Abstract
The DNA-binding hemopoietic zinc finger transcription factor GATA1 promotes terminal megakaryocyte differentiation and restrains abnormal immature megakaryocyte expansion. How GATA1 coordinates these fundamental processes is unclear. Previous studies of synthetic and naturally occurring mutant GATA1 molecules demonstrate that DNA-binding and interaction with the essential GATA1 cofactor FOG-1 (via the N-terminal finger) are required for gene expression in terminally differentiating megakaryocytes and for platelet production. Moreover, acquired mutations deleting the N-terminal 84 amino acids are specifically detected in megakaryocytic leukemia in human Down syndrome patients. In this study, we have systematically dissected GATA1 domains required for platelet release and control of megakaryocyte growth by ectopically expressing modified GATA1 molecules in primary GATA1-deficient fetal megakaryocyte progenitors. In addition to DNA binding, distinct N-terminal regions, including residues in the first 84 amino acids, promote platelet release and restrict megakaryocyte growth. In contrast, abrogation of GATA1-FOG-1 interaction leads to loss of differentiation, but growth of blocked immature megakaryocytes is controlled. Thus, distinct GATA1 domains regulate terminal megakaryocyte gene expression leading to platelet release and restrain megakaryocyte growth, and these processes can be uncoupled.
Through development and adult life tissue-restricted transcription factors, like the GATA family, coordinate differentiation and proliferation in a cell context-specific manner to ensure that appropriate numbers of terminally mature cells arise from stem/progenitor cells. In the human hemopoietic system, these processes generate ∼1010 new mature blood cells daily. Moreover, the number and mix of cells produced can be altered rapidly as required. When this process goes awry, diseases like leukemia can occur. To date, there is an incomplete understanding of how tissue-restricted transcription factors coordinate differentiation with cell cycle exit during maturation of lineage-restricted progenitor and precursor cells.
The DNA-binding zinc finger transcription factor GATA1 plays key roles in myelopoiesis. Enforced GATA1 expression specifies erythroid and megakaryocytic lineages from primary multipotential myeloid progenitors and committed myelomonocytic and lymphoid progenitors (22, 25, 27). Once the lineages are specified, continued GATA1 expression is necessary for expression of many red cell-and megakaryocyte-specific genes in terminally maturing precursor cells (48, 61, 63, 68). In mice, germ line ablation of GATA1 function results in embryonic lethality at embryonic day 11.5 (E11.5) from fatal anemia due to a block in erythroid differentiation at the proerythroblast stage accompanied by apoptosis (16, 49, 65, 66). Similarly, deletion of an upstream enhancer in the GATA1 locus (ΔneoΔHS mice), resulting in near complete loss of megakaryocyte GATA1 expression, specifically blocks terminal megakaryocyte differentiation and proplatelet formation, and mice suffer from thrombocytopenia (48, 61). However, in contrast to cell death in GATA1-null erythroid cells, immature GATA1-deficient megakaryoblasts accumulate in bone marrow (10-fold) and spleen (100-fold) and show abnormal growth when cultured in vitro. This suggests that although GATA1 is required for gene expression in both terminally maturing red cells and megakaryocytes, it interfaces with cell cycle and apoptotic machinery differently in the two cell types.
How does GATA1 execute these functions in hematopoietic cells? A number of experimental approaches including transactivation assays (31, 69), induction of partial megakaryocyte differentiation of a myeloid multipotential cell line 416B (59, 60), and rescue of erythroid differentiation in GATA1-null erythroid cells (4, 47, 67) have identified three functional domains in GATA1: two highly conserved N-terminal and C-terminal cysteine-rich zinc-binding fingers (Cys-X2-Cys-X17-Cys-NA-Cys) (12, 54) and a less well conserved N-terminal activation domain.
The C-terminal zinc finger (Cf) and adjacent C-terminal basic residues are required for high-affinity binding to all WGATAR DNA sequences (17). Deletion of Cf completely abolishes GATA1 function in all assays, suggesting that DNA binding is absolutely required (47, 60, 67). Although physical interactions between the Cf and other hematopoietic (GATA2-3, EKLF, and PU.1) and widely expressed (Sp1 and CBP) (3, 6, 9, 33, 37, 44, 71) transcriptional regulators have been reported, their importance in Cf function remains unclear.
Initial transactivation and 416B differentiation assays suggested that the N-terminal zinc finger (Nf) was dispensable for GATA1 function (31, 60). However, rescue of erythroid differentiation of GATA1-null erythroid cells and definitive erythropoiesis in transgenic mice required the Nf (47, 67). The Nf cooperates with the Cf to bind tandem WGATAR sequences (51) and binds independently to GATC DNA motifs (38). It also interacts with the proteins FOG-1, c-MYB, and STAT-3 (13, 50, 55). Structural studies show that the Nf has two surfaces: one interacts with DNA and the other binds FOG-1 (26, 30). Point mutations affecting DNA binding (R216Q) (70) or interaction with FOG-1 (e.g., V205G) (7, 8, 14, 15, 32, 39) impede terminal megakaryocyte and red cell differentiation. This results in severe thrombocytopenia accompanied by dysplastic megakaryopoiesis and variable dyserythropoietic anemia.
The functional role of the third proposed functional domain, the N terminus, is still unresolved. Reporter gene transactivation assays in fibroblasts originally defined an activation domain in the N-terminal 63 amino acids (31). However, this domain is dispensable for partial megakaryocyte differentiation of 416B cells (60) and erythroid differentiation of GATA1-null erythroid cells (67). In contrast, studies in transgenic mice suggest that the N-terminal 84 amino acids are only dispensable for erythropoiesis when the mutant protein is severalfold overexpressed (47). Discrepancies between the erythroid rescue studies may either reflect differences in the GATA1 N-terminal deletions tested or the assays employed or a combination of both.
The most convincing evidence of involvement of the N terminus in megakaryopoiesis has come from studies in children with megakaryocytic preleukaemia (transient myeloproliferative disease [TMD]) and acute megakaryoblastic leukemia (AMKL) in Down syndrome (DS) (1, 18, 19, 23, 34, 43, 64). Although the DNA mutations are varied, the functional consequences are predicted to always result in exclusive production of a shorter GATA1 protein (GATA1s) with an N-terminal 84-amino-acid truncation. DS TMD and AMKL (pre)leukemic blasts show partial megakaryocyte differentiation and exhibit altered growth. This suggests that deletion of the N-terminal domain in combination with trisomy 21 alters megakaryocyte cell fate. However, the mechanisms by which GATA1 generates appropriate numbers of megakaryocytes and coordinates terminal megakaryocyte differentiation remain unclear.
To further probe GATA1 function, we established a rescue assay of primary GATA1-deficient fetal megakaryocyte progenitors to identify domains of GATA1 required for coordinated platelet production and megakaryocyte growth. In particular, we have contrasted the role of the N terminus with the interaction of GATA1 with FOG-1.
MATERIALS AND METHODS
Mouse strains.
C57BL/6 and ΔneoΔHS mice (48) were housed according to institutional and national guidelines for humane animal care. Assays were performed on E13.5 to E14.5 fetal liver cells from wild-type C57BL/6 and ΔneoΔHS mice.
Cell staining and progenitor isolation.
To isolate Lin− CD41+ FcγRlow c-kit+ CD9+ cells, 1 × 108 to 3 × 108 single fetal liver cells were passed through a 40-μm-pore-size strainer (Becton Dickinson, San Diego, CA), resuspended in phosphate-buffered saline (PBS; Cambrex, East Rutherford, NJ)-10% fetal calf serum (FCS; PAA Laboratories GmbH, Pasching, Austria) and labeled with rat anti-mouse antibodies directed against the following: Gr-1 (monoclonal antibody [MAb] RB6-8C5), Mac-1 (MAb M1/70), Ter119, B220 (MAb RA3-6B2), Sca-1 (MAb E13-161.7) (BD Pharmingen, San Diego, CA) and interleukin-7 receptor (IL-7R) (MAb A7R34) (eBioscience, San Diego, CA). Antibody-labeled cells were removed using goat anti-rat magnetic microbeads (Miltenyi Biotec, Bergisch-Gladbach, Germany). The remaining cells were then incubated with goat anti-rat-immunoglobulin G-Tricolour antibody (equivalent to phycoerythrin [PE]-Cy5) (Caltag, Burlingame, CA) to remove residual positive cells later. After being blocked with 5% rat serum (Caltag), cells were incubated with fluorescein isothiocyanate-conjugated anti-mouse CD41 (MWReg30), PE-conjugated anti-mouse FcγR (2.4G2), allophycocyanin (APC)-conjugated anti-mouse c-kit (2B8), and biotinylated anti-mouse CD9 (KMC8) MAbs (BD Pharmingen). Cells were stained with streptavidin-APC-Cy7 (BD Pharmingen) and sorted using a MoFlow cell sorter (DAKO Cytomation, Fort Collins, CO) to isolate a Lin− CD41+ FcγRlow c-kit+ CD9+ cell population. Megakaryocyte-erythroid progenitor (MEP) and common myeloid progenitor (CMP) cell populations were sorted as previously described (52).
In vitro liquid culture analysis.
Sorted cells were resuspended in Iscove's modified Dulbecco's medium-10% fetal calf serum (FCS)-l-glutamine (Invitrogen, Carlsbad, CA), 50 U/ml penicillin, and 50 μg/ml streptomycin (Cambrex) supplemented with thrombopoietin (TPO; 1% of conditioned cell culture supernatant (58), 2 U/ml erythropoietin (EPO) (Neo Recormon; Roche, Mannheim, Germany), 10 ng/ml murine IL-3 (mIL-3), 5 ng/ml human IL-11 (huIL-11), 50 ng/ml murine stem cell factor (mSCF), 5 ng/ml murine granulocyte-macrophage colony-stimulating factor (mGM-CSF; Peprotech, Rocky Hill, NJ), and 10 ng/ml mFlt3-ligand (R&D Systems, Minneapolis, MN) to induce myeloid lineage differentiation. At day 4, cells were harvested, labeled with rat anti-mouse GPIIbIIIa-PE, GPIb-PE and GPVI-PE antibodies (B. Nieswandt, Germany) and analyzed using a Cyan Flow Cytometer (DAKO Cytomation). For every experiment, positive and negative fluorescence was distinguished by comparison with matching isotype controls. Dead cells were excluded on the basis of their Hoechst 33258 uptake (1 μg/ml; Molecular Probes, Eugene, OR).
