Abstract
The marine autotroph Aureococcus anophagefferens (Pelagophyceae) was rendered axenic in order to investigate hydrolysis rates of peptides, chitobiose, acetamide, and urea as indicators of the ability to support growth on dissolved organic nitrogen. Specific rates of hydrolysis varied between 8 and 700% of rates observed in associated heterotrophic marine bacteria.
Anthropogenic nutrient loading provides a major source of nitrogen to coastal systems and plays an important role in regulating primary production and biomass accumulation (19, 20, 24). The availability of different forms of nitrogen and their relative rates of utilization are important factors determining phytoplankton productivity and succession (2, 11). Recent studies in nutrient-enriched coastal areas indicate that while overall phytoplankton productivity may increase with availability of nitrogen, disproportionate increases in dissolved organic nitrogen (DON) versus inorganic nitrogen may significantly alter community composition and trigger harmful algal blooms (3, 5).
Aureococcus anophagefferens (Pelagophyceae), a eukaryotic picoplankton that repeatedly blooms in Long Island coastal bays (New York), forms tides that color the water brown. While algal blooms typically correlate with input of dissolved inorganic nitrogen, A. anophagefferens blooms when dissolved inorganic nitrogen (nitrate and ammonium) is near the limit of detection (15, 31). In a previous report, the increase in cell abundance of A. anophagefferens coincided with a decrease in the concentration of DON, equivalent to the observed increase in algal biomass (18). It was suggested that A. anophagefferens is able to outgrow other phytoplankton in the assemblage because of its ability to scavenge nitrogen from the labile DON pool, potentially benefiting from associations with heterotrophic bacteria (3, 18).
In order to test whether A. anophagefferens could utilize DON substrates as a source of nitrogen, it was essential to remove bacterial contaminants from the culture medium prior to physiological investigations. To characterize heterotrophic bacteria potentially associated with brown tides, the taxonomic identities and hydrolytic capabilities of three strains of heterotrophic bacteria that contaminated the initial strain of A. anophagefferens were assessed. Both A. anophagefferens and the associated bacteria were isolated during a massive brown tide in Long Island, New York.
A modification of the protocol by Cottrell and Suttle (8) was used for the preparation of an axenic culture of A. anophagefferens strain CCMP 1784 (Provasoli-Guillard Center for Culture of Marine Phytoplankton, Bigelow Laboratory, West Boothbay Harbor, Maine). The antibiotic concentrations used in the protocol were determined by defining the upper limit of tolerance of A. anophagefferens to each antibiotic. Exponentially growing cultures were exposed to the following antibiotics sequentially: penicillin G (1.0 mg ml−1), neomycin (0.25 mg ml−1), streptomycin (0.10 mg ml−1), gentamicin (0.25 mg ml−1), and penicillin G (2.0 mg ml−1). All antibiotics (purchased from Sigma Chemical) were added sequentially to the final concentrations listed above, followed by a 10-fold dilution with fresh artificial seawater (ASW) medium enriched with f/2 nutrients (13) the next day, and a 3- to 4-day recovery period from the previous antibiotic treatment. For the penicillin treatment, a sterile glucose solution (0.01% final concentration) was added to the culture 3 h before the dark period to stimulate the growth of heterotrophic bacteria. Penicillin G was then added at the onset of the dark period. With each antibiotic addition, an aliquot of the culture was taken to assess bacterial contaminants by transferring to marine broth (MB2216; Difco, Detroit, Mich.), plating on marine agar (MBA2216; Difco), and direct cell counting following staining with acridine orange. Following antibiotic treatment, three new cultures of A. anophagefferens (CCMP 1982, 1983, and 1984) were confirmed as axenic by the Provasoli-Guillard Center for Culture of Marine Phytoplankton.
