Abstract
Matrix attachment region binding proteins have been shown to play an important role in gene regulation by altering chromatin in a stage- and tissue-specific manner. Our previous studies report that SMAR1, a matrix-associated protein, regresses B16-F1-induced tumors in mice. Here we show SMAR1 targets the cyclin D1 promoter, a gene product whose dysregulation is attributed to breast malignancies. Our studies reveal that SMAR1 represses cyclin D1 gene expression, which can be reversed by small interfering RNA specific to SMAR1. We demonstrate that SMAR1 interacts with histone deacetylation complex 1, SIN3, and pocket retinoblastomas to form a multiprotein repressor complex. This interaction is mediated by the SMAR1(160-350) domain. Our data suggest SMAR1 recruits a repressor complex to the cyclin D1 promoter that results in deacetylation of chromatin at that locus, which spreads to a distance of at least the 5 kb studied upstream of the cyclin D1 promoter. Interestingly, we find that the high induction of cyclin D1 in breast cancer cell lines can be correlated to the decreased levels of SMAR1 in these lines. Our results establish the molecular mechanism exhibited by SMAR1 to regulate cyclin D1 by modification of chromatin.
The periodic movement of the cell cycle is orchestrated by programmed oscillations in the activity of cyclins, cyclin-dependent kinases (CDKs), and their target proteins, including pRb and E2F/DP1 complexes (34). In response to the mitogenic stimuli, normal cells exit G1 phase and enter S phase by assembling a D type of cyclins with respective CDK partners. Cyclin D1, a G1-phase cyclin, belongs to a family of three closely related proteins termed cyclins D1, D2, and D3. These proteins are expressed in redundant fashion in all proliferating cells and collectively control cell cycle progression along with CDK4/CDK6. Cyclin D/CDK complexes further phosphorylate retinoblastoma (Rb) protein and release an E2F transcription factor that triggers progression into S phase. The activation of CDKs is dependent on their association with cyclin partners, while inactivation is dependent on CDK inhibitors. A fine control of cyclins and CDK inhibitors is set by both transcriptional and degradation mechanisms.
A complex transcriptional regulatory mechanism has been shown to exist to coordinate the specific temporal profiles of cyclins (38). Previous studies have demonstrated that autoregulatory loops occur between CDKs and their substrate, cyclin D1 (17). Cyclin D1 is induced by several proteins in proliferative signaling and transformations, including Ras, Rac, and Stat5 (27, 41). Elevation of cyclin D1 mRNA in 50 to 70% of breast cancers, while failing to develop normal mammary glands in transgenic mice lacking both cyclin D1 alleles, associates its role in cancer as well as normal breast development (35, 47). Aberrant cyclin D1 expression in the malignancies is attributed to gene amplification, loss of transcriptional control, and stabilization (23, 40).
The molecular mechanisms involved in cyclin D1 downregulation in physiological and pathological processes have begun to be unraveled in recent years. Control of cyclin D1 is shown both at transcriptional and degradation steps. PTEN, a tumor suppressor protein, is frequently inactivated in brain, prostate, and uterine cancers. Upon overexpression, PTEN induces cell cycle arrest by reducing the levels of cyclin D1 and its decreased nuclear availability (31). In earlier reports, the tumor suppressor gene product pRb has been shown to govern the transcriptional events in the proliferating cells. Rb recruits histone deacetylase to E2F and cooperates with histone deacetylase 1 (HDAC1) to repress the E2F-regulated promoter of the gene encoding the cell cycle protein cyclin E (6). Tumor suppressor p53, a major cell cycle regulatory protein, is capable of both activating and repressing transcription in response to various stresses and genotoxic insults (44, 49). A recent report by Rocha et al. (32) shows that upon p53 induction, p52/Bcl3 activator complexes are replaced by p52/HDAC1 repressor complexes, resulting in active repression of cyclin D1 transcription (32). In another report of AT/RT (atypical teratoid and malignant rhabdoid tumors), an aggressive pediatric tumor has been shown to harbor a mutated tumor suppressor gene, INI1/hSNF5 (4). Upon reintroduction of INI1/hSNF5 in a malignant rhabdoid-derived cell line, cells were arrested at G0/G1 stage. Molecular mechanisms of cell cycle arrest correlate with the ability of INI1/hSNF5 to mediate the HDAC1-dependent transcriptional repression of the cyclin D1 gene (48). Thus, overall studies indicate the significance of histone deacetylases for regulating cell cycles by transcriptional repression and modification of the chromatin structure.
SMAR1 has been identified by virtue of its ability to bind to MARβ from a mouse T-cell expression library (8). SMAR1 shows 99% homology with BANP in humans, which has been mapped to the 16q24 locus (5). Dysregulation of the cyclin D1 gene by loss of heterozygosity (LOH) at the 16q24 locus has been well studied in breast and prostate tumors (11). Earlier we reported the interaction of SMAR1 with tumor suppressor p53 and its ability to regress the tumor in vivo (18). Further studies showed that the RS domain of SMAR1 interacts with phosphorylated p53 and stabilizes it in the nucleus (16). SMAR1 has recently been shown to interact with Cux/CDP and repress MARβ-mediated transcription (19, 20). However, the detailed role of SMAR1 in tumor regression, genes targeted by SMAR1, and the mechanism of repression exhibited by SMAR1 remain to be elucidated.
In the present study we analyzed the role of tumor suppressor SMAR1 in regulating the cell cycle and signal transduction. Ectopic expression of SMAR1 revealed the downregulation of D-type cyclins (D1 and D3) followed by downregulation of G1/S transition proteins. In the search for the mechanism exhibited by SMAR1 in repression of cyclin D1, we explored the association of SMAR1 with components of the SIN3 transcriptional corepressor complex, including HDAC1. Using chromatin immunoprecipitation (ChIP) experiments, we demonstrate cyclin D1 as a direct transcriptional target for SMAR1. Further, we report that the cyclin D1 promoter, in SMAR1 overexpressed cells, is enriched in deacetylated histones. SMAR1-mediated repression is abolished upon trichostatin A (TSA) treatment. Since amplification and dysregulation of cyclin D1 is predominantly observed in breast carcinomas, we checked the status of SMAR1 in breast cancer-derived cell lines. In the majority of the breast cancer cell lines used, elevated levels of cyclin D1 were correlated with drastically reduced levels of SMAR1. Endogenous SMAR1 knockdown studies using small interfering RNA (siRNA) increased cyclin D1 expression, which further confirms the cell line data. Our results thus provide a potential mechanism exhibited by SMAR1 in regulating cyclin D1 that, in turn, affects the cell cycle.
MATERIALS AND METHODS
Cell culture and transient transfections.
Breast cancer cell lines MCF-7, HBL-100, ZR75.30, ZR75.1, T-47D, SKBR3, MDA MB-231, and MDA-MB-468 and non-breast cancer cell lines B16F-1 and 293 were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum (Invitrogen) in the presence of 5% CO2 at 37°C. Prior to transfections, cells were seeded at a density of 1 × 106 per 30-mm dish and cultured for 24 h. One microgram of either Flag vector or Flag-SMAR1 plasmid DNA was used for transfection with Lipofectamine 2000, following the manufacturer's protocol (Invitrogen). In the case of overexpression of HDAC1, 1 μg of Flag-HDAC1 was used.
siRNA transfections.
MCF-7 cells were plated at a density of 1 × 106 on a 30-mm dish 24 h before transfection. siRNA specific for SMAR1 and control scrambled siRNA were synthesized from Ambion (SMAR1 siRNA [RS] sense, GCAGAGCAUUGACUCCAAGTT; antisense, CUUGGAGUCAAUGCUCUGCTT; scrambled siRNA sense, UACCGUAGGCAUGCAAAGCTT; antisense, AUGGCAUCCGUACGUUUCGTT; SMAR siRNA [NS] sense, GAGAAGCUAGACCUGGUCATT; antisense, UGACCAGGUCUAGCUUCUCTT). To standardize the effective concentration, various concentrations of SMAR1 siRNA and control siRNA were transfected in MCF-7 cells along with 1 μg of Flag-SMAR1 using siPort reagent following the manufacturer's instructions. Endogenous SMAR1 knockdown studies were carried out in 293 cells.
Luciferase assay.
For luciferase assays, 2 μg of cyclin D1 luciferase (CD1 Luc) or cyclin D1 mutant luciferase (CD1 mut Luc) promoter construct was transfected in 293 cells. One microgram of either Flag-SMAR1 or HDAC1 was cotransfected in cases of overexpression studies. Luciferase assays were performed 24 h posttransfection using a Luclite luminescence reporter gene assay system (Perkin Elmer). Luciferase activity was calculated using a Top count scintillation counter (Packard). Equal amounts of protein (50 μg) were used for luciferase assays, and relative light units were plotted for luciferase activity. For SMAR1 knockdown experiments, 200 nM siRNA (RS) was cotransfected along with the cyclin D1 luciferase plasmid.
Immunoblotting and antibodies.
Cells were scraped, washed with 1× PBS at different time intervals, and lysed in DIGNAM buffer. Protein concentrations were estimated using Bradford reagent (Bio-Rad). Equal amounts of protein were loaded for immunoblotting. Following sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, resolved proteins were electroblotted on polyvinylidene difluoride membranes (Amersham). The membrane was blocked overnight in Tris-buffered saline containing 0.1% Tween-20 (TBST) and 10% bovine serum albumin (BSA). The membrane was then probed with primary antibody in TBST for 2 h, followed by three 10-min TBST washes at room temperature. Incubation with the secondary antibody was done for 1 h, and three 10-min TBST washes were given prior to detection. Proteins were detected using enhanced chemiluminescence substrate (Amersham). Antibodies used in this report were for cyclin D1, cyclin D3, CDK4, p27, pRb s807/811, pStat5 (Cell Signaling), Flag antibody (Sigma), E2F1, cyclin D2, actin, total Rb, HDAC1, p107, p130, Sin3A, Sin3B, and Sin3A/Sin3B (Santa Cruz).
A purified glutathione S-transferase (GST)-fused truncation protein (400 to 548 amino acids) conjugated with appropriate adjuvant was injected in rabbits for SMAR1 antibody. Following three booster doses, the antibody titer was checked and the ascitic fluid was further purified. For antibody purification, recombinant protein A beads (Sigma) were packed onto columns and subsequently purified.
Purification of GST fusion proteins.