In vitro colony assays.
A total of 1,000 Lin− CD41+ FcγRlow c-kit+ CD9+ cells were plated in methylcellulose (M3234; Stem Cell Technologies, Vancouver, Canada) with the myeloid cytokine cocktail above. CMP (200) and MEP (1,000) cells were seeded in methylcellulose (M3234 and M3134; Stem Cell Technologies). For CMP culture, methylcellulose was supplemented with the myeloid cytokine cocktail (see above) and 2 ng/ml mIL-6 (Peprotech) instead of mGM-CSF. For MEP culture, CMP cytokine cocktail was supplemented with 30% FCS (Stem Cell Technologies), 10 μg/ml recombinant human insulin, 100 μg/ml iron-saturated human transferrin (Sigma, St. Louis, MO), 0.2 mM glutamine, and 50 U/ml penicillin-50 μg/ml streptomycin (Cambrex). Burst-forming erythroid unit and erythroid CFU colonies were either scored at day 4 (for MEP) or 6 (for CMP) of culture. All other colonies were counted at day 9. Single colonies were cytospun and stained with May-Gruenwald-Giemsa (Sigma).
In rescue experiments, 1,000 infected green fluorescent protein-positive (GFP+) sorted Lin− CD41+ FcγRlow c-kit+ CD9+ ΔneoΔHS cells were seeded into methylcellulose (M3234; Stem Cell Technology) supplemented with the same cytokines as above. Total colony number was enumerated at day 9. All colonies were checked under an inverted microscope (Olympus IX51) for GFP expression.
Real-Time RT-PCR.
GATA1 and GATA2 expression were quantitated in 5 × 103 to 5 × 105 sorted primary cells and in ∼5 × 106 retrovirally infected NIH 3T3 cells. RNA was isolated using an RNeasy Micro or Mini RNA Isolation kit (QIAGEN, Hilden, Germany), respectively. Total RNA was divided into a reverse transcribed sample (+RT) and a control without reverse transcriptase (−RT) DNase-treated RNA was reverse transcribed with either Sensiscript Kit (QIAGEN) for primary cells or Superscript II Reverse Transcriptase (Invitrogen) for NIH 3T3 cells with 10 μg/ml random primer, 800 μM deoxynucleoside triphosphate mix, 40 U of RNAseOUT, and 10 mM dithiothreitol (Sigma) in 1x buffer.
Real-Time PCR was performed as previously described (20). cDNA from erythroid (MEL) and megakaryocytic (L8057) cell lines was used to estimate the range of linearity of each probe. Real-time RT-PCR primer and probe sequences are available on request. Gene expression ratios were calculated relative to the cycle threshold (CT) value for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) according to the following mathematical equation: relative ratio = 2(CT GAPDH − CT specific primer or probe).
Cloning of wild-type and mutant GATA1 constructs.
All GATA constructs were cloned into the pMMP-mGata1/ER-IRES-GFP (where ER is endoplasmic reticulum and IRES is internal ribosome entry site) plasmid. Cloning details are available on request. Chicken GATA1 cDNA was kindly provided by Martin Zenke University of Aachen, Germany.
Generation of high-titer retrovirus.
Vesicular stomatitis virus G (VSV-G) protein-coated retroviral particles were produced by transient transfection of 293GPG stable packaging cell line, concentrated using ultracentrifugation, and titered on NIH 3T3 cells as previously described (40).
GATA1 Western blotting.
Infected harvested NIH 3T3 cells were boiled for 10 min in 4× NUPAGE LDS sample buffer (Invitrogen), loaded onto 4 to 12% NUPAGE BisTris gradient precast gels (Invitrogen), and run for 1 h at 200 V in NUPAGE MOPS (morpholinepropanesulfonic acid) running buffer (Invitrogen). Protein transfer and use of anti-mouse GATA1 antibodies (N6 or M20; Santa Cruz Biotechnology, Santa Cruz, CA) have been previously described (42).
GATA1 immunofluorescence.
Infected NIH 3T3 or ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cells were cytocentrifuged onto glass slides and fixed with 5% paraformaldehyde (Sigma), washed with 1× PBS, permeabilized in 0.1% Triton-PBS (Sigma), and stained with anti-mouse GATA1 antibody (N6; Santa Cruz Biotechnology). Images were captured using a fluorescence microscope (Olympus BX60) and Macprobe 4.3 software (Applied Imaging, Newcastle, United Kingdom). Infected ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cells were stained with ToPro-3 (20 ng/ml; Molecular Probes, Eugene, OR) for 5 min at room temperature before mounting in Vectashield (Vector Laboratories, Peterborough, United Kingdom). Confocal sections were collected using a Bio-Rad 2000 confocal system attached to an Olympus BX51 microscope (×100 objective with numerical aperture of 1.3). The average intensity of the GATA1 signal was measured on the entire nuclei of GFP+ cells (for those cells that were infected) or nuclei of noninfected cells using the Metamorph software program (Universal Imaging Corporation, Marlow, United Kingdom). For each construct tested, 100 cells were analyzed. The mean GATA1 signal intensities (±1 standard deviation [SD]) were then calculated.
EMSA.
Ninety-percent confluent B23 cells were transfected with 25 μg of plasmid DNA (wild-type and mutant GATA1 retroviral plasmids) using Lipofectamine (Invitrogen). Nuclear extracts were prepared as previously described (2). Electromobility shift assays (EMSAs) were performed as previously described (62). The oligonucleotide sequence GATCTCCGGCAACTGATAAGGATTCCCTG was used as a probe. The underlined sequence is the GATA-binding site. Cold competitor (wild-type and mutant; GATCTCCGGCAACTGTGAAGGATTCCCTG) oligonucleotides were added in 100-fold excess. One microliter of anti-mouse GATA1 antibody (N6; Santa Cruz Biotechnology) was used in supershift experiments.
Retroviral infection of ΔneoΔHS progenitors and analysis of infected cells.
E13.5 to E14.5 ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ fetal liver cells were incubated for 36 h in Iscove's modified Dulbecco's medium-10% FCS-l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin with 1% TPO-conditioned medium, 50 ng/ml mSCF, 5 ng/ml huIL-11, 5 ng/ml mGM-CSF, 10 ng/ml mIL-3 (Peprotech), 2 U/ml EPO (Roche), and 10 ng/ml mFlt3-ligand (R&D Systems). A total of 3 × 104 to 5 × 104 progenitors/sample were infected by spin inoculation with VSV-G-coated retroviral particles at a multiplicity of infection of 50 in the same medium and cytokines with 8 μg/ml polybrene (Sigma). Cells were incubated for 7 h at 37°C in 5% CO2, washed, and resuspended in the same medium and cytokines. Medium was changed after 24 h, and cytokines were switched to 1% TPO-conditioned medium, 2 ng/ml mIL-6, and 5 ng/ml huIL-11 to promote megakaryocyte maturation.
At 72 h after infection, cells were either analyzed for differentiation or stained with the DNA dye Hoechst 33342 (Molecular Probes) to evaluate the cell cycle. For differentiation analysis, dishes were scanned for proplatelet formation in GFP+ and GFP− cells under an inverted microscope (Olympus IX51). Images were captured using Openlab 3.1.5 software (Improvision, Coventry, United Kingdom) with a magnification of ×100. Cells were harvested, stained with a PE-conjugated anti-mouse GPIb antibody, and analyzed by flow cytometry. Appropriate isotype controls were used in every experiment. Hoechst 33258 (1 μg/ml; Molecular Probes) was added to distinguish between dead and live cells. To define a GPIbhigh rescue gate, wild-type fetal liver Lin− CD41+ FcγRlow c-kit+ CD9+ cells were cultured until proplatelets were shed and stained with anti-mouse GPIb-PE antibody, and GPIb expression was plotted against forward scatter. The same GPIbhigh gates were then applied to all rescue experiments. GPIb expression was analyzed on a Hoechst− and GFP+ gated cell population.
For analysis of cell cycle profiles, cells were stained with Hoechst 33342 (5 μg/ml) at 37°C for 90 min. The DNA profile was analyzed using a MoFlow flow cytometer (DAKO Cytomation). Propidium iodide (1 μg/ml) was used to discriminate between dead and live cells. The cell cycle profile was then created on a linear scale using Summit Software (DAKO Cytomation).
RESULTS
ΔneoΔHS fetal liver contains an abnormal megakaryocyte (Meg)-erythroid progenitor.
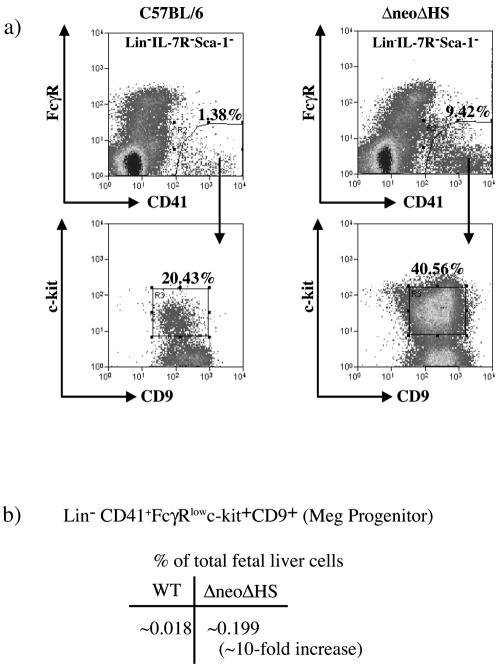
To define the differentiation block in megakaryopoiesis in ΔneoΔHS mice, myeloid progenitors were enumerated, and their hematopoietic potential was evaluated. First, to identify a fetal Meg-restricted progenitor, we modified a published protocol for isolation of an adult murine bone marrow common Meg progenitor using flow cytometry (36). This progenitor population is characterized by the immunophenotype Lin− CD41+ FcγRlow c-kit+ CD9+ (see Materials and Methods). A representative sort from ΔneoΔHS and wild-type fetal liver cells is shown in Fig. 1a. The Lin− CD41+ FcγRlow c-kit+ CD9+ cells were ∼10-fold increased in ΔneoΔHS total fetal liver compared to wild-type control (∼0.199% versus ∼0.018%) (Fig. 1b), consistent with a ∼10-fold increase in megakaryoblasts seen in histological sections in ΔneoΔHS bone marrow (48).