The phylogenetic affiliation of the bacterial contaminants was examined by amplification and sequencing of the 16S rRNA gene. Single colonies of bacteria grown on marine agar were picked to amplify 16S rRNA genes from total DNA by PCR with Taq polymerase (AmpliTaq Gold; Perkin Elmer) and two universal bacterial primers: EubF, Escherichia coli position 27 (5′-GAGTTTGATCCTGGCTCAG-3′), and EubR, E. coli position 1387 (5′-GGAACATGTGTGGCGGGCC-3′). The reaction volume of 50 μl contained 100 ng of template, 20 pmol of primer, 2.5 mM MgCl2, 50 mM KCl, 15 mM Tris-HCl (pH 8.0), 0.8 mM total nucleotide triphosphates, and 5 U of Taq Gold polymerase (Perkin Elmer Cetus). PCR amplifications were performed in a thermocycler (Stratagene 96 gradient Robocycler) using the following conditions: one step of 95°C for 10 min; 25 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and a final elongation step of 10 min at 72°C. A single product band, 1,350 bp, was observed after agarose gel electrophoresis. Both strands of the PCR products were sequenced (MWG Biotech, Germany) after purification with QIAquick PCR purification kit (Qiagen). The sequences were compared to 16S rRNA sequences in the GenBank database using BLASTN 2.1.2, available from the National Center for Biotechnology Information. The 3 sequences reported here plus 26 sequences retrieved from GenBank were aligned by using Clustal W (Genetics Computer Group package). Phylogenetic relationships were inferred by the neighbor-joining method with the Phylogenetic Analysis Using Parsimony package, version 4.0. Bootstrap analysis of neighbor-joining data (100 resamplings) was used to evaluate the tree topologies recovered for 1,617 nucleotide positions.
The capabilities of the bacteria and of A. anophagefferens to hydrolyze chitobiose and peptides were determined by using the fluorogenic substrates 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide (MUF-GlcNAc; Sigma M2133) and l-leucine-4-methylumbelliferylamide HCl (Leu-MUF; Fluka 61888) as described by Montgomery and Kirchman (21). MUF-GlcNAc and Leu-MUF were added (55 μM final concentration) to exponentially growing cultures of A. anophagefferens and bacteria in ASW medium (13) with f/2 nutrients and >1-kDa DON, recovered from West Neck Bay pore waters by ultrafiltration, as the nitrogen source. On the assumption that the MUF tag potentially prevented transport across the cell membrane, intracellular chitobiose hydrolysis activity was determined with crude cell extracts rather than whole cells. A. anophagefferens cells were harvested, resuspended in 1 ml of buffer (25 mM NaH2PO4 and 1 mM EDTA, pH 8.0), and shaken for 2 min in a bead beater (Mini-Beadbeater; Biospec Products) with an equal volume of glass beads (0.5 or 0.1 mm) to break the cells. Crude cell extract was removed from the glass beads into a 10× larger volume of buffer for MUF-GlcNAc hydrolysis determination. Urea and acetamide hydrolysis rates were assayed in the crude and soluble cell extracts of A. anophagefferens and bacteria (crude cell extract only) by adding 1 mM urea or acetamide and monitoring changes in NH4+ concentration over time. NH4+ concentration was measured as described by Grasshoff et al. (14). Protein concentration was measured with the Pierce BCA-200 protein assay kit (1856175) following the manufacturer’s instructions.
