Abstract
New cultured strains of the planctomycete division (order Planctomycetales) of the domain Bacteria related to species in the genera Gemmata and Isosphaera were isolated from soil, freshwater, and a laboratory ampicillin solution. Phylogenetic analysis of the 16S rRNA gene from eight representative isolates showed that all the isolates were members of the planctomycete division. Six isolates clustered with Gemmata obscuriglobus and related strains, while two isolates clustered with Isosphaera pallida. A double-membrane-bounded nucleoid was observed in Gemmata-related isolates but not in Isosphaera-related isolates, consistent with the ultrastructures of existing species of each genus. Two isolates from this study represent the first planctomycetes successfully cultivated from soil.
The increasing significance of planctomycetes (i.e., members of the order Planctomycetales) has been reported within the areas of evolution, molecular ecology, and cell biology (6, 24, 27, 41). The planctomycetes form a distinctive division of the domain Bacteria (20, 31, 35) characterized by budding reproduction and the absence of peptidoglycan in their cell walls (14, 19). Four cultured genera so far have been described; all are aerobic chemoheterotrophs—Planctomyces, Pirellula, Gemmata, and Isosphaera. Gemmata and Isosphaera have been described only on the basis of a single species each: Gemmata obscuriglobus and Isosphaera pallida (5, 9, 38). The diversity of the group extends beyond these to include at least two other planctomycete genera at Candidatus status (“Brocadia” and “Kuenenia”) and consisting of anaerobic ammonium-oxidizing autotrophs (32, 40).
Planctomycetes were all originally isolated from aquatic habitats as diverse as acid bogs and sewage treatment plants, and direct molecular cloning and probe studies have extended this diversity (1, 5, 9, 12, 17, 27, 28, 29, 30, 33, 43, 44). Many of the original isolates have been confirmed as planctomycetes by phylogenetic analysis of rRNA sequences (11, 42). Seven clones related to planctomycetes were reported as soil cluster II in a study of soil from the Mt. Coot-tha region; this was one of the first molecular ecology investigations of the bacterial diversity in any soil microbial community (21, 37). Subsequently, several studies reported clones of planctomycetes from clone libraries prepared from different environments (2, 10, 15, 17, 18, 32, 40). Because of the increasing evidence for the ubiquitous distribution of planctomycetes in the environment from direct cloning retrieval, isolation of such organisms in pure cultures is needed to enrich the understanding of their potential ecological role through study of their physiology and growth characteristics.
Samples used as inocula in isolation attempts were water obtained from University Lake, The University of Queensland St. Lucia Campus, Brisbane, Queensland, Australia, and the central library fountain of the same campus; soil collected from Mt. Coot-tha, Brisbane, Queensland, Australia, and Charlies Crossing Rd., Upper Coomera, Queensland, Australia; and a laboratory ampicillin solution (10 mg ml−1) which had not been filter sterilized. Primary isolation and subculture media were M1 agar (30), R2A agar (BBL, Becton Dickinson Microbiology Systems, Cockeysville, Md.), and PYGV agar (39), all with 1.5% agar, or M1 broth, 0.1% peptone in tap water, or sterile tap water; the media contained combinations of either ampicillin (200 μg ml−1), streptomycin (1,000 μg ml−1), and cycloheximide (20 to 100 μg ml−1) or penicillin G (500 μg ml−1), streptomycin (1,000 μg ml−1), and amphotericin B (Fungizone) (0.25 or 0.5 μg ml−1). Water samples were inoculated onto selective agar media; soil samples were suspended in sterile tap water before inoculation onto agar; and enrichments were inoculated by mixing an equal volume of water sample with a sterile deionized water solution of penicillin, streptomycin, and amphotericin B, each at double the concentration in selective media. The isolates have been deposited into the Australian Collection of Microorganisms (ACM) as ACM 5050 to ACM 5057. Micromanipulation was performed by using the methods and apparatus of Skerman (34) to separate a Gemmata-like isolate (strain Soil9) from a cocultured nonplanctomycete isolate after the conventional subculturing method failed to do so.
