Abstract
Efficient elimination of mitochondrial reactive oxygen species (mROS) correlates with increased cellular survival and organism life span. Detoxification of mitochondrial ROS is regulated by induction of the nuclear SOD2 gene, which encodes the manganese-dependent superoxide dismutase (MnSOD). However, the mechanisms by which mitochondrial oxidative stress activates cellular signaling pathways leading to induction of nuclear genes are not known. Here we demonstrate that release of mROS activates a signal relay pathway in which the serine/threonine protein kinase D (PKD) activates the NF-κB transcription factor, leading to induction of SOD2. Conversely, the FOXO3a transcription factor is dispensable for mROS-induced SOD2 induction. PKD-mediated MnSOD expression promotes increased survival of cells upon release of mROS, suggesting that mitochondrion-to-nucleus signaling is necessary for efficient detoxification mechanisms and cellular viability.
The release of reactive oxygen species (ROS) from the mitochondria (mROS) is intimately associated with a variety of human diseases, in addition to regulating normal cellular processes which promote aging in eukaryotes (1, 5). ROS are the physiological by-products of the mitochondrial electron transport respiratory chain, and the rates of mitochondrial superoxide and hydrogen peroxide (H2O2) production are directly related to the basal metabolic rate. In turn, mitochondrial metabolic potential is determined by the combined actions of antioxidative defenses and molecular repair mechanisms. The manganese-dependent superoxide dismutase (MnSOD) is the primary mitochondrial enzymatic defensive mechanism that converts superoxide to peroxide, which is then further degraded by catalase and peroxiredoxins (23). This ensures the efficient detoxification of ROS and protection of cells against oxidative damage to lipids, proteins, RNA, and DNA. Because MnSOD is a mitochondrial matrix protein which is encoded by a nuclear gene, SOD2, a signaling pathway or pathways must exist which relay the signal from the mitochondria, where ROS are produced, to the induction of SOD2 in the nucleus, and thus allow the efficient detoxification of mitochondria from H2O2. However, to date no specific signaling function for ROS released at the mitochondria leading to nuclear gene induction has been described.
The SOD2 gene promoter is under the control of several transcription factors, most prominently the transcription factor nuclear factor κB (NF-κB) and the Forkhead transcription factor FOXO3a (FKHRL1) (9, 10). Both proteins have been shown to be regulated by exposure of cells to extracellular H2O2 (4, 20, 22). For example, FOXO3a induces SOD2 in response to exogenous hydrogen peroxide in quiescent cells, where the protective effects of the Akt/protein kinase B (PKB) pathway are not active (10). Similarly, in response to various oxidative stress stimuli, NF-κB can be regulated either via tyrosine phosphorylation of IκBα (8) or via the canonical IκB kinase (IKK) complex. In the latter case, the serine/threonine kinase protein kinase D (PKD) promotes NF-κB activation, leading to protection of cells from oxidative stress-induced death (22). PKD is a ubiquitously expressed kinase and a member of the calcium calmodulin-dependent kinase superfamily of enzymes (15). Pools of PKD are localized at the Golgi (7, 13, 24) or alternatively can translocate from the cytosol to the plasma membrane in response to a variety of cellular stimuli (16, 17). In oxidative stress signaling, neither the cellular location nor the mechanism by which PKD promotes survival has been described. Moreover, the mechanisms by which mitochondrial ROS stimulate the induction of nuclear genes which promote mitochondrial detoxification and survival have not been explored.
MATERIALS AND METHODS
Cell culture, antibodies, and expression plasmids.
The HeLa cell line was purchased from the American Type Culture Collection (Manassas, VA) and maintained in high-glucose Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. The anti-Abl, anti-protein kinase Cδ (anti-PKCδ), anti-PKCζ, anti-PKD, anti-Akt/PKB, and anti-IKKα/β antibodies were from Santa Cruz (Santa Cruz, CA). Anti-cytochrome c (anti-CytC) was from BD Pharmingen (San Diego, CA); anti-Src was from Upstate Biotechnology (Lake Placid, NY); and anti-PKD-pS738/742, anti-Akt/PKB-pS473, and anti-IKKα-pS180/IKKβ-pS181 were from Cell Signaling Technologies (Beverly, MA). Anticatalase was from EMD Biosciences (La Jolla, CA); anti-FLAG (M2), antiactin, and antivimentin were from Sigma (St. Louis, MO); and the anti-Organelle Detector Sampler kit (anti-Bcl-2, anti-GM130, anti-integrin α2, and antinucleoporin) was from BD Biosciences (San Diego, CA). The secondary goat anti-mouse immunoglobulin G (IgG) (H+L) Cy2-conjugated and donkey anti-rat IgG (H+L) Cy3-conjugated antibodies were from Jackson Laboratories (West Grove, PA). The pY463 antibody has been described previously (22), and the antihemagglutinin (anti-HA) was purified in house as previously described (21). H2O2 (30%) was from Fisher Scientific (Pittsburgh, PA), and rotenone and diphenyleneiodonium (DPI) were from Sigma. The PKD-specific substrate peptide used was AALVRQMSVAFFFK. The MitoTracker Red dye (CM-H2XRos) was from Molecular Probes (Eugene, OR), and 4′,6′-diamidino-2-phenylindole (DAPI) was from Sigma. Superfect (QIAGEN, Valencia, CA) or TransIT HeLa Monster (Mirus, Madison, WI) was used for transient transfections according to the manufacturer's instructions. Mutagenesis was carried out using the QuickChange strategy (Stratagene), and all constructs were verified by DNA sequencing. All expression plasmids for PKD1 are based on an amino-terminal HA-tagged PKD1 in pcDNA3 and have been described previously (22). FLAG-tagged PKD2 was kindly provided by T. Seufferlein. Other expression and reporter gene plasmids were provided by J. Brugge (Src.Y527F), B. M. Burgering (SOD2 and SOD2 DBE12mut reporter), T. Maniatis (IκBα.SD), S. Ohno (PKCδ.DRA), and B. Schaffhausen (Abl p120).
Isolation of mitochondria.
