Abstract
Embryonic stem (ES) cell pluripotency and differentiation are controlled by a network of transcription factors and signaling molecules. Transcription factors such as Oct4 and Nanog are required for self-renewal and maintain the undifferentiated state of ES cells. Decreases in the expression of these factors indicate the initiation of differentiation of ES cells. Inactivation of the gene encoding the orphan nuclear receptor GCNF showed that it plays an important role in the repression of Oct4 expression in somatic cells during early embryonic development. GCNF−/− ES cells were isolated to study the function of GCNF in the down-regulation of pluripotency genes during differentiation. Loss of repression of ES cell marker genes Oct4, Nanog, Sox2, FGF4, and Stella was observed upon treatment of GCNF−/− ES cells with retinoic acid. The loss of repression of pluripotency genes is either a direct or indirect consequence of loss of GCNF. Both the Oct4 and Nanog genes are direct targets of GCNF repression during ES cell differentiation and early mouse embryonic development. In contrast Sox2 and FGF4 are indirectly regulated by GCNF through Oct4. These findings establish a central role for GCNF in the repression of pluripotency gene expression during retinoic acid-induced ES cell differentiation.
Embryonic stem (ES) cells are capable of unlimited symmetrical self-renewal and have the potential to differentiate into any cell type in the body (15, 16, 18, 26). The pluripotency of ES cells is maintained by several key regulatory factors, which establish precise patterns of gene expression and are characteristic of the undifferentiated phenotype of ES cells. The transcription factor Oct4 belongs to the POU homeodomain family and plays an essential role in the maintenance of ES cell pluripotency (4, 34, 37, 40). Oct4 is highly expressed in ES cells and embryo carcinoma (EC) cells and is down-regulated upon differentiation of either cell type (35, 42). Precise levels of Oct4 are required to sustain stem cell self-renewal, and either up- or down-regulation induces divergent developmental programs. A less than twofold increase in expression causes differentiation into primitive mesoendoderm. In contrast, repression of Oct4 induces loss of pluripotency and differentiation into trophectoderm (35).
Oct4 also plays a pivotal role during early development of embryos. Oct4 is expressed throughout the morula and inner cell mass of the blastocyst and is restricted to primordial germ cells after gastrulation. Oct4 is only expressed in the germ cell lineage in adults (4, 41, 44). Oct4-deficient embryos die at the blastocyst stage due to the loss of pluripotency of inner cell mass cells. In the absence of a true inner cell mass, trophoblast proliferation is not maintained and they differentiate into trophectoderm (33). Oct4 regulates the expression of several other genes, including Rex1, Utf1, and Sox2, of which the last was also found to be essential for ES cell maintenance (2, 36).
Recently a new transcription factor, Nanog, belonging to the ES cell regulatory network was discovered (8, 31). Nanog is a homeodomain protein that directs propagation of undifferentiated ES cells. The expression profile of Nanog is very similar to that of Oct4 in ES cells and during early embryonic development. Nanog mRNA is present in pluripotent mouse and human ES cell lines and absent from differentiated cells (6, 12). Nanog expression is restricted to the inner cell mass in preimplantation embryos and restricted to embryonic ectoderm of postimplantation embryos (21, 31). Endogenous Nanog and elevated Nanog from transgene expression act in parallel with cytokine stimulation of Stat3 to drive ES cell self-renewal, bypassing Stat3 and maintaining Oct4 levels (8, 31). Nanog-deficient inner cell mass failed to generate epiblast and only produced parietal endoderm-like cells (31). Nanog-deficient ES cells lost pluripotency and differentiated into an extraembryonic endoderm lineage (31). The level of Nanog expression is tightly correlated with the undifferentiated state of ES cells (22). These findings establish a central role for Nanog in the maintenance of ES cell pluripotency and the epiblast stage of embryonic development.
Differentiation of ES cells is accompanied by down-regulation of the whole series of transcription factors and signaling molecules that maintain the pluripotent phenotype, such as Oct4 and Nanog, and up-regulation of transcription factors involved in differentiation. Little is known about how determinants of ES cell pluripotency are regulated during differentiation of ES cells. Previous studies have shown that several members of the nuclear receptor family, including LRH-1, SF-1, GCNF, retinoic acid receptor/retinoid X receptor, and COUP TF I and II, regulate Oct4 expression by binding to its enhancer and promoter regions (3, 17, 19, 36, 42).
The orphan nuclear receptor Germ Cell Nuclear Receptor (GCNF:NR6A1) is involved in regulating early embryonic development and reproduction (10, 17, 28, 29). GCNF is a transcriptional repressor that down-regulates target gene expression through binding to response elements, which are configured as a direct repeats with no spacing between the half sites (DR0), within their promoters (9, 13, 24, 43). GCNF is predominantly expressed in germ cells in the adult (9, 28, 29) and is widely expressed in early mouse embryos (10) as well as in P19 cells (17). GCNF's expression pattern inversely correlates with that of Oct4 during mouse embryogenesis and P19 cell differentiation (17, 23). The repression of Oct4 is driven by binding of GCNF to a DR0 element located in the proximal promoter (17). In GCNF−/− embryos, Oct4 expression is not turned off efficiently in somatic cells at gastrulation, when Oct4 expression is normally restricted to the germ cell lineage, and GCNF−/− embryos subsequently die around embryonic day E10.5 (10, 17). The loss of Oct4 repression in GCNF−/− embryos suggested that GCNF might play a role in repressing Oct4 expression in ES cells upon differentiation.
To understand the role of GCNF in the regulation of ES cell differentiation, we established and characterized a GCNF−/− ES cell line. Here we report the effect of loss of GCNF function upon ES cell differentiation and the silencing of pluripotency genes during this process. Our findings suggest that GCNF plays a central role in repressing the ES cell phenotype during retinoic acid (RA)-induced differentiation through repression of pluripotency genes such as Oct4 and Nanog.
MATERIALS AND METHODS
Plasmids.
The GCNF expression vector was constructed by insertion of full-length cDNA of mouse GCNF into the pCMV-HA expression vector (Clontech, Sparks, MD). The Oct4-LUC reporter has been described previously (17). The reporter plasmid Nanog5P was generated by insertion of 2.5 kb of 5′ promoter region of the mouse Nanog gene into the pGL3-Basic luciferase reporter vector (Promega, Madison, WI) between the KpnI and XhoI sites; Nanog3P was constructed by insertion of 1.0 kb of 3′ untranslated region from the mouse Nanog gene into the pGL3-promoter luciferase reporter (Promega) at the XbaI site; and Nanog5/3P was made by insertion of 1.0 kb of 3′ untranslated region of the mouse Nanog gene into Nanog5P reporter at the XbaI site. The 2.5 kb of 5′ promoter fragment of the mouse Nanog gene was obtained from genomic DNA by PCR with primers 5P-F, AGTGGTACCAACAGTGGGTCTGAAGCCTTCGAG, and 5P-R, TCACTCGAGTGTGATGGCGAGGGAAGGGAT. The 1.0 kb of 3′ untranslated region of mouse Nanog gene was generated from genomic DNA by PCR with primers 3P-F, AGACGCTAGCAACATCTGGGCTTAAAGTCAGGGC, and 3P-R, TTAAACTAGTCCAGCTGGCATCGGTTCATC. The PCR products were sequenced to confirm the fidelity of these sequences.
Derivation of ES cell lines.
Male and female GCNF+/− mice were bred and females were checked daily for plugs. At 3.5 days of gestation, uteri were removed and flushed with ES medium containing 0.02 mM HEPES. Blastocysts were recovered with a mouth pipette and cultured for 3 days with ES medium in 60-mm petri dishes plated with gamma-irradiated confluent STO feeders. After 3 days, when blastocysts had adhered to the feeder layer, inner cell mass outgrowths were aspirated with a mouth pipette and plated individually in a single well of a 24-well plate. The next day, each well was washed twice with phosphate-buffered saline, two drops of 0.25% trypsin-EDTA (Invitrogen, Carlsbad, CA) was added for 2 min, 1 ml ES medium was added, and cells were split at 1:1. For the subsequent steps, cells were split 1:1 every 3 days. After 2 to 5 weeks, the first ES cell colonies were visible in some wells. When a well contained several well-formed colonies (around 10), cells were plated on 60-mm dishes and expanded and aliquots were frozen at −80°C. For genotyping, feeders were removed by replating two to three times on non-gelatin-coated dishes for 30 minutes, which allowed the feeder cells to adhere while most of the ES cells stayed in the suspension and were transferred into a new gelatin-coated dish for propagation.