GST-SMAR1 as well as GST(160-350) and GST(350-548) truncation clones were grown in Luria-Bertani medium with ampicillin and induced with 1 mM isopropyl-β-d-thiogalactopyranoside. Cells were resuspended in lysis buffer containing phosphate-buffered saline (PBS), Triton X-100, and protease inhibitors (Roche). After sonication, supernatant was incubated with glutathione Sepharose beads for 1 h at 4°C with gentle agitation. After three washes, each with lysis buffer and PBS, the proteins were eluted with 100 mM reduced glutathione buffer. The elute was resolved on gels, and protein bands were visualized by Coomassie staining (see Fig. S1 in the supplemental material).
In vitro binding assays.
Bead-bound GST fusion proteins (3 μg) were incubated with 293 cell lysate (300 μg) in 20 mM phosphate buffer for 10 h at 4°C. After three washes with 20 mM phosphate buffer with 0.1% Triton X-100, bound proteins were eluted in 2× sample buffer and resolved in an SDS polyacrylamide gel, membrane transferred, and probed with various antibodies. For Flag pull-down assays, Flag beads were incubated with 293 cell lysate (300 μg) overexpressing Flag-HDAC1 for 10 h. Beads were washed thrice with the dilution buffer, mixed with [35S]methionine-labeled in vitro-translated protein SMAR1 (400 μg), and incubated for 4 h. Unbound protein fractions were washed with DIGNAM buffer, and bound proteins were eluted with 2× sample buffer. Proteins were resolved on SDS polyacrylamide gels, dried for 1 h, and exposed to X-ray film.
Immunoprecipitations.
Immunoprecipitation of endogenous SMAR1, HDAC1, Sin3A, Sin3B, p107, and p130 was carried out as described below. Two-hundred micrograms of protein was diluted in 1× PBS containing 0.5% NP-40. Lysates were precleared with control immunoglobulin G (IgG) and 20 μl of protein A/G beads for 1 h at 4°C. The precleared lysates were incubated with specific antibody for 12 h and immobilized on protein A/G beads. Beads were washed thrice with 1× PBS containing 0.5% NP-40 and eluted in 2× sample buffer. Endogenous proteins were detected by Western blot analysis.
For two-step coimmunoprecipitation experiments, 300 μg of 293 cell lysate was incubated with SMAR1 antibody and bound to protein A/G beads for 6 h at 4°C. Beads were washed three times with the lysis buffer, and protein complexes were eluted with Laemmli buffer. For a second immunoprecipitation, the eluate from the first immunoprecipitation was diluted in lysis buffer and pulled with HDAC1 antibody or control IgG followed by the addition of protein A/G beads. Detection of SMAR1, Sin3A/Sin3B, and HDAC1 was done upon immunoblotting.
Semiquantitative RT-PCR.
For semiquantitative reverse transcription-PCR (RT-PCR) analysis, total RNA was isolated from cells and treated with RNase-free DNase I (Roche). For each assay, 7 μg of total RNA was subjected to reverse transcription in a 20-μl reaction mixture containing 1× random hexanucleotide mix, 1 mM deoxynucleoside triphosphates, RNase inhibitor, and MuMLV reverse transcriptase (Invitrogen). PCR was carried out in a 25-μl reaction mixture using 1 μl cDNA as template. Gene-specific primers used were SMAR1 (forward [For], GCATTGAGGCCAAGCTGCAAGCTC, and reverse [Rev], CGGAGTTCAGGGTGATGAGTGTGAC), cyclin D1 (For, CTTCCTCTCCAAAATGCCAG, and Rev, AGAGATGGAAGGGGGAAAGA), and β-actin (For, TACCACTGGCATCGTGATGGACT, and Rev, TTTCTGCATCCTGTCGGCAAT). For SMAR1 gene amplification, annealing at 63°C was used with a limiting number of cycles. The cyclin D1 gene was amplified for 30 cycles using a 55°C annealing temperature.
Real-time RT-PCR.
Real-time RT-PCR was performed by an iCycler iQ thermal cycler system (Bio-Rad) using the double-stranded DNA-specific fluorophore SYBR Green. In a 25-μl PCR, 1 μl of cDNA was amplified using 1× iQTN SYBR Green Supermix (Bio-Rad) containing 0.4 mM deoxynucleoside triphosphate mix, 1.5 mM MgCl2, 50 pmol of a forward and reverse primer mix, SYBR Green I, and 0.5 U iTaq DNA polymerase. Cycling parameters for cyclin D1 and SMAR1 were the same as those mentioned for RT-PCR analysis, except that 30 cycles was used for SMAR1. Resolution of the product of interest from nonspecific product amplification was achieved by melt curve analysis. Confirmation of a single product was checked by agarose gel analysis. Quantitation was performed with three different sets of cDNA samples. Graphs were plotted and statistical analysis was performed using Sigma Plot.
Primer used for electrophoretic mobility shift assay (EMSA) and ChIP studies.
Primers used for amplification of various regions of cyclin D1 promoter for EMSA and ChIP studies are the following. Probe I (−60 to −330), which has E2F1 and Sp1 binding site consensuses, was amplified using primers For, TAGAACAGGAAGATCGGAC, and Rev, AGGGCCGCAAACCGCGGGC. Probe II (−330 to −630), which is the SMAR1 binding sequence in the cyclin D1 promoter, was amplified by primers For, TGAAAATGAAAGAAGATGCAGTCG, and Rev, GAAACTTGCACAGGGGTTGT. Probe III (−630 to −1030), the sequence adjacent to the SMAR1 binding site, is amplified by For, CATTGGTGCTTGTGGATAG, and Rev, ATCTTGTCCTTCTAGCCTG. Probe IV, the 50-bp AT-rich MAR-like sequence within probe II region, was obtained as the custom-made oligonucleotide ATTCAATTTACACGTGTTAATGAAAATGAAAGAAGATGCAGTCGCTG and its complementary reverse. Probe V, the mutant of a 50-bp AT-rich MAR-like sequence with is probe II region, was obtained as the custom-made oligonucleotide ATTCAATCTACACGTGTCCGTGGGATGAGGGAAGATGCAGTCGCTG and its complementary reverse. Two kilobases of upstream sequence of probe II was amplified in ChIP using primers for the probe named probe VI (For, TTAAGGGCTTAACAATGGA, and Rev, AGGTGATTTCAGTTAATTT). The AT-rich sequence that was observed upstream of the probe II sequence, called probe VII, was amplified in ChIP by primers using For, TCTTTTTGTTTCCAGCAGTG, and Rev, TCTCTGTCCCCATCTGTAAA.
Electrophoretic mobility shift assay (EMSA).
For EMSA, probes were PCR labeled using [α-32P]dCTP in a 25-μl PCR. Probe I, probe II, and probe III were PCR amplified, and PCR products were then eluted from native polyacrylamide gel by a phenol-chloroform method and subsequent precipitation by 70% ethanol. Oligonucleotide labeling was done by a Klenow reaction using [α-32P]dCTP in a 20-μl reaction containing 1 mM dATG mix, Klenow buffer, and 0.5 U of Klenow (Invitrogen). Probe purification was done using a Probequant G-50 column (Amersham). Binding reactions were performed in a 10-μl total volume containing 10 mM HEPES (pH 7.9), 1 mM dithiothreitol, 50 mM KCl, 2.5 mM MgCl2, 10% glycerol, 0.5 to 1 μg double-stranded poly(dI-dC), 10 μg BSA, and 1 μg of recombinant protein. Samples were incubated for 5 min at room temperature prior to addition of radiolabeled probe. The samples were then incubated for 15 min at room temperature, and the products of binding reactions were resolved by 8% native polyacrylamide gel electrophoresis. The gels were dried under a vacuum and processed for autoradiography.
ChIP analysis.
Assays were performed using a chromatin immunoprecipitation (ChIP) assay kit (Upstate Biotechnology) following the manufacturer's instructions. Cells (1 × 106) were plated per 30-mm dish and transfected with 1 μg of Flag-vector or Flag-SMAR1 in cases of overexpression. For SMAR1 knockdown experiments, 200 nM siRNA (RS) was transfected in 293 cells. DNA-protein interactions were fixed with 1% formaldehyde at 37°C for 10 min at various time points. ChIP assays were carried out using anti-SMAR1, anti-HDAC1, anti-Sin3A, anti-Sin3B, anti-p107, anti-p130, anti-pStat5, and anti-E2F1 antibodies. Input DNA-, rabbit IgG (r-IgG)-, and mouse IgG (m-IgG)-pulled DNA served as controls for all the experiments. DNA immunoprecipitated was then subjected to 25 cycles of PCR using primers for probe II and probe III. Probes VI and VII were amplified for 26 cycles.
Histone deacetylase assay.
Flag vector and Flag-SMAR1 were overexpressed in 293 cells. Flag-SMAR1 and a Flag-associated complex were pulled using Flag antibody. To eliminate nonspecific interactions, pulled complex was washed with 20 mM phosphate buffer containing 0.1% Triton X-100. For the deacetylation assays, core histones were labeled using [3H]acetyl coenzyme A ([3H]acetyl-CoA). Core histones (2 μg) were incubated with 20 ng of p300 and 1 μl of 6.2 Ci/mmol [3H]acetyl-CoA for 15 min at 30°C. The activity of p300 was inhibited by incubating the reaction mixture for 10 min in 10 mM lysyl-CoA, p300-histone acetyltransferase inhibitor (24). The reaction mixture was further incubated with either 50 ng of recombinant HDAC1, Flag-SMAR1 complex, or Flag complex for 45 min. To inhibit HDAC activity, 200 nM TSA was added to the reaction mixture. The samples were then resolved on a 15% polyacrylamide gel and analyzed by fluorography.
RESULTS
Effect of SMAR1 overexpression on G1/S transition molecules.
