse Hoxb1 enhancer in which multimeric PH sites were shown to drive segmental gene expression (9, 10, 40, 42). The Hoxb1 enhancer appears to be a mixture of sequences endowed with both repressing and activating functions. Its activity depends on the nature of the proteins bound at the different sites, the nature of coactivators or corepressors recruited, and the specific chromatin structure. Three of the important sites bind Meinox-Pbx dimers (PM1, PM2, and R2/PM3), and the remaining two sites (R1 and R3) bind Pbx-Hoxb1 dimers. In the P19 cell culture model, upon induction with RA, the shift from repression to activation is exerted by activation of Hoxb1 via retinoic acid response elements (35), accompanied by the increase in Pbx1 expression. The R3 site appears to be the major DNA binding site for Pbx1-Hoxb1 heterodimers. Since recruitment of Hoxb1 by Pbx occurs on DNA, a site different from R3 must be required to tether Pbx1 to DNA. In this regard Meinox proteins (Meis and Prep) can serve the function of binding Pbx1 to DNA in a position- or orientation-dependent manner that may be efficiently recognized by Hoxb1. The different affinities of PM1, PM2, and R2/PM3 may be crucial in allowing or preventing Hoxb1 recruitment. If Prep1-Pbx is bound too firmly to DNA (R2/PM3), this might prevent Hoxb1 recruitment. The specific complexes bound will determine the type of coregulators to be recruited. For example, if Prep1-Pbx is bound to R2/PM3, it may recruit only corepressors. When the Prep1-Pbx-Hoxb1 complex is bound at the R3-PM2 or R3-PM1 site, coactivators might be recruited. This model also suggests that the binding activity of the R3-PM2 site for Prep-Pbx dimers may be decreased during retinoic acid induction either by binding of alternative factors or by posttranslational mechanisms that alter specific properties of Hoxb1.
Several data support this hypothesis. First, dimeric and trimeric complexes can be immunoprecipitated from cells (reference 16 and our unpublished data). Second, the timing of expression of the involved proteins during retinoic acid induction of Hoxb1 also is in line with our proposed mechanism. In fact, in P19 or NT2 cells and the absence of retinoic acid, Prep1 and Pbx1 proteins are present in the nucleus and form dimeric complexes (16). Upon addition of retinoic acid, not only is Hoxb1 itself synthesized but also Pbx1 and Meis1 are induced (16). Thus, different Meinox-Pbx dimers can be formed which might have different affinities for the different PM sites.
Role of Prep1 and other Meinox proteins in Hoxb1 expression.
The general model that emerges from this comparison of Hoxb1 and Hoxb2 regulation implies that Prep1 and/or other members of the Meinox family (Meis) are essential for cross- and autoregulatory activities mediated by Hoxb1. As shown in experiments performed in zebra fish, the down-regulation of Prep1.1 results in a major decrease in Hoxb1a expression in the hindbrain, as well as of Hoxb2 and Hoxa2, genes that are dependent on Hoxb1a (13). Moreover, the expression of dominant-negative constructs of Meis in zebra fish has a similar effect resulting in phenotypes similar to those of the lazarus/pbx mutant (55). The data presented in this paper, therefore, give molecular support to the phenotypic observations in zebra fish and show the importance of interactions between PH and PM sites to facilitate formation of the ternary Meinox-Pbx-Hoxb1 complexes and stimulate regulatory activity. This has important implications for helping to identify and define Hox-responsive target sites of in vivo relevance.
Acknowledgments
This work was supported by grants from Telethon, by the Italian Ministry for University and Research (MIUR, COFIN 2002-2005), and AIRC (Italian Association for Cancer Research), to F.B. F.C., L.M.W., and R.K. were supported by funds from the Stowers Institute for Medical Research, and S.T. was supported by a Boehringer Ingelheim Fonds Fellowship. E.F. is grateful to the EMBO (European Molecular Biology Organization) for a short-term fellowship.