Maintenance and RA differentiation of ES cells and P19 cells.
ES cells were maintained on STO feeder layers in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum tested for ES cell culture, 100 mM nonessential amino acids, 2 mM glutamine, 100 U of penicillin-streptomycin/ml (Invitrogen), and 0.55 μM β-mercaptoethanol (Sigma). The medium was changed 3 h before splitting. Cells were rinsed twice with phosphate-buffered saline, treated with 0.25% trypsin, 0.5 mM EDTA for 2 to 3 min, and split 1:5 or used for differentiation after removal of feeder cells. Around 100,000 wild-type or GCNF−/− ES cells were plated on a gelatinized 60-mm petri dish without feeder cells using ES medium containing 10−6 M all-trans-retinoic acid (RA) (Sigma). The medium was changed every day. For day 0 differentiation, cells were cultured in ES medium supplemented with 1,000 U/ml of leukemia inhibitory factor (LIF; Chemicon, Temecula, PA) instead of RA, and the medium was changed every day for 2 days.
COS1 and P19 cells were maintained with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM glutamine, and 100 U of penicillin-streptomycin/ml (Invitrogen). Differentiation of P19 cells was carried out with the same procedure as that for ES cells. CHO-K1 cells were cultured with F12 medium supplemented with 10% fetal bovine serum and 100 U of penicillin-streptomycin/ml.
Genotyping of mice and embryos.
Genotyping of the ES cell line was performed after removal of feeder cells. Genotyping of embryos was followed by whole-mount in situ hybridization. DNA was extracted from ES cells, embryos, and mouse tails with a lysis buffer (0.5% sodium dodecyl sulfate, 50 mM Tris, pH 7.5, 0.1 M NaCl, 5 mM EDTA, 0.2 mg/ml proteinase K). Genomic DNA PCR was performed following previously described conditions (10). Specific primers for Y chromosome marker were forward, CTGGAGCTCTACAGTGATGA, and reverse, CAGTTACCAATCAACACATCA. The myosin primers were forward, TTACGTTCCATCGTGGACAGCAT, and reverse, TGGGCTGGGTGTTAGTCTTAT.
Northern blot and RT-PCR.
Total RNAs from different time points of differentiated P19 and ES cells were isolated using Trizol reagent (Invitrogen, Carlsbad, CA). Northern blot analysis was performed according to protocol PT1190-1 (Clontech, Sparks, MD). Blots were hybridized with 32P-labeled cDNAs corresponding to Oct4 (bases 908 to 1260, GenBank accession number NM 013633) and Nanog (bases 507 to 1026, GenBank AY278951). To ensure equal loading of RNA samples, the blots were probed with mouse glyceraldehyde-3-phosphate dehydrogenase cDNA.
Reverse transcription was carried out with Moloney murine leukemia virus reverse transcriptase II according to the manufacturer's protocol (Invitrogen). Gene-specific primers for GCNF were forward, CTGAACAACGAACCTGTCTC, and reverse, TTGCTCTCTGAAGCCCTGTT; Oct4, forward, GGCGTTCTCTTTGGAAAGGTGTTC, and reverse, CTCGAACCACATCCTTCTCT; Nanog, forward, AAAGGATGAAGTGCAAGCGGTGG, and reverse, CTGGCTTTGCCCTGACTTTAAGC; Stella, forward, CAGCCGTACCTGTGGAGAACAAGAG, and reverse, AGCCCTGGGCCTCACAGCTT; Sox2, forward, CCTCGGATCTCTGGTCAAGT, and reverse, TGTGCGTCAGGGGCACCGTG; FGF4, forward, TATTTGCTCTCGCTACTTAGG, and reverse, ACTCCGAAGATGCTCACCACG; FGF8, forward, CAGCTCTACAGCCGCACCAGC, and reverse, TGCTCTTGGCAATTAGCTTCC; β-tubulin, forward, TCACTGTGCCTGAACTTACC, and reverse, GGAACATAGCCGTAAACTGC; and β-actin, forward, GGCCCAGAGCAAGAGAGGTATCC, and reverse, ACGCACGATTTCCCTCTCTCAGC.
Western blot analysis and immunofluorescent staining of embryoid bodies.
Total proteins were extracted with 1× passive lysis buffer (Promega) from different time points of RA-differentiated P19 or ES cells. Western blot analysis was performed according to the protocol provided by Amersham Pharmacia (RPN2209). Anti-GCNF antibody was previously described (24). Anti-Oct4 (sc-8628) and antiactin (sc-1616) antibodies and horseradish peroxidase-conjugated secondary antibodies used in Western blot assay were purchased from Santa Cruz (Santa Cruz, CA).
Embryoid bodies were formed from about 1,000 ES cells in a hanging drop on the lip of culture dish for 2 days and suspended in medium for another day. After fixation in 4% paraformaldehyde for 2 h and permeabilization with 0.2% Triton X-100 for 30 min and blocking with 10% normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA), embryoid bodies were incubated with rabbit anti-Nanog immunoglobulin G (Chemicon, Temecula, CA) and goat anti-Oct4 immunoglobulin G (Santa Cruz, CA) or normal rabbit and goat immunoglobulin G. After washing with phosphate-buffered saline, embryoid bodies were stained with fluorescein isothiocyanate-conjugated donkey anti-rabbit immunoglobulin G and Texas Red-conjugated donkey anti-goat immunoglobulin G (Jackson ImmunoResearch Laboratories, West Grove, PA). Embryoid bodies were mounted on glass slides using a Vectashield mounting medium for fluorescence (Vector Laboratories, Burlingame, CA) and examined with a Zeiss laser scanning confocal microscope (LSM510) (Carl Zeiss, Germany).
EMSA.
COS-1-overexpressed proteins P19 and ES cell nuclear extracts were extracted with 2× binding buffer (25 mM HEPES, pH 7.9, 150 mM KCl, 0.4 mM EDTA, 2 mM dithiothreitol, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail [Roche, Indianapolis, IN]). The sequence of the Oct4 DR0 probe and electrophoretic mobility shift assays (EMSAs) were performed according to the methods described previously (17). The oligodeoxynucleotide probe was end labeled with [α-32P]dCTP using Klenow (Roche) and incubated in 1× binding buffer with 0.7 μg/μl of poly(dI-dC) · poly(dI-dC) and 0.4 mM dithiothreitol for 30 min at room temperature. Specific antibody (1 μl/reaction) was used in the supershift assays and preincubated for 5 min before adding probes. The protein-DNA complexes were separated on 5% nondenaturing polyacrylamide gels in 0.5× Tris-borate-EDTA running buffer and visualized by autoradiography. The sequences of Nanog DR0 are forward, AGGCAGATTTCTGAGTTCAAGGCCAGCCTGGTCTA, and reverse, TCTGTAGACCAGGCTGGCCTTGAACTCAGAAATCT.
Chromatin immunoprecipitation (ChIP) assay.
ChIP assays were performed according to the on-line protocol provided by Upstate Company (Charlottsville, VA). Undifferentiated and RA-differentiated P19 and ES cells were treated with 1% formaldehyde. Cross-linked DNA was extracted and disrupted by sonication. The recovered DNA was amplified with gene-specific primers. Primers covering the mouse Oct4 DR0 region of proximal promoter are forward, CCTCCGTCTGGAAGACACAGGCAGATAGCG and reverse, CGAAGTCTGAAGCCAGGTGTCCAGCCATGG. Primers specific for the Nanog 5′ promoter DR0 site are forward, ACAGTGGGTCTGAAGCCTTCGAG, and reverse, CCTCCATTGCCCAGCCTGACTG. Primers for the Nanog 3′ untranslated region are F1, CCACTGAGCCATCTCACCAGC; R1, AGTTGAGTTGGTGCCCAGCATG; F2, ATGGTGGCTACTCTCGAGGATG; and R2, TCCAGCTGGCATCGGTTCATCA.