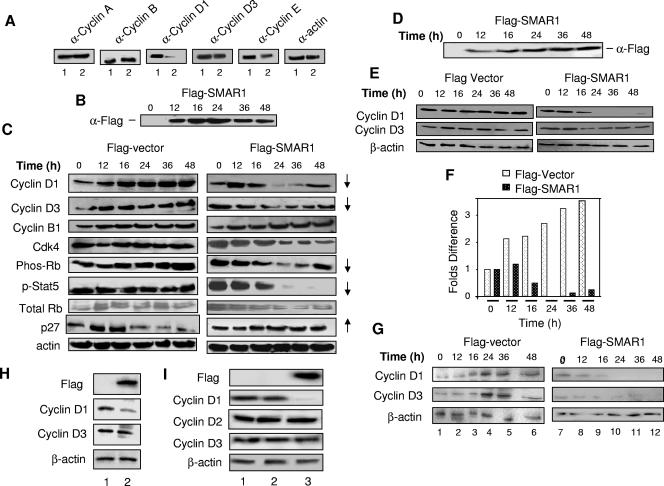
Tumor suppressor proteins have been shown to directly alter cell cycle regulation (48). Earlier we reported that SMAR1 regresses the B16-F1-induced tumor in the mouse model (18). To investigate the role of SMAR1 in regulating the cell cycle, we analyzed the expression levels of different cyclins. Flag-SMAR1 was overexpressed in asynchronous culture of B16-F1 cells and harvested at 24 h. Protein levels of cyclin D1 and cyclin E were downregulated in SMAR1-overexpressed compared to mock-transfected cells. There was no change observed in cyclins A, B1, and D3 (Fig. 1A). A human homolog of SMAR1, BANP, is shown to be located at 16q24, the locus that has gained importance as it harbors at least three breast tumor suppressor proteins. Since SMAR1 downregulated cyclin D1, whose amplification is seen in many breast cancers, we were interested to study the role of SMAR1 in human breast cancer cell lines. Thus, SMAR1 was overexpressed in non-breast cancer (293) and breast cancer (MCF-7, HBL-100, and MDA-MB-468) cell lines. Upon overexpression of SMAR1 in 293 cells, cyclin D1 and D3 downregulation started at 16 h and continued until 36 h. No change in the levels of cyclin B1 was observed. The effect of the decrease of cyclin D1 levels on the cyclin D1/CDK4 complex was verified in the same lysates by checking the phosphorylation status of Rb. As expected, a decrease in the phosphorylation of Rb (pRb s807/811) was observed from 16 to 36 h; however, the total Rb remained unaltered. A reverse pattern of p27/kip1 expression was seen in the case of SMAR1 overexpression. STAT5 transcription factor, under phosphorylated conditions, gets recruited to the cyclin D1 promoter and favors transcription (27). SMAR1 overexpression reduced the phosphorylation of Stat-5 drastically. Considerable downregulation of CDK4 was also noticed in the case of SMAR1 overexpression (Fig. 1C). Overexpression of Flag-SMAR1 in 293 cells was confirmed using Flag antibody (Fig. 1B).
FIG. 1.
SMAR1 inhibits cyclin D1 expression. (A) Expression of various cyclins was checked on mock (lane 1) and Flag-SMAR1 (lane 2) transfections in B16-F1 cells. (B and C) Western blot analysis for time course overexpression of Flag-SMAR1. Expression patterns of various cell cycle regulatory proteins were studied in a time course experiment on overexpression of Flag vector and Flag-SMAR1 in 293 cells. (D and E) Time course overexpression of Flag-SMAR1 and expression profile of cyclins D1 and D3 in MCF-7 cells. (F) Densitometric analysis of cyclin D1 expression in MCF-7 cells. (G) Expression profile of cyclins D1 and D3 in a time course experiment upon overexpression of Flag vector and Flag-SMAR1 in HBL-100 cells. (H) Expression of cyclins D1 and D3 was checked in MDA-MB-468 on mock (lane 1) and Flag-SMAR1 (lane 2) transfections. (I) Expression of cyclins D1, D2, and D3 upon stable expression of SMAR1 in MCF cells (lane 1, cells; lane 2, mock-transfected cells; lane 3, Flag-SMAR1-transfected cells).
In a similar experiment of Flag-SMAR1 overexpression in MCF-7 cells, a fivefold downregulation of cyclin D1 was observed at 16 h, and there was no detectable protein at 24 h (Fig. 1E and F). As indicated, cyclin D3 was also downregulated in MCF-7, but not drastically. Overexpression of SMAR1 was verified in MCF-7 cells by anti-Flag immunoblotting (Fig. 1D). Similar results of cyclin D1 downregulation were seen in HBL-100 cells along with a strong repression of cyclin D3 (Fig. 1G). In the case of MDA-MB-468, cyclin D1 but not cyclin D3 was downregulated (Fig. 1H). Thus, SMAR1 overexpression downregulated cyclin D1 irrespective of cell type. To further verify the effect of SMAR1, stable clones expressing Flag-SMAR1 or Flag vector in MCF-7 cells were checked for expression of cyclin D1. Transfected cells were stably selected using neomycin. Cells stably expressing SMAR1 showed downregulation of cyclin D1, but no change in cyclins D2 and D3 was observed (Fig. 1I). Thus, ectopic expression of SMAR1 downregulates cyclin D1 expression.
Expression profile of SMAR1 in various breast cancer and non-breast cancer cell lines.
Earlier studies using cancer cell lines have reported that the expression of SMAR1 is defective in the majority of cancer cell lines (18). As cyclin D1 expression is dysregulated in a majority of the breast tumors and breast cancer cell lines, we wanted to assess if the upregulation of cyclin D1 is accompanied by downregulation of SMAR1 in breast cancer cell lines.
Transcript analysis revealed that SMAR1 was reduced in MCF-7, HBL-100, MDA-MB-231/468, SK-BR3, and T-47D compared to 293. There was no detectable transcript in ZR75.1 and ZR75.30 (Fig. 2A). Further validation was done using real-time RT-PCR analysis. The graph shown in Fig. 2C was obtained by repeating the experiment thrice with different sets of cDNA samples. SMAR1 was downregulated by 12-fold in the MCF-7 cell line, while 18-fold downregulation was seen in HBL-100; 10- to 7-fold variations in MDA MB-231/468 and 3-fold differences in T-47D cells compared to 293 and B16-F1 were also seen. No amplification was seen in ZR75.1 and ZR75.30. Immunoblotting using SMAR1 antibody showed lowered protein expression in T-47D and SK-BR-3, while protein expression was undetectable in MCF-7, ZR75.1, ZR75.3, HBL-100, MDA-MB-231, and MDA-MB-468. SMAR1 expressing T-47D and SK-BR3 showed low cyclin D1 levels, while a high induction of cyclin D1 was seen in MCF-7, ZR75.1, ZR75.3, HBL-100, and MDA-MB-231/468 that showed drastically reduced levels of SMAR1 (Fig. 2B).
FIG. 2.
Expression of SMAR1 and cyclin D1 in various breast cancer and non-breast cancer cell lines. (A and C) RNA obtained from various cell lines was either undiluted or diluted 10-fold and used for reverse transcription. SMAR1, cyclin D1, and β-actin transcripts were studied using 1 μl of cDNA in RT-PCR and real time RT-PCR analyses (bar a, 293 cells; bar b, B16-F1; bar c, HBL-100; bar d, T-47D; bar e, MCF-7; bar f, MDA-MB-231; bar g, MDA-MB-468). (B) Western blot analysis of cyclin D1 and SMAR1 was performed for various cell lines.
SMAR1 represses cyclin D1 gene expression.
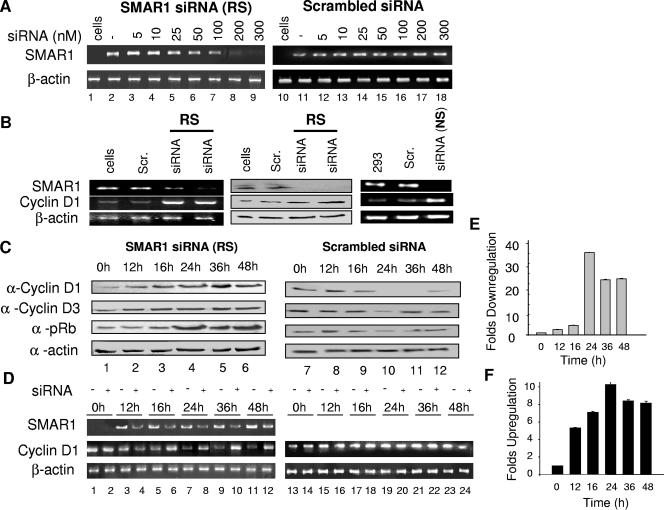
To address the role of SMAR1 in regulation of cyclin D1, we designed siRNAs corresponding to the sequence of SMAR1 as described in Materials and Methods. A scrambled siRNA sequence was used as a negative control. In the presence of 200 nM SMAR1 siRNA (RS), there was a complete knockdown of SMAR1 transcript while no change was found with control siRNA-treated samples (Fig. 3A). To further validate the results, another set of siRNA was used, siRNA (NS), at the effective concentration of 200 nM. The 293 cells were treated with 150 and 200 nM concentrations of SMAR1 siRNA (RS), SMAR1 siRNA (NS), and control siRNA for 24 h. Cells treated with siRNA or control siRNA were harvested to make total RNA and protein lysate. cDNA obtained from total RNA was subjected to PCR amplification for SMAR1, cyclin D1, and β-actin. As shown in Fig. 3B (left and right panels), downregulation of SMAR1 transcript in siRNA-treated samples was correlated with the increased cyclin D1 transcript. Since both the siRNAs targeted to SMAR1 showed increased expression of cyclin D1, further studies were carried out using SMAR1 siRNA (RS). Similarly, protein levels were verified by Western blot analysis (Fig. 3B, middle panel) where an increase in the expression of cyclin D1 was observed. β-Actin was amplified as a control in all the reactions. Thus, the cell line and siRNA data, where a lowered expression of SMAR1 was observed in relation to increased cyclin D1 expression, collectively suggest the role of endogenous SMAR1 in cyclin D1 regulation. We next examined the status of cyclin D1, cyclin D3, and pRb s807/811 upon treatment with SMAR1 siRNA. No inhibition of cyclin D1 and D3 and pRb was observed in SMAR1-overexpressing siRNA-treated cells (Fig. 3C).
FIG. 3.
SMAR1 affects cyclin D1 transcription, and SMAR1 siRNA reverses repression. (A) Optimization of SMAR1 and scrambled siRNA in MCF-7 cells by RT-PCR analysis. (B) Knockdown of endogenous SMAR1 induces cyclin D1 expression. The left-hand panel shows RT-PCR analysis of cyclin D1, SMAR1, and β-actin; in the middle panel protein levels were verified by Western blotting; and in the right-hand panel RT-PCR for SMAR1, cyclin D1, and β-actin was verified using another siRNA (NS). (C) Effect of SMAR1 and scrambled siRNA on cyclin D1, cyclin D3, and pRb protein expression upon overexpression of SMAR1 in MCF-7 cells. (D) MCF-7 cells were transfected with Flag vector (right panel) or Flag-SMAR1 (left panel). Transfected cells were either treated or untreated with SMAR1 siRNA. RNA prepared from various time points was diluted 10-fold, and cDNA obtained was used for PCR analysis of SMAR1, cyclin D1, and β-actin. (E and F) Cyclin D1 transcripts were quantified from the above-obtained cDNA by real-time RT-PCR analysis. Bar graphs represent fold changes in the transcript in the presence or absence of siRNA treatment. Scr., scrambled siRNA.