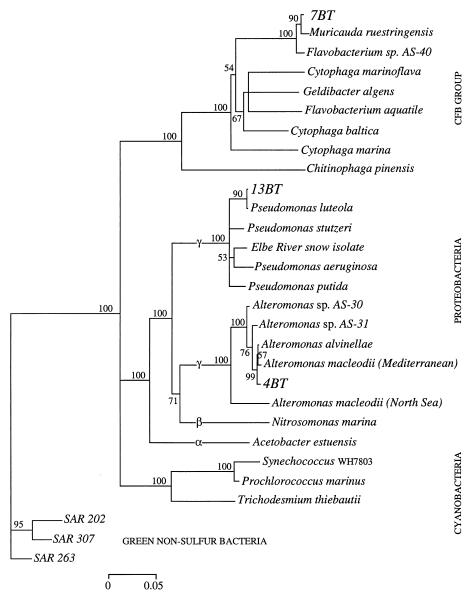
The original A. anophagefferens culture was contaminated by a number of bacterial strains that differed in their morphology, taxonomic affiliation, and hydrolytic capabilities. Comparison of growth in nonaxenic and axenic cultures demonstrated that the algal culture grew better without the associated bacteria, suggesting no direct beneficial association between A. anophagefferens and the heterotrophic bacteria. The algae grew well in established ASW medium with f/2 macronutrients and trace metals and also when nitrate was replaced by a wide variety of nitrogen sources (data not shown). Three strains of bacteria, 4BT, 7BT, and 13BT, overlapped in their hydrolytic capabilities with A. anophagefferens and may be able to outcompete it in culture, as evidenced by frequent crashes when A. anophagefferens was grown nonaxenically on DON substrates as the sole source of nitrogen. The 16S rDNA sequence of strain 7BT clustered with the Cytophaga-Flexibacter-Bacteroides group, and sequences of strains 13BT and 4BT clustered with the γ subdivision of the Proteobacteria (Fig. 1). Strain 7BT, a yellow-pigmented rod-shaped bacterium, had the closest sequence identity (98%) to the marine bacteria Flavobacterium sp. strain AS-40 and Muricauda ruestringensis, isolated from the Adriatic Sea and North Sea sediments, respectively. Strain 4BT, a nonpigmented rod-shaped bacterium, was related (98 to 99% sequence identity) to Alteromonas macleodii, Alteromonas alvinellae, and Alteromonas sp. strain AS-30/31 from the Mediterranean Sea, a hydrothermal vent community, and the Adriatic Sea, respectively (Fig. 1). Strain 13BT was the most unusual of the three isolates, growing in misshapen rods and forming nonpigmented crusts on solid agar. There was 99% sequence identity between 13BT and Pseudomonas luteola (formerly Chryseomonas luteola), suggesting that this species belongs to the Pseudomonas genus.
FIG. 1.
Phylogenetic tree based on 1,617 nucleotide positions showing relationships of 7BT, 13BT, and 4BT to representative bacterial 16S rRNA genes. The tree was inferred from nearly-complete sequences by the neighbor-joining method. The numbers of bootstrap replicates that supported the branching order, from a total of 100 replicates, are shown above the internal segments. Values below 50% are not shown. Gene sequences from the SAR cluster were used as the outgroup. The scale bar shows numbers of substitutions per nucleotide position. CFB, Cytophaga-Flexibacter-Bacteroides.
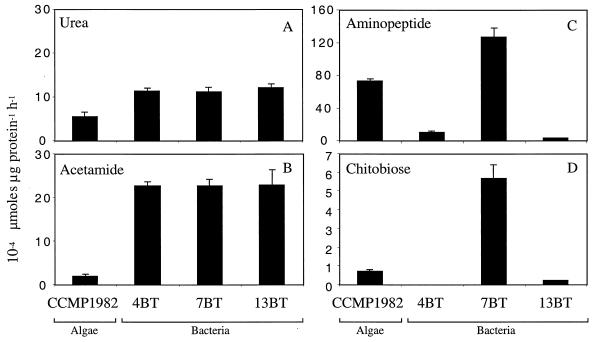
A. anophagefferens could hydrolyze all of the substrates tested, sometimes at higher rates than the associated bacteria (Fig. 2), suggesting that it can utilize a wide range of DON sources. Urease and acetamidase activities were associated predominantly with the soluble fraction (98 to 100%) of the crude cell extract, indicating that these enzymes were not membrane or cell surface bound in A. anophagefferens. The intracellular location of these enzymes is consistent with urease purified from marine picoplankton (7, 26) and acetamidase activity measured in Emiliania huxleyi (25). The specific activity of urease, 5.5 ×10−4 μmol of urea μg of protein−1 h−1, was 48% of bacterial hydrolysis rates (Fig. 2A). Field experiments demonstrate that urea flux is relatively high during brown-tide blooms (3), suggesting that it is an important source of nitrogen for A. anophagefferens. The specific rate of acetamide hydrolysis in A. anophagefferens was within the same order of magnitude as the urea hydrolysis rate (Fig. 2A and B), but acetamide hydrolysis by A. anophagefferens