For isolation of mitochondria, cells were washed twice with phosphate-buffered saline (PBS) and resuspended in 1 ml lysis buffer (20 mM HEPES-KOH, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 1 mM dithiothreitol, 250 mM sucrose) plus protease inhibitor cocktail (Sigma). Cells were homogenized with 40 strokes in a Teflon homogenizer. Lysates were centrifuged twice at 750 × g for 10 min at 4°C, and the supernatant was centrifuged at 10,000 × g for 15 min at 4°C to pellet mitochondria. Mitochondrial pellets were resuspended in lysis buffer and subjected to immunoblot analysis. Supernatants (S-100) were spun for 1 h at 10,000 × g at 4°C. Nuclear extracts were prepared as previously described (19).
Immunoblotting and immunoprecipitation.
Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 1% Triton X-100, 150 mM NaCl, 5 mM EDTA) plus protease inhibitor cocktail (Sigma) or in radioimmunoprecipitation assay buffer (for whole-cell lysates) (10 mM sodium phosphate, pH 7.2, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 150 mM NaCl, 2 mM EDTA, pH 7.4, 50 mM sodium fluoride) plus protease inhibitor cocktail (Sigma), and either lysates were used for immunoblot analysis or proteins of interest were immunoprecipitated by a 1-h incubation with the respective antibody (2 μg) followed by a 30-min incubation with protein A/G-agarose (Santa Cruz). Immune complexes were washed five times with Tris-buffered saline (TBS; 50 mM Tris-HCl, pH 7.4, 150 mM NaCl) and either resolved by SDS-polyacrylamide gel electrophoresis (PAGE) or subjected to an in vitro kinase assay.
Protein kinase assays.
PKD kinase assays were performed subsequent to immunoprecipitation of PKD (anti-PKD) and extensive washing in 20 μl kinase buffer (50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 2 mM dithiothreitol). The kinase reaction was carried out at room temperature for 20 min after adding 10 μl of kinase substrate mix (150 μM PKD-specific substrate peptide, 150 μM ATP, 10 μCi [γ-32P]ATP in kinase buffer). To terminate the kinase reaction, the samples were centrifuged and the supernatants containing the phosphorylated peptide were applied as spots to P81 phosphocellulose paper (Whatman). The papers were washed three times with 0.75% phosphoric acid and once with acetone and dried, and activity was determined by liquid scintillation counting. To verify equivalent amounts of immunoprecipitated PKD in each sample, SDS sample buffer was added to the remaining beads and the samples were resolved by SDS-PAGE.
Reporter gene assays.
Cells were transiently cotransfected with NF-κB (NF-κB-luc), FOXO3a (FHRE-luc), or SOD2 (SOD2-luc) reporter constructs (5 μg), 1 μg of pCS2-(n)β-gal, and the cDNA of interest (1 μg), using Superfect (QIAGEN). Twenty-four hours after transfection of the reporters, assays for luciferase and β-galactosidase activity were performed on total cell lysates using standard assays and measured on a luminometer. Luciferase activity was normalized to the β-galactosidase activity. Protein expression was controlled by immunoblot analysis.
RNAi.
RNA interference (RNAi) was performed as follows. To transiently silence the expression of human FOXO3a, the following oligonucleotides were cloned into the pSuper vector (2): 5′-GATCCCCGAGCTCTTGGTGGATCATCTTCAAGAGAGATGATCCACCAAGAGCTCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGAGCTCTTGGTGGATCATCTCTCTTGAAGATGATCCACCAAGAGCTCGGG −3′. To transiently silence the expression of human MnSOD, the following oligonucleotides were cloned into the pSuper vector: 5′-GATCCCCCAACCTGAACGTCACCGAGTTCAAGAGACTCGGTGACGTTCAGGTTGTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAACAACCTGAACGTCACCGAGTCTCTTGAACTCGGTGACGTTCAGGTTGGGG-3′. The pSuper constructs to silence PKD1 and PKD2 isoforms have been described previously (21, 22). HeLa cells were transfected with pSUPER or pSUPER-RNAi, using the TransIT HeLa Monster reagent (Mirus). In all experiments, the cells were transfected at 30% confluence. Transfection efficiencies (95 to 100%) were controlled using a green fluorescent protein expression vector. For reporter gene assays, genes of interest were transfected in a second transfection using Superfect after 24 h. Experiments were performed 48 h after initial transfection. Reduced expression of target proteins was evaluated by immunoblotting (PKD) or reverse transcription-PCR (RT-PCR) (FOXO3a).
Immunofluorescence.
Cells were transfected (5 μg DNA) and 24 h after transfection were plated on glass coverslips at a density of 120,000 per well in a six-well plate. The next day cells were stimulated, washed twice with PBS (RT), and fixed in 3.5% paraformaldehyde (15 min, 37°C). Following permeabilization (0.1% Triton X-100 for 10 min), cells were blocked with PBS containing 3% bovine serum albumin and 0.05% Tween 20 for 30 min at room temperature. For DAPI staining, cells were washed three times in DAPI buffer (100 mM NaCl, 10 mM EDTA, 10 mM Tris pH 7.0) and incubated for 1 h at 37°C with 0.1 μg/ml DAPI in DAPI buffer. Cells were then washed three times in DAPI buffer. The coverslips were then incubated with the primary antibody diluted in PBS-bovine serum albumin (anti-HA [rat], 1:2,000 for PKD; anti-CytC [mouse], 1:2,000) overnight at 4°C. Cells were then washed five times with PBS and then incubated with the secondary antibody diluted in PBS-bovine serum albumin (donkey anti-rat IgG Cy3 conjugated, 1:400; goat anti-mouse IgG Cy2 conjugated, 1:400) for 2 h. After extensive washes in PBS, coverslips were mounted in Gel Mount from Biomeda (Foster City, CA) and examined.
RT-PCR.