Whole-mount in situ hybridization.
Embryos from GCNF+/− female crossed with GCNF+/− males were harvested between E8.5 and E8.75 and fixed in 4% paraformaldehyde. Whole-mount in situ hybridization was carried out as described previously (17). The Nanog cDNA from an RT-PCR product were digoxigenin labeled (In Vitro Translation Kit; Promega) as cRNA probes.
Transient transfection assays.
The transient transfections were performed with Fugene 6 according to the manufacturer's protocol (Roche, Indianapolis, IN). The hemagglutinin (HA)-tagged GCNF expression vector (0, 20, 50, and 100 ng/well) and reporter DNA (200 ng/well) were cotransfected into CHO-K1 cells plated in 12-well plates (105 cells per well) for 24 h prior to transfection. Total plasmid DNAs were balanced with empty expression vector pCMV-HA. After 48 h incubation, the cells were harvested and the luciferase activity was analyzed using the luciferase assay kit (Promega) according to the manufacturer's protocols. Relative luciferase activity represents luciferase activity relative to that obtained from cotransfection of reporter plasmids and empty expression vector pCMV-HA, which was arbitrarily set at 1.
RESULTS
Induction of GCNF and formation of the TRIF complex in differentiated ES cells.
Analysis of GCNF−/− embryos in conjunction with the molecular studies in P19 cells suggested that GCNF might play a role in repressing Oct4 expression during ES cell differentiation. It has been shown that GCNF expression is induced in P19 cells upon treatment with retinoic acid (17, 23). Although P19 cells are an embryonic carcinoma cell line that has multipotent characteristics, they are different from ES cells, which are characterized as pluripotent.
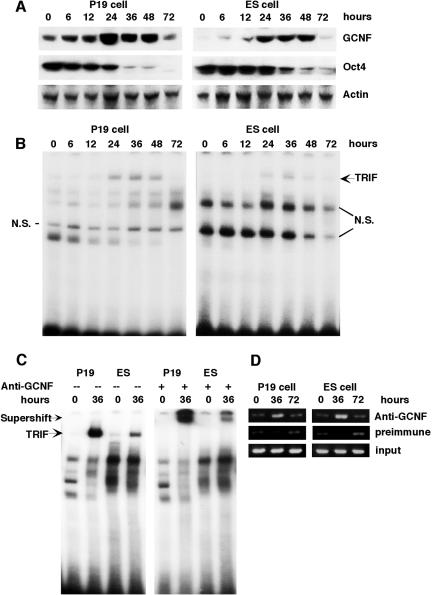
To study the role of GCNF in ES cells we wanted to first determine if GCNF could be induced in ES cells by the appropriate stimuli. Thus, we treated ES cells with RA, under conditions similar to P19 cells, and analyzed the expression patterns of GCNF and Oct4 during differentiation by Western analysis. When ES cells were treated with RA, the level of GCNF protein started to increase after 6 h of RA induction, reaching peak expression between 24 and 36 h, and dropped to undetectable levels after 3 days of RA treatment (Fig. 1A). The transient induction of GCNF during the ES cell differentiation process was similar to what was observed in P19 cells (Fig. 1A). Temporally, Oct4 protein expression was down-regulated in differentiated ES cells in the same manner as in differentiated P19 cells (Fig. 1A). In P19 cells, the expression level of Oct4 protein was dramatically decreased following 24 h of RA treatment and completely silenced after 72 h (Fig. 1A), comparatively, in differentiated ES cells, the decrease of Oct4 protein also occurred after 24 h of RA treatment. These results demonstrated that GCNF expression was RA inducible in differentiated ES cells as in P19 cells and inversely correlated with the expression of Oct4.
FIG. 1.
Expression of Oct4 and GCNF and binding of GCNF to the Oct4 promoter in differentiated P19 and ES cells. (A) The protein expression levels of GCNF and Oct4 were analyzed by using the anti-GCNF and Oct4 antibodies. The amount of total protein was normalized by β-actin, which was used as a control. (B) The TRIF complex was detected in differentiated P19 and ES cells by EMSA analysis with an Oct4 DR0 probe. (C) The TRIF complex in differentiated P19 and ES cells can be supershifted by anti-GCNF antibodies. (D). Direct binding of GCNF to DR0 site located in the endogenous Oct4 gene proximal promoter in differentiated P19 and ES cells was demonstrated by ChIP assay. P19 and ES cells were differentiated by treatment with RA for the times indicated (hours).
In P19 cells, RA-induced differentiation is accompanied by the formation of a large complex that binds to the Oct4 proximal promoter and is responsible for the repression of Oct4 expression. This complex is called the transiently retinoid-induced factor (TRIF) complex and contains GCNF in differentiated P19 cells (23). To determine whether RA induced the formation of a GCNF-containing TRIF complex in differentiated ES cells, protein extracts from RA-treated ES and P19 cells at different time points were analyzed by electrophoretic mobility shift assay for binding to the Oct4 DR0 element. In ES cells, a TRIF-like complex was induced after 24 h of treatment, increased by 36 h, and then decreased to undetectable levels after 72 h of RA treatment (Fig. 1B). This DNA binding pattern was identical to what was observed in differentiated P19 cells (Fig. 1B), and its temporal occurrence directly correlated with GCNF expression. Anti-GCNF antibodies specifically supershifted the TRIF complex in differentiated ES cells and had no effect on the Oct4 DNA binding complexes in undifferentiated ES cells containing the orphan nuclear receptor LRH-1 (Fig. 1C) (19). These results demonstrated that RA could induce the formation of a TRIF-like complex in differentiated ES cells and that like P19 cells, it contained GCNF.
The inverse expression patterns of GCNF and Oct4 in P19 and ES cells indicated that GCNF might repress the expression of Oct4 in ES cells. Although in vitro data from transient transfection assays in P19 cells support this hypothesis (17), in vivo cell autonomous evidence is still needed to prove that GCNF fulfills this function in ES cells. Based on the GCNF expression pattern in P19 and ES cells, three time points (0, 36, and 72 h) for RA treatment of P19 and ES cells were chosen for chromatin immunoprecipitation (ChIP) assays to determine if GCNF binds to the Oct4 promoters in vivo. After immunoprecipitation with anti-GCNF antibodies, the genomic DNA surrounding the DR0 site in the Oct4 proximal promoter was amplified. As shown in Fig. 1D, in the absence of RA (0 h), GCNF binding to the DR0 site in the Oct4 proximal promoter was not detected above background, although at this time point, low levels of GCNF expression were detected in P19 cells (Fig. 1A and D). After 36 h of RA-induced differentiation, a strong amplified signal was observed in both P19 and ES cells (Fig. 1D). When the expression level of GCNF dropped at 72 h, in vivo binding to the Oct4 promoter was also reduced in both P19 and ES cells (Fig. 1A and D). These results demonstrated that RA induced endogenous GCNF to bind to the DR0 of the Oct4 proximal promoter in P19 and ES cells.
Establishment of a GCNF−/− ES cell line.
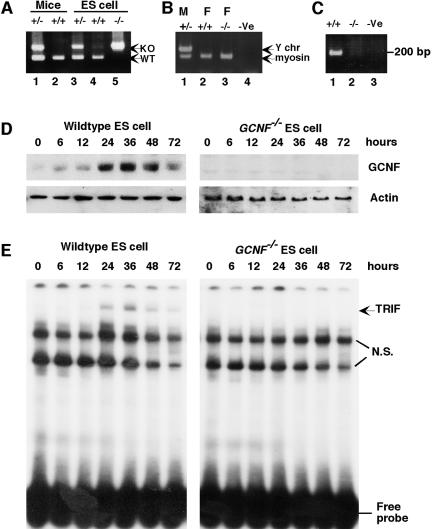
Previous results from GCNF−/− embryos (17) and the current discovery of GCNF expression in differentiated ES cells focused our interests on GCNF function in ES cells. Thus, we established a GCNF−/− ES cell lines from blastocyst outgrowths. Various ES cell lines were derived from blastocysts of GCNF+/− intercrosses and two GCNF−/− ES cell lines were isolated. Several heterozygote cell lines were also generated but not widely investigated, as GCNF+/− animals did not have a haploinsufficient phenotype (10). The genotype of the GCNF−/− ES cell line was confirmed (Fig. 2A). Most notably, 2904c was a wild-type female ES cell line, and 2412e was a GCNF−/− female ES cell line. Both the 2904c and 2412e ES cell lines are female in that they lack a Y chromosome marker (Fig. 2B), which avoids any sex-specific bias between our wild-type and GCNF−/− ES cells, for instance, differential methylation patterns.