was only 8% as efficient as that in bacteria (Fig. 2B). In bacteria and yeasts, amides are utilized as sources of both carbon and nitrogen, metabolized by aliphatic amidases (6). By contrast, phytoplankton probably use amides mainly as a source of nitrogen, and consequently the enzyme pathway may be quite different. In a preliminary investigation using degenerate primers, we were able to amplify the gene coding for urea amidohydrolase, EC 3.5.1.5, but not the gene coding for an aliphatic amidase, in A. anophagefferens (S. Nissen and G. M. Berg, unpublished data). It is possible that in A. anophagefferens, amides are metabolized by urease, similar to the enzymatic action of urease in higher plants (10, 12), or by an enzyme that is entirely different from the amidase characterized in yeasts and bacteria.
FIG. 2.
Mean nitrogen hydrolysis rates (n = 3) and 95% confidence intervals. Urea and acetamide hydrolysis was measured on crude cell extracts of bacteria and CCMP 1982. Aminopeptide and chitobiose hydrolysis was measured in whole-cell suspensions, with the exception of hydrolysis of chitobiose by CCMP 1982, which was measured in crude cell extracts.
The rate of peptide hydrolysis in A. anophagefferens was 700% higher than that in the Alteromonas-like strain 4BT but only 55% of that for the Flavobacterium-like strain 7BT (Fig. 2C). Plasma membrane-associated aminopeptidase has been reported for Chlamydomonas reinhardtii (17) and more recently for Gonyaulax polyedra (30), another bloom-forming alga, suggesting that peptide hydrolysis may be more common in algae than previously thought. The capability of A. anophagefferens to degrade peptides supports the idea that components of the high-molecular-weight DON pool are bioavailable to certain phytoplankton (18). However, peptides comprise only 1 to 2% of this pool (27), and despite rapid turnover (28, 29), it is unlikely that this pool supplies the major fraction of the nitrogen needed to support brown tides.
Another source of nitrogen to support brown tides may derive from the carbohydrate fraction of the DON pool. In the present study we observed the potential for chitobiose hydrolysis in A. anophagefferens (Fig. 2D). While chitin derivatives have not been demonstrated to be good sources of nitrogen for growth of phytoplankton in culture (1, 25), chitin concentrations and turnover rates may be relatively high and seasonally important in coastal systems (16). If A. anophagefferens is able to transport chitobiose into the cell, it is possible that chitobiose can be metabolized by a nonspecific intracellular enzyme, such as a lysozyme (4, 22, 23). It is also conceivable that A. anophagefferens is capable of scavenging nitrogen from chitin amino sugars via a deaminase. As inorganic nitrogen becomes more limited, carbohydrate-degrading bacteria similar to Flavobacterium sp. strain 7BT (9) may provide novel sources of nitrogen for growth of certain phytoplankton.
Although the exact mechanisms leading to the hydrolysis of various substrates have not been identified, our work shows that several enzymatic pathways are simultaneously active in DON-grown A. anophagefferens. Therefore, this algal species has the potential to utilize a wide range of DON compounds as inorganic nitrogen becomes depleted during the summer months. Hydrolysis rates for A. anophagefferens were within the same order of magnitude as and sometimes higher than those of the bacterial strains tested here, suggesting that the importance of DON utilization by A. anophagefferens during bloom conditions should be assessed more thoroughly.
Nucleotide sequence accession numbers.
