Abstract
To test the role of gene order in globin gene expression, mutant human β-globin locus yeast artificial chromosome constructs were used, each having one additional globin gene encoding a “marked” transcript (ɛm, γm, or βm) integrated at different locations within the locus. When a βm-globin gene was placed between the locus control region (LCR) and the ɛ-globin gene, βm-globin expression dominated primitive and definitive erythropoiesis; only βm-globin mRNA was detected during the fetal and adult definitive stages of erythropoiesis. When an Aγm-globin gene was placed at the same location, Aγm-globin was expressed during embryonic erythropoiesis and the fetal liver stage of definitive erythropoiesis but was silenced during the adult stage. The downstream wild-type γ-globin genes were not expressed. When an ɛm-globin gene was placed between the δ- and β-globin genes, it remained silent during embryonic erythropoiesis; only the LCR-proximal wild-type ɛ-globin gene was expressed. Placement of a βm-globin gene upstream of the Gγ-globin gene resulted in expression of βm-globin in embryonic cells and in a significant decrease in expression of the downstream wild-type β-globin gene. These results indicate that distance from the LCR, an inherent property of spatial gene order, is a major determinant of temporal gene expression during development.
The human β-globin locus consists of five functional globin genes, which are expressed during ontogeny in the order in which they lie on the chromosome, 5′-ɛ-Gγ-Aγ-δ-β-3′. During development, two switches occur in globin gene expression, from embryonic to fetal, coinciding with the transition from embryonic to definitive erythropoiesis, and from fetal to adult around the perinatal period. High-level expression of the β-like globin genes is dependent upon both gene-proximal and -distal cis-acting regulatory elements. The major distal regulatory element is the locus control region (LCR). It contains five DNase I-hypersensitive sites (5′HS1 to -5) (42) and is located 6 to 22 kb upstream of the ɛ-globin gene. Functions of LCRs include tissue-specific enhancement of gene expression, chromatin-opening activity, insulation of the locus from the effects of surrounding chromatin, and determination of replication timing and choice of origin used (26, 27). It is currently assumed that LCR sequences, together with their cognate transcription factors and coactivators, form an active site “hub” that interacts with gene-proximal elements and their bound transcription factors to form a transcriptosome specific for the globin gene(s) expressed at each developmental stage (8, 20, 26, 34). Less-conserved sequences between the HS cores also may function in globin gene regulation (19), perhaps by acting to constrain the complex conformation.
Because the spatial arrangement of the β-like globin genes parallels their temporal expression pattern, it has been proposed that gene order or proximity to the LCR may be a determinant of the order of globin gene expression during development. Previous studies have established an effect of either gene order or distance from the LCR (10, 18, 39, 44). Data from transgenic mice produced with α- and β-globin genes linked at various distances to the LCR demonstrated that proximity to the LCR increased the probability of interaction with the LCR (18). The effect of gene order on globin gene developmental regulation was tested by producing transgenic mice containing two tandemly arranged γ- or β-globin or γβ- and βγ-globin genes linked to a μLCR (39). These studies showed that the trans-acting environment is the primary determinant for correct developmental control but that proximity to the LCR also is involved in regulation. The effect of distance from the LCR was analyzed using a larger 70-kb β-globin locus in two linked cosmids (10). When the ɛ-globin gene was replaced with a marked β-globin (βm) gene, β-globin expression was sustained throughout development. In contrast, when the βm-globin gene was placed upstream of the δ-globin gene, βm-globin gene expression accounted for 75% of total β-globin gene expression (βm + βwt). Inversion of the whole locus relative to the LCR in β-globin locus yeast artificial chromosome (β-YAC) transgenic mice resulted in β-globin gene expression throughout development, with no ɛ-globin gene expression and limited γ-globin gene expression during primitive erythropoiesis, again suggesting that proximity to the LCR is a determinant of globin gene expression (44).
In this study we sought to further test the role of spatial gene order in the control of globin gene expression using β-YAC transgenic mice carrying constructs containing one additional globin gene encoding a “marked” transcript (ɛm, γm, or βm) integrated at a different locations within the locus. Our results indicate that distance from the LCR as determined by gene order, in addition to gene-proximal cis-acting elements, determines the appropriate developmental expression of β-like globin genes.
MATERIALS AND METHODS
YAC constructs.
A 213-kb β-YAC containing the human β-globin locus (Fig. 1) was utilized for all experiments (16, 36, 38). This β-YAC was previously estimated to be 248 kb, but with a nearly complete sequence of the human β-globin locus region now available (http://globin.cse.psu.edu) (4), the size was more accurately determined. An approximately 6-kb sequence gap still exists downstream of 3′HS1 in the latest release of the human genome sequence. The numerical coordinates used in construct descriptions below are based upon GenBank file U01317 unless indicated otherwise. All of the YAC modifications were introduced by yeast integrating plasmid (YIP)-mediated “pop-in,” “pop-out” or two-step gene replacement methods of homologous recombination as previously described (1, 31, 38). Briefly, for the “pop-in,” “pop-out” method, YIP clone DNA was linearized with the indicated restriction enzyme and transformed into spheroplasted Saccharomyces cerevisiae strain AB1380 containing the 213-kb β-YAC. Transformants were selected for uracil prototrophy on complete medium (CM), and correct recombination was determined by Southern blot hybridization analysis. Spontaneous excision of the YIP was induced by overnight growth in nonselective CM. The yeast cells were plated on 5-fluoroorotic acid (5-FOA) CM plates to select for loss of the URA3 gene residing on the YIP vector, which results in 5-FOA resistance. The two-step gene replacement protocol utilized the same selection steps as the “pop-in,” “pop-out” method, except two fragments with recombinogenic ends were used in sequential transformations. The first fragment contained the target sequence and the marked globin gene interrupted with the URA3 cassette; the second fragment was identical, but it lacked the URA3 gene. Correct recombinants were confirmed by Southern blot hybridization analysis and PCR.