Cells of isolates were negatively stained with filtered (0.22-μm-pore-size membrane) 1% uranyl acetate containing 0.4% sucrose. Methods for cryofixation, cryosubstitution, and thin sectioning are fully described elsewhere (22).
DNA was extracted from cultures grown on M1 agar by one of the following methods: (i) direct heat lysis from colony growth deposited in a PCR reaction tube, done at 98°C for 2 to 10 min, with Taq buffer (Bresatec) and primers and Taq polymerase (Bresatec) subsequently added to form a complete PCR reaction mixture; (ii) boiling cells in 200 μl of 5% Chelex 100 resin (Bio-Rad catalog no. 142-2832) and, after centrifugation, adding 4 to 5 μl of supernatant per 100 μl of PCR reaction mixture; or (iii) lysing cells in saline-EDTA with lysozyme (37°C for 30 min), followed by proteinase K-sodium dodecyl sulfate (50 to 55°C for 30 min), this step being followed by phenol-chloroform (1:1) protein denaturation and cleanup of the aqueous phase using a Prep-A-Gene kit (Bio-Rad) with 0.5 μl of the resulting DNA used per 100 μl of PCR reaction mixture.
Small-subunit (16S) ribosomal DNA (rDNA) was amplified using primer set 27f-1492r (16). PCR and cycle sequencing were performed as previously described (7, 13). The primers used for 16S rDNA sequencing of isolates were those described by Lane (16) or planctomycete-amplifying primers of our own design: 27f, 342r, 357f, 530f, 1114f, PLN945f (5′-GGGGCTCACACAAGCGGTGG), 1101r, 687r, PLN926r (5′-CCACCGCTTGTGTGAGCCCC), 1230fplanc [5′-CTGCACACGT(G/C)CTACAATG], and 1492r (primers are based on Escherichia coli numbering [3]).
16S rDNA sequences of new isolates were imported into the ARB software package (26; www.biol.chemie.tu-muenchen.de/pub/ARB/) for alignment and phylogenetic analysis. The integrity of sequence data was checked by comparing complementary stem regions determined from the secondary structure of the 16S rRNA molecule. Variable regions of the alignment were excluded from the data set by using the bacterial mask of Lane (16) within ARB. Trees were constructed by using distance matrix method DNADIST and neighbor-joining tree generation within NEIGHBOR of the PHYLIP software implemented within 2D-ANGIS (Australian National Genomic Information Service; http://morgan.angis.su.oz.au/). Bootstrap analysis was performed by using SEQBOOT and CONSENSE within PHYLIP with 100 resamplings. A pairwise similarity matrix was generated by using the ae2 alignment editor (ftp://ftp.bio.indiana.edu/molbio/unix/) and the ARB alignment for the relevant phylotypes.
Eight planctomycete-like isolates were isolated from soil, freshwater, or a laboratory antibiotic solution by using enrichment in the presence of combinations of streptomycin as a protein synthesis inhibitor, penicillin or ampicillin as a peptidoglycan synthesis-inhibiting compound, and either amphotericin B or cycloheximide as an antifungal agent. Among the eight representative isolates studied, strains JW9-3f1, JW10-3f1, CJuql4, and CJuql1 are all from a eutrophic lake (on the St. Lucia Campus of The University of Queensland); JW11-2f5 is from water from an ornamental fountain (central library fountain); JW3-8s0 is from Mt. Coot-tha soil; strain Soil9 is from soil at another site (Charlies Crossing Rd.); and C2-3 is from a laboratory ampicillin solution. Another 17 isolates derived from Mt. Coot-tha soil were found by repetitive extragenic palindromic-PCR typing to be replicates indistinguishable by band patterns from strain JW3-8s0 (unpublished data). The new isolates are all pink or red pigmented, except for strain C2-3, which is nonpigmented. Strains C2-3 and CJuql1, unlike the I. pallida type strain, which needs incubation in enriched CO2 at 42°C, can both grow in air not supplemented with CO2 at 28°C, as can all the Gemmata-like isolates. All of the new isolates, both those with close relations to Gemmata and those with closest relations to Isosphaera, possess spherical cells and reproduce by budding, as determined by phase-contrast microscopy. Morphological features are illustrated for some of these isolates in the electron micrographs of Fig. 1. Young cultures of pink Gemmata-like isolates are motile, but no swimming motility was observed in cultures of Isosphaera-like isolates. Isosphaera-like isolates did not exhibit filaments.