Cellular mRNA isolation was performed using RNA-Bee (TEL-TEST, Friendswood, TX) according to the manufacturer's instructions, and cellular mRNA was transcribed into cDNA using Superscript II (Invitrogen, Carlsbad, CA). For the transcription reaction, 1 μg oligo(dT) (18) primer (NEB, Beverly, MA) and 1 μg RNA were incubated in a total volume of 10 μl H2O at 70°C for 10 min. Buffer (5×), 40 U RNAsin (Roche, Mannheim, Germany), 200 μM deoxynucleoside triphosphate (dNTP) (NEB), 10 mM dithiothreitol, and 300 U Superscript II reverse transcriptase were then added to a total volume of 20 μl. The reaction was carried out at 45°C for 60 min and then heat inactivated at 95°C for 5 min. The resulting cDNA pool was subjected to PCR analysis using the following primers for human FOXO3a and vimentin: human FOXO3a oligonucleotides (product size, 591 bp) 5′-TTCAAGGATAAGGGCGACAG-3′ and 5′-CAGGTCGTCCATGAGGTTTT-3′ and human vimentin oligonucleotides (product size, 408 bp) 5′-CCTTGAACGCAAAGTGGAAT-3′ and 5′-GCTTCAACGGCAAAGTTCTC-3′. Reaction conditions for the PCR were 1 min of annealing at 55°C and 1 min of amplification at 72°C for 30 cycles.
Cell survival assays.
Cells were seeded in 96-well plates in cell culture media and after 24 h treated with H2O2 for 16 h. Cells were then washed twice with PBS and stained for 15 min with a crystal violet solution (0.5% crystal violet in 20% methanol). Plates were washed by rinsing the plate with water and then dried. The dye was dissolved in methanol, and optical density was measured (550 nm) on an enzyme-linked immunosorbent assay plate reader (6).
RESULTS
Mitochondrial oxidative stress activates PKD at the mitochondria.
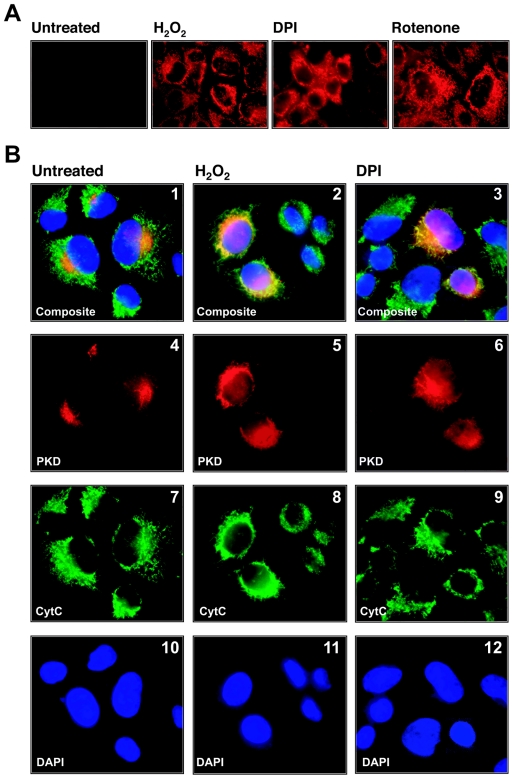
To accentuate the generation of ROS at the mitochondria, we made use of inhibitors of the mitochondrial respiratory chain, including rotenone, a mitochondrial complex I inhibitor, and DPI, which at defined doses is an inhibitor of the NADPH cytochrome P450 reductase. Both of these compounds are known to increase mitochondrial ROS (12). In HeLa cells, DPI, rotenone, and exogenous H2O2 induced the release of mROS, as measured by the increased fluorescence of the dye MitoTracker Red (CM-H2XRos), which is specifically oxidized by mitochondrial ROS (Fig. 1A). Because our recent studies have pointed to an important role for PKD in the regulation of oxidative stress responses (22), we investigated whether mROS activate PKD. We first evaluated whether the induction of mROS promotes the translocation of PKD to the mitochondria. In growing HeLa cells, wild-type PKD was primarily localized in a perinuclear region, most likely the Golgi compartment as previously reported (Fig. 1B, panel 4) (13). However, in cells treated with H2O2 or DPI, PKD partially colocalized with the mitochondria within 30 min of stimulation, as judged by the overlap in staining with the mitochondrial marker cytochrome c (Fig. 1B, panels 2 and 3).
FIG. 1.
Mitochondrial oxidative stress locates PKD to the mitochondria. (A) Cells were incubated for 15 min with the reduced MitoTracker Red (CM-H2XRos) dye. The cell culture medium then was replaced, and cells were either left untreated or stimulated with H2O2 (10 μM, 10 min), DPI (20 μM, 60 min), or rotenone (20 μM, 60 min). (B) Cells were transfected with HA-tagged PKD and 24 h after transfection seeded on glass coverslips. Cells were stimulated with H2O2 (10 μM, 10 min) or DPI (20 μM, 1 h) and stained as described in Materials and Methods (PKD, anti-HA [α-HA], red; anti-cytochrome c, [α-CytC], green; nuclei, DAPI, blue).
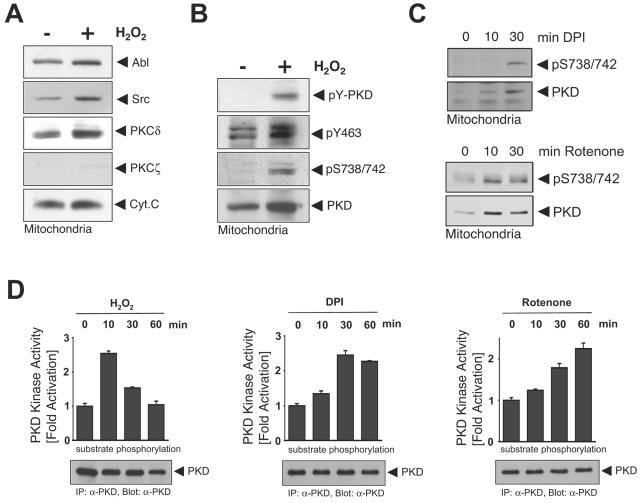
Biochemical analysis of PKD localization revealed that signal relay kinases in the PKD activation pathway, namely Src, Abl, and protein kinase Cδ (PKCδ) (22), are also localized at the mitochondria in untreated cells and further translocate in response to H2O2, consistent with published observations (Fig. 2A) (11, 14, 18). Immunoblotting for mitochondrial cytochrome c and cytoplasmic PKCζ revealed the purity of the mitochondrial preparations. Additional characterization of the mitochondrial preparations revealed that they were devoid of Golgi, plasma membranes, nuclei, and peroxisomes, as judged by immunoblotting with specific markers for each organelle (see Fig. S1 in the supplemental material). We also evaluated activation of PKD upon mROS release. In oxidative stress signaling, two signaling events control PKD activation: the first is phosphorylation of Tyr463 in the PKD pleckstrin homology (PH) domain, mediated by the tyrosine kinase Abl; this facilitates the second step, phosphorylation of PKD at the activation loop residues Ser738 and Ser742, mediated by PKCδ. In this pathway, both Abl and PKCδ are activated via Src (21, 22). Overall PKD tyrosine phosphorylation and Tyr463 and Ser738/Ser742 phosphorylation were induced at the mitochondria in response to H2O2 (Fig. 2B). mROS production also increased PKD Ser738/Ser742 phosphorylation (Fig. 2C). These data show that PKD is localized at the mitochondria upon mROS release and that it is phosphorylated at key residues which are required for its activation. This is consistent with increased PKD protein kinase activity induced by DPI, rotenone, and H2O2 in a time-dependent manner (Fig. 2D).