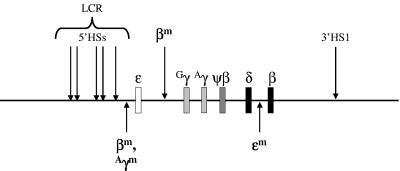
FIG. 1.
Gene order constructs. The human β-globin locus is shown as a line, the genes are indicated as rectangles, and the DNase I-hypersensitive sites are marked with arrows. The various marked globin genes and their sites of integration are shown with arrows linking them to the site of integration.
βm 5′ ɛ β-YAC.
A 4.1-kb βm-globin HpaI-XbaI fragment (GenBank coordinates 61340 to 65439) was inserted into an EcoRV site between 5′HS1 and the ɛ-globin gene at GenBank coordinate 15182 (Fig. 1). The mark in the βm-globin gene was an 8-bp ClaI linker ligated into an S1 nuclease-blunted NcoI site of the β-globin promoter at GenBank coordinate 62185 (11). A net increase of 4 bp resulted, and the linker was constructed in such a way that the initiation methionine codon was preserved. A 3,139-bp BglII-HindIII fragment (GenBank coordinates 13769 to 16908) was isolated from pHSI (gift from Q. Li) and ligated into BamHI-HindIII-cut and calf intestinal alkaline-phosphatase-treated pUC19 to produce pUC-5′ ɛ. The 4.1-kb HpaI-XbaI βm-globin fragment was isolated from pSP73βm (11), the XbaI overhang filled with E. coli polymerase I Klenow enzyme and ligated into EcoRV-cut and phosphatase-treated pUC-5′ ɛ, resulting in pUC βm 5′ ɛ. An 1,166-bp HindIII-cut and Klenow enzyme-filled URA3 fragment was isolated from YEP24-neo (gift from C. Huxley) and ligated into BamHI-cut, Klenow enzyme-filled and phosphatase-treated pUC βm 5′ ɛ to generate pUC βm::URA3 5′ ɛ. This vector was digested with PvuII to yield an 8,282-bp fragment encompassing the βm::URA3 5′ ɛ insert and two smaller fragments bearing the pUC19 plasmid vector sequence. This 8.3-kb fragment was used to transform yeast; selection was for uracil prototrophy. For the second transformation and recombination, a 2,116-bp ClaI-PstI fragment containing a portion of the βm-globin gene (GenBank coordinates 62202 to 64319) was purified from digested pUC βm 5′ ɛ; selection was for 5-FOA resistance. Wild-type and marked gene transcripts in β-YAC transgenic mice were distinguished by RNase protection analysis (RPA) or reverse transcriptase-PCR (RT-PCR) coupled with restriction fragment length polymorphism (RFLP) analysis.
βm 5′ Gγ β-YAC.
A 4.1-kb HpaI-XbaI βm fragment was inserted 5′ of the Gγ-globin gene into a BglII site at GenBank coordinate 32837 (Fig. 1). A 3,167-bp XbaI fragment 5′ to the Gγ-globin gene (GenBank coordinates 32011 to 35178) was purified from pA-36 (gift from B. Forget) and ligated into the XbaI-digested and phosphatase-treated YIP vector pRS406 (Stratagene, La Jolla, CA) to produce pRS-5′ Gγ. The 4.1-kb HpaI-XbaI βm-globin fragment was prepared as described in the preceding section and ligated into BglII-cut, Klenow enzyme-filled and phosphatase-treated pRS-5′ Gγ to produce pRS βm 5′ Gγ. This YIP was linearized with Tth111I prior to transformation of β-YAC-bearing yeast. Differential analysis of βm- and βwt-globin transcripts in murine lines was as described in the preceding section for the βm 5′ ɛ β-YAC construct.
Aγm 5′ ɛ β-YAC.
A 5.4-kb Aγm-globin SspI fragment (GenBank coordinates 38683 to 44077) was inserted at the same EcoRV site described for the βm 5′ ɛ β-YAC construct (Fig. 1). The Aγm-globin gene contains a six-base-pair deletion at +21 to +26 relative to the Aγ-globin translation start site (41). A 3,139-bp BglII-HindIII fragment (GenBank coordinates 13769 to 16908) was isolated from pHSI and ligated into BamHI-HindIII-cut and calf intestinal alkaline phosphatase-treated pBluescript KS+ (Stratagene, La Jolla, CA) to produce pBS-5′ ɛ. The 5.4-kb SspI-SalI fragment encompassing the Aγm-globin gene was isolated from pUC19 Aγm (+) (39) and ligated into EcoRV-digested and phosphatase-treated pBS-5′ ɛ to produce pBS Aγm 5′ ɛ. An 8.5-kb SalI-NotI fragment encompassing the Aγm 5′ ɛ insert was isolated from pBS Aγm 5′ ɛ and subcloned into SalI-NotI-digested and phosphatase-treated pRS406 to produce pRS Aγm 5′ ɛ. This YIP construct was linearized with PshAI (GenBank coordinate 16300) prior to transformation of yeast and recombination into the β-YAC. Differential analysis of Aγm- and Aγwt-globin transcripts in transgenic mouse lines was as described for the βm 5′ ɛ β-YAC construct.
ɛm 3′ δ β-YAC.