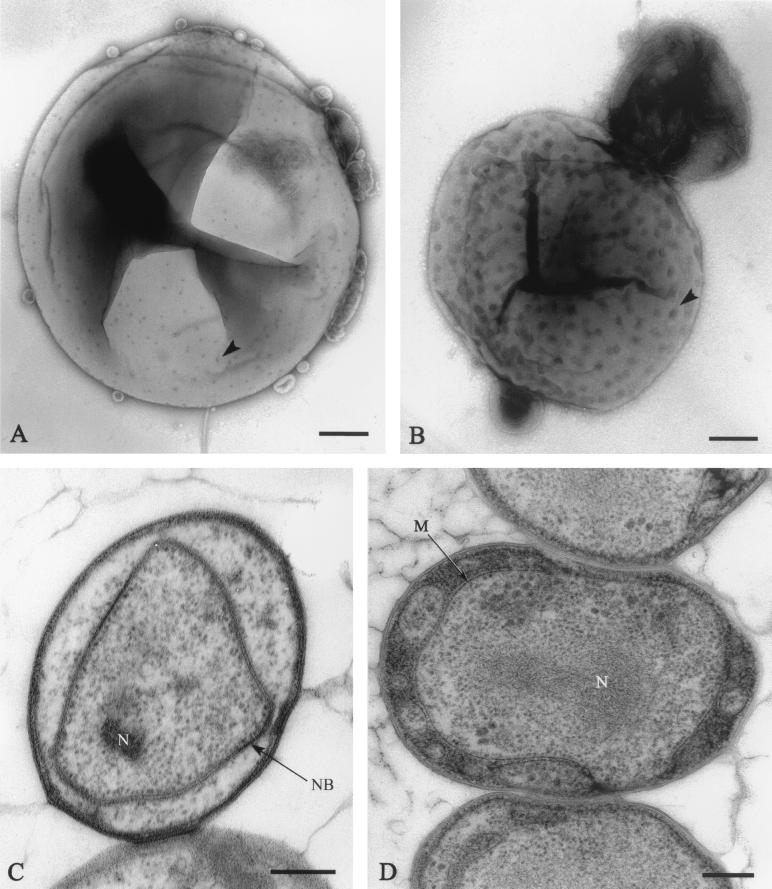
FIG. 1.
Electron micrographs of cells of new Gemmata-like and Isosphaera-like isolates. (A) Negatively stained cell of the Gemmata-like strain JW11-2f5 showing crateriform structures (arrowhead) and coccoid cell morphology. Bar marker, 200 nm. (B) Negatively stained budding cell of Isosphaera-like strain CJuql1 showing uniform crateriform structures (arrowhead) on the mother cell and coccoid cell morphology. Bar marker, 200 nm. (C) Thin section of Gemmata-like cryosubstituted cell of strain JW3-8s0 showing the double-membrane-bounded nuclear body (NB) and nucleoid (N) enclosed within it. Bar marker, 200 nm. (D) Thin section of Isosphaera-like strain C2-3 possessing a fibrillar nucleoid (N) within a cytoplasmic compartment bounded by a single membrane (M) only. Bar marker, 200 nm.
