Abstract
The transcription factors RFX and CIITA are major players in regulation of the expression of all classical and nonclassical major histocompatibility complex class II (MHC-II) genes. RFX nucleates the formation of a multiprotein complex, called the MHC-II enhanceosome, on MHC-II promoters. Assembly of this enhanceosome is an obligatory step for recruitment of the coactivator CIITA and thus for activation of MHC-II gene transcription. We have analyzed the function of the ankyrin repeat-containing protein RFXANK, which forms the heterotrimeric RFX complex together with RFX5 and RFXAP. We discovered that ANKRA2, the closest paralogue of RFXANK, can substitute for RFXANK in the activation of MHC-II genes and that this ability is mediated by its ankyrin repeat domain (ARD). This finding provided the basis for a high-resolution structure-function analysis of the ARD of RFXANK, which allowed us to map the RFX5 interaction domain and residues critical for assembly of the RFX complex. We also found that mutations in the fourth ankyrin repeat of RFXANK abolish assembly of the enhanceosome on MHC-II promoters in vivo but not in vitro, suggesting a new role of RFXANK in facilitating promoter occupation in the context of chromatin.
The function of major histocompatibility complex class II (MHC-II) molecules is crucial for the immune system. They present antigenic peptides derived from exogenous antigens to CD4-positive T cells and thereby trigger the initiation of adaptive immune responses (6). Furthermore, the interaction of immature T lymphocytes with MHC-II-positive cells in the thymus regulates key steps of the positive and negative selection processes that shape the mature T-cell repertoire (46). The central importance of these functions of MHC-II molecules is clearly manifested by the phenotype of patients suffering from MHC-II deficiency (also called the bare lymphocyte syndrome [BLS]). BLS patients fail to express MHC-II genes and consequently cannot mount efficient adaptive immune responses (11, 37).
Unlike MHC-I molecules, which are expressed on virtually all cells, the expression of MHC-II molecules is largely restricted to thymic epithelial cells and professional antigen-presenting cells, including dendritic cells, macrophages, and B cells. Other cell types can be induced to express MHC-II genes by exposure to gamma interferon or other stimuli. This tightly regulated spatial and temporal pattern of MHC-II expression is dictated mainly by the expression of a transcriptional coactivator called CIITA (14, 18, 37, 43, 44). CIITA is therefore often referred to as the “master regulator” of MHC-II genes, and much effort has consequently focused on elucidating the mechanisms by which it activates transcription. These include the induction of histone modifications, chromatin remodeling, recruitment of the general transcription machinery, and phosphorylation of RNA polymerase II (2, 4, 17, 24, 25, 41, 44, 50).
Although there is no doubt that CIITA is a key player, it is clearly not sufficient for activating MHC-II gene expression. CIITA has to be recruited to MHC-II promoters by multiple protein-protein interactions with the components of a large multiprotein complex referred to as the MHC-II enhanceosome (4, 16, 23, 37, 44, 49), which assembles on a characteristic promoter-proximal regulatory motif located ∼150 to 300 bp upstream of the transcription initiation site (1). This regulatory module is composed of four highly conserved sequence elements, called the S (or W), X, X2, and Y boxes, which are targets of unidentified S-box binding proteins, RFX, X2BP (which contains CREB), and NF-Y, respectively (21, 27, 29, 38). While NF-Y and CREB are implicated in the regulation of many genes, RFX is almost exclusively specific for MHC genes.
RFX is a trimeric complex composed of RFX5, RFXAP, and RFXANK (also called RFX-B and Tvl-1) (8, 19, 22, 31, 42). It is absolutely essential for the activation of all MHC-II genes, as is demonstrated by the fact that the absence of any one of the three RFX subunits leads to BLS (8, 22, 31, 42). The RFXANK gene is mutated in BLS complementation group B, which accounts for most known cases of the disease (22, 31, 37, 47). In RFX-deficient BLS patients, enhanceosome formation is abolished, CIITA cannot be recruited, and MHC-II expression is abrogated. Although this absolute requirement for RFX in transcriptional activation of MHC-II genes was established over 15 years ago (38), very little is known about the specific functions of the three subunits of the RFX complex. This report addresses the role of RFXANK.
RFXANK is a 260-amino-acid (aa) protein constituting the smallest subunit of the RFX complex. Its C-terminal region contains an ankyrin repeat domain (ARD) composed of four ankyrin repeats, which are well-known protein-protein interaction motifs found in numerous proteins performing diverse functions (28). RFXANK has a closely related and conserved paralogue called ANKRA2 (20, 22, 36). The conservation between RFXANK and ANKRA2 is restricted to the ARD (70% identity). Despite this homology, the function of ANKRA2 has not been linked to the expression of MHC-II genes (see Discussion).
To dissect the function of RFXANK, we designed an approach that combines our current knowledge of the three-dimensional (3-D) structure of ARD proteins with a rigorous analysis of a large panel of RFXANK mutants in a variety of in vitro assays and in a functional assay consisting of complementation of a BLS group B cell line. We first found that the ARD of ANKRA2 can functionally replace the ARD of RFXANK in the complementation of BLS cells from group B. This contributed to the design of a very precise site-directed mutagenesis analysis that permitted us to map specific residues implicated in three key functions of RFXANK. The RFXANK-RFX5 interaction domain was mapped to an unprecedented outer surface of ankyrin repeats 2 and 3. In addition, mutations within ankyrin repeats 1 and 3 were found to interfere with formation of the RFX complex without abolishing RFXANK's ability to interact separately with the other two subunits. Finally, mutations in ankyrin repeat 4 inhibited assembly of the enhanceosome complex in vivo but not in vitro, suggesting that RFXANK plays a specific role in MHC-II promoter occupation in the context of chromatin.
MATERIALS AND METHODS
Cell culture and FACS analysis.
The RFXANK-deficient BLS-1 cell line and the corresponding complemented BLS-1c cells have been described previously (15, 22, 24, 25). These cell lines and all transfectants were grown in RPMI 1640+ Glutamax I medium (Invitrogen) supplemented with 15% fetal calf serum and antibiotics. Fluorescence-activated cell sorter (FACS) analysis was performed using the following antibodies: HLA-DR monoclonal antibody 2.06 (5), HLA-DP monoclonal antibody BRAFB6 (Serotec), HLA-DQ monoclonal antibody SPVL3 (Serotec), and fluorescein isothiocyanate-conjugated rabbit anti-mouse immunoglobulin G (STAR9B; Serotec).
Transductions.
BLS-1 cells stably expressing RFXANK mutants were produced by lentiviral transduction as described previously (26, 35). Transduced cells expressing the vector-encoded mCD8 marker were stained with anti-mCD8-biotin antibody (Ly-2; PharMingen) and sorted to >95% purity with streptavidin-coated dynabeads (M-280; Dynal).
Plasmids and constructs.
Relevant sequence information for plasmids containing ANKRA2, hybrid RFXANK-ANKRA2, and wild-type and mutated RFXANK cDNAs (deletion endpoints, RFXANK-ANKRA2 fusions, and point mutations) is available on request. Wild-type and mutant RFXANK constructs contained a FLAG tag fused to the N terminus (C-terminal deletion mutants and point mutants) or C terminus (N-terminal deletion mutants). The presence of the FLAG tag was confirmed to have no influence on the function of RFXANK.
GST pull-down assays.
Wild-type and mutant RFXANK cDNAs were cloned into the pGEX4T2 plasmid (Amersham). Glutathione S-transferase (GST)-RFXANK fusion proteins were produced in B21-CodonPlus (DE3)-RIL Escherichia coli cells (Stratagene) by a 3-h induction with 0.25 M IPTG (isopropyl-β-d-thiogalactopyranoside) and were purified from the total lysate using glutathione-Sepharose Microspin modules (Amersham). For each assay, ∼10 μg of either GST or GST-RFXANK prebound to Microspin beads was mixed with 5 μl of [35S]Met-labeled in vitro-translated RFX5, RFXAP, or CIITA in 1 ml of binding buffer (50 mM Tris-HCl, pH 8.0, 150 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 5% glycerol, 1% bovine serum albumin [BSA], 1% Triton, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol [DTT]) and incubated for 30 min at 30°C. The beads were washed three times in washing buffer (binding buffer with the KCl concentration increased to 250 mM), and bound proteins were then eluted in sodium dodecyl sulfate (SDS) sample buffer, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and revealed by autoradiography.