A 3.7-kb EcoRI ɛm-globin fragment (GenBank coordinates 17482 to 21233) was inserted between the δ- and β-globin genes in a HindIII site at GenBank coordinate 59641 (Fig. 1). The mark is an eight-base-pair XhoI linker inserted in a PvuII site at GenBank coordinate 19524 (Q. Li, personal communication). A 2,595-bp EcoRI fragment encompassing sequence 3′ to the δ-globin gene (GenBank coordinates 58034 to 60629) was isolated from pδ BglII and ligated into EcoRI-digested and phosphatase-treated pUC19, in which the HindIII site had been previously ablated by digestion with HindIII, Klenow enzyme filling of the 5′ overhangs, and religation to generate pUC-3′ δ. A 3,770-bp EcoRI fragment encompassing the ɛ-globin gene was isolated from pɛ3.7 (2), Klenow enzyme filled, and ligated in HindIII-cut, Klenow enzyme-filled and phosphatase-treated pUC-3′ δ to produce pUC ɛ 3′ δ. A 1,516-bp ClaI-SpeI fragment encompassing the ɛ-globin mark was purified from pɛ3.7m (K. R. Peterson, unpublished construct) and ligated into ClaI-SpeI-cut and phosphatase-treated pUC ɛ 3′ δ to generate pUC ɛm 3′ δ. The 6,370-bp EcoRI fragment containing the entire insert from pUC ɛm 3′ δ was subcloned into EcoRI-digested and phosphatase-treated pRS406 to produce pRS ɛm 3′ δ. A PmlI site (GenBank coordinate 60366) was used to linearize this YIP vector prior to transformation of yeast and “pop-in,” “pop-out” recombination. Measurement of ɛm- and ɛwt-globin transcripts in transgenic mice was as described above for the βm 5′ ɛ β-YAC construct.
YAC purification and transgenesis.
Purification of β-YAC DNAs for microinjection was performed as described previously (16, 35-38). Briefly, pulsed-field gel electrophoresis of preparative high-mass DNA agarose plugs was used to fractionate yeast chromosomes and the β-YAC, followed by electrophoretic concentration of the YAC DNA in a 4% low-melting-point agarose gel, β-agarase digestion of the concentrated YAC DNA plugs, and filtration of the liquefied agarose solution through a 0.22-μm syringe filter in high-salt microinjection buffer. Purified and filtered YAC DNAs were microinjected into fertilized mouse oocytes (C57/Bl6) and then transferred to pseudopregnant foster mothers to produce transgenic mice. Transgenic animals were identified by PCR analysis of tail biopsy DNA. Founders were bred with nontransgenic mice to produce F1 progeny; these in turn were bred to obtain F2 staged embryos, fetuses, and adults. All structure-function analyses were carried out beginning with the F2 generation.
Structural analysis of transgene integrity and copy number determination.
Structural analysis of β-YAC transgenes was performed to assess the integrity and colinearity of transgenes as described previously (31, 38). High-molecular-weight DNA embedded in agarose was prepared, and slices were digested with SfiI. Digested DNA was fractionated by pulsed-field gel electrophoresis and transferred to a Zeta-probe charged nylon membrane (Bio-Rad, Hercules, CA) by capillary blotting. Strips representing individual lanes of the gel were cut from the blot, and each one was hybridized with one of the following 32P-radiolabeled probes: 0.7-kb PstI 5′HS3, 1.9-kb HindIII 5′HS2, 1.8-kb XbaI 5′HS1, 3.7-kb EcoRI ɛ-globin gene, 2.4-kb EcoRI fragment 3′ Aγ-globin gene, 1.0-kb EcoRV ψβ region, 2.1-kb PstI fragment 5′ δ-globin gene, 0.9-kb EcoRI-BamHI fragment 3′ β-globin gene, 1.4-kb XbaI DF10 (3′HS1), 1.9-kb BglII HPFH3, 0.5-kb HindIII H500, and 1.5-kb EcoRI-BglII HPFH6. DNA probes were labeled using the Decaprime II kit (Ambion, Austin, TX), following the manufacturer's instructions.
The presence of 5′HS4 and 5′HS5 was confirmed by Southern blot hybridization using a 32P-radiolabeled 3.2-kb BamHI fragment (Globin Gene Server human β-globin locus coordinates 215110 to 218333) as a probe (4). Digestion with SfiI (218055) and MluI (226095) produces a diagnostic 8.0-kb fragment for 5′HS5 and 5′HS4. All lines contained 5′HS4 and -5 unless otherwise indicated.
Transgene copy numbers were determined by Southern blot hybridization using LCR 5′HS3, γ-, and β-globin gene fragment probes coradiolabeled with a mouse Thy 1.1 gene fragment as described previously (15, 30). Probe-specific activity was corrected, and comparison of hybridization signal intensity between human globin transgenes and the endogenous diploid murine Thy1.1 gene gave an estimate of transgene copy number. Radioactive signals were quantitated using a phosphorimager (Packard Instruments, Meriden, CT).
RPA.