To verify the transcriptional regulatory effect of SMAR1, a time course experiment overexpressing SMAR1 in MCF-7 cells was done. Downregulation of cyclin D1 transcript was observed from 12 h and peaked at 24 h (Fig. 3D). No detectable product could be seen at 24 h; however, a very small amount of product reappeared at 36 h. To further validate observations of RT-PCR, a real-time RT-PCR analysis was performed. Fortyfold fewer transcripts were observed at 24 h in SMAR1-transfected cells (Fig. 3E). Both RT-PCR and real-time RT-PCR results were normalized using human β-actin.
The cDNAs obtained from siRNA-treated samples were subjected to RT-PCR analysis to monitor the cyclin D1 transcript profile at various time points. Transfection of SMAR1 siRNA in SMAR1-overexpressed cells reversed the silencing effect of the cyclin D1 transcript at different time points. Real-time PCR analysis was performed from the same cDNA that showed restored transcript levels in siRNA-treated samples. In real-time transcript analysis, cyclin D1 mRNA was elevated by 10-fold at 24 h (Fig. 3F). These results suggest that SMAR1 is involved in silencing cyclin D1 transcription upon overexpression in MCF-7 cells.
Mapping of the SMAR1 binding site on the cyclin D1 promoter.
To determine whether the cyclin D1 gene is the direct transcriptional target for SMAR1, we studied in vitro binding of SMAR1 on the human cyclin D1 promoter. The approximately 1-kb region with known putative transcription factor binding sites (the region from −66 to −987) of the cyclin D1 promoter was scanned for the SMAR1 binding site in gel shift assays. Either GST or GST-SMAR1 (1 μg) was used for binding studies with 0.5 and 1 μg of poly(dI-dC), using 10 ng of three radiolabeled probes, I, II, and III, in individual reactions (Fig. 4A). GST-SMAR1 showed a nucleoprotein complex with probe II against the GST control, while no detectable complexes could be seen with probes I and III (Fig. 4C to E). The affinity and specificity of the binding was then documented by cold competition experiments. One-hundred-fold-molar-excess competitor DNA was used for binding reactions. MARβ, a previously known SMAR1 binding sequence, was used as competitor DNA, while a 250-bp fragment obtained from an SK+ vector was used as nonspecific competitor DNA. Unlabeled specific competitor DNA in the binding reaction depleted the complex with GST-SMAR1, while no interference was observed using nonspecific DNA (Fig. 4F). As reported earlier, the DNA binding domain of SMAR1(350-548) showed the band shift with probe II. However, no significant complex formation was detected with the protein-interacting domain SMAR1(160-350) (Fig. 4G).
FIG. 4.
SMAR1 binds to the cyclin D1 promoter. (A) Schematic representation of the cyclin D1 promoter in which arrows indicate the primers used for amplification of probes. The solid box (probe IV) indicates the 50-bp MAR-like sequence in the cyclin D1 promoter. (B) Human (h-) and mouse (m-) MAR-like sequences obtained in the cyclin D1 promoter are shown; small letters indicate the differences in homology. Mutated bases in the oligonucleotide are underlined and in boldface. (C, D, and E) Bindings of GST and GST-SMAR1 were studied on probes I, II, and III. The first lane of each panel represents a free probe. Two lanes for every binding reaction represent 0.5 and 1 μg of poly(dI-dC) as competitor DNA. (F) A nonspecific 250-bp scrambled sequence (lanes 2 and 3) did not interfere with binding, while MARβ (lanes 4 and 5) reduced complex formation with probe II. (G) Domain specificity of SMAR1 truncations in which GST-SMAR1(350-548) (lanes 4 and 5) showed binding to probe II. (H) SMAR1 binds to a 50-bp AT-rich MAR-like region in the cyclin D1 promoter (lanes 2 and 3). MARβ in the competition reaction abolished complex formation (lanes 4 and 5). (I) IgH MAR and a scrambled 50-bp sequence could not compete for binding to probe IV. (J) Domain specificity of SMAR1 for binding to probe IV. (K) SMAR1 failed to bind to probe V (mutant oligonucleotide). In lanes 6 and 7, probe IV was used as a positive control for binding reactions. C, complex; F, free probe; Scr., scrambled siRNA; P-II, probe II.
SMAR1 is a MAR binding protein, and we searched for putative MAR-like sequences in the cyclin D1 promoter. A search using a MAR finder program showed a potential MAR-like sequence in the probe II region (Fig. 4B). We then designed the oligonucleotides for a 50-bp AT-rich sequence for EMSA. Binding reactions were performed using annealed 50-bp double-stranded oligonucleotides with either GST or GST-SMAR1. Interestingly, complex formation was observed with GST-SMAR1 with both 0.5 and 1 μg poly(dI-dC) competitor DNA (Fig. 4H). We plotted the protein saturation curve using 50 ng to 1 μg of GST-SMAR1 protein (see Fig. S2 in the supplemental material). Affinity of GST-SMAR1 to probe IV was documented using a cold probe as a competitor. SMAR1 exhibited high affinity to the 50-bp (probe IV) sequence in vitro (Kd = 0.48 to 1.4 nM) (see Fig. S2 in the supplemental material). Specificity of binding to probe IV (50 bp oligonucleotides) was further checked using another well-known 70-bp IgH MAR and scrambled 50-bp fragments as competitor DNAs. There was inhibition in complex formation upon addition of MARβ DNA; however, the 70-bp IgH MAR and scrambled 50-bp fragments did not compete for binding with the 50-bp cyclin D1 oligonucleotide (Fig. 4H and I). Thus, although SMAR1 is an AT-rich binding protein, it did not show binding to IgH MAR but exhibits primary sequence specificity to the probe II region of the cyclin D1 promoter. As seen for probe II, the DNA binding region of SMAR1(350-548) showed binding to probe IV, while no binding of the protein interacting domain of SMAR1(160-350) was observed with probe IV (Fig. 4J). To analyze the role of AT-rich sequences in SMAR1 binding, ATs were replaced by GCs. Replacement of AT by GC ablated the binding of SMAR1 to the oligonucleotidenucleotide showing the significance of the AT-rich sequence (Fig. 4B and K).
SMAR1 represses transcription from the cyclin D1 promoter.
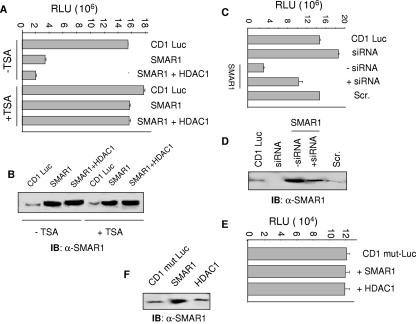
Recent reports on negative regulation of cyclin D1 transcription revealed recruitment of HDAC1 on the cyclin D1 promoter. Thus, we examined if downregulation of the cyclin D1 promoter by SMAR1 occurs by recruitment of corepressor molecules like HDACs. To verify this, the full-length cyclin D1 promoter (from −1745 to +137) with the luciferase reporter gene was transfected in 293 cells. Cells were harvested 24 h posttransfection, a time when cyclin D1 repression occurs, as checked by RT-PCR and Western blot analysis. Overexpression of SMAR1 significantly repressed the transcription by 4.6-fold from the cyclin D1 promoter (Fig. 5A). Cotransfection of SMAR1 and HDAC1 synergistically downregulated the repression by 7.6-fold, while overexpression of HDAC1 alone did not alter cyclin D1 promoter-driven luciferase activity (data not shown). Further, the activity of SMAR1-mediated repression on the cyclin D1 promoter was analyzed upon inhibition of HDAC activity using trichostatin A. SMAR1-mediated cyclin D1 repression was relieved by TSA treatment, strongly suggesting the requirement of HDAC activity for SMAR1-mediated repression of the cyclin D1 promoter. Overexpression of Flag-SMAR1 is shown by Western blotting using the same lysate (Fig. 5B).
FIG. 5.
SMAR1 represses cyclin D1 gene expression in reporter assays. (A) Cyclin D1 promoter activity was checked in 293 cells by luciferase reporter assay. Cotransfections of either Flag-SMAR1 or HDAC1 were done along with cyclin D1 luciferase (CD1 Luc) vector. In the case of TSA treatment (200 nM), cells were treated 24 h posttransfection. Relative light units (RLU) obtained were plotted. (B) SMAR1 expression was shown in the case of each transfection correlated to the bar graph. (C) In the case of siRNA treatment, siRNA was cotransfected along with cyclin D1 Luc vector, and the combination of plasmids is indicated at the top of each bar. Relative luciferase activities were calculated 24 h posttransfection by loading equal amounts of protein. (D) Western blot analysis to show expression of SMAR1 corresponding to each bar in the luciferase assay mentioned above. (E) A cyclin D1 mutant (CD1 mut-Luc) construct was made by deleting the probe II region as described in Materials and Methods. The bar graph represents the effect of SMAR1 overexpression on CD1 mut-Luc 24 h posttransfection. (F) The status of SMAR1 expression in each reaction is shown by SMAR1 Western blotting. IB, immunoblot.
The results of SMAR1-mediated cyclin D1 repression were verified by employing siRNAs specific to SMAR1. As shown in Fig. 5C, endogenous knockdown of SMAR1 increased the transcriptional activity from the cyclin D1 promoter while scrambled siRNA did not affect the transcription. Overexpression of SMAR1 along with siRNA treatment partially rescued the repression that was caused by SMAR1 (1.4-fold repression compared to 4.6-fold repression caused by SMAR1) on the cyclin D1 promoter, suggesting the role of SMAR1 in cyclin D1 promoter downregulation. The level of SMAR1 expression is shown in Fig. 5D.
Data from EMSA studies indicated the significance of the probe II region for binding of SMAR1 on the cyclin D1 promoter, thus, we made a deletion construct in which the probe II region was deleted from the full-length cyclin D1 promoter (cyclin D1 mut luc). Cotransfection of SMAR1 or HDAC1 with the cyclin D1 mut luc construct did not show any downregulation of luciferase activity (Fig. 5E). The level of SMAR1 was monitored using Western blotting (Fig. 5F). These results collectively indicate the significance of SMAR1-mediated repression of the cyclin D1 promoter.