Promoter pull-down assays.
Promoter pull-down assays with whole-cell extracts were performed essentially as described previously (23, 29). Briefly, 0.6 mg of whole-cell extracts prepared from BLS-1 cells and transfectants was incubated with a magnetic-bead-bound biotinylated HLA-DRA promoter fragment, prepared by PCR using pDRsyn (45) as a template. After a 2-h incubation in binding buffer [20 mM HEPES, pH 7.9, 100 mM KCl, 6 mM MgCl2, 1 mM DTT, 20% glycerol, 0.01% NP-40, 0.15 mg/ml poly(dI-dC) · poly(dI-dC), 0.15 mg/ml single-stranded DNA from E. coli, protease inhibitors], the beads were washed three times in washing buffer (binding buffer supplemented with 0.1% BSA) and proteins were eluted in SDS-PAGE buffer and analyzed by Western blotting using polyclonal rabbit antibodies for RFX5 and RFXAP (8) and anti-FLAG antibodies (M2; Sigma) to detect wild-type and mutant RFXANK proteins.
Coimmunoprecipitation.
Two microliters of FLAG-RFXANK, HA-RFXAP, and RFX5, all produced in HeLa cells using the Vaccinia-T7 system (10), was mixed in binding buffer (20 mM HEPES, 100 mM KCl, 0.5 mM DTT, 0.1% BSA, 0.1% NP-40, protease inhibitors) and incubated for 2 h on ice. Ten microliters of anti-FLAG M2 antibody coupled to agarose beads (Sigma) was added, and the reaction mixture was incubated for 1 h on ice. The beads were then washed three times in the same buffer, and proteins were eluted in SDS-PAGE sample buffer and analyzed by Western blotting using polyclonal rabbit antibodies for RFXANK, RFX5, and RFXAP (8, 23).
ChIP.
Chromatin immunoprecipitation (ChIP) experiments were performed as previously described (24) using polyclonal rabbit anti-RFX and anti-CIITA antisera (3, 42). Ten micrograms of cross-linked chromatin (corresponding to 1.2 × 107 cells) was used for each immunoprecipitation. Each ChIP was repeated twice with identical results. HLA-DRA promoter sequences present in the immunoprecipitates were quantified by real-time PCR using the following primers: 5′-ATTTTTCTGATTGGCCAAAGAGTAATT-3′ and 5′-AAAAGAAAAGAGAATGTGGGGTGTAA-3′. PCR was performed using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems) and a SYBR green-based kit for quantitative PCR (Eurogentec). The amounts of immunoprecipitated DNA were calculated by comparison to a standard curve generated with serial dilutions of input DNA. All PCRs were repeated at least three times.
Quantification of ANKRA2 mRNA.
Total RNAs were extracted with Trizol (Invitrogen). cDNA was synthesized from 1 μg of RNA using random hexamer primers and Superscript II reverse transcriptase (Invitrogen). Real-time PCR was then performed to quantify the amounts of ANKRA2 mRNA with the following two sets of primers, with identical results: exon2-exon 3, 5′-ACCTGGAGGTGGCATCTG-3′ and 5′-GGGTGTAGACATGCCTTACTTG-3′, and exon 4-exon 5, 5′-CTGGCTACTCGTATCGAACAAG-3′ and 5′-AGCTATTTGCCCGTGTGC-3′. PCR was performed using the iCycler iQ Real-Time PCR Detection System (Bio-Rad) and iQ SYBR green Supermix. The amounts of cDNAs were calculated by comparison to a standard curve generated with serial dilutions of a reference cDNA. All PCRs were repeated at least three times. The levels were normalized using the average values of 18S rRNA and TATA-binding protein mRNA as references. Error bars represent standard deviations from the mean obtained from three independent PCR amplifications.
3-D structure modeling.
The 3-D structure of the ankyrin repeat domain of RFXANK (amino acids 93 to 243) was modeled using the SwissModel Automated Comparative Protein Modeling Server (12, 39; http://swissmodel.expasy.org/). Four known structures of the ankyrin repeat-containing proteins GABP, myotrophin, and p18-INK4c (two independent structures) were used as templates (ExPDB database codes, 1AWCB, 2MYO_, 1IHBA, and 1BU9A).
In vitro transcription and translation.
Wild-type RFX5, HA-RFXAP, and CIITA (type IV) were cloned into pIVEX vectors (Roche; pIVEX 2.4a, 5′-His fusion, and pIVEX 2.3 and pIVEX 2.4b NdeI, 5′-His fusion, respectively) and were transcribed and translated in vitro using the TnT T7 coupled rabbit reticulocyte lysate system (Promega) in the presence of 35S-labeled methionine.
RESULTS
The ARD is sufficient for the function of RFXANK.
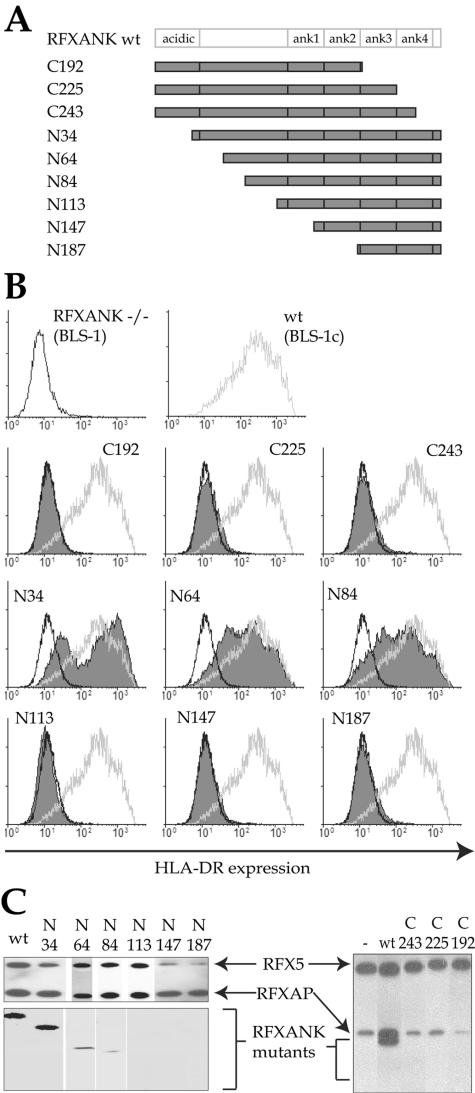
We initiated our structure-function analysis of RFXANK by generating a series of constructs encoding N-terminal and C-terminal deletion mutants (Fig. 1A). These constructs were transduced using a lentiviral vector into the BLS-1 cell line, which bears a null mutation in the RFXANK gene and thus lacks expression of MHC-II genes. The transduced cells were purified and tested by FACS for the restoration of cell surface HLA-DR expression (Fig. 1B). None of the C-terminal deletion mutants (C192, C225, and C243) were able to restore MHC-II expression. The largest N-terminal deletions (N113, N147, and N187) were also not functional. In contrast, wild-type RFXANK and shorter deletion mutants lacking only the N-terminal region of RFXANK (N34, N64, and N84) restored HLA-DR expression to wild-type levels.
FIG. 1.
Analysis of RFXANK deletion mutants. (A) Schematic representation of wild-type RFXANK and the deletion mutants used in this study. The C-terminal acidic domain and the N-terminal ankyrin repeats are indicated. The names of the mutants correspond to the end points of C- and N-terminal deletions. (B) FACS analysis of cell surface expression of HLA-DR in the BLS-1 (RFXANK−/−) B-cell line transfected stably with wild-type (wt) (BLS-1c) or mutant RFXANK constructs. The BLS-1 and BLS-1c profiles are included as a reference. Open black profiles correspond to BLS-1; open gray profiles correspond to BLS-1c; filled profiles represent cells complemented with the deletion mutants. (C) Promoter pull-down experiments performed with extracts prepared from BLS-1 cells or BLS-1 cells transfected with the indicated constructs. MHC-II enhanceosomes were assembled on HLA-DRA promoter fragments, purified, and analyzed for the presence of the three RFX subunits by Western blotting. Equal amounts of extract were used for each pull down. A partial RFX complex containing only RFX5 and RFXAP was pulled down in the absence of RFXANK (right, lane −). Functional RFXANK mutants (N34, N64, and N84) are integrated into this complex, while nonfunctional mutants (N113, N147, N187, C243, C225, and C192) are not.