Total RNA was isolated from yolk sac, fetal liver, and blood of developmentally staged transgenic F2, or later, embryos, fetuses, and adults by the method of Chomczynski and Sacchi (7) using the RNAgents total RNA isolation reagents (Promega, Madison, WI) as previously described (39). Human and murine globin mRNAs were detected by RNase protections performed as described previously (21, 39). Antisense RNA probes were prepared using the MaxiScript II kit (Ambion, Austin, TX). Template DNAs used to prepare riboprobes to measure endogenous mouse α- and ζ-globins were pT7Moα and pT7Moζ, respectively (28), and to measure human ɛ-, γ-, and β-globins, the templates were pT7Huɛ(188), pT7Aγm(170), and pT7βm, respectively (25, 43). Individual human β-like globin gene expression levels were quantitated as percentages of total human β-like globin gene expression or as percentages of murine α- and ζ-globin gene expression corrected for transgene and endogenous murine gene copy number using data collected on a Cyclone phosphorimager and OptiQuant analysis software (Packard Instruments, Meriden, CT). RPAs were done separately with each conceptus of each litter to minimize experimental error and determine variation in globin gene expression. For each construct, three transgenic lines were established, and with rare exceptions, two or three conceptuses were analyzed at each developmental stage. Quantitative data include the means and standard deviations from two or three experiments for each line. Thus, averages at each developmental stage are indicative of 4 to 9 experimental values for individual transgenic lines and 12 to 27 values for combined lines. Wild-type β-YAC lines 4 and 10 served as controls for RPA and RT-PCR RFLP (40). All experiments performed included a parallel set of wild-type β-YAC samples (data not shown). In all instances the outcome confirmed the data we have compiled in published or unpublished form over the past 12 years regarding expression levels in these lines. Wild-type β-YAC control lanes were included in each figure for comparison purposes, for size or band location indicators only, or to demonstrate proper function of riboprobes. For statistical analysis, the Student t test of comparison of means was performed, and differences were considered significant at P values of <0.025.
Semiquantitative RT-PCR.
cDNA was synthesized from 10-day embryonic yolk sac, 12-day fetal liver, 14-day fetal liver, or adult mouse blood total RNA using an oligo-dT primer (Promega Corp., Madison, WI) and Superscriptase II reverse transcriptase (Invitrogen, Carlsbad, CA). One microgram of total RNA was mixed with 0.5 μg oligo-dT and sterile water in a final volume of 11 μl. The mixture was heated to 70°C for 10 min and then chilled on ice. Once cooled, 4 μl 5× first-strand buffer, 2 μl 0.1 M dithiothreitol, 1 μl 10 mM deoxynucleoside triphosphate mix, and 1 μl RNasin (Promega Corp, Madison, WI) were added and the reaction mixture was heated at 42°C for 2 min. One μl Superscript II RT was added, and the 20-μl reaction mixture was incubated at 42°C for 50 min. RT enzyme was heat inactivated by incubation for 15 min at 70°C.
PCR was performed essentially as described by Omori et al. (32) using Biolase Taq polymerase (Bioline USA, Randolph, MA) in a 25-μl reaction mix containing 1× NH4 buffer [16 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8), 0.01% Tween-20], 2 mM MgCl2, 100 μM deoxynucleoside triphosphates, 1 μCi [α-32P]dCTP, 0.4 pmol forward primer, 0.4 pmol reverse primer, 1 U Taq polymerase, and globin gene-specific primers (Invitrogen, Carlsbad, CA). All primers were designed to cross an intron so that RNA-templated reverse-transcribed DNA amplification products could be distinguished from gene amplification products. For human β-globin and mouse α-globin, initial denaturation was at 95°C for 5 min, followed by 18 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s; final extension was at 72°C for 10 min. Separate reactions were performed for human β-globin and mouse α-globin. Parallel titration reactions showed that this cycle number was within the exponential amplification range. The primer sequences for RT-PCR were as follows: βm 5′ ɛ β-YAC or βm 5′ Gγ β-YAC β-globin forward, 5′-ACATTTGCTTCTGACACAACTG-3′, and reverse, 5′-AGGAGCCTGAAGTTCTCAG-3′. The mouse α-globin primers employed were as described previously (32). A similar scheme was used for the human ɛ-globin gene products. Amplification conditions were identical to those used for β-globin. The primer sequences for RT-PCR were as follows: ɛm 3′ δ β-YAC forward, 5′-CATATCTGCTTCCGACACAG-3′, and reverse, 5′-GGGGTAAACAACGAGGAGTC-3′. Mouse α-globin was included as a control in separate reactions.
A nonquantitative RT-PCR was employed for the γ-globins. Initial denaturation was at 95°C for 5 min, followed by 20 to 40 cycles of 1-min steps at 95°C, 60°C, and 72°C; final extension was at 72°C for 10 min. The primer sequences for RT-PCR were as follows: Aγm 5′ ɛ β-YAC γ-globin forward, 5′-ACACTCGCTTCTGGAACGT-3′, and reverse, 5′-TAGACAACCAGGAGCCTTCC-3′.
RT-PCR RFLP analysis.
RT-PCR primers were designed to amplify regions of the wild-type and marked globin gene products that were distinguished by RFLP analysis. Following semiquantitative RT-PCR, 5 μl of the RT-PCR products were digested overnight with restriction enzymes and fractionated on an 8% native polyacrylamide gel, which was dried and subjected to phosphorimager analysis (Typhoon, GE Healthcare, Waukesha, WI). In the βm-globin gene constructs, the normal NcoI site was replaced with a ClaI site by virtue of introduction of the mark. Thus, the βwt-globin product was digested with NcoI but not ClaI, and the converse was true for the βm-globin product. For the βm 5′ ɛ β-YAC and βm 5′ Gγ β-YAC mice, the full-length PCR product was 370 bp. Digestion of the wild-type product with NcoI produced 320-bp plus 50-bp bands, whereas the βm-globin product was not digested. ClaI digested the βm-globin product but not the βwt-globin product to produce 324-bp plus 50-bp fragments. All experiments were run twice using the samples indicated in the figures.