FIG. 1.
Isolation of a Lin− CD41+ FcγRlow c-kit+ CD9+ progenitor cells from mouse E13.5 to E14.5 fetal liver. (a) Flow cytometric analysis of Lin−IL-7R−Sca-1− wild-type and ΔneoΔHS fetal liver cells labeled with CD41-fluorescein isothiocyanate, FcγR-PE, c-kit-APC, CD9-biotin, and streptavidin-APC-Cy7. Sorting gates for isolation of Lin− CD41+ FcγRlow c-kit+ CD9+ cells are shown. (b) The table shows the average frequencies of Lin− CD41+ FcγRlow c-kit+ CD9+ population in wild-type and ΔneoΔHS fetal liver from nine different sorting experiments. Note that only 10% of total fetal liver cells are Lin−.
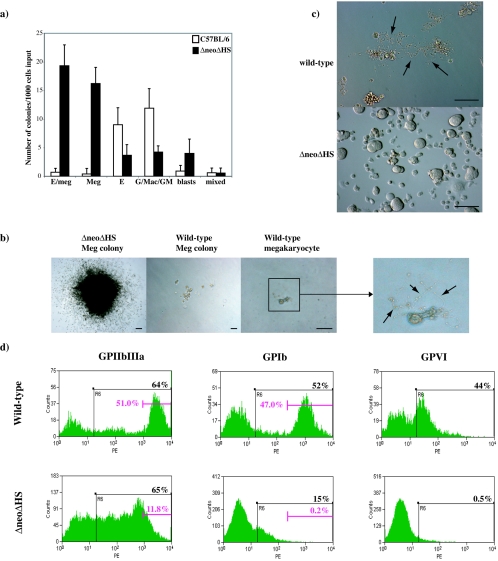
To assess the functionality of this population, purified cells were grown in in vitro colony assays and liquid culture. For colony assays, 1,000 cells were plated in methylcellulose containing TPO, EPO, SCF, IL-3, IL-11, Flt3-ligand, and GM-CSF. The average colony numbers of at least three independent experiments are shown in Fig. 2a. ΔneoΔHS cells had a plating potential of ∼5% and gave rise to mainly erythroid-Meg (E/Meg) and Meg colonies (∼70% of total colonies). E/Meg and Meg colonies were abnormally large, macroscopic, and composed of tens of thousands of cells that never shed proplatelets (Fig. 2b). The remaining colonies were erythroid, granulocyte and/or macrophage (G/Mac/GM), BLAST, and mixed myeloid (GEMM) (Fig. 2a). In contrast, Lin− CD41+ FcγRlow c-kit+ CD9+ wild-type cells gave rise to strikingly few Meg and E/Meg colonies, but, instead, the majority of cells differentiated into single mature megakaryocytes with proplatelet extensions (Fig. 2b, right panel) within 2 days of culture (60 to 70 megakaryocytes per 1,000 cells). Wild-type cells exhibit a lower plating potential (∼2%) with E, G/Mac/GM, BLAST, and GEMM colonies present (Fig. 2a). The few Meg colonies observed were normal-sized (much smaller compared to those from ΔneoΔHS cells) and formed proplatelets (Fig. 2b). Therefore, wild-type and ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cell populations have quite different hematopoietic potentials.
FIG. 2.
Clonogenic potential and liquid culture analysis of isolated Lin− CD41+ FcγRlow c-kit+ CD9+ progenitor cells from wild-type and ΔneoΔHS fetal livers. (a) Mean number (±1 SD) of day 9 colonies determined by culturing 1,000 Lin− CD41+ FcγRlow c-kit+ CD9+ progenitors in methylcellulose with Epo, Tpo, IL-3, IL-11, SCF, Flt3-ligand, and GM-CSF (n = 3 independent experiments). Samples were plated in triplicates. G/Mac/GM, combined number of granulocyte, macrophage, and mixed granulocyte/macrophage colonies; blasts, BLAST cell colonies; mixed, colonies with multilineage composition. (b) Morphology of a typical megakaryocyte colony derived from ΔneoΔHS (left panel) and wild-type (middle panel) progenitors at day 9 of culture (original magnification, ×40). Scale bar, 100 μm. The majority of wild-type Lin− CD41+ FcγRlow c-kit+ CD9+ cells differentiated into megakaryocytes with proplatelet extensions within 2 days in culture (right panel; original magnification, ×100). Scale bar, 100 μm. An enlarged view in the adjacent panel further to the right shows the proplatelet extensions (black arrows). (c) Photographs of wild-type (top) and ΔneoΔHS (bottom) Lin− CD41+ FcγRlow c-kit+ CD9+ day 4 liquid cultures (original magnification, ×100). Scale bar, 100 μm. Wild-type cells formed proplatelets (black arrows); ΔneoΔHS cells accumulated and never shed proplatelets. (d) Analysis of GPIIbIIIa, GPIb, and GPVI expression by flow cytometry at day 4 in liquid cultures of sorted wild-type (top panels) and ΔneoΔHS (bottom panels) Lin− CD41+ FcγRlow c-kit+ CD9+ fetal liver cells.
To evaluate proplatelet formation, sorted Lin− CD41+ FcγRlow c-kit+ CD9+ cells were cultured in medium containing TPO, EPO, IL-3, IL-11, SCF, mFlt3-ligand, and GM-CSF. Within 3 days, most wild-type cells shed proplatelets and expressed high levels of the megakaryocyte maturation markers GPIIbIIIa, GPIb, and GPVI (Fig. 2c and d). In contrast, cultured ΔneoΔHS cells accumulated without proplatelet formation (Fig. 2c). Expression of late-stage megakaryocyte maturation markers, GPIb and GPVI, was either very low or absent, and though 65% of cells expressed GPIIbIIIa, only 11.8% exhibited high-level expression (Fig. 2d). In conclusion, we have identified an abnormal, bi-potential E/Meg progenitor that accumulates in ΔneoΔHS but not wild-type fetal liver and displays a block in terminal maturation and has altered growth properties.
CMP and MEP cell populations in ΔneoΔHS mice.
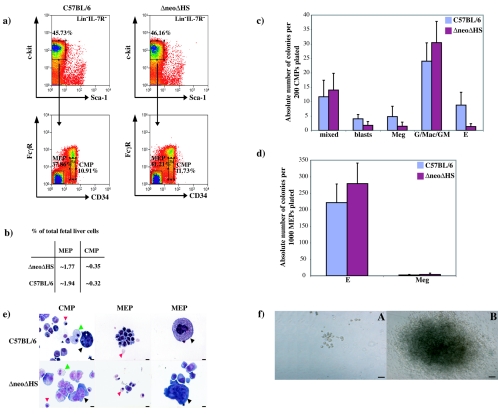
To determine if abnormal progenitors could be detected earlier in myelopoiesis, published sorting protocols (52) were used to isolate fetal liver CMP and MEP populations. Representative fluorescence-activated cell sorting (FACS) plots of these isolated populations are shown in Fig. 3a. The frequency of CMP and MEP within total fetal liver cells was similar (Fig. 3b). Myeloid colony-forming potential from at least three independent experiments is depicted in Fig. 3c and d. A decrease in erythroid colonies was detected in ΔneoΔHS compared to wild-type CMP, whereas the frequency of all other colony types was similar. Morphological analysis confirmed wild-type and ΔneoΔHS fetal liver CMP gave rise to all myeloid cell types (Fig. 3e). This was confirmed by FACS analysis from in vitro liquid cultured CMP (data not shown). As expected, the MEP population only matured into erythroid cells (Ter119+) and megakaryocytes (GPIIbIIIa+) (Fig. 3d and e and data not shown). The most important difference between ΔneoΔHS and wild-type CMP and MEP colonies was that ΔneoΔHS Meg colonies were abnormally large and failed to shed proplatelets, reminiscent of colonies seen from Lin− CD41+ FcγRlow c-kit+ CD9+ ΔneoΔHS cells (compare Fig. 3f with Fig. 2b).
FIG. 3.
Isolation and colony-forming potential of wild-type and ΔneoΔHS mouse fetal liver CMP and MEP populations. (a) FACS-sorted Lin− CD34+ FcγRlow c-kit+ Sca-1− (CMP) or Lin− CD34− FcγRlow c-kit+ Sca-1− (MEP) cells from wild-type (left panels) and ΔneoΔHS (right panels) fetal liver cells are shown. (b) The average frequency of CMP and MEP populations within total fetal liver from nine different sorting experiments is summarized in the table. Note that Lin− cells comprise only 10% of the total fetal liver population. (c and d) Mean numbers of colonies (±1 SD) enumerated from panel c (200 CMP) or panel d (1,000 MEP) are shown from three independent experiments. All samples were analyzed in triplicates. The nature of the colonies is as shown in Fig. 2. (e) May-Gruenwald-Giemsa-stained cells from pooled wild-type (top panels) or ΔneoΔHS (bottom panels) colonies. CMPs gave rise to red cells (red arrowhead), myeloid cells (green arrowhead) and megakaryocytes (black arrowhead). MEPs differentiated into red cells (red arrowhead) of all maturation stages. Megakaryocytes (black arrowhead) from wild-type but not ΔneoΔHS MEP show normal terminal maturation. Scale bar, 10 μm. (f) Wild-type CMP and MEP gave rise to normal megakaryocyte colonies (frame A) whereas ΔneoΔHS CMP and MEP formed abnormal, very large and hyperproliferative megakaryocyte colonies (frame B). Scale bar, 100 μm.