To corroborate these findings with biochemical support, we performed a promoter pull-down assay which we developed previously to assess the assembly of a stable higher-order enhanceosome complex containing RFX, NF-Y, CREB, and the S-box binding factor (23, 29). Briefly, DNA fragments containing the HLA-DRA promoter region were incubated with extracts from BLS-1 cells complemented with wild-type or mutant RFXANK. DNA-bound enhanceosome complexes were purified, eluted, and analyzed by Western blotting for the presence of RFX (Fig. 1C). The results show that both wild-type RFXANK and the short N-terminal deletion mutants (N34, N64, and N84) can be incorporated into the enhanceosome complex. In contrast, the longer N-terminal deletion mutants and the C-terminal deletion mutants are not incorporated. The C-terminal region containing the ARD (aa 84 to 260) is thus essential and sufficient for the function of RFXANK. These findings are in agreement with previous studies demonstrating the importance of the ARD of RFXANK (7, 32-34).
In addition to the four ankyrin repeats, an additional region located between aa 84 and 113 is required for the function of RFXANK. These residues could potentially mediate a specific function of RFXANK. We do not favor this possibility, because they are not conserved in ANKRA2, which can complement the lack of RFXANK (see below). We hypothesize that residues located within this region play a structural role in stabilizing the 3-D structure of the ARD. This is supported by the fact that residues 104 to 112 are predicted to fold into a helix adjacent to the ARD (see Fig. 3A).
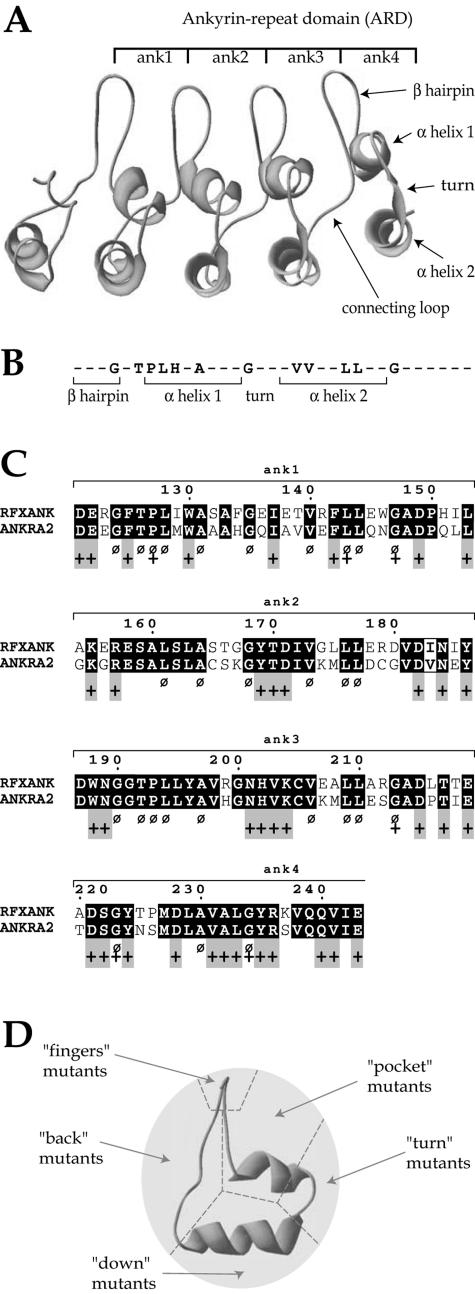
FIG. 3.
Selection of residues mutated in RFXANK. (A) Computer-generated three-dimensional model of the ankyrin repeat domain of RFXANK. The four ankyrin repeats are shown. Each consists of a β-hairpin and two anti-parallel α-helices. (B) Consensus ankyrin repeat sequence. Residues highly conserved between ankyrin repeat-containing proteins are indicated. Nonconserved residues are represented by dashes. (C) Alignment of the ankyrin repeat domains from RFXANK and ANKRA2. Residues conserved between the two proteins are highlighted in black. Residues that are highly conserved between many different ankyrin repeat proteins, and that were for this reason excluded from our selection, are indicated by a Ø sign. Residues predicted to be solvent accessible are indicated by a + sign. Gray boxes indicate residues that are accessible and conserved between RFXANK and ANKRA2 but not conserved in other ankyrin proteins. These residues were mutated in this study. (D) Residues selected for mutagenesis were divided into five groups according to their positions in the ankyrin repeat structure. A side view of the structure is shown. “Finger” mutants are located in the β-hairpin, “pocket” mutants in the ankyrin “groove,” “turn” mutants in the loop connecting the two α-helices, “down” mutants in the downward surface of the external α-helix, and “back” mutants in the loop connecting two adjacent ankyrin repeats.
ANKRA2 can substitute functionally for RFXANK.
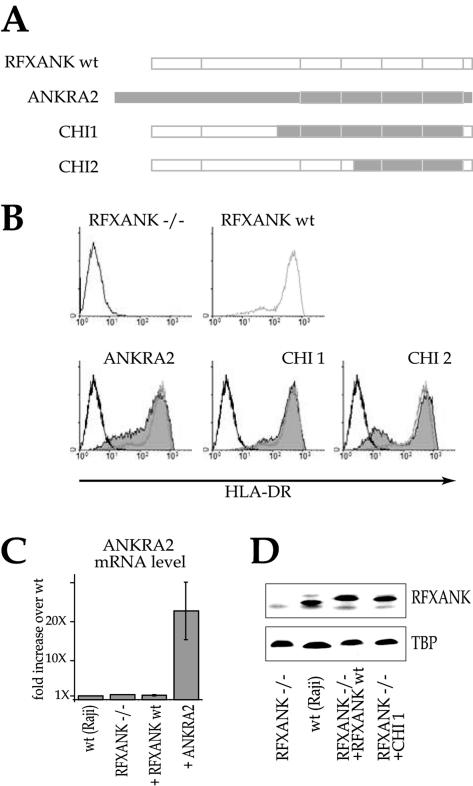
The ANKRA2 gene was previously identified as being the closest paralogue of human RFXANK (22). Like RFXANK, ANKRA2 contains a C-terminal ARD composed of four ankyrin repeats. We exploited the homology between RFXANK and ANKRA2 to gain insight into the respective importance of conserved versus divergent amino acids in the ARD of RFXANK. We created two chimeric constructs (CHI1 and CHI2) in which either all four or only the three most homologous ankyrin repeats of RFXANK were replaced with the corresponding regions from ANKRA2 (Fig. 2A). These constructs were assayed for the ability to restore HLA-DR expression in BLS-1 cells. Both chimeric constructs could replace RFXANK (Fig. 2B). These results imply that the function of RFXANK is not affected by amino acid changes at positions that differ between RFXANK and ANKRA2. In contrast, residues that are conserved between the two proteins must be critical for the function of RFXANK.
FIG. 2.
ANKRA2 can substitute functionally for RFXANK. (A) Schematic representations of RFXANK, ANKRA2, and the chimeric fusion constructs used to complement the BLS-1 (RFXANK−/−) cell line. (B). Stable transfectants were analyzed by FACS for HLA-DR cell surface expression as in Fig. 1. (C) Steady-state mRNA levels for ANKRA2 were determined by quantitative real-time reverse transcription-PCR for wild-type (wt) B cells, RFXANK−/− cells, RFXANK−/− cells complemented with cDNA for RFXANK, and RFXANK−/− cells complemented with cDNA for ANKRA2. The error bars indicate standard deviations. (D) Protein levels of RFXANK (in total extracts from RFXANK−/− cells, wild-type cells, and RFXANK−/− cells complemented with RFXANK) and the RFXANK-ANKRA2 fusion protein (in extracts from RFXANK−/− cells complemented with the CHI1 fusion) were determined by Western blotting. The level of TATA-binding protein (TBP) was used as a loading control.