FIG. 2.
Isolation and characterization of GCNF−/− ES cells. (A) Genotyping of isolated ES cells from blastocyst outgrowths. Genomic DNA extracted from mouse tails was genotyped as controls (lanes 1 and 2). ES cells in lane 3, 4, and 5 were isolated from the same litter, corresponding to ES cell lines 2904d, 2904c, and 2412e, respectively. (B) Gender analysis of ES cell lines 2904d (lane 1), 2904c (lane 2), and 2412e (lane 3) and the PCR negative control (lane 4) was determined using the Y chromosome-specific primers and the myosin-specific primers as a control. (C) GCNF mRNA was analyzed by RT-PCR with DNA-binding domain-specific primers (10). Total RNAs were extracted from RA-differentiated cells and reverse-transcribed into cDNA (2904c, lane 1; 2412e, lane 2; and RT control, lane 3). (D) Western blot analysis of GCNF expression in GCNF−/− ES cells; 20 μg of total proteins extracted from RA-differentiated ES cells at different time points were analyzed. (E) EMSA analysis of the TRIF complex in the GCNF−/− ES cells using the Oct4 DR0 as a probe. Protein samples were the same as in panel D.
We first wanted to confirm the absence of GCNF mRNA and protein in our GCNF−/− ES cell line. No GCNF mRNA was detected in RA-differentiated 2412e GCNF−/− ES cells by RT-PCR (Fig. 2C). Similarly, no GCNF protein was detected in differentiated GCNF−/− ES cells by Western blot (Fig. 2D). In contrast, GCNF protein is highly expressed after 24 to 48 h of RA treatment of GCNF+/+ ES cells and decreases after 72 h of RA treatment (Fig. 2D). EMSA analysis further confirmed the loss of the GCNF TRIF complex in differentiated GCNF−/− ES cells (Fig. 2E). The latter result not only confirmed that GCNF was present in the TRIF complex but also clearly showed that the formation of the complex was GCNF dependent.
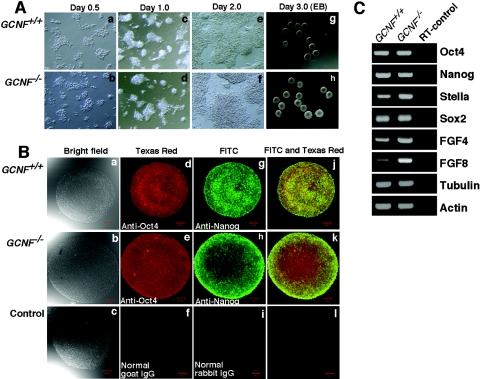
Although the GCNF−/− cell line that we generated had lost GCNF expression, we wanted to ensure that it displayed the characteristics of an embryonic stem cell line. When GCNF−/− cells were cultured as a monolayer on gelatinized culture dishes in the presence of LIF, they tended to aggregate to form colonies similar to wild-type ES cells. The GCNF−/− cells had a proliferative rate similar to that of wild-type ES cells (Fig. 3A, panels a to f). GCNF−/− ES cells also formed embryoid bodies when cultured in hanging drops (Fig. 3A, panels g to h). Immunofluorescent staining demonstrated that GCNF−/− ES cells in the embryoid bodies coexpressed high levels of the pluripotent marker genes Oct4 and Nanog (Fig. 3B, panels a to l). Analysis of a small panel of pluripotent marker genes was examined in monolayer cultured ES cells by RT-PCR. The GCNF−/− ES cells expressed all the marker genes tested at levels comparable to wild-type ES cells (Fig. 3C). Clearly, the GCNF−/− ES cell line that was isolated maintained ES cell features in the undifferentiated state even though the expression of GCNF was lost.
FIG. 3.
ES cell phenotype of GCNF−/− ES cells. (A) Morphology of GCNF−/− ES cells cultured under the control of LIF as monolayer (panels a to f) and embryoid bodies (EB, panels g to h). (B) Immunofluorescent staining of embryoid bodies. Embryoid bodies were incubated with rabbit anti-Nanog and goat anti-Oct4 antibodies and visualized by fluorescein isothiocyanate-labeled donkey anti-rabbit and Texas Red-labeled donkey anti-goat secondary antibodies. The images were taken under bright field (panels a to c), fluorescein isothiocyanate filter (panels d to f), and Texas Red filter (panels g to i) and merged images from fluorescein isothiocyanate and Texas Red filters (panels j to l) using a Zeiss laser scanning confocal microscope (LSM510). (C) RT-PCR analysis of pluripotent marker gene expression in GCNF−/− ES cells cultured as a monolayer for 2 days in the presence of LIF. Wild-type ES cells were cultured and analyzed as controls.
Oct4 expression in GCNF−/− ES cells.
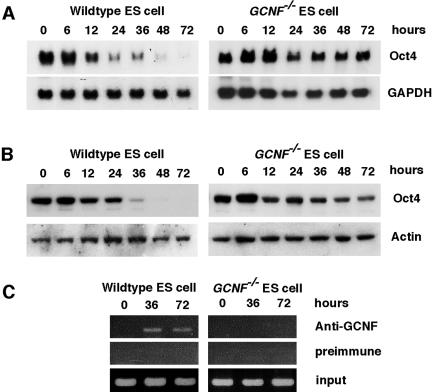
The expression of the Oct4 gene in differentiated GCNF−/− ES cells was studied. Northern blot results showed that in wild-type ES cells, Oct4 mRNA levels started to decrease after 12 h of RA treatment and kept decreasing to undetectable levels at 72 h (Fig. 4A). In contrast, in GCNF−/− ES cells after 12 h of RA treatment, Oct4 mRNA was expressed at higher levels than in undifferentiated ES cells and then subsequently was maintained at somewhat lower levels from 24 h to 72 h without being repressed (Fig. 4A). Western blot analysis also showed that the Oct4 protein levels were maintained in differentiated GCNF−/− ES cells, in contrast to wild-type ES cells, where Oct4 protein levels dropped to undetectable levels after 48 h of RA treatment (Fig. 4B). Thus, in GCNF−/− ES cells, there is a cell autonomous loss of Oct4 repression upon differentiation with RA.
FIG. 4.
Loss of repression of Oct4 gene expression in differentiated GCNF−/− ES cells. (A) Oct4 expression was detected by Northern blot analysis in RA-differentiated ES cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression was detected to control for loading amounts and integrity of the RNAs. (B) Oct4 protein was detected by Western blot analysis in RA-differentiated ES cells and β-actin was used as a loading and integrity control. (C) Direct binding of GCNF to the DR0 element located in the proximal promoter of the endogenous Oct4 gene in RA-differentiated wild-type and GCNF−/− ES cells was analyzed by ChIP assay. Preimmunized serum immunoglobulin G was used as a negative control.
To determine if the loss of repression was a direct effect of GCNF mediated by binding to the Oct4 promoter in vivo in ES cells, we used the GCNF-dependent ChIP assay. The results confirmed the loss of GCNF binding to Oct4 DR0 in the proximal promoter in GCNF−/− ES cells (Fig. 4C). These results demonstrated that loss of GCNF expression resulted in loss of Oct4 repression in differentiated GCNF−/− ES cells and revealed that GCNF plays a pivotal role in the down-regulation of Oct4 expression during differentiation of ES cells.
Loss of repression of Nanog and other ES cell genes in RA-treated GCNF−/− ES cells.