SMAR1 associates with HDAC1, SIN3, and pocket Rbs.
It has been suggested that histone deacetylation could be one of the mechanisms by which repressor proteins (25, 45) mediate transcriptional repression. First we analyzed the effect of TSA in SMAR1-mediated cyclin D1 repression. Flag-SMAR1 was overexpressed in MCF-7 cells following TSA treatment. As seen in Fig. 6A (lane 2), the repressive effect of SMAR1 was eliminated upon TSA treatment. To get further insights into SMAR1-mediated repression, we studied the association of SMAR1 with specific HDACs. In the in vitro interaction studies, GST-SMAR1 was immobilized on GST beads, and 300 μg of 293 cell lysate was incubated for 10 h to check interaction with HDAC1. As shown in Fig. 6B, endogenous HDAC1 interacts with GST-SMAR1. In a similar experiment, overexpressed Flag-HDAC1 was immobilized on Flag beads to which in vitro-translated [35S]methionine-labeled SMAR1 was added and incubated for 4 h. Detection of 35S-labeled SMAR1 in Flag-HDAC1-pulled samples further confirmed the in vitro interaction between SMAR1 and HDAC1 (Fig. 6C). Domain specificity of SMAR1 interaction with HDAC1 was delineated using GST-SMAR1(160-350) and GST-SMAR1(350-548). As shown in Fig. 6B, the protein-interacting domain GST-SMAR1(160-350) showed interaction with HDAC1 while GST-SMAR1(350-548) did not support the interaction.
FIG. 6.
SMAR1 interacts with HDAC1, SIN3, and pocket Rbs. (A) MCF-7 cells transiently transfected with Flag-SMAR1 were treated with TSA 48 h posttransfection. Addition of TSA reversed the repression of cyclin D1 (lane 2). (B) 293 cell extract was incubated with GST, GST-SMAR1, GST(160-350), and GST(400-548) as indicated. The bound fraction from the glutathione Sepharose beads and input cell extract were analyzed for HDAC1 by Western blot analysis. (C) Flag-HDAC1-bound beads were incubated with in vitro-translated 35S-labeled SMAR1. The bound fraction along with the input was detected by autoradiography. (D) Cell extracts from 293 cells overexpressing either Flag-SMAR1, Flag-SMAR1(160-350), or Flag-SMAR1(400-548) were subjected to immunoprecipitation with anti-Flag, anti-HDAC1, or control antibody (preimmune) as indicated for detection of interactions with HDAC1 and Flag-SMAR1, respectively. (E) 293 cell extracts were immunoprecipitated using SMAR1 antibody and control IgG (preimmune) as indicated and were analyzed by HDAC1 antibody for checking the endogenous interaction of SMAR1 and HDAC1. (F) GST pull-down assays for detection of Sin3A/Sin3B were performed by incubating 293 cell extracts to GST-SMAR1 or GST protein. Bound fractions were analyzed by Western blotting as indicated. 293 cell extracts were immunoprecipitated (IP) with SMAR1 antibody or 293 cell extracts overexpressing Flag-SMAR1 were immunoprecipitated with Flag antibody, and they were analyzed for Sin3A/Sin3B by Western blotting. (G) The procedures for the two-step coimmunoprecipitation are outlined in the graphic on the left. 293 cell lysate was used for the first immunoprecipitation using SMAR1 antibody and protein A/G beads. The complex was eluted followed by the second step of coimmunoprecipitation with anti-HDAC1 antibody or control IgG. Protein samples were then analyzed by Western blotting separately with anti-SMAR1, anti-HDAC1, and anti-Sin3A/Sin3B. (H) GST pull-down assays for detection of p107 and p130 were performed by incubating 293 cell lysate with either GST-SMAR1 or GST protein, and the bound fraction was analyzed by Western blotting as indicated. Flag-SMAR1 overexpressing 293 cell lysate was immunoprecipitated by Flag antibody and was analyzed for p107 and p130 by Western blotting. Pre, preimmune; IB, immunoblot.
To analyze whether SMAR1 directly interacts with HDAC1, coimmunoprecipitation studies were done using 293 lysate or 293 lysate overexpressing Flag-SMAR1. As shown in Fig. 6D (IP: α-SMAR1 for endogenous interaction and IP: α-Flag upon overexpression) and E (IP: α-HDAC1 for endogenous interaction), SMAR1 associated with HDAC1 in endogenous as well as in overexpressed conditions. Domain specificity of SMAR1 and HDAC1 interaction was performed by immunoprecipitations upon transfection of Flag-tagged SMAR1 truncations SMAR1(160-350) and SMAR1(350-548). Consistent with GST pulldown assay data, SMAR1(160-350) showed interaction with HDAC1 (Fig. 6D, middle panel).
To assess the corepressor complex associated with SMAR1 and HDAC1, coimmunoprecipitations were carried out using 293 lysate or 293 lysate that overexpresses Flag-SMAR1. SMAR1- or Flag-immunoprecipitated samples revealed the presence of endogenous mSin3A and mSin3B proteins, as also shown in GST pulldown assays (Fig. 6F, IP: α-SMAR1 for endogenous interaction and IP: α-Flag upon overexpression). The specificity of the interaction in every immunoprecipitation is shown by IgG controls (preimmune control). To further examine whether SMAR1, HDAC1, Sin3A, and Sin3B could form the ternary complex, we performed two-step coimmunoprecipitation studies using 293 cell lysate. In the first immunoprecipitation reaction, SMAR1 antibody was used to pull SMAR1, and the associated complex was eluted in Laemmli buffer. The eluate was then immunoprecipitated with either HDAC1 antibody or control IgG, followed by Western blot analysis to detect Sin3A and Sin3B. As shown in Fig. 6G (right panel), Sin3A and Sin3B were present in the final immunoprecipitate but not in the control immunoprecipitate, indicating SMAR1, HDAC1, Sin3A, and Sin3B exist as a complex. These results, along with immunoaffinity purification results (see Fig. S3 in the supplemental material), indicate SMAR1 is associated with the HDAC1/SIN3 complex. Further nuclear colocalization of SMAR1, HDAC1, Sin3A, and Sin3B were visualized using confocal studies (see Fig. S4 in the supplemental material).
Tumor suppressor Rb regulates the transcriptional events important for cell proliferation. Withdrawal of the cell cycle due to inhibition of E2F-regulated genes was observed upon binding of pRb to E2F species. pRb, along with related pocket proteins p107 and p130, utilizes different mechanisms to elicit this effect. GST pull-down assays and immunoprecipitations showed the interaction of SMAR1 with p107 and p130 (Fig. 6H). These results suggest that SMAR1 forms a multiprotein complex by associating with HDAC1, SIN3, and pocket Rbs.
SMAR1 associates with the corepressor complex at the cyclin D1 promoter locus.
To determine if SMAR1 is directly recruited to the cyclin D1 promoter in vivo and its correlation to the HDAC1 recruitment, we performed ChIP assays in 293 cells and MCF-7 cells. As shown in Fig. 7A (left panel), the recruitment of both SMAR1 and HDAC1 along with Sin3A/Sin3B was observed on the probe II region in 293 cells under endogenous conditions, suggesting the occupancy of the cyclin D1 promoter by SMAR1. However, much less amplification of probe II was observed in p107- and p130-immunoprecipitated DNA. In a similar experiment of chromatin immunoprecipitation using SMAR1 and HDAC1 antibody in MCF-7 cells, there was no amplification of the probe II region from immunoprecipitated DNA in endogenous conditions (Fig. 7A, right panel). Lack of probe II amplification in MCF-7 cells in endogenous conditions is attributed to undetectable levels of SMAR1 protein in this cell line (as shown in Fig. 2C). Thus, we overexpressed SMAR1 in MCF-7 cells and studied the recruitment of SMAR1 and the associated HDAC1 protein complex on probe II and probe III regions of the cyclin D1 promoter. As shown in Fig. 7A (right panel), recruitment of SMAR1 and HDAC1 was observed upon overexpression of SMAR1. SMAR1-immunoprecipitated DNA at different time points of transfections from 293 and MCF-7 cells were subjected to PCR amplification using primers designed for probe II and probe III regions. Interestingly, time-dependent recruitment of SMAR1 was observed at the probe II region of the cyclin D1 promoter (Fig. 7B), while there was no amplification of probe III (data not shown), which was correlated to EMSA results. In both the cell lines, binding of SMAR1 peaked at 24 h to 48 h. Since 293 cells express significant levels of SMAR1 protein, we could see the endogenous levels of SMAR1 occupying probe II at 0 h, while no endogenous SMAR1 was observed in MCF-7 (Fig. 7B). These results thus indicated the time-dependent recruitment of SMAR1 (upon SMAR1 overexpression) on the cyclin D1 promoter that peaks at 24 h, which is correlated with drastic downregulation of transcript and protein levels. To check for the functionality of SMAR1 with HDAC1, SIN3, and pocket Rbs on the cyclin D1 promoter, ChIP assays were performed. Immunoprecipitated DNA samples from 293 and MCF-7 at the 24-h time point were subjected to PCR using primers for probe II and probe III. We found specific interaction of HDAC1, Sin3A, Sin3B, p107, and p130 on probe II (Fig. 7C). Amplification of probe III was seen with p107- and p130-pulled fractions, while HDAC1-pulled fractions failed to show amplification. Phospho-Stat and E2F1 transcription factor binding on the cyclin D1 promoter was also studied. A low amplification of probe II was observed in E2F1-pulled DNA, while no amplification could be detected for p-Stat5 (Fig. 7C, left and right panels). As a positive control, input DNA was used while nonspecific amplification was monitored by using mouse IgG-, rabbit IgG-, and no-antibody-pulled DNA samples. These results are consistent with the direct recruitment of HDAC1, SIN3, and pocket Rb complexes by SMAR1 on cyclin D1, which is responsible for the observed repressive effects.
FIG. 7.