FIG. 2.
Mitochondrial oxidative stress activates PKD at the mitochondria. (A to C) Cells were stimulated with H2O2 (10 μM, 10 min), DPI (20 μM), or rotenone (10 μM) as indicated. Mitochondrial fractions were prepared, and lysates were resolved by SDS-PAGE and immunoblotted against the indicated proteins. All results are typical of three independent experiments. (D) Cells were stimulated with H2O2 (10 μM), DPI (20 μM), or rotenone (10 μM) for the indicated times. PKD was immunoprecipitated (IP), and a PKD substrate kinase assay was performed. Expression of PKD was determined by immunoblotting with anti-PKD (α-PKD; bottom panels).
ROS-stimulated PKD activation induces SOD2 and MnSOD.
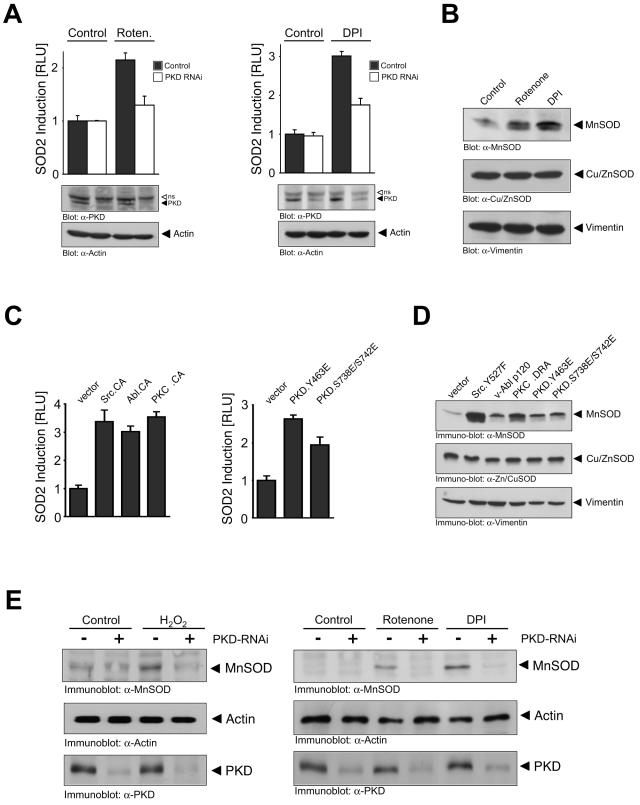
Although MnSOD detoxifies mitochondrial ROS, it is actually encoded by a nuclear gene, SOD2. To investigate whether activation of PKD by mROS is linked to SOD2 induction, we transfected cells with a SOD2-luciferase reporter and induced mitochondrial oxidative stress. Stimulation of cells with rotenone and DPI led to SOD2 promoter activation (Fig. 3A). Importantly, this was dependent on PKD because RNAi-mediated silencing of PKD blunted SOD2 induction (Fig. 3A). Moreover, increased SOD2 induction in response to mROS was concomitant with increased levels of MnSOD protein (Fig. 3B). Activated alleles of signal relay kinases which activate PKD also stimulated SOD2 induction and MnSOD expression when transiently transfected in HeLa cells (Fig. 3C and D). Similarly, constitutively active PKD alleles also increased SOD2 reporter activity and MnSOD protein expression (Fig. 3C and D). Finally, induction of MnSOD protein in cells exposed to H2O2, DPI, or rotenone was also attenuated in cells transfected with PKD RNAi (Fig. 3E). Thus, mitochondrial ROS activate PKD, which in turn is required for MnSOD expression. Note that in these and all other RNAi experiments, a combination of RNAi targeting PKD1 and PKD2 was used. The specificity of PKD1 RNAi to silence PKD1 but not PKD2 expression, and conversely the ability of PKD2 RNAi to target PKD2 but not PKD1, was verified by expression of tagged alleles of PKD1 and PKD2 with each RNAi either separately or in combination (see Fig. S2 in the supplemental material).
FIG. 3.
ROS-stimulated PKD activation induces SOD2 and MnSOD. (A) Cells were transfected with vector control (pSuper) or PKD1/2 RNAi (pSuper PKD1/pSuper PKD2) for 24 h. Then cells were transfected a second time with reporter constructs and 8 h after transfection stimulated with rotenone (Roten.; 10 μM, 16 h) or DPI (1 μM, 16 h). Reporter gene assays were performed to measure SOD2 gene reporter transcriptional activity (SOD2-luciferase reporter plasmid) or β-galactosidase activity. Error bars represent standard deviation. Protein expression was controlled by immunoblotting against PKD (anti-PKD [α-PKD]) and actin (antiactin [α-Actin]). The open arrow indicates a nonspecific (ns) band detected by the anti-PKD antibody. (B) Cells were stimulated with rotenone (10 μM, 8 h) or DPI (20 μM, 8 h), and lysates were immunoblotted against MnSOD (anti-MnSOD [α-MnSOD), Cu/ZnSOD (anti-Cu/ZnSOD [α-Cu/ZnSOD]), or vimentin (antivimentin [α-Vimentin]). (C) Cells were transfected with the SOD2 or β-galactosidase reporters and either vector alone or active alleles of Src (Src.Y527F, Src.CA), Abl (v-Abl p120, Abl.CA), PKCδ (PKCδ.DRA, PKCδ.CA), or PKD (PKD.Y463E or PKD.S738E/S742E). After 16 h, luciferase and β-Gal reporter gene assays were performed. Protein expression was controlled by immunoblot analysis (not shown). (D) Cells were transfected with active alleles of Src (Src.Y527F), Abl (v-Abl p120), PKCδ (PKCδ.DRA), or PKD (PKD.Y463E or PKD.S738E/S742E). After 16 h, lysates were analyzed for MnSOD (anti-MnSOD), Cu/ZnSOD (anti-Cu/ZnSOD), or vimentin (antivimentin) expression. (E) Cells were transfected with vector control (pSuper) or PKD RNAi (pSuper PKD1/2) for 48 h and then treated with H2O2 (16 h, 1 μM), rotenone (5 μM), or DPI (10 μM). MnSOD expression and PKD silencing by RNAi were analyzed by immunoblot analysis using anti-MnSOD or anti-PKD antibodies. Analysis of actin (antiactin) expression served as a loading control.