REFERENCES
- 1.Azpiazu, N., and G. Morata. 1998. Functional and regulatory interactions between Hox and extradenticle genes. Genes Dev. 12:261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bel-Vialar, S., N. Itasaki, and R. Krumlauf. 2002. Initiating Hox gene expression in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 129:5103-5115. [DOI] [PubMed] [Google Scholar]
- 3.Berthelsen, J., J. Vandekerkhove, and F. Blasi. 1996. Purification and characterization of UEF3, a novel factor involved in the regulation of the urokinase and other AP-1 controlled promoters. J. Biol. Chem. 271:3822-3830. [DOI] [PubMed] [Google Scholar]
- 4.Berthelsen, J., V. Zappavigna, E. Ferretti, F. Mavilio, and F. Blasi. 1998. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 17:1434-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthelsen, J., V. Zappavigna, F. Mavilio, and F. Blasi. 1998. Prep1, a novel functional partner of Pbx proteins. EMBO J. 17:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burglin, T. 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25:4173-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burglin, T. R., and G. Ruvkun. 1992. New motif in PBX genes. Nat. Genet. 1:319-320. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, S. B. 1995. Homeotic genes and the evolution of arthropods and chordates. Nature 376:479-485. [DOI] [PubMed] [Google Scholar]
- 9.Chan, S.-K., L. Jaffe, M. Capovilla, J. Botas, and R. S. Mann. 1994. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 78:603-615. [DOI] [PubMed] [Google Scholar]
- 10.Chan, S.-K., and R. S. Mann. 1996. A structural model for a homeotic protein extradenticle-DNA complex accounts for the choice of HOX protein in the heterodimer. Proc. Natl. Acad. Sci. USA 93:5223-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, S.-K., H.-D. Ryoo, A. Gould, R. Krumlauf, and R. Mann. 1997. Switching the in vivo specificity of a minimal HOX-responsive element. Development 124:2007-2014. [DOI] [PubMed] [Google Scholar]
- 12.Chang, C.-P., W.-F. Shen, S. Rozenfeld, H. J. Lawrence, C. Largman, and M. L. Cleary. 1995. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 9:663-674. [DOI] [PubMed] [Google Scholar]
- 13.De Florian, G., N. Tiso, E. Ferretti, F. Blasi, M. Bortolussi, and F. Argenton. 2004. Prep1.1 has essential and unique genetic functions in hindbrain development and neural crest cells differentiation. Development 131:613-627. [DOI] [PubMed] [Google Scholar]
- 14.Di Rocco, G., A. Gavalas, H. Popperl, R. Krumlauf, F. Mavilio, and V. Zappavigna. 2001. The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 auto-regulatory enhancer function. J. Biol. Chem. 276:20506-20515. [DOI] [PubMed] [Google Scholar]
- 15.Di Rocco, G., F. Mavilio, and V. Zappavigna. 1997. Functional dissection of a transcriptionally active, target-specific Hox-Pbx complex. EMBO J. 16:3644-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferretti, E., H. Marshall, H. Pöpperl, M. Maconochie, R. Krumlauf, and F. Blasi. 2000. Segmental expression of Hoxb2 in r4 requires two distinct sites that facilitate cooperative interactions and ternary complex formation between Perp, Pbx and Hox proteins. Development 127:155-166. [DOI] [PubMed] [Google Scholar]
- 17.Gould, A., A. Morrison, G. Sproat, R. A. H. White, and R. Krumlauf. 1997. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 11:900-913. [DOI] [PubMed] [Google Scholar]
- 18.Itasaki, N., S. Bel-Vialar, and R. Krumlauf. 1999. “Shocking” developments in chick embryology: electroporation and in ovo gene expression. Nat. Cell Biol. 1:E203-E207. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs, Y., C. A. Schnabel, and M. L. Cleary. 1999. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol. Cell. Biol. 19:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawagoe, H., R. K. Humphries, A. Blair, H. J. Sutherland, and D. E. Hogge. 1999. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia 13:687-698. [DOI] [PubMed] [Google Scholar]