GenBank accession numbers for the sequenced bacteria are AF386740 (strain 7BT), AF386741 (strain 13BT), and AF386742 (strain 4BT).
Acknowledgments
This work was supported by grants from the Suffolk County Department of Health Services, Office of Ecology, the Alexander Von Humboldt Foundation, and the Deutsche Forschungsgemeinschaft.
We thank Helen Murray-Tobin for help with preparation of the axenic culture and Uwe Rabsch for help with logistics.
REFERENCES
- 1.Antia, N. J., B. R. Berland, D. J. Bonin, and S. Y. Maestrini. 1975. Comparative evaluation of certain organic and inorganic sources of nitrogen for phototrophic growth of marine microalgae. J. Mar. Biol. Assoc. U. K. 55:519–539. [Google Scholar]
- 2.Berg, G. M., P. M. Glibert, N. O. G. Jørgensen, M. Balode, and I. Purina. 2001. Variability in inorganic and organic nitrogen uptake associated with riverine nutrient input in the Gulf of Riga, Baltic Sea. Estuaries 24:204–214. [Google Scholar]
- 3.Berg, G. M., P. M. Glibert, M. W. Lomas, and M. Burford. 1997. Organic nitrogen uptake and growth by the chrysophyte Aureococcus anophagefferens during a brown tide event. Mar. Biol. 129:377–387. [Google Scholar]
- 4.Cabib, E. 1987. The synthesis and degradation of chitin. Adv. Enzymol. 59:59–101. [DOI] [PubMed] [Google Scholar]
- 5.Carlsson, P., H. Edling, and C. Bechemin. 1998. Interactions between a marine dinoflagellate (Alexandrium catenella) and a bacterial community utilizing riverine humic substances. Aquat. Microb. Ecol. 16:65–80. [Google Scholar]
- 6.Chebrou, H., F. Bigey, A. Arnaud, and P. Galzy. 1996. Study of the amidase signature group. Biochim. Biophys. Acta 1298:285–293. [DOI] [PubMed] [Google Scholar]
- 7.Collier, J. L., B. Brahamsha, and B. Palenik. 1999. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology 145:447–459. [DOI] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and C. A. Suttle. 1995. Production of axenic cultures of Micromonas pusilla (Pracinophyceae) using antibiotics. J. Phycol. 29:385–387. [Google Scholar]
- 9.Cottrell, M. T., D. N. Wood, L. Yu, and D. L. Kirchman. 2000. Selected chitinase genes in cultured and uncultured marine bacteria in the α- and γ-subclasses of the Proteobacteria. Appl. Environ. Microbiol. 66:1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon, N. P., P. W. Riddles, C. Gazzola, R. L. Blakeley, and B. Zerner. 1980. Jack bean urease (EC 3.5.1.5). V. On the mechanism of action of urease on urea, formamide, acetamide, N-methylurea, and related compounds. Can. J. Biochem. 58:1335–1344. [DOI] [PubMed] [Google Scholar]
- 11.Eppley, R. W., and B. J. Peterson. 1979. Particulate organic matter flux and planktonic new production in the deep ocean. Nature 282:677–680. [Google Scholar]
- 12.Fishbein, W. 1977. Formamide: the minimum-structure substrate for urease. Biochim. Biophys. Acta 484:433–442. [DOI] [PubMed] [Google Scholar]
- 13.Goldman, J. C., and J. J. McCarthy. 1978. Steady state growth and ammonium uptake of a fast-growing marine diatom. Limnol. Oceanogr. 23:695–703. [Google Scholar]
- 14.Grasshoff, K., M. Ehrhardt, and K. Kremling. 1999. Methods of seawater analysis. Wiley-VCH Verlag GmbH, Weinheim, Germany.