SMAR1 recruits the repressor complex at the cyclin D1 promoter. Chromatin immunoprecipitation was performed as described in Materials and Methods. (A) Endogenous recruitment of SMAR1, HDAC1, Sin3A/Sin3B, p107, and p130 on the cyclin D1 promoter probe II and III region was studied in the 293 cell line (left panel). SMAR1 protein is undetectable in MCF-7 cells, thus ChIP for recruitment of SMAR1 on the probe II and III region was studied in untransfected MCF-7 (0 h) or cells transfected (24 h) with Flag-SMAR1 (right panel); two lanes for each time point indicate two different concentrations of templates (1 and 0.5 μl, respectively) used for amplification of immunoprecipitated DNA. (B) Recruitment of SMAR1 on the cyclin D1 promoter upon overexpression of Flag-SMAR1 in a time course experiment from the time of transfection was studied in 293 and MCF-7 cells (left and right panels, respectively). Chromatin was immunoprecipitated at various time points, and pulled DNA was amplified using primers for probe II. Densitometric analysis for SMAR1 recruitment in each cell line is represented in the bar graph. (C) 293 and MCF-7 cells were transfected with 1 μg Flag-SMAR1, and cells were cross-linked 24 h posttransfection. ChIP assay using various antibodies against the proteins associated with SMAR1 was performed in 293 and MCF-7 cells (left and right panels, respectively), and immunoprecipitated DNA was amplified using primers for probe II and probe III. For every PCR, 1 and 0.5 μl of the template was used for amplification. Ab, antibody.
SMAR1-associated HDAC1 deacetylates histones in vitro.
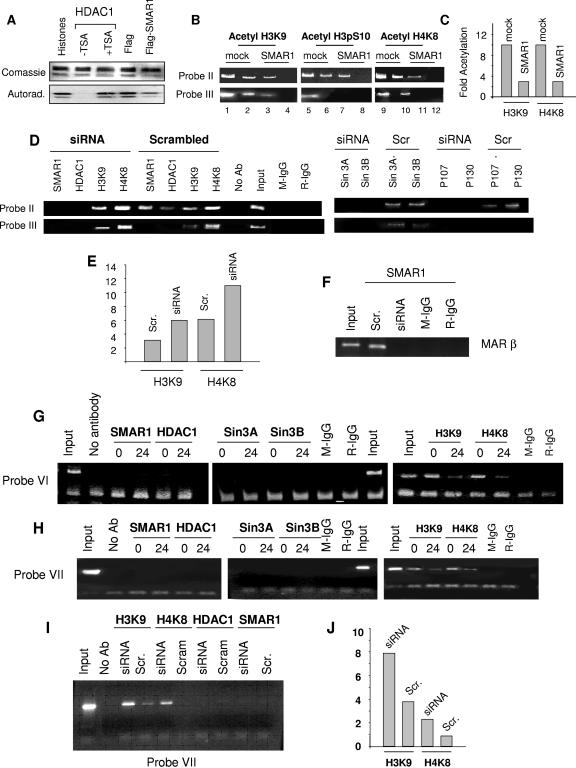
Since our results demonstrate SMAR1 interaction with HDAC1 on the cyclin D1 promoter, we wanted to analyze the functionality of this association. Samples overexpressing Flag-SMAR1 were subjected to Flag pull-down assays and directly assayed for HDAC1 activity. Labeled histones were incubated with either recombinant HDAC1 or Flag-pulled samples. Upon fluorography, we observed significant HDAC activity associated with the Flag-SMAR1-pulled complex (Fig. 8A). A positive control using recombinant HDAC1 showed strong deacetylase activity. However, in a reaction of recombinant HDAC1 along with TSA treatment, we did not observe deacetylase activity. The ability of SMAR1-associated complex to deacetylate the histones suggested a possible role of SMAR1 in deacetylating the cyclin D1 promoter.
FIG. 8.
Histone modifications at the cyclin D1 promoter locus. (A) HDAC activity was determined in Flag-immunoprecipitated fractions obtained from Flag or Flag-SMAR1 overexpressed 293 cells upon incubation with labeled histones. (B) In ChIP assays, chromatin was immunoprecipitated by using antibodies against H3K9, H4K8, and H3 p-Ser-10. The probe II and probe III region of the cyclin D1 promoter was amplified from pulled chromatin. For every PCR, 1 and 0.5 μl of the template was used for amplification. (C) Densitometric analysis of the acetylation status of the cyclin D1 promoter locus (probe II). (D) Endogenous SMAR1 was depleted using SMAR1 siRNA (RS) and studied for recruitment of SMAR1, HDAC1, Sin3A/SinB, p107, and p130. Further histone modifications were studied upon depletion of SMAR1 as indicated using the probe II and probe III region of the cyclin D1 promoter. (E) Densitometric analysis of the acetylation status of the cyclin D1 promoter locus (probe II) upon depletion of endogenous SMAR1. (F) MAR β was amplified as the positive control from SMAR1-pulled samples in siRNA (RS) and scrambled siRNA-treated cells. No amplification of MARβ in siRNA (RS)-treated cells further confirms the depletion of SMAR1. (G) An additional SMAR1 binding site and associated repressor molecule binding were checked on the probe VI region of the cyclin D1 promoter in 293 cells (0 h) and 293 cells transfected with Flag-SMAR1 (24 h) (left and middle panel, respectively), while the status of histone modifications on probe VI were studied in 293 cells (0 h) and cells overexpressing Flag-SMAR1 (24 h) (left panel). (H) An additional SMAR1 binding site and associated repressor molecule binding were checked on the probe VII (a MAR-like region 5 kb upstream) region of the cyclin D1 promoter in 293 cells (0 h) and 293 cells transfected with Flag-SMAR1 (24 h) (left and middle panels, respectively), while the status of histone modifications on probe VI were studied in 293 cells (0 h) and cells overexpressing Flag-SMAR1 (24 h). Histone modifications were studied at the probe VII (5 kb upstream) region of the cyclin D1 promoter in 293 cells (0 h) and 293 cells transfected with Flag-SMAR1 (24 h) (left panel). (I) Histone modifications and recruitment of SMAR1 and HDAC1 were checked at probe VII (5 kb upstream) of the cyclin D1 promoter upon depletion of endogenous SMAR1 from 293 cells. (J) Densitometric analysis of acetylation status of the cyclin D1 promoter locus (probe VII) upon depletion of endogenous SMAR1 from 293 cells. Autorad., autoradiography; Scr, scrambled siRNA.
SMAR1-recruited complex deacetylates histones in vivo.
Since SMAR1-associated complex deacetylated the acetylated histones, we monitored the acetylation status of the cyclin D1 promoter by ChIP analysis using acetylated H3K9 and H4K8 antibodies. We observed that probe II chromatin was acetylated both at H3K9 and H4K8 in mock- compared to SMAR1-transfected cells, and the ratio of deacetylation varied by threefold at 24 h (Fig. 8B and C). H3 Ser-10 has been shown to be phosphorylated upon K9 acetylation in active chromatin. In SMAR1-overexpressed cells, the phosphorylation status was reduced by threefold at 24 h (Fig. 8B, middle panel). To check whether this effect was restricted to probe II, where SMAR1 and HDAC1 are recruited, the probe III region was also analyzed (Fig. 8B and C). A similar status of deacetylation was observed with probe III, suggesting deacetylation is not restricted to the probe II region.
Endogenous depletion of SMAR1 increased acetylation of histones at the cyclin D1 promoter.
Since the expression of cyclin D1 was increased in 293 cells upon knockdown of endogenous SMAR1, we further verified the endogenous SMAR1-mediated recruitment of HDAC1, SIN3, p107, and p130 to the cyclin D1 promoter by using siRNA specific for SMAR1. We depleted the endogenous SMAR1 from 293 cells using siRNA. As shown in Fig. 8D, we do not observe either SMAR1, HDAC1, Sin3A/Sin3B, p107, or p130 recruitment on probe II upon siRNA treatment, while cells treated with scrambled siRNA showed recruitment of both SMAR1 and HDAC1 on the probe II region. The SMAR1 binding site MARβ was amplified from the same set of samples that served as positive controls (Fig. 8F). We further analyzed the status of histone acetylation on the cyclin D1 promoter in siRNA-treated and -untreated cells using acetyl-H3K9 and -H4K9 antibodies. Acetylation of H3K9 and H4K8 was increased by twofold in siRNA treatment compared to the scrambled fragments on the probe II and III region of the cyclin D1 promoter, indicating the reversal of deacetylation mediated by endogenous SMAR1 (Fig. 8D and E).
SMAR1 directs the histone modifications at a distance.
After examining the role of SMAR1 in histone modifications at the cyclin D1 promoter region (both on probes II and III), we next analyzed whether SMAR1 directs the chromatin remodeling at a distance. Our studies mapped SMAR1 binding to a MAR-like consensus (probe II) in the cyclin D1 promoter region, thus, we searched for a putative MAR consensus 10 kb upstream of the cyclin D1 promoter. Although the EMSA studies have shown that SMAR1 specifically binds and exhibits high affinity (Kd = 0.48 to 1.4 nM) to probe IV in the cyclin D1 promoter, we further studied whether SMAR1 has any additional binding site on the MAR-like consensus observed in the nearby region of the cyclin D1 promoter. We designed a set of primers for ChIP studies of 3 kb, where there was no MAR consensus, and of the 5 kb upstream of the promoter, where we observed a MAR-like region (designated probes VI and VII, respectively). Chromatin immunoprecipitation using SMAR1 antibody in 293 cells revealed no amplification of the probe VI and VII region, suggesting that SMAR1 does not have an additional binding site and specifically binds to probe II (Fig. 8G and H, left panels). ChIP was done using three sets of primers spanning 1-kb regions around the MAR stretch observed, and the results were consistent with the probe VII data represented in Fig. 8H. We then studied the recruitment of HDAC1, Sin3A, and Sin3B in the probe VI and VII region. As shown in Fig. 8G and H (middle panels), we did not observe recruitment of any of the above-mentioned factors.
Since overexpression of SMAR1 deacetylated the histones in the probe II and III region and depletion of SMAR1 increased acetylation in this region, we studied whether SMAR1 controls the histone acetylation status at a distance. Therefore, we immunoprecipitated chromatin from 293 cells and 293 cells overexpressing SMAR1 (24 h) using acetyl-H3K9 and acetyl-H4K8 antibodies. Decreased amplification of the probe VI and VII region in SMAR1-overexpressing cells indicated deacetylation of the probe VI and VII region compared to 293 cells alone (Fig. 8G and H, respectively, right panels).
To further prove the role of SMAR1 in modifying histones, at a 5-kb distance ChIP assays were performed upon depletion of endogenous SMAR1. Depletion of endogenous SMAR1 in 293 cells showed an increased amplification of the probe VII region by twofold, reflecting an increased acetylation. These results suggest that the SMAR1-mediated repressive effect spreads over at least the 5-kb region that has been studied (Fig. 8I and J).