GATA1 and GATA2 expression levels in ΔneoΔHS mice.
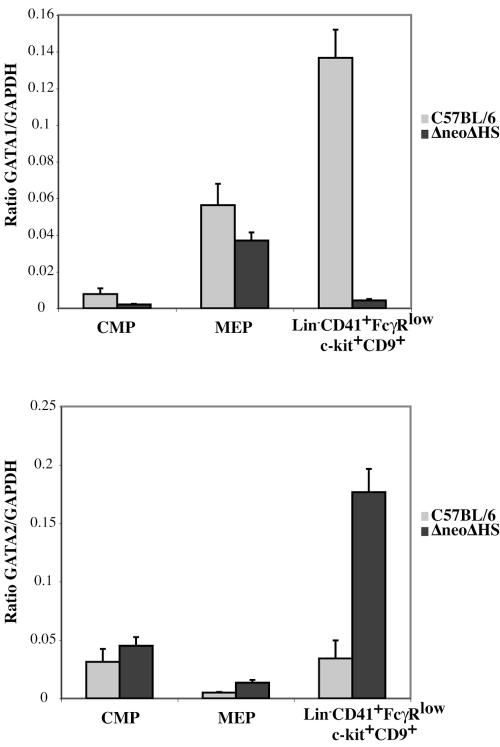
To correlate the phenotype of wild-type and ΔneoΔHS CMP, MEP, and Lin− CD41+ FcγRlow c-kit+ CD9+ cells with GATA1 and GATA2 expression levels, quantitative TaqMan real-time RT-PCR was performed (Fig. 4). GATA1 and GATA2 mRNA levels were similar in both genotypes in CMP and MEP cells. In contrast, GATA1 expression was ∼10-fold decreased and GATA2 levels were about fourfold increased in ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cells compared to wild-type counterparts. These data are in accord with previously published data showing GATA1 levels are ∼5% of normal in arrested ΔneoΔHS megakaryoblasts (48).
FIG. 4.
Expression levels of GATA1 and GATA2 in sorted progenitor populations. TaqMan real-time RT-PCR quantitation of GATA1 (top panel) and GATA2 (bottom panel) mRNA expression normalized to GAPDH is shown in FACS-sorted wild-type and ΔneoΔHS fetal liver CMP, MEP, and Lin− CD41+ FcγRlow c-kit+ CD9+ progenitor populations.
GATA1 structure/function study by retroviral rescue of ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cells.
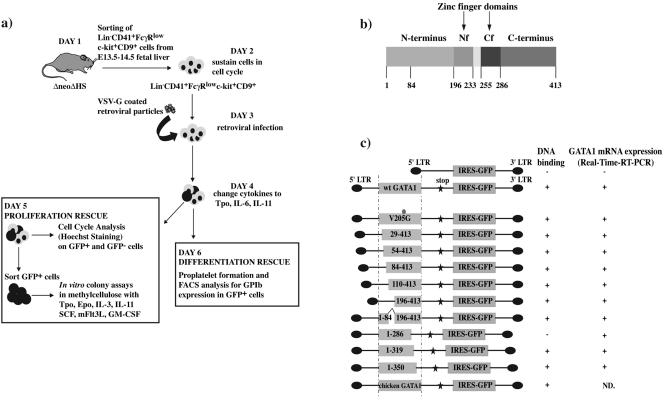
Given the abnormal megakaryocyte differentiation potential and low level of expressed GATA1 in ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cells, we infected this population with VSV-G pseudotyped wild-type and mutant GATA1 retroviral particles to (i) localize functionally important sequences within the N terminus of mGATA1 protein, (ii) contrast function of N terminus sequences with sequences mediating interaction with FOG-1, and (iii) determine if other GATA1 domains are required for platelet release/expression of GPIb and restoration of normal megakaryocyte growth. An overview of the experimental protocol is shown in Fig. 5a.
FIG. 5.
Outline of rescue experiments and phenotypic characterization of wild-type and mutant GATA1 VSV-G coated retroviral particles. (a) Outline of rescue experiments of GATA1-deficient progenitors by retroviral infection of wild-type and mutant GATA1 molecules. (b) Modular structure of wild-type murine GATA1. (c) Schematic representation of GATA1 constructs tested. Wild-type and mutant GATA1 constructs (box) were linked to an artificial stop codon (star) and IRES-GFP (box) and expressed under the control of retroviral long terminal repeat (LTR) sequences. The dot in the V205G construct indicates the location of the V205G mutation within the GATA1 molecule. Summary of in vitro DNA binding properties (analyzed by EMSA) of different GATA1 proteins and mRNA expression (determined by TaqMan real-time RT-PCR) in infected NIH 3T3 cells is shown on the right. ND, not done.
The modular structure of mGATA1 and mutant GATA1 constructs tested are shown in Fig. 5b and c. Viral GATA1 mRNA and protein expression in infected NIH 3T3 cells was analyzed by TaqMan real-time RT-PCR, immunofluorescence, and Western blot analysis. All mGATA1 constructs expressed GATA1 mRNA, and expression was not detected in IRES-GFP control vector-infected cells (Fig. 5c). Western blot analysis confirmed appropriate GATA1 protein expression in NIH 3T3 cells and B23 cells (see Fig. S1a in the supplemental material, upper and lower panels, respectively). Constructs expressing wild-type (wt) GATA1 and V205G proteins (that disrupt interaction with FOG-1) and proteins with deletions in GATA1 spanning residues 29 to 413, 54 to 413, 1 to 286, 1 to 319, and 1 to 350 were detected almost exclusively in the nucleus (see Fig. S1b, M, N, O, P, R, S, and T in the supplemental material). In contrast, uninfected and IRES-GFP (control vector)-infected NIH 3T3 cells did not show GATA1 signal (see Fig. S1b, K and L, in the supplemental material). Because of epitope deletion (the epitope is located around amino acid 84), mutant GATA1 proteins spanning residues 84 to 413 (see Fig. S1b, Q, in the supplemental material), 110 to 413, 196 to 413, and residues 1 to 84 added to residues 196 to 413 (1 to 84^196 to 413) (data not shown) could not be tested by immunofluorescence. All constructs expressed GFP (see Fig. S1b in the supplemental material and data not shown). The ability of constructs to bind DNA in EMSAs is summarized in Fig. 5c. All constructs containing the C-terminal zinc finger bound DNA in vitro, except the construct spanning amino acid residues 1 to 286. This construct does not contain basic residues C-terminal to the C-finger required for in vitro DNA binding (17).
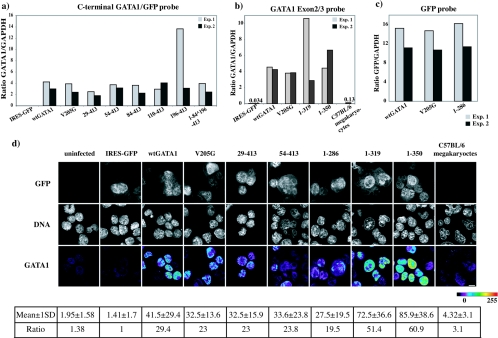
To document exogenous (viral) wild-type and mutant GATA1 mRNA expression in infected Lin− CD41+ FcγRlow c-kit+ CD9+ cells, cDNA was prepared from FACS-sorted GFP+ progenitor cells infected with different GATA1 constructs. Levels of exogenous wild-type and mutant GATA1 mRNAs were determined by TaqMan real-time RT-PCR using primers/probe spanning either the C-terminal end of the mGata1 gene and the GFP gene (Fig. 6a) or exon 2 and 3 in the mGata1 gene (Fig. 6b) or the GFP gene alone (Fig. 6c). The choice of primers/probe was dependent on the sequences present in the mutant GATA1 mRNA species. Results from two independent experiments (experiment 1 and 2) demonstrate some variation in the level of exogenous GATA1 mRNA expressed, but for most samples this was less than 1.5-fold. To help interpret results of rescue assays with mutant GATA1 molecules, we compared the level of exogenous GATA1 mRNA in infected cells to the physiological level of endogenous GATA1 mRNA in mature noninfected wild-type megakaryocytes (Fig. 6b). Exogenous GATA1 mRNAs were significantly overexpressed (between 20- and 40-fold) compared to the endogenous level of GATA1 mRNA in mature normal megakaryocytes.
FIG. 6.
Expression analysis of wild-type and mutant GATA1 mRNA and protein in infected ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cells. (a to c) GATA1 mRNA expression quantitated by TaqMan real-time RT-PCR from retrovirally infected ΔneoΔHS fetal Lin− CD41+ FcγRlow c-kit+ CD9+ cells using either the C-terminal GATA1-GFP probe (a) or the GATA1 Exon2/3 probe (b) or the GFP probe (c). Purified wild-type C57BL/6 megakaryocytes (C57BL/6) were analyzed to compare the physiological expression level of GATA1 with retrovirally expressed GATA1. mRNA from two independent cell sortings and infections are shown for each construct (experiment 1, gray; experiment 2, black). Relative mRNA ratios were normalized with respect to GAPDH mRNA. (d) Micrographs showing the level of GATA1 protein expressed in ΔneoΔHS fetal Lin− CD41+ FcγRlow c-kit+ CD9+ cells infected with different GATA1 constructs. Panels show GFP expression (upper panel), ToPro-3 staining (DNA, middle panel), and GATA1 protein signal (lower panel). GATA1 expression is displayed in pseudo-color (scale bar below) to show the level of GATA1 in different infected cells. The numbers below each panel show the mean GATA1 protein signal intensity ± 1 SD/fixed cell volume when 350 to 400 cells where analyzed. To compare results between panels, the mean signal intensity in all the panels has been normalized with respect to the mean signal intensity in the IRES-GFP panel.