The ɛm 3′ δ β-YAC samples were similarly processed, except that XhoI was used to distinguish the ɛwt- and ɛm-globin RT-PCR products. The 164-bp ɛwt-globin product was not digested with XhoI, whereas the 172-bp ɛm-globin product was cut into 155-bp and 17-bp fragments.
For the Aγm-globin construct, following RT-PCR, portions of the PCR product were subjected to overnight digestion with appropriate restriction enzymes, fractionated on a 2 to 3% agarose gel, and visualized by ethidium bromide staining. DdeI digestion was used to analyze the Aγm- and Aγwt-globin gene products. The Aγwt-globin RT-PCR product from Aγm 5′ ɛ β-YAC mice was cut with DdeI into 130-bp plus 19-bp fragments, whereas the Aγm-globin RT-PCR product remained as the full-length 159-bp fragment.
RESULTS
The goal of our studies was to analyze the effect of gene order on the temporal expression of the human β-like globins using β-YAC transgenic mice. Towards this purpose, four modified β-YAC constructs were made (Fig. 1). Each of the constructs contained an additional marked β-like globin gene inserted in a nonnative location within a 213-kb β-YAC. For the construct βm 5′ ɛ β-YAC, a βm-globin gene was inserted 1.6 kb 3′ to 5′HS1 and 4.4 kb 5′ to the ɛ-globin gene CAP site. The βm 5′ Gγ β-YAC construct bore a βm-globin gene inserted 1.7 kb 5′ to the Gγ-globin CAP site. For the Aγm 5′ ɛ β-YAC construct, an Aγm-globin gene was inserted at the same location as the βm-globin gene in the βm 5′ ɛ β-YAC construct. The ɛm 3′ δ β-YAC construct contained an ɛm-globin gene inserted 4.3 kb 3′ to δ-globin exon 2 and 2.5 kb 5′ to the β-globin CAP site.
An adult β-globin gene located proximally to the LCR is preferentially expressed throughout development.
Three βm 5′ ɛ β-YAC transgenic lines were established. Each line harbored at least one intact human β-globin locus transgene extending from 5′HS5 through the β-globin gene enhancer (see Fig. S1 in the supplemental material). Expression studies were done by RNase protection assay using embryonic, fetal, and adult erythroid cells of F2 progeny of the transgenic lines.
As shown in Fig. 2, the βm-globin gene was expressed throughout development, beginning as early as detectable (day 8) and continuing through adulthood. ɛ-Globin mRNA was not detected and γ-globin gene expression was reduced during embryonic erythropoiesis. At day 10 postconception, the γ-globin level was 15% of total human globin expression [γ/(γ+β)] versus 90% in wild-type β-YAC transgenic mice [γ/(ɛ+γ)].
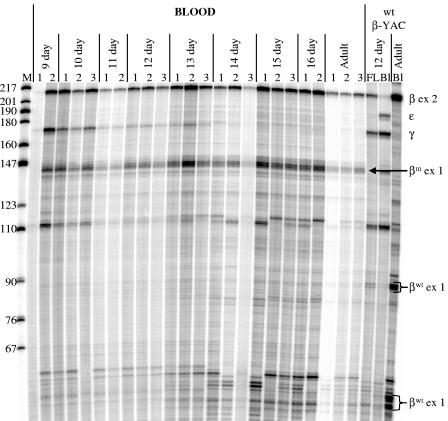
FIG. 2.
Developmental profile of β-like globin gene expression in βm 5′ ɛ β-YAC transgenic mice. Total RNA was isolated from the blood of staged fetuses and subjected to RNase protection analysis. Representative data from line 1 are shown. The days after conception are shown at the top of the autoradiograph. The numbers indicate individuals of the same developmental stage. Antisense RNA probes for human ɛ-, γ-, and β-globin mRNAs were utilized. Protected fragments are displayed on the right. The protected fragment sizes are as follows: human β-globin exon 2 (β ex 2), 205 bp; ɛ-globin exon 2 (ɛ), 188 bp; γ-globin exon 2 (γ), 170 bp; marked β-globin exon 1 (βm ex 1), 145 bp; wild-type β-globin exon 1 (βwt ex 1), 91 and 50 bp. The β-globin antisense probe hybridizes with exons 1 and 2 of both the βwt- and βm-globin mRNAs. Thus, the exon 2 protected fragment is the sum of these two gene products, and the exon 1 protected fragments distinguish the two mRNAs. The marker (M) in the left lane is a radiolabeled MspI digest of plasmid pBR322; fragment sizes are shown in base pairs. FL, fetal liver; Bl, blood; wt β-YAC, wild-type 213-kb human β-globin locus YAC transgenic mouse line 4 (42). Lines 2 and 3 showed an identical phenotype.
Mean γ-globin expression in the day 10 yolk sac for all lines (nine embryos), calculated as a percentage of murine α-like globin genes and corrected for endogenous and transgene copy numbers, was 30.7% ± 21.5% compared to 59.9 ± 2.4% (P < 0.025) in wild-type β-YAC lines. No γ-globin mRNA was detected in the liver during fetal definitive erythropoiesis by RNase protection assay or antibody staining for globin chains (data not shown), although γ-globin mRNA was detected in the peripheral circulation, which still contains high numbers of embryonic erythroblasts, through day 14 (Fig. 2). Only βm-globin was detected during adult definitive erythropoiesis; no wild-type β-globin mRNA was observed.