When viewed by transmission electron microscopy after negative staining with uranyl acetate, all of the isolates display crateriform structures on their cell walls, as illustrated for the Gemmata-like strain JW11-2f5 and the Isosphaera-like strain CJuql1 in Fig. 1A and B. These structures are electron-dense circular regions of negative stain accumulation characteristic of planctomycetes and were evenly distributed over the cell surface, whether the strain proved to be Gemmata-like or Isosphaera-like by 16S rRNA sequence analysis. Thin-sectioned cells of strains JW3-8s0, JW9-3f1, JW10-3f1, JW11-2f5, Soil9, and CJuql4 (Gemmata-like, as determined by 16S rRNA sequence relationships; see below) all display a nuclear body containing the nucleoid itself and surrounded by an envelope consisting of two membranes, as illustrated for representative strain JW3-8s0 in Fig. 1C. They also possess at the cell rim a peripheral electron-dense region bounded by an inner intracytoplasmic membrane, which divides the cell into two compartments, one with ribosomes and one without them. The central ribosome-containing region is the one which contains the nuclear body compartment. Even strain JW10-3f1, which is relatively distant from G. obscuriglobus but still clusters within the Gemmata group, possesses a nuclear body. Compartmentalization was seen for cells of C2-3 and CJuql1 (Isosphaera-like as determined by 16S rRNA sequence relationships; see below); however, their compartments differ in appearance from those of the Gemmata group, as illustrated for Isosphaera-like strain C2-3 in Fig. 1D. There are no double-membrane-bounded nuclear bodies in the Isosphaera-like strains. The nucleoid is contained within an internal compartment bounded by a single membrane only.
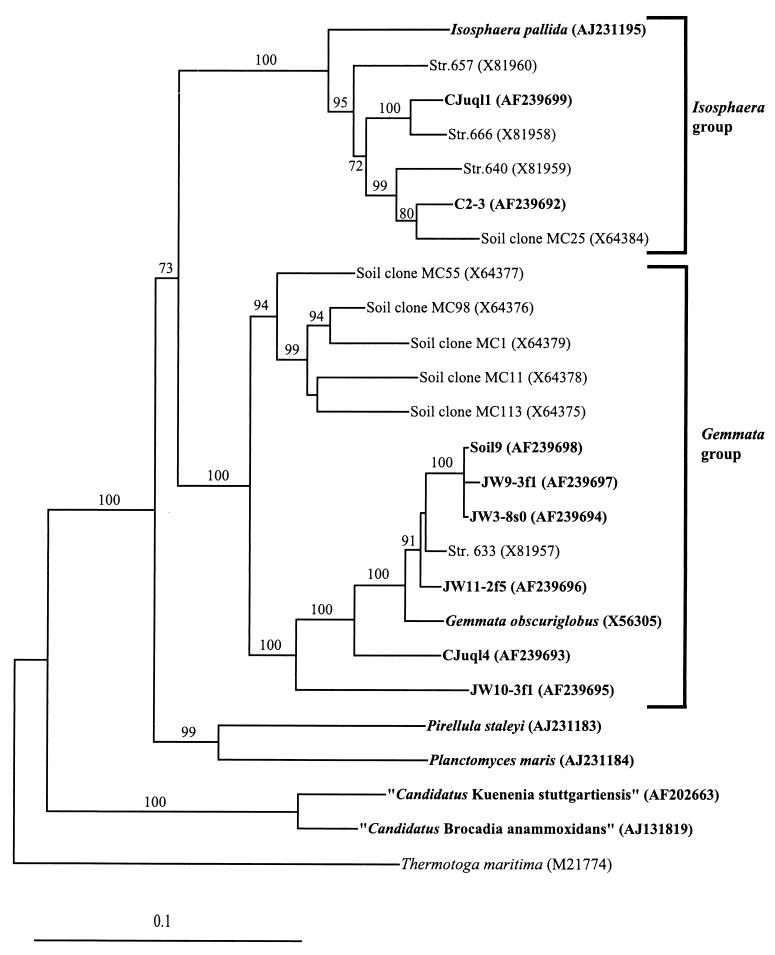
Nearly complete 16S rRNA gene sequences (1,447 to 1,511 bp) were obtained for the eight new isolates, and their phylogenetic relationship to existing planctomycete isolates and cloned sequences from uncultured members of native microbial communities was assessed. Six of the eight new isolates, JW11-2f5, JW9-3f1, Soil9, JW3-8s0, CJuql4, and JW10-3f1, clustered within the Gemmata group defined by G. obscuriglobus. Isolates C2-3 and CJuql1 clustered within the Isosphaera group defined by I. pallida (Fig. 2). Similarity values between the Gemmata-related strains and G. obscuriglobus and the Isosphaera-related strains and I. pallida are consistent with phylogenetic clusters. The Gemmata-like strain JW11-2f5 is the most similar to G. obscuriglobus at 96.4%, while JW9-3f1, Soil9, and JW3-8s0 are greater than 99% similar to each other and range from 94.3 to 95.1% in sequence similarity to G. obscuriglobus. Strains CJuql4 and JW10-3f1 display the lowest level of similarity to G. obscuriglobus at 92.2 and 86.6%, respectively. The Isosphaera-like strains C2-3 and CJuql1 display only 89.4 and 89.8% similarity to I. pallida but cluster with the Mt. Coot-tha soil clone MC25 and the northern Germany compost water strain 666, respectively. Most of the planctomycete signature nucleotides for 16S rRNA that are characteristic of a given genus group (7), either the Gemmata group or the Isosphaera group, are also present in the new isolates. Isolates clustered with the Gemmata group all possess in their 16S rRNA gene a 10-base sequence insertion between E. coli positions 998 and 999, characteristic of the group (21).