The complementation of MHC-II expression by ANKRA2 in RFXANK−/− B cells could mean that ANKRA2 is normally absent or expressed at insufficient levels in these cells. We therefore measured the expression of ANKRA2 in wild-type B cells, RFXANK−/− cells, and the mutant cells complemented with RFXANK or ANKRA2 (Fig. 2C). The wild-type, RFXANK−/−, and RFXANK-complemented cells express only low levels of ANKRA2 mRNA. Complementation with the ANKRA2 cDNA increases its expression >20-fold compared to the three other cell lines. We therefore conclude that the endogenous level of ANKRA2 is insufficient to support expression of MHC-II genes in the absence of RFXANK.
The expression levels of RFXANK and the chimeric RFXANK-ANKRA2 proteins could be compared because they share the N terminus, which is recognized by our anti-RFXANK antibody. Both wild-type RFXANK and the CHI1 chimera were expressed at levels that are practically identical to the level of RFXANK in wild-type cells (Fig. 2D).
Generation of a panel of RFXANK mutants.
A combined experimental and computer-assisted approach was designed to select amino acid residues that are likely to mediate the function of RFXANK (Fig. 3). First, since the ARD of ANKRA2 can replace that of RFXANK, only residues that are conserved between these two proteins were retained as potential candidates (Fig. 3C). Second, we generated a computer model of the ARD of RFXANK (Fig. 3A). Residues predicted to be buried within the hydrophobic core were considered to be unlikely to mediate specific protein-protein interactions because they are probably important for maintaining the overall molecular structure of the ARD. We consequently included only residues that were predicted to be solvent accessible (Fig. 3C). Third, we excluded residues that are conserved between ankyrin repeats found in many different proteins (consensus sequence in Fig. 3B and C). These residues map mainly to the two well-conserved α-helices and are likely to be involved in maintaining the conserved structure of the domain. In summary, three criteria were used to select candidate residues that are likely to mediate the specific function(s) of RFXANK: residues that are conserved between RFXANK and ANKRA2, residues that are accessible at the surface of the protein, and residues that are not generally conserved between most ankyrin repeat proteins. Residues meeting all three criteria (Fig. 3C) were selected for site-directed mutagenesis. Mutations introduced at these residues were divided into five groups (pocket, finger, turn, bottom, and back mutants) on the basis of their localization within the ankyrin repeat structure (Fig. 3D). In each group, mutants affecting either one or several repeats were generated. A mutant affecting a defined surface of a single ankyrin repeat contained one, two, three, or four amino acid substitutions, depending on how many residues within their surfaces met the selection criteria (see Fig. 4 to 7 for details of the amino acid changes made). Mutants were named using a letter to indicate the localization of the mutations (D for down, F for finger, B for back, T for turn, and P for pocket), followed by one or more numbers referring to the ankyrin repeats containing the mutations (1, 2, 3, and 4 for the four repeats). All mutants were assayed for the ability to restore MHC-II expression in the BLS-1 complementation assay.
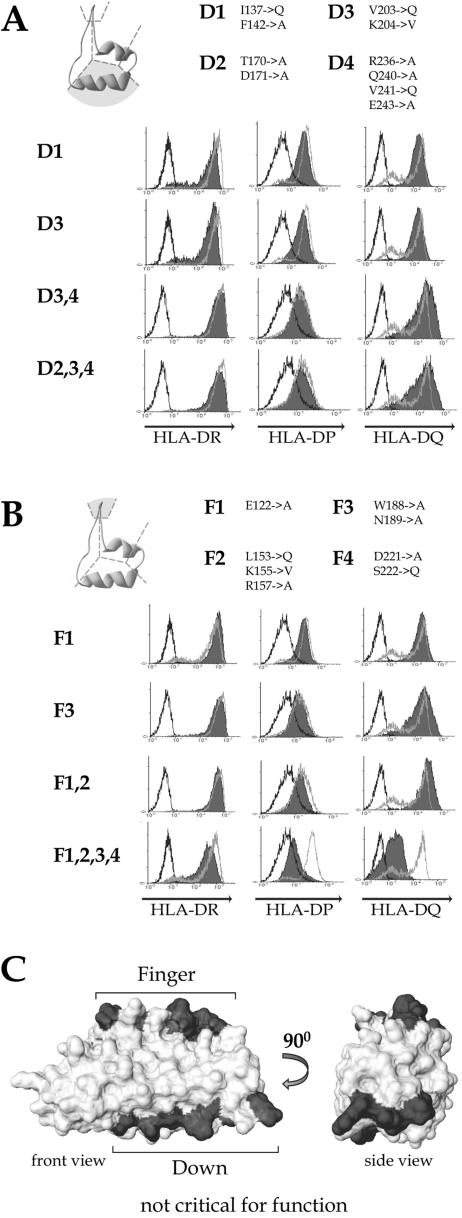
FIG. 4.
Mutations in the external α-helix and β-hairpins have no effect on RFXANK function. Mutations affecting the outer α-helix (“down” mutants) (A) and β-hairpins (“finger” mutants) (B) were analyzed in the complementation assay as described in the legend to Fig. 1B. The amino acid changes made in each mutant are listed at the top of each panel. A side view of an ankyrin repeat is shown schematically in the top left corner. The region that was mutated is shaded. (C) A space-filling model of the RFXANK ARD is shown so that the β-hairpins point upward and the external α-helices downward. The regions mutated in down and finger mutants are shaded in gray.
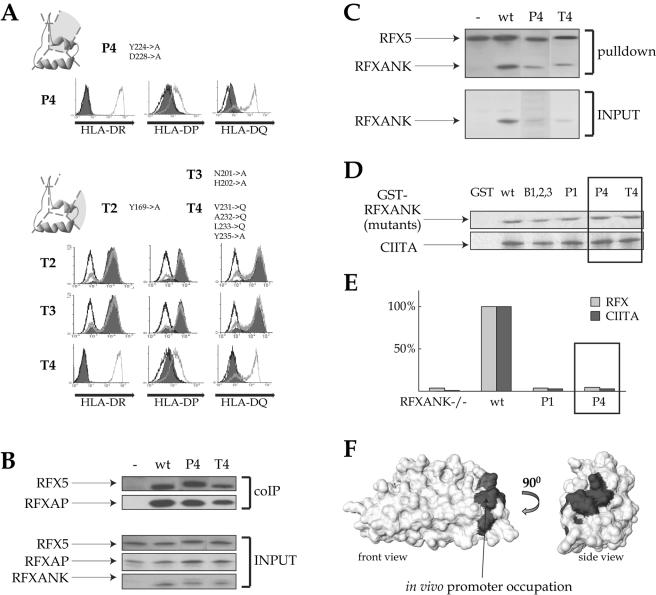
FIG. 7.
The fourth ankyrin repeat of RFXANK is necessary for in vivo promoter occupation. Mutations in the pocket of the fourth ankyrin repeat (P4) and in the turns of repeats 2, 3, and 4 (T2, T3, and T4) were analyzed in the complementation (A), promoter pull-down (B), coimmunoprecipitation (coIP) (C), and GST pull-down (D) assays as described in the legends to Fig. 1 and 5. (E) ChIP experiments with antibodies directed against RFX5 (light-gray bars) and CIITA (dark-gray bars) were performed in BLS-1 (RFXANK−/−) cells complemented with wild-type RFXANK and the P1 or P4 mutant. Noncomplemented BLS-1 cells were used as a negative control. Immunoprecipitates were analyzed for the presence of promoter sequences from the HLA-DRA gene. Signals observed in wild-type cells were set at 100%. A representative of three ChIP experiments is shown. (F) Space-filling model of the RFXANK ARD. The surface required for in vivo promoter occupation is marked in gray.
Mutations in the β-hairpins and outer α-helices do not affect RFXANK function.
Four mutants containing amino acid substitutions in the outer α-helices (down mutants D1, D3, D3,4, and D2,3,4) and four containing changes in the β-hairpins (finger mutants F1, F3, F1,2, and F1,2,3,4) were tested for the ability to restore MHC-II expression in BLS-1 cells. All four down mutants restored expression of the three HLA isotypes to wild-type levels (Fig. 4A), indicating that the outer α-helical surface of RFXANK does not engage in functionally important interactions. Of the four finger mutants, three restored normal MHC-II expression (Fig. 4B). Only the combined F1,2,3,4 mutant, which has a total of eight mutated residues affecting the β-hairpins of all four ankyrin repeats, had a partial effect. F1,2,3,4 restored expression of HLA-DR to nearly wild-type levels but did not completely restore the expression of HLA-DP and -DQ (Fig. 4B). The fact that mutations in the β-hairpin motifs of the ankyrin repeat domain do not destroy the function of RFXANK was unexpected, as these regions were previously proposed to mediate an interaction with RFXAP (33) (see Discussion and Fig. 8).