Semiquantitative RT-PCR coupled with restriction enzyme digestion analysis (RT-PCR RFLP) was performed using primers for the β-globin 5′ untranslated region (UTR) and exon 1/exon 2 boundary. cDNAs were synthesized from adult blood RNA and amplified by PCR, and the PCR products were digested overnight with restriction enzymes ClaI or NcoI. These data confirmed that only the βm-globin gene was expressed in adult mice carrying the βm 5′ ɛ β-YAC construct (Fig. 3). This analysis also demonstrated that the wild-type β-globin gene was not expressed at any stage of development. ClaI treatment showed complete digestion of the 370-bp fragment, whereas NcoI did not digest the PCR product at all, demonstrating that all of the β-globin mRNA synthesis in these lines was βm-globin. Furthermore, βwt-globin protected fragments were not observed in Fig. 2, nor were they observed in RNase protection assays using only a β-globin exon 1 antisense riboprobe to exclusively detect and distinguish βm- and βwt-globin mRNAs (data not shown).
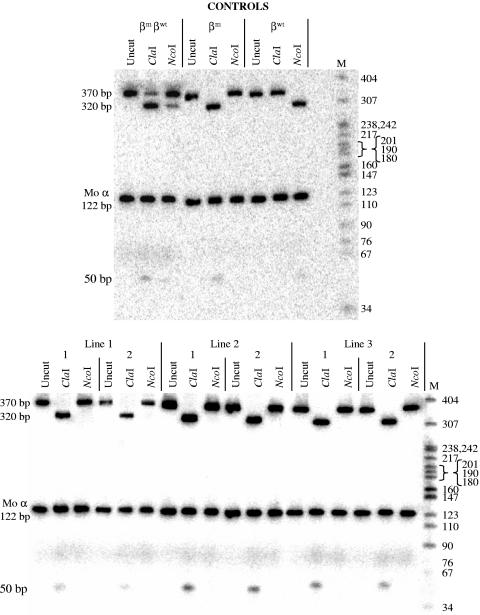
FIG. 3.
Semiquantitative RT-PCR RFLP analysis of β-globin expression in βm 5′ ɛ β-YAC transgenic mouse lines. RT-PCRs were performed to amplify the 5′ end of the β-globin transcript in adult mouse blood total RNA samples from βm 5′ ɛ β-YAC lines 1, 2, and 3 (See Materials and Methods). Representative data from two adult individuals for each line are shown. The 4-bp insertion in the βm-globin gene created a diagnostic ClaI restriction enzyme site and destroyed an NcoI site. ClaI digestion of the βm-globin produces 324-bp and 50-bp fragments, whereas NcoI does not cut (βm control lane). The reverse occurs for wild-type β-globin; NcoI produces 320-bp and 50-bp fragments, whereas ClaI does not cut (βwt control lane). Both enzymes digest proportionately to gene product levels as shown in the βmβwt control lane, where the majority of expression in these μLCRβmβwt mice is βm-globin. Top panel, controls: βmβwt, μLCRβmβwt transgenic mouse adult blood (39); βm, μLCRβmAγ transgenic mouse adult blood (39); βwt, wild-type 213-kb β-YAC transgenic mouse line 4 adult blood (40). Bottom panel: βm 5′ ɛ β-YAC lines 1, 2, and 3. Digest fragment sizes are shown on the left, along with the location and size of the mouse α-globin product used to normalize our data. Molecular weight marker (M) sizes are listed on the right side of the panels.
Midlocus localization of an adult β-globin gene alters its expression pattern.
Three βm 5′ Gγ β-YAC transgenic lines were established. All lines contained at least one β-globin locus copy that encompassed the LCR and the βm-globin gene (see Fig. S2 in the supplemental material). However, line 3 (Δβwt) contained a deletion of the normally located βwt-globin gene but contained sequences through the ψβ-globin gene. Globin gene expression was measured by RPA in embryonic, fetal, and adult erythroid cells of F2 animals.
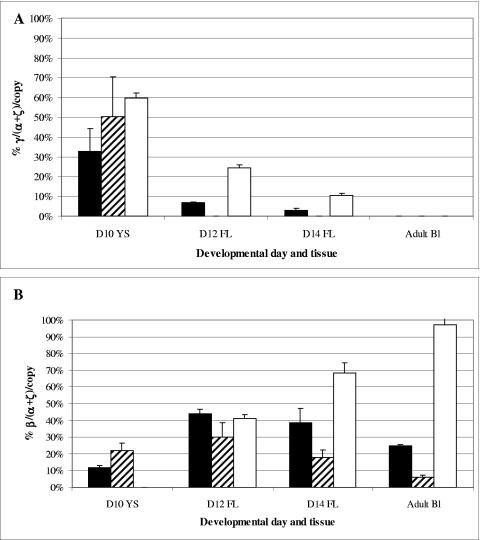
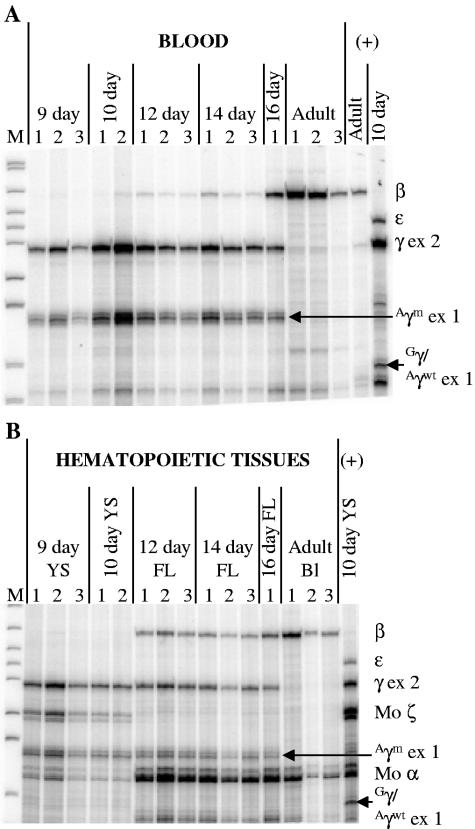
Representative results by RNase protection assay are shown in Fig. 4. βm-Globin expression was detected in the embryonic yolk sac, and expression continued through definitive erythropoiesis in both fetal liver and adult blood (Fig. 4B). Nearly all of the β-globin expressed was βm-globin, although βwt-globin expression was detectable in day 14 fetal liver and adult blood by RPA using only a β-globin antisense riboprobe to reduce background (Fig. 4C). In the full-length β-YAC mice (lines 1 and 2), mean ɛ-globin expression in the day 10 yolk sac was 2.5-fold higher than in wild-type β-YAC lines (P < 0.01), whereas γ-globin expression was 45% lower than in control mice (P < 0.01) (Table 1; Fig. 5A). During day 12 and 14 fetal definitive erythropoiesis, mean γ-globin expression was approximately 28% of the wild-type β-YAC expression level (Fig. 5A; Table 1).