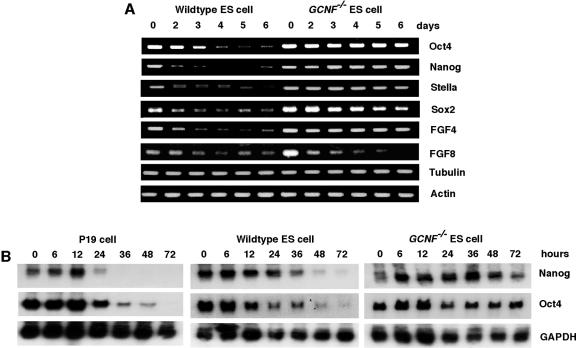
Even though Oct4 is one of the determinants of ES cell pluripotency and it is directly regulated by GCNF, other pluripotency factors, like Nanog, appear to act independently. The expression of pluripotency genes was further investigated during RA-induced differentiation of wild-type and GCNF−/− ES cells by semiquantitative RT-PCR. The expression of the pluripotency factors Oct4, Nanog, Stella, Sox2, and FGF4 was analyzed in a 6-day time course of RA treatment. FGF8 was also analyzed as a control along with tubulin and actin. The RT-PCR has been repeated at least twice using RNAs originating from two distinct differentiation experiments.
Surprisingly, the RT-PCR results demonstrated that the expression of not only Oct4 mRNA was maintained in differentiated GCNF−/− ES cells, but also other pluripotency genes, Nanog, Stella, Sox2, and FGF4 responded in a manner similar to Oct4 in differentiated GCNF−/− ES cells, that is, loss of repression (Fig. 5A). All of these pluripotency factors were highly expressed in undifferentiated ES cells and decreased to undetectable levels upon differentiation of ES cells, while, in GCNF−/− ES cells, all of them maintained their expression during 6 days of RA differentiation. The loss of repression of pluripotency genes was specific during the differentiation process, as the expression of FGF8 was still repressed in GCNF−/− ES cells in a manner similar to wild-type ES cells. The housekeeping genes for β-tubulin V and β-actin were used as controls to ensure relatively similar total RNA inputs. All of the RT-PCR fragments were sequenced to confirm their gene-specific sequences.
FIG. 5.
Analysis of the expression of ES cell pluripotency factors in RA-differentiated wild-type and GCNF−/− ES cells. (A) Comparison of undifferentiated and differentiated ES cell marker gene expression in wild-type and GCNF−/− ES cells by RT-PCR analysis. The number on the top of the figure indicates days of RA treatment. (B) Nanog expression was detected by Northern blot analysis over 72 h of differentiation of P19 and wild-type and GCNF−/− ES cells with RA. The number on the top of the figure indicates the hours of RA treatment.
Because Nanog has been shown to be essential for the maintenance of pluripotency of the epiblast and ES cells, we analyzed the expression of Nanog in greater detail. To confirm the loss of repression of the Nanog gene in GCNF−/− ES cells, the expression of Nanog was further studied by Northern blot analysis. Considering the early induction of GCNF in P19 and ES cells, a shorter time course of RA induction was used. The results showed that Nanog was highly expressed in undifferentiated ES cells and P19 cells (Fig. 5B). Coincident with GCNF induction from 24 to 48 h (Fig. 1), Nanog mRNA was dramatically decreased after 1 day of RA treatment in ES and P19 cells and dropped to undetectable levels after 2 days of treatment in wild-type ES cells. In contrast, Nanog gene expression was maintained at relatively high levels in GCNF−/− ES cells in the presence of RA (Fig. 5B). The repression of Nanog inversely correlated with induction of GCNF expression in P19 and ES cells. Thus, we predicted that GCNF might directly regulate the expression of Nanog gene in P19 and ES cells.
GCNF directly represses Nanog expression via binding to DR0 sites in the gene.
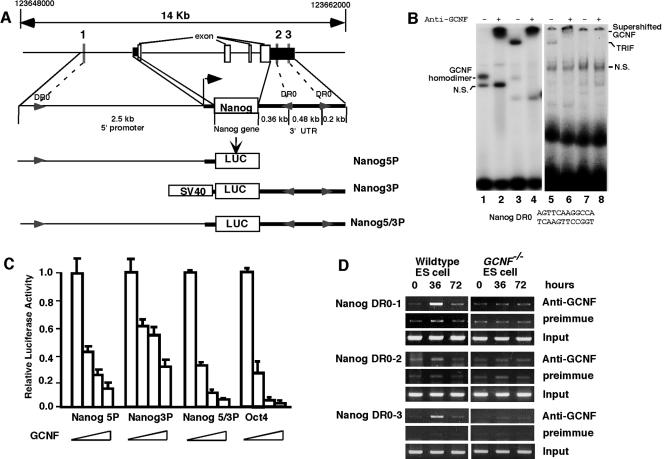
To determine if Nanog is a direct target of GCNF repression, the regulatory regions of the Nanog gene were analyzed for DR0 elements. From the published literature, a set of GCNF response elements (DR0) were used (Table 1) to generate a hidden markov model (HMM) using Hmmer v0.2.32 and used to search the entire mouse genome (14, 19, 25). Three candidate DR0 elements were identified in 14 kb of genomic DNA spanning the entire Nanog gene. One is located 2.5 kb upstream from the ATG start site and the other two DR0s were located in 1 kb of the 3′ untranslated region (Fig. 6A). The sequences of these three DR0s are identical (AGTTCAAGGCCA) and the orientation is indicated in Fig. 6A.
TABLE 1.
List of known GCNF response elements
| Gene | Sequence | Reference |
|---|---|---|
| Bone morphogenic protein 15 (BMP15) | AGGCCAAGTTCA | 29 |
| Bone morphogenic protein 15 (BMP15) | AGTTCAAGGTAA | 29 |
| Growth differentiation factor 9 (GDF9) | TGGTCAAGTACT | 29 |
| Growth differentiation factor 9 (GDF9) | CAGCCAAGGTCA | 29 |
| Growth differentiation factor 9 (GDF9) | AGTTTAAGGCCA | 29 |
| Homeobox D 11 (HOXD11) | ATGTCAAGGTCA | Unpublished data |
| Octamer-binding transcription factor (Oct4) | AGGTCAAGGCTA | 17 |
| Protamine 1 (PRM1) | AGTTCAAGGTCA | 24 |
| Protamine 2 (PRM2) | AGGTCAAGTTCC | 24 |
| Lactoferrin | AGGTCAAGGCGA | A. Jetten, personal communication |
| Synthetic GCNF binding oligonucleotide | AG(G/T)TCAAG(G/T)TCA | 43 |
FIG. 6.
Direct repression of Nanog expression by GCNF binding to DR0 elements in the Nanog gene. (A) Localization of three DR0 elements in the promoter and 3′ untranslated region of the mouse Nanog gene and description of the Nanog luciferase reporter constructs. Arrows indicate the DR0 elements and their relative orientations. (B) Analysis of binding of GCNF to the Nanog DR0 sequence by EMSA. GCNF-overexpressing COS1 extracts (lanes 1 and 2) or RA-differentiated P19 cell extracts (lanes 3 and 4) or GCNF+/+ ES cell extracts (lanes 5 and 6) or GCNF−/− ES cell extracts (lanes 7 and 8) were incubated with the Nanog DR0 probe in the absence or presence of anti-GCNF antibodies. (C) GCNF dose-dependently repressed the Nanog reporter gene expression in transiently transfected CHO-K1 cells; 200 ng/well of three Nanog-Luc reporters, Nanog5P, Nanog3P, and Nanog5/3P, and Oct4-Luc as a positive control were cotransfected with different amounts of HA-GCNF expression vector (0, 20, 50, and 100 ng/well). (D) Direct binding of GCNF to the endogenous Nanog gene DR0 elements was demonstrated by ChIP assay with GCNF antibodies. GCNF+/+ and GCNF−/− ES cells were differentiated by treatment with RA for the times indicated (hours).
Binding of GCNF to the Nanog DR0s was tested by EMSA. The results showed that COS1-overexpressed HA-tagged GCNF could bind to this element as a homodimer and the binding was specifically supershifted by the anti-GCNF antibody (Fig. 6B, lanes 1 to 2). When RA-differentiated P19 and ES cell protein extracts were incubated with the Nanog DR0 probe, a slow-migrating TRIF complex was detected that could be supershifted by the anti-GCNF antibody (Fig. 6B, lanes 3 to 6). When the GCNF−/− ES cell extracts were used, the ES cell complex disappeared (Fig. 6B, lanes 7 to 8). This complex was identical to the GCNF TRIF complex detected using the Oct4 DR0 (AGGTCAAGGCTA) as a probe (Fig. 1B). Thus, GCNF can bind directly to the Nanog DR0 elements.