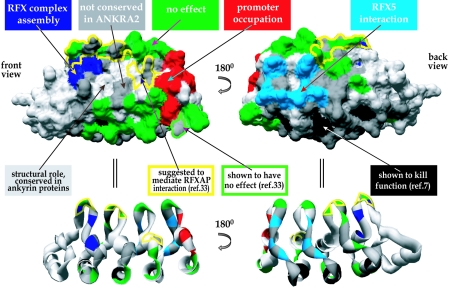
FIG. 8.
Model representing all known functions and interaction surfaces in RFXANK. Front and back views of the space-filling (top) and ribbon (bottom) models of RFXANK's ARD are marked with different colors representing different mutations and surfaces analyzed in this and previous studies. Residues implicated in RFX complex assembly (Fig. 6) are marked in dark blue, residues important for promoter occupation in vivo (Fig. 7) are marked in red, and those mediating interactions with RFX5 (Fig. 5) are marked in light blue. Residues at which the mutations had no effect (Fig. 4) are shown in green. Dark gray was used for residues which are not conserved in ANKRA2 and were therefore not analyzed. Light gray indicates residues which were not analyzed because they either are conserved in different ankyrin-containing proteins (and are therefore unlikely to mediate specific functions of RFXANK) or are inaccessible at the protein surface. Light gray was also used for the helix located adjacent to the ARD (leftmost part in the front view). Mutations of the residues marked in black were shown previously to be deleterious for RFXANK function (7). Since these mutations lie in the conserved α-helices, they probably disrupt the structure of the ARD. The yellow contour indicates residues suggested previously to mediate interactions with RFXAP (33). The same study showed that mutations of the residues marked with a green contour have no effect.
Mapping of the RFXANK-RFX5 interaction surface.
Four mutants containing substitutions in the “back” surface of the ankyrin repeat domain (residues located in the loops connecting adjacent repeats) were generated. Three of these mutants have changes in single ankyrin repeats (B1, B2, and B3). The fourth is a combined mutant containing changes in repeats 1, 2, and 3 (B1,2,3). Only B1 was fully functional in the complementation assay (Fig. 5A). Mutations in repeats 2 and 3 (B2 and B3) led to a partial reduction in the ability to restore MHC-II expression (Fig. 5A). The combined B1,2,3 mutant was completely inactive (Fig. 5A). Taken together, these results indicate that the surface defined by the B2 and B3 mutations (back of ankyrin repeats 2 and 3) is critical for the function of RFXANK.
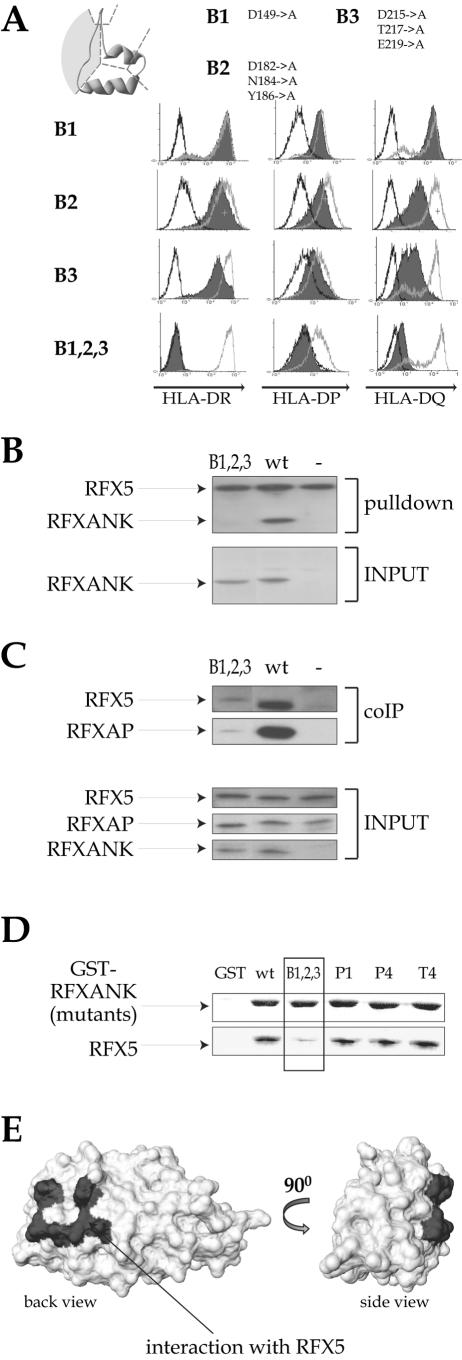
FIG.5.
Identification of the surface that mediates the interaction with RFX5. (A) Mutations located in the loops connecting two adjacent ankyrin repeats (“back” mutants) were analyzed in the complementation assay as described in the legend to Fig. 1B. (B) Promoter pull-down experiments with equal amounts of extracts prepared from BLS-1 cells complemented with either the B1,2,3 mutant or wild-type (wt) RFXANK were performed as described in the legend to Fig. 1C. Non complemented BLS-1 cells were used as a negative control (−). Input extracts were analyzed for the presence of RFXANK. The amounts of the other subunits of the RFX complex were equal in all input extracts (data not shown). (C) Vaccinia-T7-produced Flag-RFXANK proteins (wild-type or B1,2,3 mutant), RFX5 and RFXAP, were incubated together to form RFX complexes. Anti-Flag antibody was then used to immunoprecipitate RFXANK, and the presence of RFX5 and RFXAP in the immune complexes was analyzed by Western blotting. No RFXANK was added in the control lane (−). Inputs were analyzed in the bottom panels. (D) Equal amounts of glutathione-Sepharose bound GST or GST fusions containing wild-type and mutant RFXANK were incubated with in vitro-translated 35S-labeled RFX5 and washed. Bound proteins were fractionated by SDS-PAGE and analyzed by Coomassie blue staining (for GST fusions; top) or autoradiography (for RFX5; bottom). (E) Space-filling model of the RFXANK ARD. The RFXANK-RFX5 interaction domain is marked in gray.
Three biochemical assays were performed to determine why the back mutants destroy the function of RFXANK. To determine whether these mutations affect the ability of RFXANK to be integrated into the enhanceosome complex, we performed a promoter pull-down assay with extracts prepared from the transfected BLS-1 cells. The B1,2,3 mutant was unable to associate with the HLA-DRA promoter in this assay (Fig. 5B). We next checked by means of coimmunoprecipitation experiments whether B1,2,3 retained its ability to form the RFX complex. Coimmunoprecipitation of B1,2,3 with the other two subunits (RFX5 and RFXAP) of the RFX complex was strongly reduced (Fig. 5C). Finally, we performed GST pull-down assays to determine whether the back mutations interfere with the interaction of RFXANK with RFX5 or RFXAP. We found that the B1,2,3 mutant was strongly affected in its ability to interact with RFX5 (Fig. 5D). As positive controls, we used wild-type RFXANK and a selection of other nonfunctional mutants affecting different surfaces of RFXANK (see below). The loss of interaction with RFX5 is specific, as B1,2,3 interacts normally with both RFXAP and CIITA (Fig. 6C and 7D). Taken together, these results demonstrate that the “back” surfaces of ankyrin repeats 2 and 3 of RFXANK interact with RFX5 (Fig. 5E).
FIG. 6.