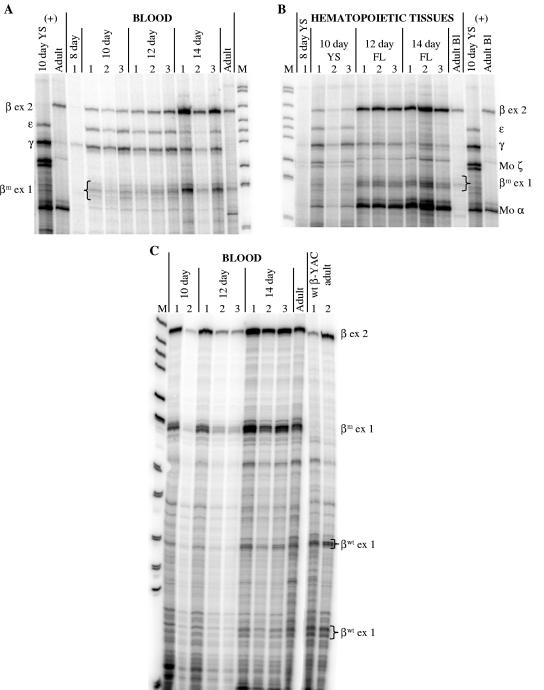
FIG. 4.
Globin gene expression during development in βm 5′ Gγ β-YAC transgenic mice. RPAs of samples from line 1 are shown. (A) Human β-like globin expression in blood. (B) Expression of human and murine globin genes in hematopoietic tissues. (C) βm- versus βwt-globin mRNA expression in blood. Labeling is as for Fig. 2. Developmental days are indicated above the images; β-like globin gene fragments are indicated on the right or left side, as are markers. The protected fragment sizes are the same as for Fig. 2 with the addition of mouse α-globin (Mo α), 128 bp, and mouse ζ-globin (Mo ζ), 151-bp doublet. YS, yolk sac; +, wild-type 213-kb β-YAC transgenic mouse line 4 (40) positive controls.
TABLE 1.
Human globin mRNA levels per copy of transgene and copy of endogenous murine α- and ζ-globin in βm 5′ Gγ β-YAC transgenic mice and wild-type β-YAC control mice
| β-YAC line | % of murine α-plus ζ-globin mRNA (mean ± SD)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| ɛ-Globin mRNA during embryonic erythropoiesis; day 10 yolk sac | γ-Globin mRNA during embryonic erythropoiesis; day 10 yolk sac | γ-Globin mRNA during definitive erythropoiesis
|
β-Globin mRNA during definitive erythropoiesis
|
|||||
| Day 12 liver | Day 14 liver | Day 10 yolk sac | Day 12 liver | Day 14 liver | Adult blood | |||
| 1 | 10.8 ± 2.2 | 40.8 ± 12.4 | 7.1 ± 0.2 | 2.1 ± 0.4 | 12.5 ± 2.7 | 42.2 ± 4.9 | 32.5 ± 2.8 | 25.4 ± 0.0 |
| 2 | 15.5 ± 6.2 | 24.6 ± 7.2 | 6.8 ± 0.6 | 3.5 ± 0.3 | 11.1 ± 2.7 | 46.1 ± 0.6 | 44.7 ± 2.9 | 23.9 ± 5.3 |
| 3 (Δβwt) | 25.0 ± 4.9 | 50.2 ± 20.2 | 0 | 0 | 21.9 ± 4.7 | 30.2 ± 8.3 | 17.9 ± 4.4 | 6.3 ± 1.2 |
| Mean for lines 1, 2 | 13.1 ± 3.3 | 32.7 ± 11.4 | 6.9 ± 0.2 | 2.8 ± 1.0 | 11.8 ± 1.0 | 44.1 ± 2.8 | 38.6 ± 8.6 | 24.1 ± 0.1 |
| Mean for wt linesa | 5.3 ± 0.3 | 59.9 ± 2.4 | 24 ± 0.6 | 10.5 ± 1.0 | 0 | 41.2 ± 2.3 | 68.2 ± 6.2 | 97.4 ± 18.6 |
wt, wild-type β-YAC transgenic lines.
FIG. 5.