Binding of GCNF to DR0 sequences is known to cause repression of target genes (13). Thus, the promoter and 3′ untranslated regions of the mouse Nanog gene were amplified from genomic DNA by PCR and inserted either 5′ or 3′ of the luciferase reporter gene, respectively. Three reporter plasmids were constructed with either the promoter or 3′ untranslated region individually, as shown in Fig. 6A. The repression effect of GCNF on these reporters was examined by cotransfection of an HA-GCNF expression vector and reporters into CHO-K1 cells, which do not express endogenous GCNF. The results are shown in Fig. 6C.
The Nanog5P luciferase reporter activity was significantly reduced with increasing amounts of transfected GCNF expression vector; 20 ng of GCNF expression plasmid caused 60% reduction in Nanog5P luciferase activity, 50 ng of vector led to about 75% reduction, and 100 ng of the GCNF vector resulted in 85% reduction of reporter activity. Comparatively, the same amount of GCNF plasmid produced less repression of the Nanog3P reporter activity than those of the Nanog5P reporter, probably due to the strength of the simian virus 40 promoter. Strikingly, the repression effect of GCNF on the combined Nanog5/3P reporter activity was comparable to the repression of Oct4 reporter activity by GCNF; 20 ng of GCNF expression vector generated around 65 to 75% reduction of Nanog5/3P and Oct4 reporter luciferase activities and 100 ng of GCNF plasmid almost silenced both of their activities (more than 95% reduction). Thus, transfected GCNF can dose dependently repress Nanog promoter activity, and the binding of GCNF to the 3′ untranslated region of the Nanog gene also increased the reduction of promoter activity.
To examine binding of endogenous GCNF to the Nanog gene DR0 elements in ES cells, GCNF-dependent ChIP assays were used. Three ChIP assay primer sets were designed around the three Nanog DR0 elements (Fig. 6A). The pattern of binding of endogenous GCNF to the Nanog DR0s was similar to that observed on the Oct4 DR0 in wild-type differentiated ES cells (Fig. 5D). After induction for 36 h with RA, GCNF obviously bound to Nanog DR0-1 located 2.5 kb upstream of the transcriptional start site and DR0-3 in the 3′ untranslated region. Comparatively, the binding of GCNF to the DR0-2 element was weaker than to DR0-1 and DR0-3, and potentially DR0-2 may not be functional in vivo. In GCNF−/− ES cells, all GCNF-dependent binding was lost. Thus, direct binding of GCNF to at least two of the Nanog DR0 elements resulted in repression of Nanog gene expression in differentiated ES cells.
Loss of repression of Nanog expression in GCNF−/− embryos.
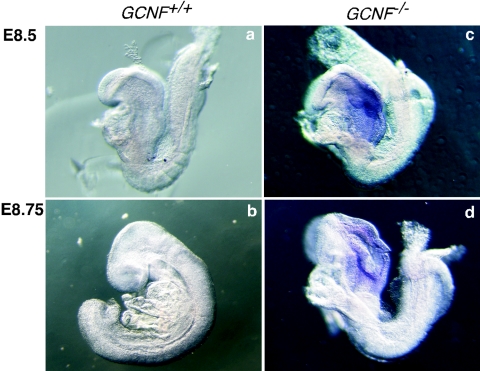
To determine if the loss of repression of the Nanog gene in GCNF−/− ES cells was physiologically relevant, we analyzed the expression of Nanog in GCNF−/− embryos (Fig. 7). In GCNF−/− embryos, expression of Oct4 was not restricted to primordial germ cells after gastrulation but remained expressed in somatic cells, such as in the neural folds and in the posterior of the embryos (17). In wild-type embryos, the Nanog gene is repressed in the entire embryo at E8.5 to E8.75 (21), however, in the GCNF−/− embryos, Nanog mRNA was clearly detectable in the neural folds. This result confirmed that the loss of GCNF function leads to loss of repression of Nanog expression in the somatic cells of gastrulating embryos, which agrees with the results obtained from the GCNF−/− ES cells.
FIG. 7.
Loss of repression of Nanog gene expression in GCNF−/− embryos. Whole-mount in situ hybridization with the Nanog cRNA probe was used to detect Nanog expression in embryos derived from GCNF+/− crosses. Expression of Nanog was analyzed in wild-type and GCNF−/− embryos at E8.5 and E8.75. Repression of Nanog gene expression after gastrulation is lost in GCNF−/− embryos.
DISCUSSION
Maintenance of pluripotence and induction of differentiation are contradictory states that are carefully balanced in ES cells and in embryonic development by transcription factors and other signaling factors. Important genes specifically expressed in undifferentiated ES cells are termed pluripotency genes or ES cell marker genes, and include Oct4, Nanog, Stella, Sox2, FGF4, BMP4, Stat3, UTF1, and Rex1 (12). The expression of these genes is required for the self-renewal of ES cells and function to maintain pluripotence. Upon differentiation of ES cells, the expression of these pluripotency genes is immediately repressed, which indicates the loss of pluripotency of the ES cells. There are two ways to reduce gene expression, either passively by loss of activation or actively by induction of repression.
Our previous studies demonstrated that the orphan nuclear receptor GCNF is a key regulator of Oct4 gene expression in P19 cells, which are embryonic carcinoma cells, as well as during embryonic development (17). Here we show that GCNF is also induced in the form of a TRIF complex in ES cells, which is physiologically relevant, and GCNF expression inversely correlates with Oct4 expression during RA-induced differentiation of ES cells. This finding suggested that although P19 and ES cells are derived from different stages of embryonic development, the former from epiblast and the later from blastocyst, GCNF has a similar function in the regulation of Oct4 gene expression in both cell systems. Since GCNF appears vital to the control of pluripotency gene expression, we established a GCNF−/− ES cell line. Strikingly, analysis of the GCNF−/− ES cell line revealed that many of ES cell marker genes are not repressed during RA-induced differentiation, including Oct4, Nanog, Sox2, FGF4, and Stella. Thus, GCNF plays a cell-autonomous role in the repression of pluripotency genes. As an essential regulator of pluripotency genes, the question is whether GCNF plays a direct or indirect role in the repression of these genes.
For Oct4 as a determinant of ES cell fate, it is clear that GCNF represses its expression through binding to a DR0 site in its proximal promoter during differentiation and gastrulation (17, 23). We also demonstrated that GCNF could directly affect the expression of the Nanog gene in differentiated P19 cells, ES cells, and developing embryos. Nanog is required for the maintenance of pluripotency in mouse epiblast and ES cells (8, 31). We found that Nanog was also expressed in undifferentiated P19 cells and its expression was turned off faster than that of Oct4, and also faster than it is turned off in ES cells (Fig. 5B). We have three pieces of evidence (EMSA, transient transfection assay, and ChIP assay) that demonstrate that GCNF directly represses the expression of the Nanog gene through binding to at least two of the three DR0 elements in the upstream and downstream regulatory regions of the Nanog gene. Loss of Nanog repression in GCNF−/− embryos is strong evidence for the effect of GCNF on Nanog gene expression (Fig. 6).
Sox2 is a transcription factor that is also involved in early embryonic development and the maintenance of pluripotence, as well as playing a role in neuronal differentiation (2, 27, 36). Although Sox2 has the same expression pattern as Oct4 in the GCNF−/− ES cells, we failed to find evidence that GCNF directly regulates its expression. In fact, our genomewide analysis of DR0 elements showed that the mouse Sox2 gene lies in a 35-kb genomic region devoid of DR0 elements (data not shown). Further analysis identified a degenerate DR0-like sequence 7 kb upstream from the transcriptional start site, however, GCNF failed to bind to this sequence using EMSA (data not shown). In addition, GCNF had no repression function on a reporter gene driven by this region of the Sox2 loci (data not shown). Thus, it seems more likely that the failure to repress Sox2 gene expression in GCNF−/− ES cells during RA treatment is an indirect consequence of GCNF inactivation. The loss of Sox2 repression in differentiated GCNF−/− ES cells is probably a direct consequence of the maintenance of Oct4 expression because previous reports have shown that Oct4 regulates Sox2 expression by binding to a conserved POU binding site in the Sox2 enhancer region (1, 7, 38).