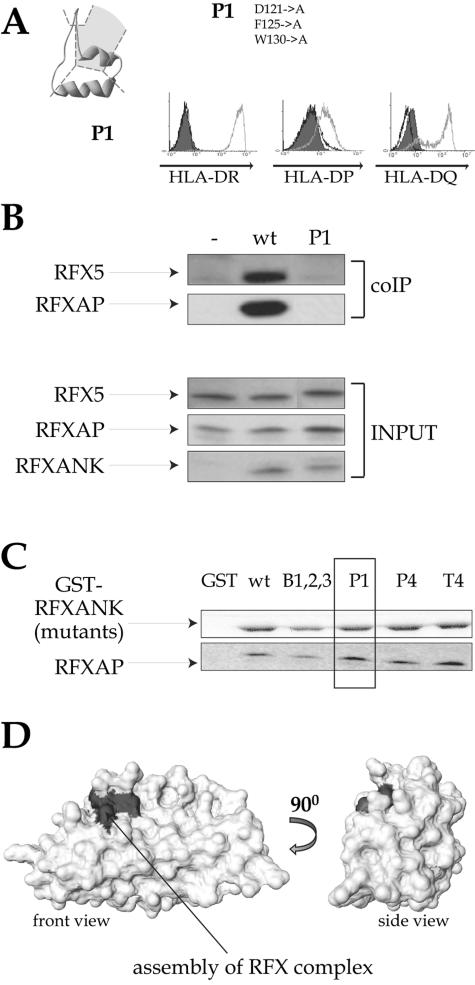
Mutations in the pocket of ankyrin repeat 1 abolish assembly of the RFX complex. The RFXANK mutant containing substitutions in the ankyrin groove region of repeat 1 (pocket mutant P1) was analyzed in the complementation (A), coimmunoprecipitation (B), and GST pull-down (C) assays as described in the legends to Fig. 1 and 5. wt, wild type. (D) Space-filling model of the RFXANK ARD. The surface implicated in RFX complex assembly is marked in gray.
The pocket of ankyrin repeat 1 is critical for assembly of the RFX complex.
The pocket mutant P1 was found to be unable to activate MHC-II expression in the transduced BLS-1 cells (Fig. 6A). P1 contains mutations in the ankyrin groove situated between the β-hairpin and the first α-helix of repeat 1. Coimmunoprecipitation experiments demonstrated that the P1 mutant is strongly affected in its ability to be integrated into the RFX complex (Fig. 6B). Surprisingly, GST pull-down experiments demonstrated that this assembly defect is not due to an abrogation of the interaction of RFXANK with RFX5 (Fig. 5D) or RFXAP (Fig. 6C). These findings indicate that the stable trimeric RFX complex cannot form despite the fact that the P1 mutant can interact separately with each of the other two components of the RFX complex. This apparent contradiction is addressed in Discussion.
The fourth ankyrin repeat of RFXANK is critical for promoter occupation in vivo.
The last mutants we analyzed contained mutations in the ankyrin “groove” (P4) or “turn” (T4) of the fourth ankyrin repeat. Neither mutant was able to correct the lack of MHC-II expression in BLS-1 cells (Fig. 7A). As was done with the previous mutants, we performed promoter pull-down assays, coimmunoprecipitation experiments, and GST pull-down assays to attempt to define the function that is defective in P4 and T4. Both mutants associated normally with RFXAP and RFX5 to form the RFX complex (Fig. 7B). P4 and T4 were also integrated into the enhanceosome complex formed on the HLA-DRA promoter (Fig. 7C). Finally, they interacted normally with RFXAP and RFX5 in GST pull-down assays (Fig. 5D and 6C). These RFXANK mutants can therefore interact normally with the other subunits of RFX and the other components of the enhanceosome in vitro, although they cannot activate the expression of MHC-II genes in vivo.
RFXANK has been shown to interact with CIITA (13, 16, 33, 49). We therefore performed GST pull-down experiments to check whether this interaction might be lost in the P4 and T4 mutants. Both mutants interacted with CIITA as efficiently as wild-type RFXANK (Fig. 7D).
Finally, we performed quantitative ChIP to determine whether the mutations in repeat 4 affect the ability of the enhanceosome complex to form on the HLA-DRA promoter in vivo. Chromatins were isolated from BLS-1 cells and BLS-1 cells transfected stably with wild-type RFXANK or the P4 mutant. Anti-RFX5 and anti-CIITA antibodies were used to precipitate chromatin fragments, and real-time PCR was used to quantify the HLA-DRA promoter fragments present in the immunoprecipitates. As demonstrated previously (24, 25), the absence of RFXANK results in a complete lack of HLA-DRA promoter occupation, indicated by the fact that no association of RFX5 and CIITA with the promoter is observed in BLS-1 cells (Fig. 7E). In cells complemented with wild-type RFXANK, the association of RFX and CIITA with the promoter is restored. In contrast, RFX and CIITA were not found at the promoter in cells complemented with the P4 mutant. This loss of promoter occupation is as severe as that observed when the cells are complemented with a mutant (P1) that cannot be integrated into the RFX complex. In conclusion, although the P4 mutant is integrated normally into RFX and can support enhanceosome assembly in vitro, it fails to be recruited to MHC-II promoters in vivo. This surprising finding suggests that repeat 4 performs a function that is important for formation of the MHC-II enhanceosome complex in vivo in the context of a native chromatin environment (Fig. 7F).
DISCUSSION
Elucidation of the genetic defects that are responsible for BLS has led to the discovery of four transcription factors that are highly specific for MHC-II genes (37). One of these factors, CIITA, has been the subject of intense research, and its function has become much clearer at the molecular level. For the remaining three factors, only very limited information is available regarding their contribution to the activation of MHC-II gene expression. In this study, we set out to elucidate the role of RFXANK. We took advantage of the availability of a B-cell line derived from a BLS patient (BLS-1) having a null mutation in the RFXANK gene. Functional complementation of BLS-1 cells with a series of RFXANK mutants, coupled with various complementary biochemical assays, was used to gain insight into key structural requirements and mechanistic aspects of the function of RFXANK.
The fact that RFXANK-deficient BLS patients exhibit an almost complete absence of MHC-II expression implies that RFXANK cannot be replaced by a functionally redundant factor in vivo. It thus came as a surprise that MHC-II expression can be restored in BLS-1 cells by ANKRA2. Our analysis shows that ANKRA2 is expressed at insufficient levels in B cells to support MHC-II expression in the absence of RFXANK. However, ANKRA2 is expressed widely in many tissues (36). Thus, the possibility that ANKRA2 could contribute to the activation of MHC-II expression in other cell types or under particular physiological conditions cannot be excluded. This possibility raises the interesting idea that two forms of the RFX complex might coexist in certain cells. These could differ with respect to their affinities or specificities for different MHC-II promoters or have different activation potentials. Importantly, the fact that increasing the expression of ANKRA2 to levels similar to those observed for RFXANK in wild-type cells is sufficient to activate MHC-II expression demonstrates that the two proteins have equal capacities to activate transcription. To determine which of the two proteins plays a dominant role in different cell types will require a comparison of the expression levels of RFXANK and ANKRA2 and quantification of MHC-II promoter occupation by these two factors in various different primary MHC-II-positive cells, particularly thymic epithelial cells and different types of resting and activated antigen-presenting cells.
Although ANKRA2 was initially identified as the closest homologue of RFXANK, subsequent studies suggested that its principal function might in fact be unrelated to MHC-II expression. It was shown to interact with megalin, a member of the LDL-receptor superfamily, which is involved in the endocytosis of a variety of metabolites by kidney epithelial cells (36). This interaction was found to be mediated by the ARD of ANKRA2. We show here that the same domain is responsible for its ability to substitute for RFXANK. In this context, it is interesting that RFXANK has also been implicated in two very different processes. In addition to its well-established role in the regulation of MHC-II expression, RFXANK has been suggested to be implicated in Raf-1 signaling. The interaction of RFXANK with Raf-1 was found to be mediated by its ankyrin repeat domain (19). It remains to be determined whether both RFXANK and ANKRA2 indeed have alternative roles in very different cellular processes or if the aforementioned findings merely reflect a certain level of promiscuity in the interactions mediated by ankyrin repeat-containing proteins, which are often able to interact with many cellular partners.