FIG. 2.
Evolutionary distance tree derived from comparative analysis of 16S rDNAs from freshwater and soil isolates and reference strains of the order Planctomycetales. Database accession numbers are shown in parentheses after species, strain, or clone names. Bootstrap values of greater than 70% from 100 bootstrap resamplings from the distance analysis are presented at nodes. Thermotoga maritima was used as an outgroup. Isolates from this study and representative named species of the planctomycetes are indicated in bold. The scale bar represents 0.1 nucleotide substitution per nucleotide position.
We have demonstrated for the first time that planctomycetes, specifically Gemmata-like representatives of this division (as illustrated by strains JW3-8s0 and Soil9), can be isolated in pure cultures from a soil habitat. Planctomycetes were previously isolated from soil only by use of 16S rRNA gene libraries (2, 15, 21) and in situ hybridization experiments (43). The type species of the genus, G. obscuriglobus, was isolated from freshwater (5), and other Gemmata-like isolates were isolated from leakage water from a compost heap (42). Of particular interest is the isolation of a Gemmata-related strain, JW3-8s0, from soil of Mt. Coot-tha. This isolation confirms the occurrence of culturable Gemmata strains in soil from the same site as was used in one of the first molecular 16S rRNA gene direct cloning studies of a soil habitat (21); in that study, several 16S rDNA clones were shown to be most closely related to G. obscuriglobus within the planctomycetes. These results are consistent with those of other studies using direct molecular ecology or hybridization methods to demonstrate that planctomycetes may be widely distributed soil inhabitants (4, 15, 17, 43, 44). Isolation in this study depended on the use of the β-lactam antibiotic penicillin or ampicillin, the aminoglycoside protein synthesis-inhibiting antibiotic streptomycin, and antifungal compounds; in one case, isolation occurred from an ampicillin solution habitat, consistent with known antibiotic sensitivities for planctomycetes and their cell wall composition (14, 19).
The planctomycete genera Gemmata and Isosphaera are at present described only on the basis of a single species each. In this study, we have isolated additional strains which are closely related to existing members of these genera. This study thus supplements previous work in which strains closely related to Gemmata and Isosphaera were isolated from various aquatic habitats (30, 42). The latter study (42) demonstrated the isolates within the Schlesner collection (30) to include a further Gemmata-like strain (strain 633) from leakage water from a compost heap and Isosphaera-like strains (strains 640, 657, and 666) also from leakage water from a compost heap. Isosphaera-like strains were also isolated from activated sludge in another study (25).