DISCUSSION
Progression of normal cells into malignant ones involves the genetic alteration of several classes of genes, including proto-oncogenes, tumor suppressor genes, and DNA damage repair genes (33). Alteration of any one of these genes results in loss of control on cell cycle restriction points, and hence uncontrolled proliferation occurs. Cyclin D1 (bcl1 or PRAD-1), a G1 cyclin, is a proto-oncogene product that is overexpressed or amplified in numerous malignancies (3, 39). Unlike other cyclins, the expression of cyclin D1 is regulated by transcriptional induction and translational and posttranscriptional modification (10, 13). In the present study, we report tumor suppressor SMAR1 as a transcriptional regulator of the cyclin D1 gene.
In earlier studies (18), we provided evidence of tumor suppressor function of SMAR1 in B16-F1-induced melanoma and its repressor function by interaction with Cux/CDP. However, the mechanism by which SMAR1 exhibits tumor suppressor and repressor functions is unknown. To understand SMAR1-mediated cell cycle regulation, we overexpressed SMAR1 in an asynchronous population of B16-F1 cells and checked for the status of various cyclins. A drastic downregulation of cyclin D1 and lowered levels of cyclins D3 and E was also observed. In this report we demonstrate that SMAR1 represses cyclin D1 gene expression by acting as a transcriptional repressor. As a consequence of reduction of cyclin D1 and CDK4, we have shown that pRb is dephosphorylated specifically at the residues that are phosphorylated by the cyclin D/CDK4 complex but not cyclin E or the cyclin A/CDK4 complex. Mitogen-activated protein kinase and Ras-mediated induction of cyclin D1 transcription occurs through binding of p-Stat to the promoter (27). In assignment to downregulation of cyclin D1, we find that the phosphorylation status of Stat5 is also downregulated upon overexpression of SMAR1. To study whether the effect of SMAR1 on cyclin D1 expression is cell type specific, we screened cell types of different origin. Consistent with the B16-F1 cell line data, SMAR1 downregulated cyclin D1 in 293, MCF-7, MDA-MB-468, and HBL-100 cell lines upon transient and stable transfection. Cyclin D3 was also downregulated in 293, MCF-7, and HBL-100. However, there was no effect in B16-F1 and MDA-MB-468 cell lines exhibiting cell type specificity. Cell cycle analysis upon overexpression of SMAR1 in MCF-7 cells showed G1/S arrest (see Fig. S5 in the supplemental material) that suggest its effect on the cell cycle.
Existing data on the MAR-associated proteins in relation to tumor regression suggest their repressor role in breast tumors. MAR binding proteins SAFB1 and SAFB2 are implicated as repressors of estrogen receptors in breast cancers. Similarly, HET is also shown to be a repressor of hsp27 in breast cancers (29, 37). Since SMAR1 showed repression of the cyclin D1 gene and cyclin D1 dysregulation is implicated in breast tumors, further studies were carried out in breast cancer lines.
SMAR1 is a DNA binding protein, and the major mechanism by which it exhibits its tumor suppression function is probably by regulating the transcription of cyclin D1. In agreement with this hypothesis, we found downregulation of cyclin D1 transcript upon overexpression of SMAR1 in a time course experiment. Depletion of SMAR1 by siRNA reverted the silencing effect, confirming the role of SMAR1 in cyclin D1 repression. EMSA studies further prove that SMAR1 acts as a repressor by directly binding to a MAR consensus in the cyclin D1 promoter. Since cyclin D1 downregulation was seen in mouse and human cell lines, MAR consensus homology for mouse and human cyclin D1 promoters was calculated. Although the entire promoter sequence is not conserved, 93% homology was seen in the MAR-like region. Thus, we presume that a common mechanism is exhibited by SMAR1 in repressing the cyclin D1 promoter in mouse and human cell lines.
Matrix-associated proteins have been shown to act as either transcriptional repressors or activators (7, 15). It has been suggested that repressor proteins like Cux/CDP and SATB1 exist in complex with HDACs in case of transcriptional repression (25, 45). At least three different kinds of repressor complexes are shown to exist, namely, SIN3, Mi2-NURD, and CoREST (1, 2, 14, 46). The repressor matrix attachment region binding protein SATB1 has been shown to interact with the SIN3/NURD complex to remodel the interleukin-2-Rα promoter locus (45). We checked for the association of SMAR1 with HDAC1 and associated complex proteins, and our results suggest interaction of SMAR1 with HDAC1 and SIN3. In this report we also show that SMAR1 interacts with HDAC1 through SMAR1(160-350), and the promoter binding domain is restricted to SMAR1(350-548). A deacetylase assay using SMAR1 further confirms functionality of SMAR1-associated repressor complexes.
pRb was shown to control expression of many cell cycle regulatory genes (6). Binding of pRb to E2F along with HDAC and pocket Rbs is well documented (12, 26). SMAR1 overexpression reduced the phosphorylation status of pRb, indicating the SMAR1-associated repressor complex might involve Rb-related proteins. In accordance with the existing literature, we found the involvement of pocket Rb proteins p130 and p107 in the SMAR1-associated repressor complex. Although we observed the interaction of pocket Rb proteins in the SMAR1-associated complex, Western analysis data from an Rb-null cell line (MDA-MB-468) and Rb-inactive cell lines (293 and HBL-100) suggest that the Rb is dispensable for SMAR1-mediated cyclin D1 repression.
A body of experimental data indicates that HDACs essentially repress transcription through deacetylation of histone tails, resulting in local modification of chromatin structure (42, 43). Regulation of gene expression towards activation or inhibition appears to correlate with the differential status of H3 and H4 acetylation (22, 36). Similarly, acetylation of H3 is directly correlated with phosphorylation of H3 p-Ser-10 (9, 28). To address this issue, we first confirmed the recruitment of the SMAR1-associated repressor complex on the cyclin D1 promoter. We found the deacetylation of H3 and H4 over the 5-kb sequence upstream of the cyclin D1 promoter region studied, indicating that the SMAR1-recruited complex on cyclin D1 represses the transcription by deacetylating histones at the promoter locus. Increases in the acetylation status of histones over the cyclin D1 promoter and the 5-kb upstream sequence upon endogenous SMAR1 depletion by siRNA treatment further confirm the role of SMAR1 in modulating the chromatin at that locus. Reduction of phosphorylation of H3 p-Ser-10 is explainable as it is directly correlated with H3K9 status. Earlier, SATB1 was shown to regulate chromatin structure and histone modifications over long distances (7). Although we have not studied the interaction of SMAR1 with chromatin remodeling complexes like the NURD complex, our data showing modification of histones over the range of the 5 kb studied without additional SMAR1 binding sites on the cyclin D1 promoter suggest the possible involvement of remodeling complexes. Taken together, SMAR1 can tether HDAC1 and associated complexes on the cyclin D1 promoter. We investigated if SMAR1 uses the deacetylase activity to mediate repression and its sensitivity to TSA. The abrogation of SMAR1-mediated deacetylation of H3 and H4 upon treatment with TSA further confirms the requirement of HDAC1 for chromatin modulation at the promoter locus (data not shown).
Our work has addressed the signaling pathway upon SMAR1 overexpression and the mechanism of repression mediated by SMAR1 to achieve this function. Since our data suggest SMAR1 as a cyclin D1 regulatory protein and cyclin D1 is overexpressed in breast cancers, we verified the levels of these proteins in breast cancer cell lines. Elevated levels of cyclin D1 can be reasoned as a consequence of reduced levels of SMAR1 in breast cancer cell lines, and SMAR1 knock-out experiments using siRNA-increased cyclin D1 expression suggest the same. Binding of endogenous SMAR1 from 293 cells to probe II in supershift experiments (see Fig. S5 in the supplemental material) and occupancy of the cyclin D1 promoter by SMAR1 and HDAC1 in ChIP assays further support the hypothesis, as we do not see any binding of SMAR1 or recruitment in MCF-7 compared to 293 in endogenous conditions. This is attributed to 12-fold less transcript levels of SMAR1 in MCF-7 and no detectable amount of the protein. The human homolog of SMAR1, BANP, is located at the 16q24 locus. At least three different genes located at this locus have been shown to play an important role in controlling breast cancer (21, 30). A loss of heterozygosity at this region is implicated in breast cancers. However, we did not find correlation of SMAR1 with respect to LOH and non-LOH cell lines. Our finding reports two cell lines, ZR75.1 and ZR75.30, having LOH that do not express SMAR1 and are null for SMAR1. Thus, our studies demonstrate tumor suppressor SMAR1 as a candidate repressor protein controlling cyclin D1 gene expression, and we hypothesize that reduced levels of SMAR1 in breast cancer cell lines might be one of the reasons for dysregulation of cyclin D1.
Supplementary Material
Acknowledgments
We thank R. G. Pestell, Lombardi Comprehensive Cancer Research Centre, for kindly providing us the cyclin D1 promoter-luciferase construct. We thank S. L. Schreiber and Bradely Bernstein, Howard Hughes Medical Institute, Cambridge, Mass., for providing the Flag-HDAC1 construct. We also thank G. C. Mishra, Director, NCCS, for providing us all support.