To further investigate the two issues of overexpression and variation in the level of exogenous GATA1 protein in infected progenitors, we performed immunofluorescence assays to quantitate the level of GATA1 protein expression (Fig. 6d). These data confirm that the GATA1 protein was ∼6- to 20-fold overexpressed in infected cells compared to wild-type megakaryocytes. In addition, there was about a two- to threefold variation in the mean level of GATA1 expression between the different constructs. However, given that the standard deviation in protein expression is ∼50% of the mean signal intensity and given the “semiquantitative” nature of the analysis, these data have to be interpreted with caution. In summary, overexpression of mutant mRNA and protein is an important caveat in interpreting results from the rescue assay (see below and Discussion).
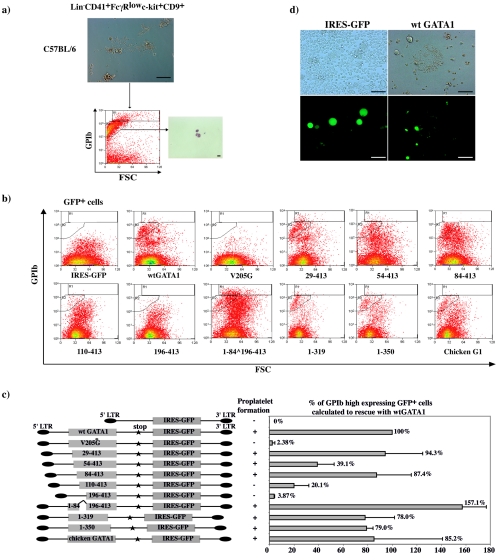
The GATA1 N terminus is indispensable for megakaryocyte differentiation.
To define GATA1 domains required for megakaryocyte differentiation, we assayed the ability of mutant molecules to rescue proplatelet formation from ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cells. To quantitatively measure GPIb-expressing proplatelets and megakaryocyte fragments, we developed a flow cytometric assay. First, wild-type E13.5 to E14.5 fetal liver sorted Lin− CD41+ FcγRlow c-kit+ CD9+ cells were cultured until proplatelet formation was observed. The culture was harvested, stained with a PE-conjugated anti-mouse GPIb antibody, and analyzed using flow cytometry (Fig. 7a). Two GPIbhigh expression gates (R1 and R2), containing megakaryocyte fragments and proplatelets, were then used in all subsequent rescue experiments. The extent of differentiation was analyzed as a function of GPIb expression in these gates in Hoechst− GFP+ cells (Fig. 7b). Average viability of rescued cells was 80 to 90%. Detection of a GPIbhigh population always correlated with proplatelet formation in culture (Fig. 7c and d).
FIG. 7.
Rescue of terminal megakaryocyte differentiation of ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cells by GATA1 proteins. (a) Photograph of liquid culture from wild-type Lin− CD41+ FcγRlow c-kit+ CD9+ cells used to set analysis gates for megakaryocyte GPIb expression (top). FACS plot of GPIb expression plotted against the forward scatter of cells/proplatelets (bottom). GPIbhigh expression gates (R1+R2) were set. On the right, sorted events from R2 were cytospun and stained with May-Gruenwald-Giemsa to reveal proplatelet and megakaryocyte fragments. Scale bar, 10 μm. (b) Flow cytometric analysis of GPIb expression in the R1 and R2 rescue gate when Lin− CD41+ FcγRlow c-kit+ CD9+ cells were infected with different constructs as indicated. (c) GATA1 constructs as described in the legend of Fig. 5c (left). To control for interexperimental variation, infection with constructs expressing no GATA1 protein (IRES-GFP construct) and wt GATA1 protein were performed in every rescue experiment. In each rescue experiment, the percentage of rescue by wt GATA1 was set to 100%, and all other mutant rescues were normalized with respect to that. The graph (right) represents the mean (±1 SD) of three independent cell sortings and infections for each construct. A summary of whether proplatelet formation was observed for each construct is shown to the left. (d) Microscopic analysis of IRES-GFP and wt GATA1 infected ΔneoΔHS Lin−CD41+ FcγRlow c-kit+ CD9+ cells 72 h after infection. The upper frames show fields by phase-contrast microscopy (original magnification, ×100). Lower frames show GFP fluorescence in the same fields. Scale bar, 100 μm.
Differentiation rescue experiments by GATA1 molecules were repeated on three independently isolated progenitor cell populations. Representative FACS plots of one experiment are shown in Fig. 7b and summarized in Fig. 7c. Expected results were obtained when wt GATA1, control GFP-expressing vector (IRES-GFP), V205G (defective for interaction with FOG-1), and the DNA-binding defective mutant (construct 1-286) were tested. Proplatelet formation and GPIbhigh population was seen with rescue with wt GATA1 but not the other constructs. These results validate the assay. They also suggest that even though the V205G mutant is overexpressed 30-fold, the GATA1-FOG-1 interaction is likely to be significantly impaired (Fig. 7b and c and data not shown). N-terminal deletion up to the first 84 amino acids did not markedly influence differentiation rescue. With further deletion to amino acid 110 (construct 110-413), the extent of rescue dropped to ∼20% of wt GATA1. When the complete N terminus was deleted (construct 196-413), the rescue potential decreased to less than 5% (Fig. 7b and c). Unexpectedly, when the first 84 amino acids were directly linked to GATA1 residues 196 to 413, a GPIbhigh population was restored, and proplatelet formation was detected in culture (Fig. 7b and c). This suggests that amino acids in the first 84 residues aid platelet release in the context of the GATA1 zinc fingers and C terminus. Deletion of the GATA1 C terminus to amino acid 319 (constructs 1-319 and 1-350) did not reduce differentiation rescue. Lastly, chicken GATA1 protein rescued differentiation, suggesting conservation of this function between the two species.
Thus, in addition to domains necessary for DNA-binding and FOG-1 interaction, two regions in the GATA1 N terminus are likely to be required for rescue of terminal megakaryocyte differentiation and proplatelet formation: (i)between amino acids 84 to 110 and (ii) residues in the first 84 amino acids when attached to GATA1 zinc fingers and C terminus.
Domains of GATA1 protein required to restrain growth of abnormal GATA1 deficient colonies.
To map GATA1 domains required to restrain megakaryocyte growth, FACS-sorted GFP+ ΔneoΔHS progenitors were subjected to colony assays and cell cycle analysis (Fig. 8a and b). All colonies expressed GFP (data not shown) and were large abnormal megakaryocyte colonies identical to uninfected ΔneoΔHS Meg colonies (Fig. 2b). Cells infected with control vector and the DNA-binding defective mutant (construct 1-286) formed ∼84 colonies/1,000 cells (Fig. 8a and data not shown). In contrast, wild-type GATA1- and the V205G mutant-infected cells yielded low colony numbers (Fig. 8a). Similarly, when the first 29 or 54 amino acids were deleted (constructs 29-413 and 54-413), the colony number was still low and comparable to wild-type GATA1-infected cells. With further N-terminal deletion, to residue 84 (construct 84-413), colony numbers increased 8- to 10-fold and peaked when the whole N terminus was removed (construct 196-413) (Fig. 8a). Interestingly, addition of amino acids 1 to 84 to GATA1 zinc fingers and C terminus (construct 1-84 ^ 196-413) reduced colony numbers (Fig. 8a). Deletions in the C terminus had no effect on colony formation (constructs 1-319 and 1-350) (Fig. 8a). When ΔneoΔHS cells were infected with chicken GATA1, ∼18 colonies/1,000 cells were formed. Thus, chicken GATA1 reduced abnormal colony numbers (∼80 colonies to ∼18 colonies) by ∼80% compared to mouse GATA1 (∼80 colonies to ∼3 colonies) (Fig. 8a).
FIG. 8.
Abrogation of abnormal growth from ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cells. (a) Mean number (±1 SD) of abnormal colonies present at day 9 from 1,000 GFP+ cells infected with different retroviral constructs from two independent sorting and infection experiments. (b) Flow cytometric cell cycle analysis of ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ cells infected with either vector (IRES-GFP; top) frames or wt GATA1 (bottom) at 24 h and 72 h of culture. GFP− (uninfected, black line) and the GFP+ (infected, green solid line) profiles were overlaid. Hoechst fluorescence was analyzed on a linear scale. At 72 h after infection the sub-G1 peak in wt GATA1-infected cells was sorted and analyzed morphologically. The upper frame shows May-Gruenwald-Giemsa staining; the lower frame illustrates benzidine staining (hemoglobinized cells stain a brown color). Scale bar, 10 μm.
In summary, in addition to DNA binding, N-terminal residues after amino acid 54 are required to dampen the growth of cells from ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ progenitors. In contrast, the GATA1 C terminus may not be necessary for this function.
To evaluate the cell cycle status of control vector- and wild-type GATA1-rescued cells, we performed Hoechst staining 24, 48, and 72 h after infection in GFP+ and GFP− cells (Fig. 8b, 48 h, and data not shown). No differences in cell cycle profiles were detected in control vector- and wild-type GATA1-rescued cells 24 h after infection (Fig. 8b). At 48 h, the percentage of cells in G2/M in the GFP+ population of wild-type GATA1-infected cells decreased slightly (data not shown). At 72 h after infection, GFP+ and GFP− cells infected with control vector showed similar cell cycle profiles (Fig. 8b). In contrast, dramatic changes were seen in cells infected with wt GATA1. Here, the percentage of GFP+ cells present in the original G1 and G2/M peaks was reduced compared to the GFP− (uninfected) population (Fig. 8b). GFP+ cells in the original G1 and G2/M peak are likely to be Lin− CD41+ FcγRlow c-kit+ CD9+ cells that were unable to fully differentiate into proplatelets at 72 h as Lin− CD41+ FcγRlow c-kit+ CD9+ progenitors successfully rescued by wt GATA1 would have already differentiated into proplatelets and platelets by this time point. Since platelets do not contain a nucleus, they are undetectable in the Hoechst profile.