Of great interest also is the finding that six newly isolated strains with a close relationship to Gemmata, as demonstrated by 16S rDNA phylogenetic analysis and by the presence of Gemmata-specific signature nucleotide sequences in their 16S rDNAs, all possess membrane-bounded nuclear bodies, that is, nucleoid regions bounded by two membranes, analogous to that described for G. obscuriglobus (8). This finding is consistent with the apparent correlation between ultrastructural features and the phylogenetic position of a species within the planctomycetes, noted previously for Pirellula species (7, 22). Of particular interest from this perspective is the finding that even the isolate within the Gemmata group most distantly related to G. obscuriglobus, strain JW10-3f1, displays a clear nuclear body. The compartmentalization of the cells of the Gemmata-like and Isosphaera-like strains described here is also consistent with that which has been described for these genera in detail elsewhere (8, 23, 24).
According to 16S rRNA sequence analysis, six of the isolates clustered within the Gemmata group, while two clustered within the Isosphaera group. On the basis of the criteria of Stackebrandt and Goebel (36), according to which two organisms related at 16S rRNA sequence homology values of less than 97.5% are unlikely to be members of the same species, each of the new Gemmata-like isolates appears to represent a species separate from the reference strain, G. obscuriglobus. The Gemmata group and the Isosphaera group are clearly both quite phylogenetically diverse, perhaps to an even greater extent than indicated in a previous culture-based study (42). The new isolates are consistent with the 16S rRNA phylogenetic profile and diversity of the family Planctomycetaceae reported in previous studies (7, 42). The isolate from Mr. Coot-tha soil, JW3-8s0, clustered closer to G. obscuriglobus (UQM 2246) and strain 633, a Gemmata-like isolate from northern Germany (42), than to the Gemmata-like MC clones. Both the similarity matrix and the phylogenetic tree showed that JW9-3f1, Soil9, and JW3-8s0 may be members of the same species, as their similarity values are over 99%. The fact that sequences of these soil isolates differ from those of all MC clones may indicate that the diversity of planctomycetes in soil is wider than molecular cloning methods could detect and emphasizes the need for culturing as well as direct PCR retrieval for studying microbial diversity; however, this fact may also reflect differences in community composition at the same site at different times.
This study has established the validity of selective enrichment with antibiotics based on known properties of cultured planctomycetes for the isolation of planctomycetes from soil and water. In addition, it has demonstrated that culturable planctomycetes and members of the Gemmata group in particular exist in a soil habitat previously known to harbor planctomycetes only via evidence from direct cloning studies. It has also established (i) that significant 16S rRNA structural characteristics of the Gemmata group of planctomycetes known previously from only one cultured strain and cloned sequences are conserved throughout the group and (ii) that the ultrastructural feature of cell compartmentalization involving a membrane-bounded nucleoid is correlated with the phylogenetic distinction of the Gemmata group of planctomycetes and is present in Gemmata isolates from different habitats. The planctomycetes are now recognized as being ubiquitous in the environment. Furthermore, several studies indicate that these organisms may make up a significant proportion of the total microbial population (4, 27, 43).
The discovery of planctomycetes in new ecological niches and of new planctomycetes highlights the fact that the order Planctomycetales is a ubiquitously distributed and highly diverse order within the domain Bacteria that may harbor organisms which play a much more significant role in the environment than first imagined with regard to the cycling of organic and inorganic compounds. The development in this study of simple methods for the isolation of planctomycetes from soil should allow more extensive studies of the distribution, physiology, and ecological role of this distinct group of the domain Bacteria in microbial communities of agricultural and environmental significance.
Nucleotide sequence accession numbers.
Nucleic acid sequences from this study have been deposited in the GenBank database under accession numbers AF239692 to AF239699.
Acknowledgments
We thank AMRAD Discovery Technologies Pty Ltd. for graduate student stipend support to J.W., Australian Postgraduate Award Scheme for stipend support for C.I., and AMRAD Discovery Technologies Pty Ltd. and the Australian Research Council for grant funding for planctomycete research in the laboratory of J.A.F.
We thank Philip Hugenholtz for discussions on the use of ARB and Chris Hayward for editorial assistance to J.W.