- 21.Kessel, M., and P. Gruss. 1991. Homeotic transformations of murine prevertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67:89-104. [DOI] [PubMed] [Google Scholar]
- 22.Keynes, R., and R. Krumlauf. 1994. Hox genes and regionalization of the nervous system. Annu. Rev. Neurosci. 17:109-132. [DOI] [PubMed] [Google Scholar]
- 23.Knoepfler, P. S., Q. Lu, and M. P. Kamps. 1996. Pbx-1 Hox heterodimers bind DNA on inseparable half-sites that permit intrinsic DNA binding specificity of the Hox partner at nucleotides 3′ to a TAAT motif. Nucleic Acids Res. 24:2288-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krumlauf, R. 1994. Hox genes in vertebrate development. Cell 78:191-201. [DOI] [PubMed] [Google Scholar]
- 25.Li, X., A. Veraksa, and W. McGinnis. 1999. A sequence motif distinct from Hox binding sites controls the specificity of a Hox response element. Development 126:5581-5589. [DOI] [PubMed] [Google Scholar]
- 26.Lu, Q., and M. P. Kamps. 1996. Structural determinants within Pbx1 that mediate cooperative DNA binding with pentapeptide-containing HOX proteins: proposal for a model of a PBX1-HOX-DNA complex. Mol. Cell. Biol. 16:1632-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lumsden, A., and R. Krumlauf. 1996. Patterning the vertebrate neuraxis. Science 274:1109-1115. [DOI] [PubMed] [Google Scholar]
- 28.Maconochie, M., R. Krishnamurthy, S. Nonchev, P. Meier, M. Manzanares, P. Mitchell, and R. Krumlauf. 1999. Regulation of Hoxa2 in cranial neural crest cells involves members of the AP-2 family. Development 126:1483-1494. [DOI] [PubMed] [Google Scholar]
- 29.Maconochie, M., S. Nonchev, M. Studer, S.-K. Chan, H. Pöpperl, M.-H. Sham, R. Mann, and R. Krumlauf. 1997. Cross-regulation in the mouse HoxB complex: the expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes Dev. 11:1885-1896. [DOI] [PubMed] [Google Scholar]
- 30.Mann, R. S., and M. Affolter. 1998. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 8:423-429. [DOI] [PubMed] [Google Scholar]
- 31.Mann, R. S., and S.-K. Chan. 1996. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 12:258-262. [DOI] [PubMed] [Google Scholar]
- 32.Manzanares, M., S. Bel-Vialer, L. Ariza-McNaughton, E. Ferretti, H. Marshall, M. K. Maconochie, F. Blasi, and R. Krumlauf. 2001. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involves auto and cross-regulatory mechanisms. Development 128:3595-3607. [DOI] [PubMed] [Google Scholar]
- 33.Manzanares, M., S. Cordes, L. Ariza-McNaughton, V. Sadl, K. Maruthainar, G. Barsh, and R. Krumlauf. 1999. Conserved and distinct roles of kreisler in regulation of the paralogous Hoxa3 and Hoxb3 genes. Development 126:759-769. [DOI] [PubMed] [Google Scholar]
- 34.Manzanares, M., P. Trainor, S. Nonchev, L. Ariza-McNaughton, J. Brodie, A. Gould, H. Marshall, A. Morrison, C.-T. Kwan, M.-H. Sham, D. G. Wilkinson, and R. Krumlauf. 1999. The role of kreisler in segmentation during hindbrain development. Dev. Biol. 211:220-237. [DOI] [PubMed] [Google Scholar]
- 35.Marshall, H., M. Studer, H. Pöpperl, S. Aparicio, A. Kuroiwa, S. Brenner, and R. Krumlauf. 1994. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature 370:567-571. [DOI] [PubMed] [Google Scholar]
- 36.Mathis, L., P. M. Kulesa, and S. E. Fraser. 2001. FGF receptor signalling is required to maintain neural progenitors during Hensen's node progression. Nat. Cell Biol. 3:559-566. [DOI] [PubMed] [Google Scholar]
- 37.McGinnis, W., and R. Krumlauf. 1992. Homeobox genes and axial patterning. Cell 68:283-302. [DOI] [PubMed] [Google Scholar]
- 38.Moens, C. B., and V. E. Prince. 2002. Constructing the hindbrain: insights from the zebrafish. Dev. Dyn. 224:1-17. [DOI] [PubMed] [Google Scholar]