S.R. is a Senior Research Fellow of CSIR, Government of India. This work was supported by grants from the Department of Biotechnology (DBT) and the Department of Science and Technology (DST), New Delhi, India.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahringer, J. 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16:351-356. [DOI] [PubMed] [Google Scholar]
- 2.Andres, M. E., C. Burger, M. J. Peral-Rubio, E. Battaglioli, M. E. Anderson, J. Grimes, J. Dallman, N. Ballas, and G. Mandel. 1999. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 96:9873-9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartkova, J., J. Lukas, M. Strauss, and J. Bartek. 1995. Cyclin D1 oncoprotein aberrantly accumulates in malignancies of diverse histogenesis. Oncogene 10:775-778. [PubMed] [Google Scholar]
- 4.Biegel, J. A., J. Y. Zhou, L. B. Rorke, C. Stenstrom, L. M. Wainwright, and B. Fogelgren. 1999. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 59:74-79. [PubMed] [Google Scholar]
- 5.Birot, A., L. Duret, L. Bartholin, B. Santalucia, I. Tigaud, J. Magaud, and J. Rouault. 2000. Identification and molecular analysis of BANP. Gene 253:189-196. [DOI] [PubMed] [Google Scholar]
- 6.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 7.Cai, S., H. J. Han, and T. Kohwi-Shigematsu. 2003. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34:42-51. [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhyay, S., R. Kaul, A. Charest, D. Housman, and J. Chen. 2000. SMAR1, a novel, alternatively spliced gene product, binds the scaffold/matrix-associated region at the T cell receptor beta locus. Genomics 68:93-96. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905-915. [DOI] [PubMed] [Google Scholar]
- 10.Choi, Y. H., S. J. Lee, P. Nguyen, J. S. Jang, J. Le, M. L. Wu, E. Takano, M. Maki, P. A. Henkart, and J. B. Trepel. 1997. Regulation of cyclin D1 by calpain protease. J. Biol. Chem. 272:28479-28484. [DOI] [PubMed] [Google Scholar]
- 11.Cleton-Jansen, A. M., D. F. Callen, R. Seshadri, S. Goldup, B. Mccallum, J. Crawford, J. A. Powell, C. Settasatian, H. van Beerendonk, E. W. Moerland, V. T. Smit, W. H. Harris, R. Millis, N. V. Morgan, D. Barnes, C. G. Mathew, and C. J. Cornelisse. 2001. Loss of heterozygosity mapping at chromosome arm 16q in 712 breast tumors reveals factors that influence delineation of candidate regions. Cancer Res. 61:1171-1177. [PubMed] [Google Scholar]
- 12.Corbeil, H. B., and P. E. Branton. 1997. Characterization of an E2F-p130 complex formed during growth arrest. Oncogene 15:657-668. [DOI] [PubMed] [Google Scholar]
- 13.Diehl, J. A., M. Cheng, M. F. Roussel, and C. J. Sherr. 1998. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12:3499-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guschin, D., P. A. Wade, N. Kikyo, and A. P. Wolffe. 2000. ATP-dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry 39:5238-5245. [DOI] [PubMed] [Google Scholar]
- 15.Herrscher, R. F., M. H. Kaplan, D. L. Lelsz, C. Das, R. Scheuermann, and P. W. Tucker. 1995. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 9:3067-3082. [DOI] [PubMed] [Google Scholar]
- 16.Jalota, A., K. Singh, L. Pavithra, R. Kaul-Ghanekar, S. Jameel, and S. Chattopadhyay. 2005. Tumor suppressor SMAR1 activates and stabilizes p53 through its arginine-serine-rich motif. J. Biol. Chem. 280:16019-16029. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, D. G., K. Ohtani, and J. R. Nevins. 1994. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 8:1514-1525. [DOI] [PubMed] [Google Scholar]
- 18.Kaul, R., S. Mukherjee, F. Ahmed, M. K. Bhat, R. Chhipa, S. Galande, and S. Chattopadhyay. 2003. Direct interaction with and activation of p53 by SMAR1 retards cell-cycle progression at G2/M phase and delays tumor growth in mice. Int. J. Cancer. 103:606-615. [DOI] [PubMed] [Google Scholar]
- 19.Kaul-Ghanekar, R., S. Jalota, L. Pavithra, P. Tucker, and S. Chattopadhyay. 2004. SMAR1 and Cux/CDP modulate chromatin and act as negative regulators of TCR beta enhancer (E beta). Nucleic Acids Res. 16:4862-4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaul-Ghanekar, R., S. Majumdar, A. Jalota, N. Gulati, N. Dubey, B. Saha, and S. Chattopadhyay. 2005. Abnormal V(D)J recombination of T cell receptor β locus in SMAR1-transgenic mice. J. Biol. Chem. 10:1074. [DOI] [PubMed] [Google Scholar]
- 21.Kochetkova, M., O. L. McKenzie, A. J. Bais, J. M. Martin, G. A. Secker, R. Seshadri, J. A. Powell, S. J. Hinze, A. E. Gardner, H. E. Spendlove, N. J. O'Callaghan, A. M. Cleton-Jansen, C. Cornelisse, S. A. Whitmore, J. Crawford, G. Kremmidiotis, G. R. Sutherland, and D. F. Callen. 2002. CBFA2T3 (MTG16) is a putative breast tumor suppressor gene from the breast cancer loss of heterozygosity region at 16q24.3. Cancer Res. 62:4599-4604. [PubMed] [Google Scholar]
- 22.Kouzarides, T. 1999. Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev. 9:40-48. [DOI] [PubMed] [Google Scholar]
- 23.Lamb, J., S. Ramaswamy, H. L. Ford, B. Contreras, R. V. Martinez, F. S. Kittrell, C. A. Zahnow, N. Patterson, T. R. Golub, and M. E. Ewen. 2003. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell 114:323-334. [DOI] [PubMed] [Google Scholar]
- 24.Lau, O. D., T. K. Kundu, R. E. Soccio, S. Ait-Si-Ali, E. M. Khalil, A. Vassilev, A. P. Wolffe, Y. Nakatani, R. G. Roeder, and P. A. Cole. 2000. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell 5:589-595. [DOI] [PubMed] [Google Scholar]
- 25.Li, S., L. Moy, N. Pittman, G. Shue, B. Aufiero, E. J. Neufeld, N. S. LeLeiko, and M. J. Walsh. 1999. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J. Biol. Chem. 274:7803-7815. [DOI] [PubMed] [Google Scholar]
- 26.Luo, Q., J. Li, B. Cenkci, and L. Kretzner. 2004. Autorepression of c-myc requires both initiator and E2F-binding site elements and cooperation with the p107 gene product. Oncogene 23:1088-1097. [DOI] [PubMed] [Google Scholar]
- 27.Matsumura, T., H. Kitamura, H. Wakao, H. Tanaka, K. Hashimoto, C. Albanese, J. Downward, R. G. Pestell, and Y. Kanakura. 1999. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 18:1367-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowak, J., and V. G. Corces. 2000. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes Dev. 14:3003-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oesterreich, S. 2003. Scaffold attachment factors SAFB1 and SAFB2: innocent bystanders or critical players in breast tumorigenesis? J. Cell. Biochem. 90:653-661. [DOI] [PubMed] [Google Scholar]
- 30.Powell, J. A., A. E. Gardner, A. J. Bais, S. J. Hinze, E. Baker, S. Whitmore, J. Crawford, M. Kochetkova, H. E. Spendlove, N. A. Doggett, G. R. Sutherland, D. F. Callen, and G. Kremmidiotis. 2002. Sequencing, transcript identification, and quantitative gene expression profiling in the breast cancer loss of heterozygosity region 16q24.3 reveal three potential tumor-suppressor genes. Genomics 80:303-310. [DOI] [PubMed] [Google Scholar]
- 31.Radu, A., V. Neubauer, T. Akagi, H. Hanafusa, and M. M. Georgescu. 2003. PTEN induces cell cycle arrest by decreasing the level and nuclear localization of cyclin D1. Mol. Cell. Biol. 23:6139-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocha, S., A. M. Martin, D. W. Meek, and N. D. Perkins. 2003. p53 represses cyclin D1 transcription through downregulation of Bcl-3 and inducing increased association of the p52 NF-κB subunit with histone deacetylase 1. Mol. Cell. Biol. 23:4713-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 34.Sherr, C. J. 2000. Cell cycle control and cancer. Harvey Lect. 96:73-92. [PubMed] [Google Scholar]
- 35.Sicinski, P., J. L. Donaher, S. B. Parker, T. Li, A. Fazeli, H. Gardner, S. Z. Haslam, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621-630. [DOI] [PubMed] [Google Scholar]
- 36.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 37.Townson, S. M., T. Sullivan, Q. Zhang, G. M. Clark, C. K. Osborne, A. V. Lee, and S. Oesterreich. 2000. HET/SAF-B overexpression causes growth arrest and multinuclearity and is associated with aneuploidy in human breast cancer. Clin. Cancer Res. 6:3788-3796. [PubMed] [Google Scholar]
- 38.Watanabe, G., C. Albanese, R. J. Lee, A. Reutens, G. Vairo, B. Henglein, and R. G. Pestell. 1998. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol. Cell. Biol. 18:3212-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinstat-Saslow, D., M. J. Merino, R. E. Manrow, J. A. Lawrence, R. F. Bluth, K. D. Wittenbel, J. F. Simpson, D. L. Page, and P. S. Steeg. 1995. Overexpression of cyclin D mRNA distinguishes invasive and in situ breast carcinomas from non-malignant lesions. Nat. Med. 1:1257-1260. [DOI] [PubMed] [Google Scholar]
- 40.Westerheide, S. D., M. W. Mayo, V. Anest, J. L. Hanson, and A. S. Baldwin. 2001. The putative oncoprotein Bcl-3 induces cyclin D1 to stimulate G(1) transition. Mol. Cell. Biol. 21:8428-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westwick, J. K., Q. T. Lambert, G. J. Clark, M. Symons, L. Van Aelst, R. G. Pestell, and C. J. Der. 1997. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol. Cell. Biol. 17:1324-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolffe, A. P. 1996. Histone deacetylase: a regulator of transcription. Science 272:371-372. [DOI] [PubMed] [Google Scholar]
- 43.Urnov, F. D., J. Yee, L. Sachs, T. N. Collingwood, A. Bauer, H. Beug, Y. B. Shi, and A. P. Wolffe. 2000. Targeting of N-CoR and histone deacetylase 3 by the oncoprotein v-erbA yields a chromatin infrastructure-dependent transcriptional repression pathway. EMBO J. 19:4074-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, L., R. B. Corcoran, J. W. Welsh, D. Pennica, and A. J. Levine. 2000. WISP is a Wnt-1- and beta-catenin-responsive oncogene. Genes Dev. 14:585-595. [PMC free article] [PubMed] [Google Scholar]
- 45.Yasui, D., M. Miyano, S. Cai, P. Varga-Weisz, and T. Kohwi-Shigematsu. 2002. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419:641-645. [DOI] [PubMed] [Google Scholar]
- 46.You, A., J. K. Tong, C. M. Grozinger, and S. L. Schreiber. 2001. CoREST is an integral component of the Co. Proc. Natl. Acad. Sci. USA 98:1454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Z. K., K. P. Davies, J. Allen, L. Zhu, R. G. Pestell, D. Zagzag, and G. V. Kalpana. 2002. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol. Cell. Biol. 22:5975-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao, R., K. Gish, M. Murphy, Y. Yin, D. Notterman, W. H. Hoffman, E. Tom, D. H. Mack, and A. J. Levine. 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14:981-993. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.