REFERENCES
- 1.Bauld, J., and J. T. Staley. 1976. Planctomyces maris sp. nov.: a marine isolate of the Planctomyces-Blastocaulis group of budding bacteria. J. Gen. Microbiol. 97:45–55. [Google Scholar]
- 2.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosius, J., M. L. Palmer, J. P. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derakshani, M., T. Lukow, and W. Liesack. 2001. Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 67:623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franzmann, P. D., and V. B. D. Skerman. 1984. Gemmata obscuriglobus, a new genus and species of budding bacteria. Antonie Leeuwenhoek 50:261–268. [DOI] [PubMed] [Google Scholar]
- 6.Fuerst, J. A. 1995. The planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology 141:1493–1506. [DOI] [PubMed] [Google Scholar]
- 7.Fuerst, J. A., H. G. Gwilliam, M. Lindsay, A. Lichanska, C. Belcher, J. E. Vickers, and P. Hugenholtz. 1997. Isolation and molecular identification of planctomycete bacteria from postlarvae of giant tiger prawn, Penaeus monodon. Appl. Environ. Microbiol. 63:254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuerst, J. A., and R. I. Webb. 1991. Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. USA 88:8184–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovannoni, S. J., E. Schabtach, and R. W. Castenholz. 1987. Isosphaera pallida, gen. and comb. nov., a gliding, budding eubacterium from hot springs. Arch. Microbiol. 147:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray, J. P., and R. P. Herwig. 1996. Phylogenetic analysis of the bacterial communities in marine sediments. Appl. Environ. Microbiol. 62:4049–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griepenburg, U., N. Ward-Rainey, S. Mohamed, H. Schlesner, H. Marxen, F. A. Rainey, E. Stackebrandt, and G. Auling. 1999. Phylogenetic diversity, polyamine pattern and DNA base composition of members of the order Planctomycetales. Int. J. Syst. Bacteriol. 49:689–696. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch, P., and M. Muller. 1985. Planctomyces limnophilus sp.nov., a stalked and budding bacterium from fresh water. Syst. Appl. Microbiol. 6:276–280. [Google Scholar]
- 13.Hugenholtz, P., E. Stackebrandt, and J. A. Fuerst. 1994. A phylogenetic analysis of the genus Blastobacter with a view to its future reclassification. Syst. Appl. Microbiol. 130:200–205. [Google Scholar]
- 14.König, H., H. Schlesner, and P. Hirsch. 1984. Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch. Microbiol. 138:200–205. [Google Scholar]
- 15.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 6:3614–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rRNA sequencing, p.115–175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., Chichester, United Kingdom.
- 17.Lee, S. Y., J. Bollinger, D. Bezdicek, and A. Ogram. 1996. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62:3787–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liesack, W., P. H. Janssen., F. A. Rainey, N. L. Ward-Rainey, and E. Stackebrandt. 1997. Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques, p.375–439. In J. D. Van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, Inc., New York, N.Y.
- 19.Liesack, W., H. König, H. Schlesner, and P. Hirsch. 1986. Chemical composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella/Planctomyces group. Arch. Microbiol. 145:361–366. [Google Scholar]
- 20.Liesack, W., R. Söller, T. Steward, H. Haas, S. Giovannoni, and E. Stackebrandt. 1992. The influence of tachytelically (rapidly) evolving sequences on the topology of phylogenetic trees—intrafamily relationships and the phylogenetic position of Planctomycetaceae as revealed by comparative analysis of 16S ribosomal RNA sequences. Syst. Appl. Microbiol. 15:357–362. [Google Scholar]
- 21.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsay, M. R., R. I. Webb, and J. A. Fuerst. 1997. Pirellulosomes: a new type of membrane-bounded cell compartment in planctomycete bacteria of the genus Pirellula. Microbiology 143:739–748. [DOI] [PubMed] [Google Scholar]