PKD regulates the SOD2 gene via NF-κB.
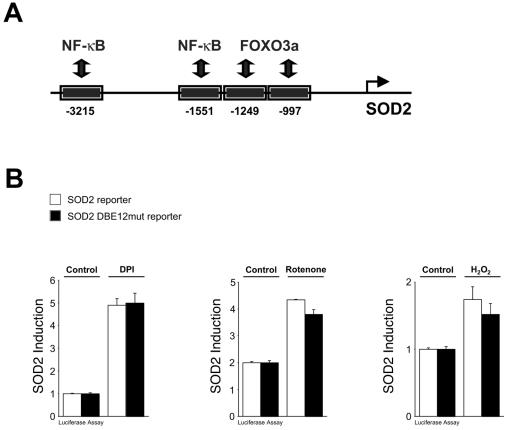
We next turned our attention to the mechanisms by which PKD relays the signal from the mitochondria to the induction of the nuclear SOD2 gene. We focused on two key transcription factors, FOXO3a and NF-κB, which have been shown to modulate SOD2 induction under different cellular conditions (Fig. 4A) (9, 10). In serum-starved cells, FOXO3a activates SOD2 in response to extracellular H2O2 (10). However, in growing cells FOXO3a activity is maintained in an inactive state in a phosphatidylinositol 3-kinase (PI 3-K)/Akt/PKB-dependent manner (3). Under normal growth conditions, we found that FOXO3a is mostly cytoplasmic (data not shown), consistent with previous reports (3). Treatment of cells with exogenous H2O2 stimulated Akt/PKB activation, as judged by Ser473 phosphorylation, and this occurred in a PI 3-K-dependent manner (see Fig. S3 in the supplemental material). When we compared the SOD2 reporter construct with a SOD2 reporter mutated in the FOXO3a-binding sites (SOD2 DBE12mut), we found that DPI, rotenone, and H2O2 all induced SOD2 promoter activation to the same extent in both reporters (Fig. 4B). This indicates that mitochondrial oxidative stress activates the SOD2 gene in a manner which is independent of FOXO3a. Therefore, FOXO3a is dispensable for mROS-induced induction of the SOD2 gene.
FIG. 4.
Mitochondrial oxidative stress regulates the SOD2 gene via NF-κB. (A) Schematic overview of FOXO3a and NF-κB binding sites in the SOD2 promoter (9, 10). (B) Cells were transfected with reporter constructs (SOD2-luciferase reporter plasmid or SOD2 DBE12mut-luciferase reporter plasmid) and 8 h after transfection stimulated with DPI (1 μM, 16 h), rotenone (20 μM, 16 h), or H2O2 (500 nM, 16 h). Reporter gene assays were performed to measure SOD2 gene reporter transcriptional activity or β-galactosidase activity (normalization). Error bars represent standard deviation.
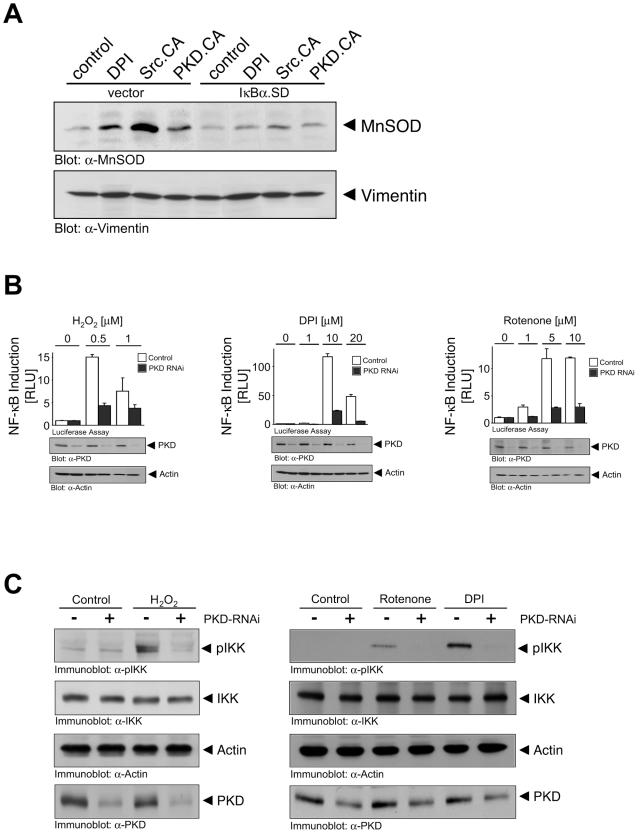
Since FOXO3a is not required for SOD2 induction upon release of mROS, we next turned our attention to NF-κB. First, we used a superdominant IκBα allele (IκBα.SD) and found that it abolished MnSOD expression in response to mROS as well as by expression of constitutively active Src and PKD (Fig. 5A). Similarly, H2O2, rotenone, and DPI increased NF-κB transcriptional activity, and this was dependent on PKD, as demonstrated with PKD RNAi (Fig. 5B). H2O2, DPI, and rotenone also stimulated an increase in the phosphorylation of IKK at the activation loop Ser180/Ser181 residues, indicative of the activation of the IKK complex (Fig. 5C). This was dependent on PKD because, in each case, IKK phosphorylation was blocked in cells transduced with PKD RNAi. Therefore, mitochondrial ROS production stimulates PKD, which promotes NF-κB activation, leading to SOD2 gene induction and MnSOD expression.