Ankyrin repeats are one of the most common protein-protein interaction motifs and are found in numerous proteins having diverse functions, including cytoskeletal organization, cell cycle control, differentiation, and transcriptional regulation (28). Their characteristic L-shaped structure (sometimes described as a “hand”) is formed by two short antiparallel α-helices connected by a short loop and a β-hairpin structure extending at an almost 90° angle from one of the α-helices (in the “hand” analogy, the β-hairpins form the fingers and the α-helices correspond to the palm). At the level of their primary sequences, ankyrin repeats exhibit a marked degree of conservation, particularly in the hydrophobic core of the two α-helices and at residues located at the α-helix-β-hairpin interface. These conserved regions are believed to be necessary to maintain the three-dimensional structure of the motif. Ankyrin repeats are always found in tandem arrays of 2 to more than 20 repeats. The extended modular structure of the ankyrin domain is maintained by hydrophobic interactions between the helical domains and hydrogen bonds between the β-hairpins of the neighboring repeats (28). In our mutagenesis approach, we were careful to avoid mutations that might disrupt the conformation of the ARD by altering conserved residues implicated in maintaining the structure of each ankyrin repeat unit or residues involved in stabilizing the interactions between adjacent repeats. The maintenance of the structural integrity of our mutants is supported by the fact that they are all expressed stably in the transfected cells (Fig. 5 and 7 and data not shown).
Our analysis demonstrates that mutations in the surface composed of the outer α-helices of the four ankyrin repeats (“down” mutants) do not have any deleterious effects. The same is true for mutations in the β-hairpins (“finger” mutants). The latter is surprising, because an earlier study reported that mutations in the β-hairpins abolish the interaction of RFXANK with RFXAP in GST pull-down assays (Fig. 8) (33). However, the mutations introduced in that report were very severe (three or four adjacent alanine substitutions). Rather than affecting a single protein-protein interaction interface, these drastic mutations may have disrupted the overall structure of the ARD. This is in fact quite likely, because several of the residues that were mutated in the earlier study are predicted to form hydrogen bonds that stabilize the three-dimensional structure of the ARD (G124, E156, D187, and G190).
Our results demonstrate that the interaction of RFXANK with RFX5 is mediated by residues located in ankyrin repeats 2 and 3. These residues map to the loops that connect the outer α-helix of the repeat with the β-hairpin of the next repeat and are found at the “back” of the ankyrin domain. This is a very unusual interaction surface for ankyrin repeat domains. Most of the known contacts between ankyrin repeat-containing proteins and their partners are mediated by a combined surface composed of the β-hairpin and first α-helix (the ankyrin “groove”) (28). An interesting exception to this rule is represented by the Saccharomyces cerevisiae transcription factor Swi6, in which the “back” of the ankyrin repeat domain engages in an intramolecular interaction with the transcription activation domain to suppress its activity (9, 40). This example confirms that the back surface of an ARD may indeed engage in protein-protein interactions. However, the RFXANK-RFX5 interaction is the first example of an intermolecular interaction mediated by the back surface of the ARD.
In addition to the RFX5 interaction domain, we show here that mutations lying in a second noncontiguous surface of the RFXANK ARD prevent formation of the RFX complex. These mutations are located in the first ankyrin repeat (P1 mutant). This mutant can interact separately with both RFXAP and RFX5 in GST pull-down assays, yet it is not able to form the trimeric RFX complex. One possible explanation is that this mutation alters the geometry of the interaction of RFXANK with one of the subunits (possibly RFXAP) in such a way that the third partner (RFX5) cannot interact with the incorrectly assembled dimer. This would be consistent with the observation that a more drastic mutation consisting of alanine substitutions at four consecutive residues in the first ankyrin repeat disrupts the RFXANK-RFXAP interaction (33). Alternatively, the P1 mutation might prevent a conformational change required to accommodate the assembly of all three subunits. The latter seems a likely scenario, because formation of the trimeric RFX complex is highly cooperative—individual interactions between any two isolated subunits are weak, while the trimer is very stable.
Mutations in the fourth ankyrin repeat (P4 or T4) do not abolish formation of RFX (as demonstrated by coimmunoprecipitation and GST pull-down experiments) or assembly of the enhanceosome complex on the HLA-DRA promoter in vitro (as demonstrated by the promoter pull-down assays). A previous report also showed that a mutation similar to P4 does not interfere with binding of RFX to DNA in vitro (48). Despite these findings, these mutants cannot support enhanceosome formation in vivo. We verified the possibility that the nuclear import of RFXANK is inhibited by the mutation of repeat 4. This does not appear to be the case, since the nuclear/cytoplasmic distribution of RFXANK is unchanged in cells expressing the P4 mutant (data not shown). This is consistent with the observation that RFXANK does not seem to contain a specific nuclear localization signal required for active nuclear import (30). Taken together, these observations lead us to suggest that these mutations of repeat 4 cancel an as yet unknown function required only in the context of chromatin. What could this function be? Several possibilities come to mind. Since RFXANK was shown to interact with NF-Y (16), it is possible that the mutation of repeat 4 alters enhanceosome assembly in such a way that it cannot bind effectively to nucleosomal DNA. Alternatively, RFXANK might be necessary to overcome the repressive influence of chromatin by interacting with nucleosomes or by recruiting chromatin-remodeling or -modifying factors. This is an attractive possibility, because it is known that the enhanceosome contributes to chromatin modification at some MHC-II promoters (25). This would be the first example of the involvement of an ankyrin repeat-containing protein in chromatin dynamics.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation, the Comission pour la Technologie et de l'Innovation, and NovImmune.
REFERENCES
- 1.Benoist, C., and D. Mathis. 1990. Regulation of major histocompatibility complex class II genes: X, Y and other letters of the alphabet. Annu. Rev. Immunol. 8:681-715. [DOI] [PubMed] [Google Scholar]
- 2.Beresford, G. W., and J. M. Boss. 2001. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat. Immunol. 2:652-657. [DOI] [PubMed] [Google Scholar]
- 3.Bontron, S., C. Ucla, B. Mach, and V. Steimle. 1997. Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol. Cell. Biol. 17:4249-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boss, J. M., and P. E. Jensen. 2003. Transcriptional regulation of the MHC class II antigen presentation pathway. Curr. Opin. Immunol. 15:105-111. [DOI] [PubMed] [Google Scholar]
- 5.Charron, D. J., and H. O. McDevitt. 1979. Analysis of HLA-D region-associated molecules with monoclonal antibody. Proc. Natl. Acad. Sci. USA 76:6567-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cresswell, P. 1994. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 12:259-293. [DOI] [PubMed] [Google Scholar]
- 7.DeSandro, A. M., U. M. Nagarajan, and J. M. Boss. 2000. Associations and interactions between bare lymphocyte syndrome factors. Mol. Cell. Biol. 20:6587-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand, B., P. Sperisen, P. Emery, E. Barras, M. Zufferey, B. Mach, and W. Reith. 1997. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 16:1045-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foord, R., I. A. Taylor, S. G. Sedgwick, and S. J. Smerdon. 1999. X-ray structural analysis of the yeast cell cycle regulator Swi6 reveals variations of the ankyrin fold and has implications for Swi6 function. Nat. Struct. Biol. 6:157-165. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griscelli, C., B. Lisowska-Grospierre, and B. Mach. 1993. Combined immunodeficiency with defective expression in MHC class II genes, p. 141-154. In F. S. Rosen and M. Seligman (ed.), Immunodeficiencies. Harwood Academic Publishers, Chur, Switzerland.