- 23.Lindsay, M. R., R. I. Webb, H. Hosmer, and J. A. Fuerst. 1995. Effects of fixative and buffer on morphology and ultrastructure of a freshwater planctomycete, Gemmata obscuriglobus. J. Microbiol. Methods 21:45–54. [Google Scholar]
- 24.Lindsay, M. R., R. I. Webb, M. Strous, M. S. M. Jetten, M. K. Butler, R. J. Forde, and J. A. Fuerst. 2001. Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175:413–429. [DOI] [PubMed] [Google Scholar]
- 25.Liu J. R., C. A. McKenzie, E. M. Seviour, R. I. Webb, L. L. Blackall, C. P. Saint, and R. J. Seviour. 2001. Phylogeny of the filamentous bacterium ‘Nostocoida limicola’ III from activated sludge. Int. J. Syst. Evol. Microbiol. 51:195–202. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554–568. [DOI] [PubMed] [Google Scholar]
- 27.Neef, A., R. Amann, H. Schlesner, and K.-H. Schleifer. 1998. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257–3266. [DOI] [PubMed] [Google Scholar]
- 28.Schlesner, H. 1986. Pirella marina sp. nov., a budding, peptidoglycan-less bacterium from brackish water. Syst. Appl. Microbiol. 8:177–180. [Google Scholar]
- 29.Schlesner, H. 1989. Planctomyces brasiliensis sp.nov., halotolerant bacterium from a salt pit. Syst. Appl. Microbiol. 12:159–161. [Google Scholar]
- 30.Schlesner, H. 1994. The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst. Appl. Microbiol. 17:135–145. [Google Scholar]
- 31.Schlesner, H., and E. Stackebrandt. 1986. Assignment of the genera Planctomyces and Pirella to a new family Planctomycetaceae fam. nov. and description of the order Planctomycetales ord. nov. Syst. Appl. Microbiol. 8:174–176. [Google Scholar]
- 32.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. Jetten, J. W. Metzger, K.-H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93–106. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt, J. M. 1978. Isolation and ultrastructure of freshwater strains of Planctomyces. Curr. Microbiol. 1:65–70. [Google Scholar]
- 34.Skerman, V. B. D. 1968. A new type of micromanipulator and microforge. J. Gen. Microbiol. 54:287–297. [DOI] [PubMed] [Google Scholar]
- 35.Stackebrandt, E., A. Fischer, P. Hirsch, T. Roggentin, and H. Schlesner. 1986. The phylogeny of an ancient group of budding peptidoglycan-less eubacteria: the genera Planctomyces and Pirella. Endocytobiosis Cell Res. 3:29–40. [Google Scholar]
- 36.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849. [Google Scholar]
- 37.Stackebrandt, E., W. Liesack, and B. M. Goebel. 1993. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 7:232–236. [DOI] [PubMed] [Google Scholar]
- 38.Staley, J. T., and J. A. Fuerst. 1989. Budding and/or appendaged bacteria, p.1890–1961. In J. T. Staley, M. P. Bryant, N. Pfenning, and J. G. Holt (ed.), Bergey’s manual of systematic bacteriology, vol. 3. The Williams & Wilkins Co., Baltimore, Md.
- 39.Staley, J. T., J. A. Fuerst, S. Giovannoni, and H. Schlesner. 1992. The order Planctomycetales and the genera Planctomyces, Pirellula, Gemmata and Isosphaera, p.3710–3731. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed., vol. IV. Springer-Verlag, New York, N.Y.
- 40.Strous, M., J. A. Fuerst, E. H. M. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. M. Jetten. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446–449. [DOI] [PubMed] [Google Scholar]
- 41.Vergin, K. L., E. Urbach, J. L. Stein, E. F. DeLong, B. D. Lanoil, and S. J. Giovannoni. 1998. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64:3075–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward, N., F. A. Rainey, E. Stackebrandt, and H. Schlesner. 1995. Unraveling the extent of diversity within the order Planctomycetales. Appl. Environ. Microbiol. 61:2270–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zarda, B., D. Hahn, A. Chatzinotas, W. Schönhuber, A. Neef, R. I. Amann, and J. Zeyer. 1997. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch. Microbiol. 168:185–192. [Google Scholar]
- 44.Zhou, J., M. E. Davey, J. B. Figueras, E. Rivkina, D. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913–3919. [DOI] [PubMed] [Google Scholar]