Furthermore, an obvious sub-G1 peak is seen in GFP+ cells (Fig. 8b). To determine the nature of cells in sub-G1, events in this peak were sorted by FACS onto glass slides. Morphologically, these cells were erythroid normoblasts that stained positive with benzidine and thus express hemoglobin (Fig. 8b). Therefore, we suggest that exogenous wt GATA1 expression can direct ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ progenitors to differentiate into red cells as well as megakaryocytes in liquid culture. This is not surprising as colony assays demonstrate that this progenitor has mixed erythroid and megakaryocyte potential (Fig. 2a). A possible explanation for the appearance of this mature erythroid population as a sub-G1 peak is that during erythroid differentiation chromatin compaction is likely to limit accessibility of DNA to intercalating dyes, such as Hoechst (10). As Hoechst staining was performed on a mixed cell population, we were unable to examine the cell cycle status of megakaryocytes alone.
Level of GATA2 expression and rescue of megakaryocyte differentiation and proliferation.
As GATA2 is required for proliferating progenitor cells (53) and GATA2 mRNA levels are elevated in ΔneoΔHS Lin−CD41+ FcγRlow c-kit+ CD9+ progenitors (Fig. 4), we asked if the ability of GATA1 mutants to restrain abnormal megakaryocyte colony formation correlated with suppression of high-level GATA2 expression. We assayed GATA2 mRNA levels by TaqMan quantitative RT-PCR in sorted GFP+ ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ progenitors 48 h after infection with GATA1 constructs (Fig. 9). GATA2 mRNA deceased by ∼2.5-fold when cells were infected with wt GATA1. In contrast, GATA2 levels did not fall appreciably when progenitors were infected with either V205G or the GATA1 DNA-binding mutant (construct 1-286). These findings are consistent with published data suggesting that repression of GATA2 by GATA1 requires GATA1 to bind DNA and interact with FOG-1 (28, 41). Moreover, GATA2 levels are higher in cells infected with GATA1 proteins that lack N-terminal residues 84 to 196. This suggests that these residues are required either directly or indirectly to repress GATA2 expression. Separately, these data support the hypothesis that suppression of GATA2 expression is not required for controlling proliferation of ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ progenitors or, indeed, for proplatelet formation (construct 1-84 ^ 196-413). However, there are two caveats to these findings. First, we have only assayed GATA2 mRNA and not protein expression. Second, we have conducted our studies in populations of cells; thus, it is unclear if low GATA2 levels correlate with suppression of proliferation and terminal differentiation in any one cell.
FIG. 9.
GATA2 expression levels in Lin− CD41+ FcγRlow c-kit+ CD9+ cells infected with GATA1 constructs. Relative GATA2 mRNA expression levels (normalized with respect to GAPDH mRNA) are shown in ΔneoΔHS fetal Lin− CD41+ FcγRlow c-kit+ CD9+ cells, 48 h after retroviral infection with different GATA1 constructs. The table summarizes data from the experiments shown in Fig. 7 and 8.
DISCUSSION
Characterization of the megakaryocyte defect in ΔneoΔHS mice.
GATA1 has an essential role in restraining megakaryocyte growth and promoting terminal differentiation leading to platelet release (see the introduction). However, a systematic study of GATA1 domains required for these processes had not been undertaken. Our work involved rescue of ΔneoΔHS primary fetal cells with different GATA1 mutant molecules. We adopted this approach for three reasons. First, primary cells are subject to physiologic growth control and complete terminal maturation. Second, as ΔneoΔHS cells only express 5 to 10% of wild-type GATA1 mRNA in megakaryocyte progenitors (Fig. 4), function of exogenous GATA1 can be studied with little confounding full-length endogenous GATA1 activity. Third, we wished to probe the function of the N-terminal GATA1 domain, which is deleted in DS TMD and AMKL (see introduction). As DS TMD and AMKL are initiated in fetal life and since previous studies had suggested functional differences between fetal and adult progenitors (21, 29, 46, 56), it was important to study fetal and not adult cells. One important caveat is that retroviral GATA1 mRNA and protein were significantly overexpressed in infected ΔneoΔHS progenitors, and this may have altered the stoichiometry of critical transcription factor interactions.
Compared to wild-type, ΔneoΔHS mice accumulate an abnormal Lin− CD41+ FcγRlow c-kit+ CD9+ population with erythroid-megakaryocytic potential (Fig. 1 and 2). When cultured in methylcellulose or medium, this population does not release proplatelets or express cell surface markers associated with terminal megakaryocyte maturation at a high level and produces abnormally large colonies of arrested megakaryocyte precursors (Fig. 2). Thus, the population recapitulates the phenotype of GATA1 megakaryocyte deficiency. Rescue of this population with wild-type GATA1 promoted terminal megakaryocyte and red cell differentiation in liquid culture (Fig. 7b, c, and d and 8b) and loss of large abnormal colony formation (Fig. 8a). Thus, reintroduction of wt GATA1 alters the cell fate of ΔneoΔHS Lin− CD41+ FcγRlow c-kit+ CD9+ progenitors. In liquid culture GATA1 does this, at least in part, by directing differentiation that is likely to be accompanied by cell cycle exit (Fig. 8b). However, we cannot rule out that other cell fate options may also be triggered by exogenous GATA1 expression.
In contrast to the Lin− CD41+ FcγRlow c-kit+ CD9+ compartment, no major differences were observed in numbers of fetal CMP and MEP cells between wild-type and ΔneoΔHS mice. In addition, ΔneoΔHS CMP and MEP cells terminally differentiated normally into all lineages, except the megakaryocytic. These findings correlate well with where the GATA1 expression defect is seen in ΔneoΔHS compared to wild-type myeloid progenitors (Fig. 4). In addition, these data suggest the −3.5 enhancer, deleted in ΔneoΔHS mice, has a nonredundant role in directing GATA1 expression to Lin− CD41+ FcγRlow c-kit+ CD9+ cells and differentiating megakaryocytes.
Role of GATA1-FOG-1 interaction.
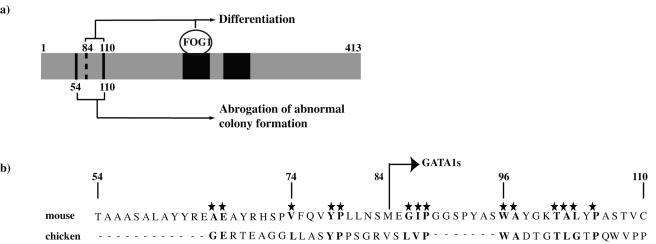
Rescue with different GATA1 molecules revealed different roles for a critical residue required for GATA1-FOG-1 interaction compared to amino acids in the N terminus (Fig. 10a). Whereas interaction with FOG-1 is critical for terminal differentiation (proplatelet formation and GPIb expression), it is not required to abrogate growth (prevent abnormal colony formation). The critical role of FOG-1 in terminal maturation in our assay is consistent with previous studies of human patients and germ line mutant mice (7, 8, 14, 15, 32, 39). The mechanisms by which GATA1-FOG-1 interaction directs terminal maturation are likely to be multiple. First, GATA1-FOG-1 interaction may facilitate productive interactions with other transcription factors required for megakaryocyte gene expression, for example, the megakaryocyte regulator Fli-1 (63). Second, GATA1-FOG-1 complexes have been proposed to anchor distant cis elements regulating erythroid globin gene expression (57). A similar situation may be operative at megakaryocyte gene loci. Third, some GATA1-FOG-1 target genes are likely to be other transcription factors and regulators of megakaryocyte cell fate, e.g., p45NF-E2, FOG-1, GATA1 and c-mpl (61, 63; B. Guyot and P. Vyas, unpublished data). Thus, their increased expression would generate a feed-forward loop of gene expression promoting terminal megakaryocyte differentiation.
FIG. 10.
Alignment of N-terminal mouse and chicken GATA1 amino acid sequence. (a) Summary of the sequence requirements within mouse GATA1 for coordinated terminal megakaryocyte differentiation and growth arrest. (b) Alignment of N-terminal mouse and chicken GATA1 amino acids between murine residues 54 to 110. The stars and bold letters indicate conserved amino acid residues between those two species. The location of the translation start site of GATA1s in mGATA1 is shown. This is equivalent to the translation start of GATA1s that is expressed in DS TMD and AMKL.
However, the finding that regulation of growth is FOG-1 independent is novel. Previous studies of mice and human patients with mutations in GATA1 residues required to interact with FOG-1 have suggested that loss of GATA1-FOG-1 interaction leads to increased megakaryocyte numbers (7, 8, 14, 15, 32, 39). However, the effects of abrogating GATA1-FOG-1 interaction on megakaryocyte growth have not been documented. As mentioned above, one caveat is that the V205G mutant GATA1 construct was significantly overexpressed. Given the small number of progenitors (∼103) available for study after infection, we could not study whether the GATA1-FOG-1 interaction had occurred. However, absence of differentiation in V205G-infected cells would argue against a productive GATA1-FOG-1 interaction. At the very least, our data show a stricter requirement for GATA1-FOG-1 interaction for terminal megakaryocyte differentiation compared to restricting growth of abnormal ΔneoΔHS colonies.
Role of the N terminus.
In contrast to the findings with the V205G mutant, residues in the N terminus are required for control of both megakaryocyte differentiation and growth (Fig. 10a). The region between residues 54 to 110 is required to dampen growth. For differentiation, the situation is more complex. Two domains between residues 1 to 110 are functional in proplatelet and GPIb expression rescue assays. The data are most consistent with two independent N-terminal domains promoting proplatelet release: one between residues 84 to 110 (Fig. 7c, compare constructs 84-413 and 110-413) and one between residues 1 to 84 (Fig. 7c, compare constructs 1-84^ 196-413 and 196-413).