- 12.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 13.Hake, S. B., K. Masternak, C. Kammerbauer, C. Janzen, W. Reith, and V. Steimle. 2000. CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation. Mol. Cell. Biol. 20:7716-7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harton, J. A., and J. P. Ting. 2000. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol. Cell. Biol. 20:6185-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hume, C. R., L. A. Shookster, N. Collins, R. O'Reilly, and J. S. Lee. 1989. Bare lymphocyte syndrome: altered HLA class II expression in B cell lines derived from two patients. Hum. Immunol. 25:1-11. [DOI] [PubMed] [Google Scholar]
- 16.Jabrane-Ferrat, N., N. Nekrep, G. Tosi, L. Esserman, and B. M. Peterlin. 2003. MHC class II enhanceosome: how is the class II transactivator recruited to DNA-bound activators? Int. Immunol. 15:467-475. [DOI] [PubMed] [Google Scholar]
- 17.Landmann, S., J. M. Waldburger, K. Masternak, A. Muhlethaler-Mottet, and W. Reith. 2003. CIITA and the MHCII enhanceosome in the regulation of MHCII expression. Curr. Genom. 4:343-363. [Google Scholar]
- 18.LeibundGut-Landmann, S., J. M. Waldburger, M. Krawczyk, L. A. Otten, T. Suter, A. Fontana, H. Acha-Orbea, and W. Reith. 2004. Specificity and expression of CIITA, the master regulator of MHC class II genes. Eur. J. Immunol. 34:1513-1525. [DOI] [PubMed] [Google Scholar]
- 19.Lin, J. H., A. Makris, C. McMahon, S. E. Bear, C. Patriotis, V. R. Prasad, R. Brent, E. A. Golemis, and P. N. Tsichlis. 1999. The ankyrin repeat-containing adaptor protein Tvl-1 is a novel substrate and regulator of Raf-1. J. Biol. Chem. 274:14706-14715. [DOI] [PubMed] [Google Scholar]
- 20.Long, A. B., and J. M. Boss. 2005. Evolutionary conservation and characterization of the bare lymphocyte syndrome transcription factor RFX-B and its paralogue ANKRA2. Immunogenetics 56:788-797. [DOI] [PubMed] [Google Scholar]
- 21.Mantovani, R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239:15-27. [DOI] [PubMed] [Google Scholar]
- 22.Masternak, K., E. Barras, M. Zufferey, B. Conrad, G. Corthals, R. Aebersold, J. C. Sanchez, D. F. Hochstrasser, B. Mach, and W. Reith. 1998. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat. Genet. 20:273-277. [DOI] [PubMed] [Google Scholar]
- 23.Masternak, K., A. Muhlethaler-Mottet, J. Villard, M. Zufferey, V. Steimle, and W. Reith. 2000. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 14:1156-1166. [PMC free article] [PubMed] [Google Scholar]
- 24.Masternak, K., N. Peyraud, M. Krawczyk, E. Barras, and W. Reith. 2003. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 25.Masternak, K., and W. Reith. 2002. Promoter-specific functions of CIITA and the MHC class II enhancesosome in transcriptional activation. EMBO J. 21:1379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matheux, F., A. Ikinciogullari, D. A. Zapata, E. Barras, M. Zufferey, F. Dogu, J. R. Regueiro, W. Reith, and J. Villard. 2002. Direct genetic correction as a new method for diagnosis and molecular characterization of MHC class II deficiency. Mol. Ther. 6:824-829. [DOI] [PubMed] [Google Scholar]
- 27.Moreno, C. S., G. W. Beresford, P. Louis-Plence, A. C. Morris, and J. M. Boss. 1999. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity 10:143-151. [DOI] [PubMed] [Google Scholar]
- 28.Mosavi, L. K., T. J. Cammett, D. C. Desrosiers, and Z. Y. Peng. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13:1435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muhlethaler-Mottet, A., M. Krawczyk, K. Masternak, C. Spilianakis, A. Kretsovali, J. Papamatheakis, and W. Reith. 2004. The S box of MHC class II promoters is a key determinant for recruitment of the transcriptional coactivator CIITA. J. Biol. Chem. 279:40529-40535. [DOI] [PubMed] [Google Scholar]
- 30.Nagarajan, U. M., A. B. Long, M. T. Harreman, A. H. Corbett, and J. M. Boss. 2004. A hierarchy of nuclear localization signals governs the import of the regulatory factor X complex subunits and MHC class II expression. J. Immunol. 173:410-419. [DOI] [PubMed] [Google Scholar]
- 31.Nagarajan, U. M., P. Louis-Plence, A. DeSandro, R. Nilsen, A. Bushey, and J. M. Boss. 1999. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity 10:153-162. [DOI] [PubMed] [Google Scholar]
- 32.Nagarajan, U. M., A. Peijnenburg, S. J. Gobin, J. M. Boss, and P. J. van den Elsen. 2000. Novel mutations within the RFX-B gene and partial rescue of MHC and related genes through exogenous class II transactivator in RFX-B-deficient cells. J. Immunol. 164:3666-3674. [DOI] [PubMed] [Google Scholar]
- 33.Nekrep, N., M. Geyer, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. Analysis of ankyrin repeats reveals how a single point mutation in RFXANK results in bare lymphocyte syndrome. Mol. Cell. Biol. 21:5566-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nekrep, N., N. Jabrane-Ferrat, and B. M. Peterlin. 2000. Mutations in the bare lymphocyte syndrome define critical steps in the assembly of the regulatory factor X complex. Mol. Cell. Biol. 20:4455-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peretti, M., J. Villard, E. Barras, M. Zufferey, and W. Reith. 2001. Expression of the three human major histocompatibility complex class II isotypes exhibits a differential dependence on the transcription factor RFXAP. Mol. Cell. Biol. 21:5699-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rader, K., R. A. Orlando, X. Lou, and M. G. Farquhar. 2000. Characterization of ANKRA, a novel ankyrin repeat protein that interacts with the cytoplasmic domain of megalin. J. Am. Soc. Nephrol. 11:2167-2178. [DOI] [PubMed] [Google Scholar]
- 37.Reith, W., and B. Mach. 2001. The bare lymphocyte syndrome and the regulation of mhc expression. Annu. Rev. Immunol. 19:331-373. [DOI] [PubMed] [Google Scholar]
- 38.Reith, W., S. Satola, C. Herrero Sanchez, I. Amaldi, B. Lisowska-Grospierre, C. Griscelli, M. R. Hadam, and B. Mach. 1988. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell 53:897-906. [DOI] [PubMed] [Google Scholar]
- 39.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sedgwick, S. G., I. A. Taylor, A. C. Adam, A. Spanos, S. Howell, B. A. Morgan, M. K. Treiber, N. Kanuga, G. R. Banks, R. Foord, and S. J. Smerdon. 1998. Structural and functional architecture of the yeast cell-cycle transcription factor swi6. J. Mol. Biol. 281:763-775. [DOI] [PubMed] [Google Scholar]
- 41.Spilianakis, C., A. Kretsovali, T. Agalioti, T. Makatounakis, D. Thanos, and J. Papamatheakis. 2003. CIITA regulates transcription onset viaSer5-phosphorylation of RNA Pol II. EMBO J. 22:5125-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steimle, V., B. Durand, E. Barras, M. Zufferey, M. R. Hadam, B. Mach, and W. Reith. 1995. A novel DNA binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome). Genes Dev. 9:1021-1032. [DOI] [PubMed] [Google Scholar]
- 43.Steimle, V., L. A. Otten, M. Zufferey, and B. Mach. 1993. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency. Cell 75:135-146. [PubMed] [Google Scholar]
- 44.Ting, J. P., and J. Trowsdale. 2002. Genetic control of MHC class II expression. Cell 109(Suppl.):S21-S33. [DOI] [PubMed] [Google Scholar]
- 45.Tsang, S. Y., M. Nakanishi, and B. M. Peterlin. 1990. Mutational analysis of the DRA promoter: cis-acting sequences and trans-acting factors. Mol. Cell. Biol. 10:711-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viret, C., and C. A. J. Janeway. 1999. MHC and T cell development. Rev. Immunogenet. 1:91-104. [PubMed] [Google Scholar]
- 47.Wiszniewski, W., M. C. Fondaneche, N. Lambert, K. Masternak, C. Picard, L. Notarangelo, K. Schwartz, J. Bal, W. Reith, C. Alcaide, B. de Saint, A. Fischer, and B. Lisowska-Grospierre. 2000. Founder effect for a 26-bp deletion in the RFXANK gene in North African major histocompatibility complex class II-deficient patients belonging to complementation group B. Immunogenetics 51:261-267. [DOI] [PubMed] [Google Scholar]
- 48.Wiszniewski, W., M. C. Fondaneche, P. Louise-Plence, A. Prochnicka-Chalufour, F. Selz, C. Picard, F. Le Deist, J. F. Eliaou, A. Fischer, and B. Lisowska-Grospierre. 2003. Novel mutations in the RFXANK gene: RFX complex containing in-vitro-generated RFXANK mutant binds the promoter without transactivating MHC II. Immunogenetics 54:747-755. [DOI] [PubMed] [Google Scholar]
- 49.Zhu, X. S., M. W. Linhoff, G. Li, K. C. Chin, S. N. Maity, and J. P. Ting. 2000. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol. Cell. Biol. 20:6051-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zika, E., and J. P. Ting. 2005. Epigenetic control of MHC-II: interplay between CIITA and histone-modifying enzymes. Curr. Opin. Immunol. 17:58-64. [DOI] [PubMed] [Google Scholar]