FGF4 is a growth factor that had been previously described to be regulated by synergistic interaction of Sox2 and Oct4 (2, 20). Loss of Sox2 and Oct4 repression in GCNF−/− ES cells maintains expression of FGF4 in these ES cells during RA-induced differentiation. Stella is reported to be a pluripotent marker gene expressed in cells of the germ lineage, preimplantation embryos, and ES cells of the inner cell mass, similar to the Oct4 gene expression pattern (5, 11, 39). Interestingly, in the human genome, Stella is clustered with the Nanog and GDF3 genes on chromosome 12p13, which are all highly expressed in human testicular germ cell tumors (11). Maintained expression of Stella in differentiated GCNF−/− ES cells suggests that GCNF, Oct4, or Nanog may directly regulate the expression of Stella, which is currently unknown.
The expression of several ES cell marker genes was maintained in the absence of GCNF in RA-treated ES cells, which raised the possibility that RA-induced differentiation of the ES cells was impaired. However, analysis of the expression of other ES cell marker genes, such as FGF8, showed the same pattern of repression in wild-type and mutant cells (Fig. 5A). Analysis of the FGF8 promoter also showed that there was no GCNF binding sites, and GCNF could not repress FGF8 promoter activity. Even though the GCNF−/− ES cells lost the repression of pluripotency genes, RA treatment still induced differentiation because the neural marker gene Nestin was induced (data not shown). We conclude that sustained expression of Oct 4 and Nanog is directly caused by the loss of GCNF repression, while the loss of repression of FGF4 and Sox2 is an indirect result of the loss of repression of Oct4.
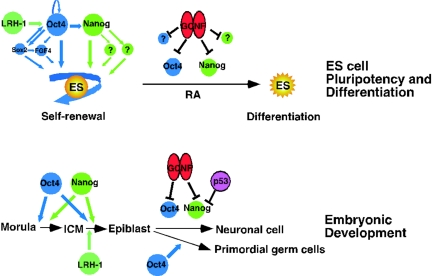
Based on our findings from experimentation with ES cell RA-induced differentiation and embryonic development, we have established a model for GCNF regulation of pluripotent ES cell marker genes Oct4 and Nanog and early development of embryos shown in Fig. 8. In ES cells, induction of GCNF expression facilitates differentiation through inhibition of the pluripotent state by repression of important regulatory genes, such as Oct4 and Nanog. The significance of our findings is greatly increased by recent demonstrations that Oct4 and Sox2 regulate Nanog gene expression and also autoregulate Oct4 expression (27, 36). Thus, ES cell self-renewal and pluripotence are maintained by positive feedforward and feedback loops that maintain the expression of Oct4, Nanog, and Sox2. Thus, loss of repression of Oct4 and Nanog cannot occur passively and requires active repression mediated by GCNF binding to the Oct4 and Nanog genes (Fig. 8).
FIG. 8.
Model for regulation of pluripotency gene expression by GCNF in ES cells and embryos. Expression of Oct4 (blue) and Nanog (green) maintains self-renewal of ES cells. Oct4 also activates expression of other pluripotency genes, such as Sox2 and FGF4. The target genes of Nanog are currently unknown (?). Upon RA treatment, induction of GCNF (red) and p53 (magenta) represses the expression of Oct4, Nanog, and other unknown genes facilitating the differentiation of ES cells. GCNF also represses the expression of Oct4 and Nanog in the somatic cells of developing embryos, such as neuronal cells, during gastrulation and thus indirectly restricts their expression to primordial germ cells. LRH-1 (aquamarine) activates the expression of Oct4 in undifferentiated ES cells and the epiblast of embryos.
In addition, it was recently reported that the tumor suppressor p53 suppresses Nanog gene expression in ES cells by recruitment of phosphorylated p53 protein into the murine Sin3a complex and subsequently binds to the promoter region of the Nanog gene (30). Whether GCNF and p53 functionally interact to repress Nanog gene expression is currently unknown. Another nuclear receptor, LRH-1, maintains the expression of Oct4 in undifferentiated ES cells and at the epiblast stage of mouse embryos (19). LRH-1 and GCNF bind to the same element in the Oct4 proximal promoter but have antagonistic effects on transcription of the gene, suggesting a reciprocal regulatory model (Fig. 8).
In the developing embryo, GCNF indirectly restricts the expression of Oct4 and Nanog to primordial germ cells after gastrulation by repressing expression of these genes in somatic cells. Inactivation of GCNF expression in embryos leads to maintained Oct4 and Nanog expression in some but not all somatic cells after gastrulation (17) (Fig. 7). The repression of Oct4 and Nanog expression in some somatic cells of GCNF−/− embryos might be indicative of additional factors involved in the repression of these genes in specific cell types or lineages. Alternatively, the repression of the Oct4 and Nanog genes in some somatic cells of the GCNF−/− embryos might reflect passive loss of activation of these genes, as expression of their regulators is in turn switched off. Other factors involved in regulating Oct4 expression are Oct4 itself, Sox2, SP1, RAR, SF1, and LRH-1 (3, 19, 36).
Interestingly, GCNF−/− embryos gastrulate and eventually die around E10.5, due to cardiovascular defects. The death of the GCNF−/− embryos occurs at a stage later than would be predicted for maintained expression of the pluripotency factors Oct4 and Nanog. Thus, it is possible that while Oct4 and Nanog maintain pluriopotence, they cannot block differentiation and patterning at gastrulation, which is consistent with the observations in RA-induced differentiation of the GCNF−/− ES cells. However, a caveat to this argument is the ectopic expression of Oct4 and Nanog proteins in the GCNF−/− embryos has not been established.
In terms of regulating pluripotence, ES cell self-renewal and differentiation are coordinately regulated upon differentiation. GCNF plays an important role in inhibiting the pluripotent phenotype by actively repressing the expression of the important regulatory factors that are high in the hierarchy of maintenance of pluripotence. Repression of Oct4 and Nanog in turn leads to loss of repression of downstream pluripotency genes that are lower in the regulatory hierarchy. Thus, it would be predicted that novel genes that contain DR0 elements that are directly regulated by GCNF are likely to be high in the hierarchy of pluripotency regulation.
The function of GCNF in regulating pluripotent gene expression implies a potential application in the treatment of human cancer cells which reexpress preimplantation embryonic genes, such as Oct4 (32). The major implication for the important position of GCNF in regulating the hierarchy of pluripotency genes lies in the fact that GCNF in an orphan nuclear receptor that holds the potential to be a ligand regulated transcription factor. A GCNF antagonist would be predicted to inhibit the repression of pluripotency genes, leading to maintenance of pluripotency, ES cell self-renewal, and propagation, thus facilitating ES cell culture and derivation of new ES cell lines. This an exciting possibility for the future of stem cell therapies based on ES cell manipulation that needs to be explored.
Acknowledgments
This work was supported by U54 HD07495 and R01 DK73524 to A.J.C.
We thank Thomas Zwaka for critical reading of the manuscript.