- 15.Keller, A. A., and R. L. Rice. 1989. Effects of nutrient enrichment on natural populations of the brown tide phytoplankton Aureococcus anophagefferens (Chrysophyceae). J. Phycol. 25:636–646. [Google Scholar]
- 16.Kirchman, D. L., and J. White. 1999. Hydrolysis and mineralization of chitin in the Delaware Estuary. Aquat. Microb. Ecol. 18:187–196. [Google Scholar]
- 17.Langheinrich, U. 1995. Plasma membrane-associated aminopeptidase activities in Chlamydomonas reinhardtii and their biochemical characterization. Biochim. Biophys. Acta 1249:45–57. [DOI] [PubMed] [Google Scholar]
- 18.LaRoche, J., R. Nuzzi, R. Waters, K. Wyman, P. G. Falkowski, and D. W. R. Wallace. 1997. Brown tide blooms in Long Island’s coastal waters linked to interannual variability in groundwater flow. Global Change Biol. 3:101–114. [Google Scholar]
- 19.Mallin, M. A., H. W. Paerl, J. Rudek, and P. W. Bates. 1993. Regulation of estuarine primary production by watershed rainfall and river flow. Mar. Ecol. Prog. Ser. 93:199–203. [Google Scholar]
- 20.Malone, T. C., L. H. Crocker, S. E. Pike, and B. W. Wendler. 1988. Influences of river flow on the dynamics of phytoplankton production in a partially stratified estuary. Mar. Ecol. Prog. Ser. 48:235–249. [Google Scholar]
- 21.Montgomery, M. T., and D. L. Kirchman. 1993. Estimating degradation rates of chitin in aquatic samples, p.597–600. In P. Kemp et al. (ed.), Current methods in aquatic microbial ecology. Lewis, Chelsea, Mich.
- 22.Mulisch, M. 1993. Chitin in protistan organisms. distribution, synthesis and deposition. Eur. J. Protistol. 29:1–18. [DOI] [PubMed] [Google Scholar]
- 23.Muzzarelli, R. A. A. 1977. Chitin. Pergamon Press, New York, N.Y.
- 24.Paerl, H. W., J. Rudek, and M. A. Mallin. 1990. Stimulation of phytoplankton production in coastal waters by natural rainfall inputs: nutritional and trophic implications. Mar. Biol. 107:247–254. [Google Scholar]
- 25.Palenik, B., and S. E. Henson. 1997. The use of amides and other organic nitrogen sources by the phytoplankton Emiliania huxleyi. Limnol. Oceanogr. 42:1544–1551. [Google Scholar]
- 26.Palinska, K. A., T. Jahns, R. Rippka, and N. Tandeau de Marsac. 2000. Prochlorococcus marinus strain PCC 9511, a picoplanktonic cyanobacterium, synthesizes the smallest urease. Microbiology 146:3099–3107. [DOI] [PubMed] [Google Scholar]
- 27.Pantoja, S., and C. Lee. 1999. Molecular weight distribution of proteinaceous material in Long Island Sound sediments. Limnol. Oceanogr. 44:1323–1330. [Google Scholar]
- 28.Pantoja, S., and C. Lee. 1999. Peptide decomposition by extracellular hydrolysis in coastal seawater and salt marsh sediment. Mar. Chem. 63:273–291. [Google Scholar]
- 29.Pantoja, S., C. Lee, and J. F. Marecek. 1997. Hydrolysis of peptides in seawater and sediment. Mar. Chem. 57:25–40. [Google Scholar]
- 30.Sankievicz, D., and P. Colepicolo. 1999. A new member of the leucyl aminopeptidase family purified and identified from a marine unicellular algae. Biochem. Biophys. Res. Commun. 262:557–561. [DOI] [PubMed] [Google Scholar]
- 31.Smayda, T. J., and T. A. Villareal. 1989. An extraordinary, noxious brown tide in Narragansett Bay. I. The organism and its dynamics, p.129–132. In T. Okaichi, D. M. Anderson, and T. Nemoto (ed.), Red tides: biology, environmental science, and toxicology. Elsevier Science Publishing Co., Inc., New York, N.Y.