FIG. 5.
PKD regulates the SOD2 gene via NF-κB. (A) Cells were transfected with vector control or superdominant IκBα and active Src or active PKD as indicated for 16 h or stimulated with DPI (10 μM). After 16 h, lysates were analyzed for MnSOD (anti-MnSOD [α-MnSOD]) or vimentin (antivimentin [α-Vimentin]) expression. (B) Cells were transfected with vector control (pSuper) or PKD1/2 RNAi (pSuper PKD1/pSuper PKD2) for 24 h. Cells were transfected in a second transfection with reporter constructs (NF-κB-luc, β-galactosidase) and 8 h after transfection stimulated with H2O2, rotenone, or DPI as indicated for 16 h. Reporter gene assays were performed to measure NF-κB reporter activity or β-galactosidase activity. Error bars represent standard deviation. Protein expression was controlled by immunoblotting against PKD (anti-PKD [α-PKD]) and actin (antiactin [α-Actin]). (C) Cells were transfected with vector control (pSuper), or PKD RNAi (pSuper PKD1/2) for 48 h and then treated with H2O2 (10 min, 10 μM), rotenone (60 min, 20 μM), or DPI (60 min, 20 μM) and immunoblotted for phospho-IKK and IKK. Actin protein levels and PKD knockdown by RNAi were analyzed by immunoblot analysis using antiactin or anti-PKD antibodies.
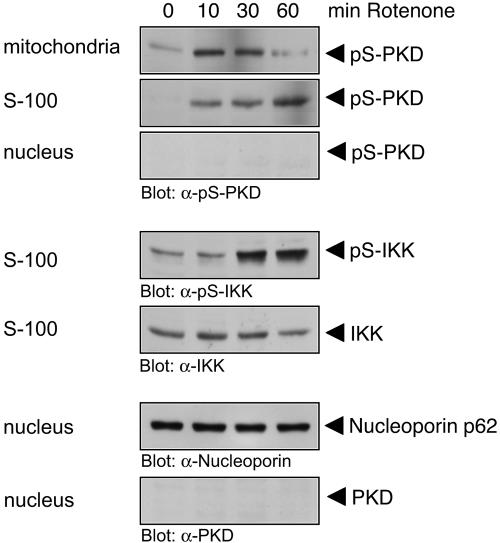
We also determined the kinetics of PKD and IKK phosphorylation and their localization upon induction of mROS. Upon exposure of cells to rotenone, phosphorylation of PKD at the mitochondria was evident within 10 min of stimulation, but at later times (60 min) phospho-PKD was found in the cytoplasmic S-100 fraction (Fig. 6). This was concomitant with the appearance of phosphorylated IKK also in the S-100 fraction at later times. However, we did not detect any phospho-IKK in the mitochondrial fractions (data not shown). Moreover, there was no detectable localization of PKD in the nuclear fraction (Fig. 6). This suggests that translocation and activation of PKD at the mitochondria relay the signal to phosphorylation of cytoplasmic IKK, leading to NF-κB activation and ultimately, SOD2 induction.
FIG. 6.
Correlation of mitochondrial PKD activation with IKK activation. Cells were stimulated with rotenone (10 μM) over time as indicated. Mitochondrial, cytosolic (S-100), or nuclear fractions were prepared, and lysates were resolved by SDS-PAGE and immunoblotted against the indicated proteins. α-, anti-.
PKD and MnSOD control cellular survival in response to mitochondrial oxidative stress.
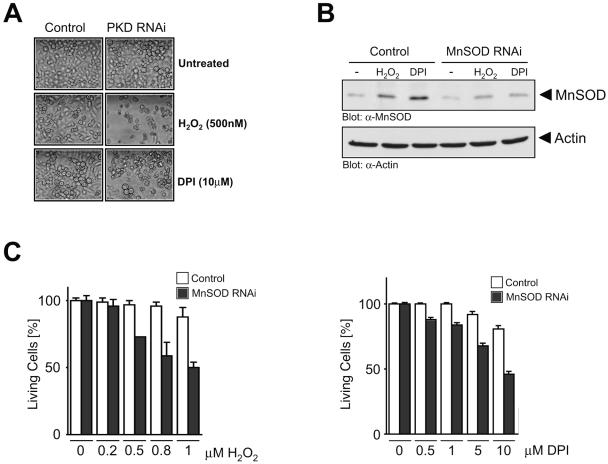
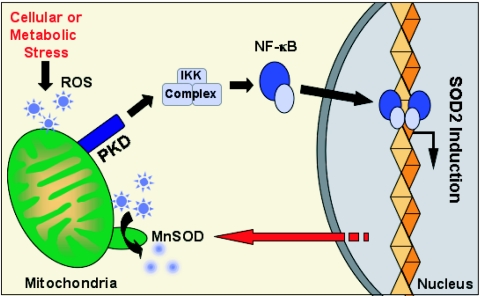
Finally, we determined the importance of the MnSOD induction by the PKD/NF-κB pathway for cellular responses to mitochondrial oxidative stress. Exposure of cells to low doses of extracellular H2O2, as well as DPI, had no effect on cell death, likely due to the induction of MnSOD (Fig. 7A). However, the same doses of H2O2 and DPI promoted significant cell death in cells transfected with PKD RNAi (Fig. 7A). To further implicate MnSOD in the survival response, we used an MnSOD-specific RNAi and found that this significantly reduced MnSOD expression under all conditions (Fig. 7B). Importantly, cell death resulting from exposure of cells to increasing concentrations of H2O2 and DPI was also significantly increased in cells in which MnSOD expression was reduced with RNAi (Fig. 7C). Thus, induction of MnSOD by the PKD pathway provides a survival advantage which allows cells to escape the damaging effects of mROS. The net effect is that when mitochondria produce lethal doses of ROS, PKD- and NF-κB-dependent induction of MnSOD allows efficient detoxification, thus promoting increased cell survival. We therefore propose a model, depicted in Fig. 8, in which PKD acts as a molecular integrator of mitochondrial oxidative stress responses by inducing NF-κB, which in turn modulates the induction of MnSOD, effectively promoting cellular survival. In this model, the FOXO3a transcription factor is dispensable for SOD2 induction under conditions in which mROS are released.