REFERENCES
- 1.Ambrosetti, D. C., C. Basilico, and L. Dailey. 1997. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 17:6321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avilion, A. A., S. K. Nicolis, L. H. Pevny, L. Perez, N. Vivian, and R. Lovell-Badge. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17:126-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnea, E., and Y. Bergman. 2000. Synergy of SF1 and RAR in activation of Oct-3/4 promoter. J. Biol. Chem. 275:6608-6619. [DOI] [PubMed] [Google Scholar]
- 4.Boiani, M., S. Eckardt, H. R. Scholer, and K. J. McLaughlin. 2002. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 16:1209-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowles, J., R. P. Teasdale, K. James, and P. Koopman. 2003. Dppa3 is a marker of pluripotency and has a human homologue that is expressed in germ cell tumours. Cytogenet Genome Res. 101:261-265. [DOI] [PubMed] [Google Scholar]
- 6.Calhoun, J. D., R. R. Rao, S. Warrenfeltz, R. Rekaya, S. Dalton, J. McDonald, and S. L. Stice. 2004. Transcriptional profiling of initial differentiation events in human embryonic stem cells. Biochem. Biophys. Res. Commun. 323:453-464. [DOI] [PubMed] [Google Scholar]
- 7.Catena, R., C. Tiveron, A. Ronchi, S. Porta, A. Ferri, L. Tatangelo, M. Cavallaro, R. Favaro, S. Ottolenghi, R. Reinbold, H. Scholer, and S. K. Nicolis. 2004. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J. Biol. Chem. 279:41846-41857. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643-655. [DOI] [PubMed] [Google Scholar]
- 9.Chen, F., A. J. Cooney, Y. Wang, S. W. Law, and B. W. O'Malley. 1994. Cloning of a novel orphan receptor (GCNF) expressed during germ cell development. Mol. Endocrinol. 8:1434-1444. [DOI] [PubMed] [Google Scholar]
- 10.Chung, A. C., D. Katz, F. A. Pereira, K. J. Jackson, F. J. DeMayo, A. J. Cooney, and B. W. O'Malley. 2001. Loss of orphan receptor germ cell nuclear factor function results in ectopic development of the tail bud and a novel posterior truncation. Mol. Cell. Biol. 21:663-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, A. T., R. T. Rodriguez, M. S. Bodnar, M. J. Abeyta, M. I. Cedars, P. J. Turek, M. T. Firpo, and R. A. Reijo Pera. 2004. Human STELLAR, NANOG, and GDF3 genes are expressed in pluripotent cells and map to chromosome 12p13, a hotspot for teratocarcinoma. Stem Cells 22:169-179. [DOI] [PubMed] [Google Scholar]
- 12.Constantinescu, S. 2003. Stemness, fusion and renewal of hematopoietic and embryonic stem cells. J. Cell. Mol. Med. 7:103-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooney, A. J., G. C. Hummelke, T. Herman, F. Chen, and K. J. Jackson. 1998. Germ cell nuclear factor is a response element-specific repressor of transcription. Biochem. Biophys. Res. Commun. 245:94-100. [DOI] [PubMed] [Google Scholar]
- 14.Eddy, S. 2003. HMMER: profile HMMs for protein sequence analysis. Bioinformatics 14:755-563. [Google Scholar]
- 15.Evans, M., and S. Hunter. 2002. Source and nature of embryonic stem cells. C. R. Biol. 325:1003-1007. [DOI] [PubMed] [Google Scholar]
- 16.Evans, M. J., and M. H. Kaufman. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154-156. [DOI] [PubMed] [Google Scholar]
- 17.Fuhrmann, G., A. C. Chung, K. J. Jackson, G. Hummelke, A. Baniahmad, J. Sutter, I. Sylvester, H. R. Scholer, and A. J. Cooney. 2001. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev. Cell 1:377-387. [DOI] [PubMed] [Google Scholar]
- 18.Gardner, R. L., and F. A. Brook. 1997. Reflections on the biology of embryonic stem (ES) cells. Int. J. Dev. Biol. 41:235-243. [PubMed] [Google Scholar]
- 19.Gu, P., B. Goodwin, A. C.-K. Arthur C.-K. Chung, X. Xu, D. Wheeler, R. P. Price, C. Galardi, L. Peng, A. M. Latour, B. H. Koller, J. Gossen, S. A. Kliewer, and A. J. Cooney. 2005. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol. Cell. Biol. 25:3492-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanna, L. A., R. K. Foreman, I. A. Tarasenko, D. S. Kessler, and P. A. Labosky. 2002. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 16:2650-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hart, A. H., L. Hartley, M. Ibrahim, and L. Robb. 2004. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev. Dyn. 230:187-198. [DOI] [PubMed] [Google Scholar]
- 22.Hatano, S. Y., M. Tada, H. Kimura, S. Yamaguchi, T. Kono, T. Nakano, H. Suemori, N. Nakatsuji, and T. Tada. 2005. Pluripotential competence of cells associated with Nanog activity. Mech. Dev. 122:67-79. [DOI] [PubMed] [Google Scholar]
- 23.Heinzer, C., U. Susens, T. P. Schmitz, and U. Borgmeyer. 1998. Retinoids induce differential expression and DNA binding of the mouse germ cell nuclear factor in P19 embryonal carcinoma cells. Biol. Chem. 379:349-359. [DOI] [PubMed] [Google Scholar]
- 24.Hummelke, G. C., M. L. Meistrich, and A. J. Cooney. 1998. Mouse protamine genes are candidate targets for the novel orphan nuclear receptor, germ cell nuclear factor. Mol. Reprod. Dev. 50:396-405. [DOI] [PubMed] [Google Scholar]
- 25.Karolchik, D. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31:51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, Y., and Y. Tsunoda. 1993. Totipotency and pluripotency of embryonic nuclei in the mouse. Mol. Reprod. Dev. 36:276-278. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda, T., M. Tada, H. Kubota, H. Kimura, S. Y. Hatano, H. Suemori, N. Nakatsuji, and T. Tada. 2005. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 25:2475-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lan, Z. J., P. Gu, X. Xu, and A. J. Cooney. 2003. Expression of the orphan nuclear receptor, germ cell nuclear factor, in mouse gonads and preimplantation embryos. Biol. Reprod. 68:282-289. [DOI] [PubMed] [Google Scholar]
- 29.Lan, Z. J., P. Gu, X. Xu, K. J. Jackson, F. J. DeMayo, B. W. O'Malley, and A. J. Cooney. 2003. GCNF-dependent repression of BMP-15 and GDF-9 mediates gamete regulation of female fertility. EMBO J. 22:4070-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, T., C. Chao, S. Saito, S. J. Mazur, M. E. Murphy, E. Appella, and Y. Xu. 2005. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 7:165-171. [DOI] [PubMed] [Google Scholar]
- 31.Mitsui, K., Y. Tokuzawa, H. Itoh, K. Segawa, M. Murakami, K. Takahashi, M. Maruyama, M. Maeda, and S. Yamanaka. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631-642. [DOI] [PubMed] [Google Scholar]
- 32.Monk, M., and C. Holding. 2001. Human embryonic genes re-expressed in cancer cells. Oncogene 20:8085-8091. [DOI] [PubMed] [Google Scholar]
- 33.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379-391. [DOI] [PubMed] [Google Scholar]
- 34.Niwa, H. 2001. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct. Funct. 26:137-148. [DOI] [PubMed] [Google Scholar]
- 35.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 36.Okumura-Nakanishi, S., M. Saito, H. Niwa, and F. Ishikawa. 2005. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 280:5307-5317. [DOI] [PubMed] [Google Scholar]
- 37.Pan, G. J., Z. Y. Chang, H. R. Scholer, and D. Pei. 2002. Stem cell pluripotency and transcription factor Oct4. Cell Res. 12:321-329. [DOI] [PubMed] [Google Scholar]
- 38.Remenyi, A., K. Lins, L. J. Nissen, R. Reinbold, H. R. Scholer, and M. Wilmanns. 2003. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17:2048-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitou, M., S. C. Barton, and M. A. Surani. 2002. A molecular programme for the specification of germ cell fate in mice. Nature 418:293-300. [DOI] [PubMed] [Google Scholar]
- 40.Scholer, H. R., R. Balling, A. K. Hatzopoulos, N. Suzuki, and P. Gruss. 1989. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 8:2551-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholer, H. R., A. K. Hatzopoulos, R. Balling, N. Suzuki, and P. Gruss. 1989. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 8:2543-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoorlemmer, J., A. van Puijenbroek, M. van Den Eijnden, L. Jonk, C. Pals, and W. Kruijer. 1994. Characterization of a negative retinoic acid response element in the murine Oct4 promoter. Mol. Cell. Biol. 14:1122-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan, Z. H., A. Medvedev, T. Hirose, H. Gotoh, and A. M. Jetten. 1997. Characterization of the response element and DNA binding properties of the nuclear orphan receptor germ cell nuclear factor/retinoid receptor-related testis-associated receptor. J. Biol. Chem. 272:10565-10572. [DOI] [PubMed] [Google Scholar]
- 44.Yoshimizu, T., N. Sugiyama, M. De Felice, Y. I. Yeom, K. Ohbo, K. Masuko, M. Obinata, K. Abe, H. R. Scholer, and Y. Matsui. 1999. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 41:675-684. [DOI] [PubMed] [Google Scholar]