The mechanisms by which N-terminal residues control growth and promote terminal maturation are unclear. They may be required for either critical protein-protein interactions or proper folding of GATA1 protein. To date, interaction with only the critical megakaryocyte transcription factor RUNX1 (24) has been mapped to the N terminus (amino acids 1 to 84) by coimmunoprecipitation assays of transfected proteins in heterologous cell lines (11). However, in that study RUNX1 independently interacted with both the N- and C-terminal GATA1 domains. Given that our nonfunctional N-terminal mutants had an intact C terminus that could have interacted with RUNX1, the importance of GATA1-RUNX1 interaction in mediating N-terminal GATA1-directed megakaryocyte differentiation is unclear. Further studies to characterize the full complement of protein interactions with the GATA1 N-terminal region are in progress (45). Separately, structural studies of GATA1 coupled with identification of functionally inactive N-terminal point mutants will provide insight into the role of the N terminus. Given the conservation of function between mouse and chicken GATA1 in our assays, the relative lack of sequence conservation (Fig. 10b) is perhaps both a surprise and of help in selecting amino acids for further study.
Function of GATA1s in DS TMD and AMKL.
To a large extent, GATA1s (residues 84 to 413) retains the ability to direct terminal megakaryocyte maturation. This is in agreement with expression profiling data of DS AMKL blasts, which have a distinctive mRNA profile composed of many GATA1 megakaryocyte target genes (5). Though GATA1s permits significant megakaryocyte maturation, it is inefficient at restricting the growth of immature megakaryocyte precursors from an abnormal Lin− CD41+ FcγRlow c-kit+ CD9+ progenitor population, consistent with the proliferative phenotype seen in DS TMD. Thus, to some extent, there is an uncoupling of coordinated terminal differentiation and growth arrest.
What implications do these results have for the role of GATA1s in DS TMD and AMKL? First, though our results have been obtained in normal karyotype murine cells rather than human cells with trisomy 21, the proproliferative nature of GATA1s in our assays suggests aspects of GATA1s biology in DS TMD and AMKL could be unraveled in a wild-type mouse model. Second, there are several possible reasons why the TMD clone extinguishes (see reference 1 for discussion), and this study suggests one reason may be that GATA1s permits terminal megakaryocyte differentiation. Third, there has been debate about whether abnormal expansion of immature megakaryocytes in TMD and AMKL is due to simply loss of wild-type GATA1 function or whether GATA1s has unique oncogenic properties. In contrast, our data suggest yet a third hypothesis, namely that GATA1s behaves as a hypomorphic GATA1 allele.
To conclude, we have localized defined, overlapping, but possibly distinct, regions in the GATA1 N terminus required for terminal megakaryocyte differentiation leading to platelet release and control of immature precursor growth. In contrast, though interaction with FOG-1 is strictly required for differentiation, it may be dispensable for growth control. The challenge now is to identify key N-terminal residues and unveil the mechanisms by which these residues restrain growth and facilitate differentiation.
Additional note.
While the manuscript was under review, two other publications have addressed the role of the GATA1 N terminus in regulating murine megakaryocyte terminal differentiation and growth and support many of the findings outlined here (29, 35).
Supplementary Material
Acknowledgments
We thank Jon Frampton and Marella de Bruijn for their advice; Hedia Chagraoui, Boris Guyot, and Marella de Bruijn for reading the manuscript; Isla Hamlett for help with Western blot analysis; Martin Zenke for chicken GATA1 cDNA; and all members of the Vyas and Porcher laboratories for their support.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahmed, M., A. Sternberg, G. Hall, A. Thomas, O. Smith, A. O'Marcaigh, R. Wynn, R. Stevens, M. Addison, D. King, B. Stewart, B. Gibson, I. Roberts, and P. Vyas. 2004. Natural history of GATA1 mutations in Down syndrome. Blood 103:2480-2489. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blobel, G. A., T. Nakajima, R. Eckner, M. Montminy, and S. H. Orkin. 1998. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. USA 95:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blobel, G. A., M. C. Simon, and S. H. Orkin. 1995. Rescue of GATA-1-deficient embryonic stem cells by heterologous GATA-binding proteins. Mol. Cell. Biol. 15:626-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourquin, J.-P., C. Langebrake, A. Subramanian, X. Li, D. Reinhardt, O. Bernard, C. M. Zwaan, H. Hasle, M. Schrappe, T. R. Golub, and S. H. Orkin. 2004. A molecular signature of the transcription gactor GATA1 in acute megakaryoblastic leukemia of childhood. Blood 104S:1117. [Google Scholar]
- 6.Boyes, J., P. Byfield, Y. Nakatani, and V. Ogryzko. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594-598. [DOI] [PubMed] [Google Scholar]
- 7.Chang, A. N., A. B. Cantor, Y. Fujiwara, M. B. Lodish, S. Droho, J. D. Crispino, and S. H. Orkin. 2002. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc. Natl. Acad. Sci. USA 99:9237-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crispino, J. D., M. B. Lodish, J. P. MacKay, and S. H. Orkin. 1999. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell 3:219-228. [DOI] [PubMed] [Google Scholar]
- 9.Crossley, M., M. Merika, and S. H. Orkin. 1995. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15:2448-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darzynkiewicz, Z., F. Traganos, L. Staiano-Coico, J. Kapuscinski, and M. R. Melamed. 1982. Interaction of rhodamine 123 with living cells studied by flow cytometry. Cancer Res. 42:799-806. [PubMed] [Google Scholar]
- 11.Elagib, K. E., F. K. Racke, M. Mogass, R. Khetawat, L. L. Delehanty, and A. N. Goldfarb. 2003. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 101:4333-4341. [DOI] [PubMed] [Google Scholar]
- 12.Evans, T., and G. Felsenfeld. 1989. The erythroid-specific transcription factor eryf1: a new finger protein. Cell 58:877-885. [DOI] [PubMed] [Google Scholar]
- 13.Ezoe, S., I. Matsumura, K. Gale, Y. Satoh, J. Ishikawa, M. Mizuki, S. Takahashi, N. Minegishi, K. Nakajima, M. Yamamoto, T. Enver, and Y. Kanakura. 2005. GATA transcription factors inhibit cytokine-dependent growth and survival of a hematopoietic cell line through the inhibition of STAT3 activity. J. Biol. Chem. 280:13163-13170. [DOI] [PubMed] [Google Scholar]
- 14.Freson, K., K. Devriendt, G. Matthijs, A. Van Hoof, R. De Vos, C. Thys, K. Minner, M. F. Hoylaerts, J. Vermylen, and C. Van Geet. 2001. Platelet characteristics in patients with X-linked macrothrombocytopenia because of a novel GATA1 mutation. Blood 98:85-92. [DOI] [PubMed] [Google Scholar]
- 15.Freson, K., G. Matthijs, C. Thys, P. Marien, M. F. Hoylaerts, J. Vermylen, and C. Van Geet. 2002. Different substitutions at residue D218 of the X-linked transcription factor GATA1 lead to altered clinical severity of macrothrombocytopenia and anemia and are associated with variable skewed X inactivation. Hum. Mol. Genet. 11:147-152. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara, Y., C. P. Browne, K. Cunniff, S. C. Goff, and S. H. Orkin. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 93:12355-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghirlando, R., and C. D. Trainor. 2003. Determinants of GATA-1 binding to DNA: the role of non-finger residues. J. Biol. Chem. 278:45620-45628. [DOI] [PubMed] [Google Scholar]
- 18.Groet, J., S. McElwaine, M. Spinelli, A. Rinaldi, I. Burtscher, C. Mulligan, A. Mensah, S. Cavani, F. Dagna-Bricarelli, G. Basso, F. E. Cotter, and D. Nizetic. 2003. Acquired mutations in GATA1 in neonates with Down's syndrome with transient myeloid disorder. Lancet 361:1617-1620. [DOI] [PubMed] [Google Scholar]
- 19.Gurbuxani, S., P. Vyas, and J. D. Crispino. 2004. Recent insights into the mechanisms of myeloid leukemogenesis in Down syndrome. Blood 103:399-406. [DOI] [PubMed] [Google Scholar]
- 20.Guyot, B., V. Valverde-Garduno, C. Porcher, and P. Vyas. 2004. Deletion of the major GATA1 enhancer HS 1 does not affect eosinophil GATA1 expression and eosinophil differentiation. Blood 104:89-91. [DOI] [PubMed] [Google Scholar]
- 21.Hegyi, E., M. Nakazawa, N. Debili, S. Navarro, A. Katz, J. Breton-Gorius, and W. Vainchenker. 1991. Developmental changes in human megakaryocyte ploidy. Exp. Hematol. 19:87-94. [PubMed] [Google Scholar]
- 22.Heyworth, C., S. Pearson, G. May, and T. Enver. 2002. Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J. 21:3770-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hitzler, J. K., J. Cheung, Y. Li, S. W. Scherer, and A. Zipursky. 2003. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood 101:4301-4304. [DOI] [PubMed] [Google Scholar]
- 24.Ichikawa, M., T. Asai, T. Saito, S. Seo, I. Yamazaki, T. Yamagata, K. Mitani, S. Chiba, S. Ogawa, M. Kurokawa, and H. Hirai. 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 10:299-304. [DOI] [PubMed] [Google Scholar]
- 25.Iwasaki, H., S. Mizuno, R. A. Wells, A. B. Cantor, S. Watanabe, and K. Akashi. 2003. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity 19:451-462. [DOI] [PubMed] [Google Scholar]
- 26.Kowalski, K., R. Czolij, G. F. King, M. Crossley, and J. P. Mackay. 1999. The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG. J. Biomol. NMR 13:249-262. [DOI] [PubMed] [Google Scholar]