FIG. 7.
PKD and MnSOD control cellular survival in response to mitochondrial oxidative stress. (A) Cells were transfected with vector control (pSuper) or PKD RNAi (pSuper PKD1/2) for 48 h. Cells were treated with DPI (10 μM) or H2O2 (500 nM) for 16 h and photographed. Silencing of PKD protein was measured by immunoblotting (see Fig. S5 in the supplemental material). (B) Cells were transfected with vector control (pSuper) or MnSOD RNAi (pSuper MnSOD) for 48 h and stimulated with DPI (10 μM) or H2O2 (1 μM) for 16 h. MnSOD induction and silencing were analyzed by immunoblotting. (C) Cells were transfected with vector control (pSuper) or MnSOD RNAi (pSuper MnSOD) for 48 h. Cells were treated with increasing concentrations of DPI or H2O2 as indicated for 16 h. Living cells were analyzed by crystal violet staining. Error bars represent standard deviation. α-, anti-.
FIG. 8.
Proposed model for the mitochondrion-to-nucleus signaling, regulation of SOD2 gene expression, and mitochondrial detoxification in response to mROS.
DISCUSSION
The production of reactive oxygen species, such as superoxide and H2O2, at the mitochondria has been intimately linked to a variety of human diseases, as well as the normal physiological processes which promote aging in eukaryotes (1, 5). Rates of mitochondrial ROS production are directly related to the basal metabolic rate. In turn, mitochondrial metabolic potential is determined by the combined actions of antioxidative defenses and molecular repair mechanisms. The conversion of superoxide to H2O2 is mediated by MnSOD. Because MnSOD is a mitochondrial protein which is encoded by a nuclear gene, SOD2, we reasoned that a specific signaling pathway must exist which relays the signal from the mitochondria where ROS are produced, to the induction of SOD2 in the nucleus, and thus allows the efficient detoxification of superoxide. To date no mitochondrion-to nucleus signaling function for ROS released at the mitochondria has been described. Here, we provide evidence for such a pathway which modulates this signal relay to increase cell survival upon mitochondrial ROS production.
To accentuate the generation of ROS at the mitochondria, we made use of inhibitors of the mitochondrial respiratory chain, including rotenone (a mitochondrial complex I inhibitor) and DPI (an inhibitor of the NADPH cytochrome P450 reductase) (12). Because our recent studies have pointed to an important role for PKD in the regulation of oxidative stress responses (21, 22), we investigated whether mitochondrial oxygen radicals induce mitochondrion-to-nucleus signaling via PKD. We found that PKD is localized to the mitochondria in cells exposed to both exogenous and mitochondrial ROS (Fig. 1). Moreover, under the same conditions, PKD is potently phosphorylated and activated (Fig. 1). Upon exposure of cells to ROS, two events control PKD activation: the first is phosphorylation of Tyr463 in the PKD PH domain, mediated by the tyrosine kinase Abl, and the second is phosphorylation of PKD at the activation loop residues Ser738 and Ser742, mediated by PKCδ. In this pathway, both Abl and PKCδ are activated by Src. Consistent with published observations (11, 14, 18), we found translocation of Src, Abl, and PKCδ to the mitochondria. Moreover, phosphorylation of PKD at Tyr463 and Ser738/Ser742 occurs at this organelle specifically upon mitochondrial ROS release (Fig. 2). Importantly, we did not detect any nuclear localization of PKD under normal or mROS-stimulated conditions (Fig. 6). Instead, our data indicate that translocation and activation of PKD occur at the mitochondria upon release of mROS. Subsequently, PKD may dissociate from mitochondria in an activated state and phosphorylate substrates such as those which participate in IKK activation. The precise mechanisms which control PKD localization at the mitochondria and its accessibility to substrates have not yet been elucidated.
We next turned our attention to the mechanisms by which PKD relays the mitochondrial signal to the induction of the nuclear SOD2 gene. We focused on two key transcription factors, FOXO3a and NF-κB, which have been shown to modulate SOD2 induction under other cellular conditions. The redox-regulated transcription factor, FOXO3a, which has been implicated in SOD2 gene activation in senescent cells, is maintained in the cytoplasm of growing cells and is not directly involved in SOD2 induction (Fig. 4B). This is consistent with previous reports which have shown that under normal growing conditions, FOXO3a, a proapoptotic transcription factor, is retained in the cytoplasm in an Akt/PKB-dependent manner (3). It is, however, worth noting that under normal cellular growth conditions, FOXO3a may play a negative role in SOD2 induction, because silencing with RNAi alone induced the SOD2 reporter (see Fig. S4 in the supplemental material). This suggests a negative regulatory role for FOXO3a in the induction of SOD2 in growing cells. However, under conditions of elevated mROS release, FOXO3a is clearly not required, since neither removal of the FOXO3a binding sites on the SOD2 reporter nor silencing FOXO3a with RNAi increased mROS-induced SOD2 induction. Instead, our data indicate that upon increased mitochondrial ROS release, PKD activates NF-κB, which in turn is required for the induction of SOD2, and this translates into increased MnSOD expression. The net effect is that when mitochondria produce lethal doses of ROS, PKD-dependent induction of MnSOD promotes efficient detoxification, thus promoting increased cell survival. We therefore propose a model, depicted in Fig. 8, in which PKD acts as a central integrator of mitochondrial oxidative stress responses, such that NF-κB modulates the induction of MnSOD and promotes survival.
We propose that the production of ROS at the mitochondria initiates a mitochondrion-to-nucleus signaling pathway in which PKD plays a critical signaling function. PKD translocates to and is activated at the mitochondria. Excessive mROS production leads to PKD-dependent activation of NF-κB, which translocates to the nucleus and induces SOD2, leading to the expression of MnSOD and subsequent accumulation at the mitochondria. The subsequent detoxification of ROS results in increased cellular survival, such that deregulation of any components of this pathway results in increased cell death. These observations may have consequences for a number of human diseases and pathologies which have been causally linked to mitochondrial oxidative stress. Similarly, the normal processes of aging of eukaryotic cells and organisms depend on mitochondrial ROS production, and thus PKD may represent a viable target for modulating life span.