- 39.Muramatsu, T., Y. Mizutani, Y. Ohmori, and J.-I. Okumura. 1997. Comparison of three non-viral transfection methods for foreign gene expression in early chicken embryos in ovo. Biochem. Biophys. Res. Commun. 230:376-380. [DOI] [PubMed] [Google Scholar]
- 40.Pöpperl, H., M. Bienz, M. Studer, S. K. Chan, S. Aparicio, S. Brenner, R. S. Mann, and R. Krumlauf. 1995. Segmental expression of Hoxb1 is controlled by a highly conserved autoregulatory loop dependent upon exd/Pbx. Cell 81:1031-1042. [DOI] [PubMed] [Google Scholar]
- 41.Prince, V. E., C. B. Moens, C. B. Kimmel, and R. K. Ho. 1998. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development 125:393-406. [DOI] [PubMed] [Google Scholar]
- 42.Ryoo, H. D., T. Marty, F. Casares, M. Affolter, and R. S. Mann. 1999. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development 126:5137-5148. [DOI] [PubMed] [Google Scholar]
- 43.Serpente, P., S. Tumpel, N. B. Ghyselinck, K. Niederreither, L. M. Wiedemann, P. Dolle, P. Chambon, R. Krumlauf, and A. P. Gould. 2005. Direct crossregulation between retinoic acid receptor {beta} and Hox genes during hindbrain segmentation. Development 132:503-513. [DOI] [PubMed] [Google Scholar]
- 44.Sham, M. H., C. Vesque, S. Nonchev, H. Marshall, M. Frain, R. Das Gupta, J. Whiting, D. Wilkinson, P. Charnay, and R. Krumlauf. 1993. The zinc finger gene Krox-20 regulates Hoxb-2 (Hox2.8) during hindbrain segmentation. Cell 72:183-196. [DOI] [PubMed] [Google Scholar]
- 45.Shen, W. F., S. Rozenfeld, H. J. Lawrence, and C. Largman. 1997. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J. Biol. Chem. 272:8198-8206. [DOI] [PubMed] [Google Scholar]
- 46.Simeone, A., D. Acampora, L. Arcioni, P. W. Andrews, E. Boncinelli, and F. Mavilio. 1990. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature 346:763-766. [DOI] [PubMed] [Google Scholar]
- 47.Smith, K. S., Y. Jacobs, C. P. Chang, and M. L. Cleary. 1997. Chimeric oncoprotein E2a-Pbx1 induces apoptosis of hematopoietic cells by a p53-independent mechanism that is suppressed by Bcl-2. Oncogene 14:2917-2926. [DOI] [PubMed] [Google Scholar]
- 48.Streit, A., A. J. Berliner, C. Papanayotou, A. Sirulnik, and C. D. Stern. 2000. Initiation of neural induction by FGF signalling before gastrulation. Nature 406:74-78. [DOI] [PubMed] [Google Scholar]
- 49.Studer, M., A. Gavalas, H. Marshall, L. Ariza-McNaughton, F. Rijli, P. Chambon, and R. Krumlauf. 1998. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development 125:1025-1036. [DOI] [PubMed] [Google Scholar]
- 50.Studer, M., A. Lumsden, L. Ariza-McNaughton, A. Bradley, and R. Krumlauf. 1996. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature 384:630-635. [DOI] [PubMed] [Google Scholar]
- 51.Studer, M., H. Pöpperl, H. Marshall, A. Kuroiwa, and R. Krumlauf. 1994. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science 265:1728-1732. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dijk, M. A., and C. Murre. 1994. extradenticle raises the DNA binding specificity of homeotic selector gene products. Cell 78:617-624. [DOI] [PubMed] [Google Scholar]
- 54.Vesque, C., M. Maconochie, S. Nonchev, L. Ariza-McNaughton, A. Kuroiwa, P. Charnay, and R. Krumlauf. 1996. Hoxb-2 transcriptional activation by Krox-20 in vertebrate hindbrain requires an evolutionary conserved cis-acting element in addition to the Krox-20 site. EMBO J. 15:5383-5396. [PMC free article] [PubMed] [Google Scholar]
- 55.Waskiewicz, A. J., H. A. Rikhof, and C. B. Moens. 2002. Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev. Cell 3:723-733. [DOI] [PubMed] [Google Scholar]
- 56.Whiting, J., H. Marshall, M. Cook, R. Krumlauf, P. W. J. Rigby, D. Stott, and R. K. Allemann. 1991. Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev. 5:2048-2059. [DOI] [PubMed] [Google Scholar]
- 57.Wilson, D. S., and C. Desplan. 1999. Structural basis of Hox specificity. Nat. Struct. Biol. 6:297-300. [DOI] [PubMed] [Google Scholar]