- 27.Kulessa, H., J. Frampton, and T. Graf. 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9:1250-1262. [DOI] [PubMed] [Google Scholar]
- 28.Letting, D. L., Y. Y. Chen, C. Rakowski, S. Reedy, and G. A. Blobel. 2004. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc. Natl. Acad. Sci. USA 101:476-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Z., F. J. Godinho, J. H. Klusmann, M. Garriga-Canut, C. Yu, and S. H. Orkin. 2005. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat. Genet. 37:613-619. [DOI] [PubMed] [Google Scholar]
- 30.Liew, C. K., R. J. Simpson, A. H. Kwan, L. A. Crofts, F. E. Loughlin, J. M. Matthews, M. Crossley, and J. P. Mackay. 2005. Zinc fingers as protein recognition motifs: structural basis for the GATA-1/friend of GATA interaction. Proc. Natl. Acad. Sci. USA 102:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, D., and S. Orkin. 1990. Transcriptional activation and DNA-binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4:1886-1898. [DOI] [PubMed] [Google Scholar]
- 32.Mehaffey, M. G., A. L. Newton, M. J. Gandhi, M. Crossley, and J. G. Drachman. 2001. X-linked thrombocytopenia caused by a novel mutation of GATA-1. Blood 98:2681-2688. [DOI] [PubMed] [Google Scholar]
- 33.Merika, M., and S. H. Orkin. 1995. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol. Cell. Biol. 15:2437-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mundschau, G., S. Gurbuxani, A. S. Gamis, M. E. Greene, R. J. Arceci, and J. D. Crispino. 2003. Mutagenesis of GATA1 is an initiating event in Down syndrome leukemogenesis. Blood 101:4298-4300. [DOI] [PubMed] [Google Scholar]
- 35.Muntean, A. G., and J. D. Crispino. 28April2005. Differential requirements for the activation domain and FOG-interaction surface of GATA-1 in megakaryocyte gene expression and development. Blood 10.1182/blood-2005-02-0551. [DOI] [PMC free article] [PubMed]
- 36.Nakorn, T., T. Miyamoto, and I. Weissman. 2003. Characterization of mouse clonogenic megakaryocyte progenitors. Proc. Natl. Acad. Sci. USA 100:205-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nerlov, C., E. Querfurth, H. Kulessa, and T. Graf. 2000. GATA-1 interacts with the myeloid PU. 1 transcription factor and represses PU.1-dependent transcription. Blood 95:2543-2551. [PubMed] [Google Scholar]
- 38.Newton, A., J. Mackay, and M. Crossley. 2001. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J. Biol. Chem. 276:35794-35801. [DOI] [PubMed] [Google Scholar]
- 39.Nichols, K. E., J. D. Crispino, M. Poncz, J. G. White, S. H. Orkin, J. M. Maris, and M. J. Weiss. 2000. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat. Genet. 24:266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ory, D. S., B. A. Neugeboren, and R. C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 93:11400-11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pal, S., A. B. Cantor, K. D. Johnson, T. B. Moran, M. E. Boyer, S. H. Orkin, and E. H. Bresnick. 2004. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc. Natl. Acad. Sci. USA 101:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porcher, C., E. C. Liao, Y. Fujiwara, L. I. Zon, and S. H. Orkin. 1999. Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development 126:4603-4615. [DOI] [PubMed] [Google Scholar]
- 43.Rainis, L., D. Bercovich, S. Strehl, A. Teigler-Schlegel, B. Stark, J. Trka, N. Amariglio, A. Biondi, I. Muler, G. Rechavi, H. Kempski, O. A. Haas, and S. Izraeli. 2003. Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood 102:981-986. [DOI] [PubMed] [Google Scholar]
- 44.Rekhtman, N., F. Radparvar, T. Evans, and A. I. Skoultchi. 1999. Direct interaction of hematopoietic transcription factors PU. 1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13:1398-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez, P., E. Bonte, J. Krijgsveld, K. Kolodziej, B. Guyot, A. Heck, P. Vyas, E. de Boer, F. Grosveld, and J. Strouboulis. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24:2354-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwinger, W., M. Benesch, H. Lackner, R. Kerbl, M. Walcher, and C. Urban. 1999. Comparison of different methods for separation and ex vivo expansion of cord blood progenitor cells. Ann. Hematol. 78:364-370. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu, R., S. Takahashi, K. Ohneda, J. D. Engel, and M. Yamamoto. 2001. In vivo requirements for GATA-1 functional domains during primitive and definitive erythropoiesis. EMBO J. 20:5250-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shivdasani, R. A., Y. Fujiwara, M. A. McDevitt, and S. H. Orkin. 1997. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 16:3965-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi, S., K. Onodera, H. Motohashi, N. Suwabe, N. Hayashi, N. Yanai, Y. Nabesima, and M. Yamamoto. 1997. Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J. Biol. Chem. 272:12611-12615. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi, T., N. Suwabe, P. Dai, M. Yamamoto, S. Ishii, and T. Nakano. 2000. Inhibitory interaction of c-Myb and GATA-1 via transcriptional co-activator CBP. Oncogene 19:134-140. [DOI] [PubMed] [Google Scholar]
- 51.Trainor, C. D., R. Ghirlando, and M. A. Simpson. 2000. GATA zinc finger interactions modulate DNA binding and transactivation. J. Biol. Chem. 275:28157-28166. [DOI] [PubMed] [Google Scholar]
- 52.Traver, D., T. Miyamoto, J. Christensen, J. Iwasaki-Arai, K. Akashi, and I. Weissman. 2001. Fetal liver myelopoiesis occurs through distinct, prospectively isolatable progenitor subsets. Blood 98:627-635. [DOI] [PubMed] [Google Scholar]
- 53.Tsai, F.-Y., G. Keller, F. C. Kuo, M. J. Weiss, J.-Z. Chen, M. Rosenblatt, F. Alt, and S. H. Orkin. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371:221-226. [DOI] [PubMed] [Google Scholar]
- 54.Tsai, S. F., D. I. Martin, L. I. Zon, A. D. D'Andrea, G. G. Wong, and S. H. Orkin. 1989. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature 339:446-451. [DOI] [PubMed] [Google Scholar]
- 55.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujiwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 56.Vainchenker, W., J. Guichard, and J. Breton-Gorius. 1979. Growth of human megakaryocyte colonies in culture from fetal, neonatal, and adult peripheral blood cells: ultrastructural analysis. Blood Cells 5:25-42. [PubMed] [Google Scholar]
- 57.Vakoc, C. R., D. L. Letting, N. Gheldof, T. Sawado, M. A. Bender, M. Groudine, M. J. Weiss, J. Dekker, and G. A. Blobel. 2005. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17:453-462. [DOI] [PubMed] [Google Scholar]
- 58.Villeval, J. L., K. Cohen-Solal, M. Tulliez, S. Giraudier, J. Guichard, S. A. Burstein, E. M. Cramer, W. Vainchenker, and F. Wendling. 1997. High thrombopoietin production by hematopoietic cells induces a fatal myeloproliferative syndrome in mice. Blood 90:4369-4383. [PubMed] [Google Scholar]
- 59.Visvader, J., and J. M. Adams. 1993. Megakaryocytic differentiation induced in 416B myeloid cells by GATA-2 and GATA-3 transgenes or 5-azacytidine is tightly coupled to GATA-1 expression. Blood 82:1493-1501. [PubMed] [Google Scholar]
- 60.Visvader, J. E., M. Crossley, J. Hill, S. H. Orkin, and J. M. Adams. 1995. The C-terminal zinc finger of GATA-1 or GATA-2 is sufficient to induce megakaryocytic differentiation of an early myeloid cell line. Mol. Cell. Biol. 15:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vyas, P., K. Ault, C. W. Jackson, S. H. Orkin, and R. A. Shivdasani. 1999. Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood 93:2867-2875. [PubMed] [Google Scholar]
- 62.Vyas, P., M. A. McDevitt, A. B. Cantor, S. G. Katz, Y. Fujiwara, and S. H. Orkin. 1999. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development 126:2799-2811. [DOI] [PubMed] [Google Scholar]
- 63.Wang, X., J. D. Crispino, D. L. Letting, M. Nakazawa, M. Poncz, and G. A. Blobel. 2002. Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J. 21:5225-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wechsler, J., M. Greene, M. A. McDevitt, J. Anastasi, J. E. Karp, M. M. Le Beau, and J. D. Crispino. 2002. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 32:148-152. [DOI] [PubMed] [Google Scholar]
- 65.Weiss, M. J., G. Keller, and S. H. Orkin. 1994. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1-embryonic stem cells. Genes Dev. 8:1184-1197. [DOI] [PubMed] [Google Scholar]
- 66.Weiss, M. J., and S. H. Orkin. 1995. Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc. Natl. Acad. Sci. USA 92:9623-9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weiss, M. J., C. Yu, and S. H. Orkin. 1997. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 17:1642-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Welch, J. J., J. A. Watts, C. R. Vakoc, Y. Yao, H. Wang, R. C. Hardison, G. A. Blobel, L. A. Chodosh, and M. J. Weiss. 2004. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104:3136-3147. [DOI] [PubMed] [Google Scholar]
- 69.Yang, H. Y., and T. Evans. 1992. Distinct roles for the two cGATA-1 finger domains. Mol. Cell. Biol. 12:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu, C., K. K, Niakan, M. Matsushita, G. Stamatoyannopoulos, S. H. Orkin, and W. H. Raskind. 2002. X-linked thrombocytopenia with thalassemia from a mutation in the amino finger of GATA-1 affecting DNA binding rather than FOG-1 interaction. Blood 100:2040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, P., G. Behre, J. Pan, A. Iwama, N. Wara-Aswapati, H. S. Radomska, P. E. Auron, D. G. Tenen, and Z. Sun. 1999. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU. 1. Proc. Natl. Acad. Sci. USA 96:8705-8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.