Supplementary Material
Acknowledgments
We thank T. Seufferlein, T. Maniatis, J. Brugge, B. M. Burgering, B. Schaffhausen, and S. Ohno for generously providing expression plasmids. We are grateful to S. T. Gray for providing the MnSOD and Cu/ZnSOD antibodies. We also thank members of the BIDMC/HMS joint Friday meeting and members of the Toker laboratory for discussions and advice.
This work was supported by grants from the National Institutes of Health (CA075134 to A.T.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Barnham, K. J., C. L. Masters, and A. I. Bush. 2004. Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 3:205-214. [DOI] [PubMed] [Google Scholar]
- 2.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 3.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 4.Brunet, A., L. B. Sweeney, J. F. Sturgill, K. F. Chua, P. L. Greer, Y. Lin, H. Tran, S. E. Ross, R. Mostoslavsky, H. Y. Cohen, L. S. Hu, H. L. Cheng, M. P. Jedrychowski, S. P. Gygi, D. A. Sinclair, F. W. Alt, and M. E. Greenberg. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011-2015. [DOI] [PubMed] [Google Scholar]
- 5.Finkel, T., and N. J. Holbrook. 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408:239-247. [DOI] [PubMed] [Google Scholar]
- 6.Grell, M., E. Douni, H. Wajant, M. Lohden, M. Clauss, B. Maxeiner, S. Georgopoulos, W. Lesslauer, G. Kollias, K. Pfizenmaier et al. 1995. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 83:793-802. [DOI] [PubMed] [Google Scholar]
- 7.Hausser, A., G. Link, L. Bamberg, A. Burzlaff, S. Lutz, K. Pfizenmaier, and F. J. Johannes. 2002. Structural requirements for localization and activation of protein kinase C mu (PKC mu) at the Golgi compartment. J. Cell Biol. 156:65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imbert, V., R. A. Rupec, A. Livolsi, H. L. Pahl, E. B. Traenckner, C. Mueller-Dieckmann, D. Farahifar, B. Rossi, P. Auberger, P. A. Baeuerle, and J. F. Peyron. 1996. Tyrosine phosphorylation of I kappa B-alpha activates NF-kappa B without proteolytic degradation of I kappa B-alpha. Cell 86:787-798. [DOI] [PubMed] [Google Scholar]
- 9.Kim, H. P., J. H. Roe, P. B. Chock, and M. B. Yim. 1999. Transcriptional activation of the human manganese superoxide dismutase gene mediated by tetradecanoylphorbol acetate. J. Biol. Chem. 274:37455-37460. [DOI] [PubMed] [Google Scholar]
- 10.Kops, G. J., T. B. Dansen, P. E. Polderman, I. Saarloos, K. W. Wirtz, P. J. Coffer, T. T. Huang, J. L. Bos, R. H. Medema, and B. M. Burgering. 2002. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419:316-321. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., A. Bharti, N. C. Mishra, D. Raina, S. Kharbanda, S. Saxena, and D. Kufe. 2001. Targeting of the c-Abl tyrosine kinase to mitochondria in the necrotic cell death response to oxidative stress. J. Biol. Chem. 276:17281-17285. [DOI] [PubMed] [Google Scholar]
- 12.Li, N., K. Ragheb, G. Lawler, J. Sturgis, B. Rajwa, J. A. Melendez, and J. P. Robinson. 2003. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 278:8516-8525. [DOI] [PubMed] [Google Scholar]
- 13.Liljedahl, M., Y. Maeda, A. Colanzi, I. Ayala, J. Van Lint, and V. Malhotra. 2001. Protein kinase D regulates the fission of cell surface destined transport carriers from the trans-Golgi network. Cell 104:409-420. [DOI] [PubMed] [Google Scholar]
- 14.Majumder, P. K., N. C. Mishra, X. Sun, A. Bharti, S. Kharbanda, S. Saxena, and D. Kufe. 2001. Targeting of protein kinase C delta to mitochondria in the oxidative stress response. Cell Growth Differ. 12:465-470. [PubMed] [Google Scholar]
- 15.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 298:1912-1934. [DOI] [PubMed] [Google Scholar]
- 16.Marklund, U., K. Lightfoot, and D. Cantrell. 2003. Intracellular location and cell context-dependent function of protein kinase D. Immunity 19:491-501. [DOI] [PubMed] [Google Scholar]
- 17.Matthews, S. A., T. Iglesias, E. Rozengurt, and D. Cantrell. 2000. Spatial and temporal regulation of protein kinase D (PKD). EMBO J. 19:2935-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyazaki, T., L. Neff, S. Tanaka, W. C. Horne, and R. Baron. 2003. Regulation of cytochrome c oxidase activity by c-Src in osteoclasts. J. Cell Biol. 160:709-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller, G., M. Ayoub, P. Storz, J. Rennecke, D. Fabbro, and K. Pfizenmaier. 1995. PKC zeta is a molecular switch in signal transduction of TNF-alpha, bifunctionally regulated by ceramide and arachidonic acid. EMBO J. 14:1961-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemoto, S., and T. Finkel. 2002. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science 295:2450-2452. [DOI] [PubMed] [Google Scholar]
- 21.Storz, P., H. Doppler, and A. Toker. 2004. Protein kinase Cδ selectively regulates protein kinase D-dependent activation of NF-κB in oxidative stress signaling. Mol. Cell. Biol. 24:2614-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storz, P., and A. Toker. 2003. Protein kinase D mediates a stress-induced NF-kappaB activation and survival pathway. EMBO J. 22:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace, D. C. 2001. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found. Symp. 235:247-266. [DOI] [PubMed] [Google Scholar]
- 24.Yeaman, C., M. I. Ayala, J. R. Wright, F. Bard, C. Bossard, A. Ang, Y. Maeda, T. Seufferlein, I. Mellman, W. J. Nelson, and V. Malhotra. 2004. Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat. Cell Biol. 6:106-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.