Human γ- and β-globin mRNA levels in βm 5′ Gγ β-YAC transgenic mice during embryonic and definitive erythropoiesis. γ- or β-globin gene expression levels were quantitated from RNase protection analyses of RNAs derived from the indicated tissues at the developmental stages shown by phosphorimaging. Levels were calculated as percentages of murine α- and ζ-globin gene expression corrected for transgene and endogenous murine gene copy number. The average and standard deviation for all individuals within all lines are plotted on the x axis. The number of conceptuses tested for each genotype is indicated in Materials and Methods; the range was from 9 to 27. (A) γ-Globin mRNA expression. (B) β-Globin mRNA expression. Black bars, full-length βm 5′ Gγ β-YAC lines 1 and 2; hatched bars, βm 5′ Gγ Δβwt β-YAC line 3; white bars, wild-type β-YAC line 4 (40). Other abbreviations are as described in previous figure legends.
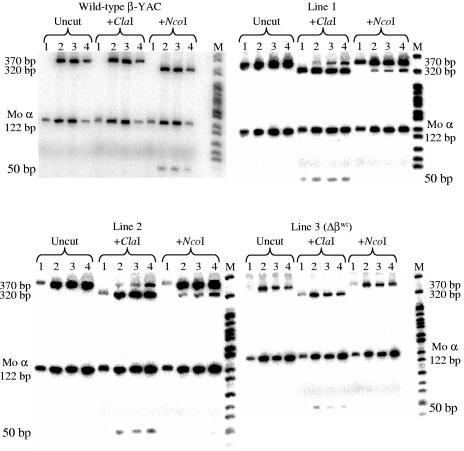
In the day 10 yolk sac of the full-length lines (1 and 2), 12% of the human globin mRNA was β-globin (Fig. 5; Table 1). At day 12 in the fetal liver, the total β-globin mRNA (βwt+βm) was similar to that of the wild-type β-YAC mice. Thereafter, β-globin mRNA declined to 56% of the control level in day 14 fetal liver and to 25% in adult blood. To confirm the pattern and level of βm- versus βwt-globin mRNA expression, semiquantitative RT-PCR RFLP was performed as described in Materials and Methods. For these experiments, cDNAs were synthesized from day 10 embryonic yolk sac, day 12 fetal liver, day 14 day fetal liver, and adult blood RNA. Wild-type β-globin PCR product was digested by NcoI in wild-type β-YAC transgenic mice that do not have a βm-globin gene but not by ClaI (Fig. 6, top left panel, wild-type β-YAC mice). The reverse was true for the βm-globin product in βm 5′ Gγ β-YAC mice lacking the βwt-globin gene, which was digested by ClaI but not by NcoI [Fig. 6, bottom right panel, βm 5′ Gγ β-YAC mouse line 3 (Δβwt)]. In βm 5′ Gγ β-YAC mouse lines 1 and 2, which contain both the βm- and βwt-globin genes, only βm-globin expression was observed in the day 10 yolk sac, but both βwt- and βm-globins were detected in day 12 fetal liver, day 14 fetal liver, and adult blood (Fig. 6, top right and bottom left panels, βm 5′ Gγ β-YAC mouse lines 1 and 2). βwt-globin constituted 7.3% ± 0.7% of total human β-globin gene expression [βwt/(βm+βwt)] in day 12 fetal liver, 10.6% ± 1.2% in day 14 fetal liver, and 16.1% ± 2.2% in adult blood (means and standard deviations of lines 1 and 2 combined, two conceptuses per line, duplicate experiments).
FIG. 6.
Semiquantitative RT-PCR RFLP analysis of β-globin gene expression in βm 5′ Gγ β-YAC transgenic mice. RT-PCRs were performed for wild-type β-YAC and three βm 5′ Gγ β-YAC mouse lines. Total mRNAs from 10 day embryonic yolk sac (lanes 1), 12-day fetal liver (lanes 2), 14-day fetal liver (lanes 3), and adult blood (lanes 4) were analyzed using primers for the 5′ UTR and exon 1/exon 2 boundary of the β-globin gene (See Materials and Methods). The full-length product is 370 bp. Digestion of the wild-type β-globin product with NcoI produces 320-bp and 50-bp fragments, whereas the βm-globin product is not digested. ClaI digests the βm-globin product, but not the βwt-globin product, to produce 324-bp and 50-bp fragments. Top panel: left, wild-type 213-kb β-YAC transgenic mouse line 4 (Wild-type β-YAC [40]); right, βm 5′ Gγ β-YAC mouse line 1 (Line 1). Bottom panel: left, βm 5′ Gγ β-YAC line 2 (Line 2); right, βm 5′ Gγ Δβwt-globin β-YAC mouse line 3, which bears a deletion of the normally located wild-type β-globin gene [Line 3 (Δβwt)]. NcoI completely digested the βwt-globin product, whereas ClaI did not, in the wild-type β-YAC mouse line. ClaI digested the βm-globin PCR product, whereas NcoI did not, in βm 5′ Gγ Δβwt-globin β-YAC line 3.
For line 3 (Δβwt) mice, which carried a β-globin locus lacking the wild-type β-globin gene, ɛ-globin expression was 4.7-fold higher than that for controls, but γ-globin expression was essentially the same as for wild-type β-YAC mice (Table 1; Fig. 5A). γ-Globin gene expression was not detected during fetal or adult definitive erythropoiesis. βm-Globin gene expression in the yolk sac was twofold higher than that for lines 1 and 2 (P < 0.01), suggesting that the absence of the βwt-globin gene resulted in a higher level of expression from the βm-globin gene (Fig. 5B; Table 1). However, β-globin expression was consistently lower than in lines 1 and 2 during all stages of definitive erythropoiesis (Table 1; Fig. 5B).
Together, these data suggest that the LCR-proximal βm-globin gene is expressed exclusively early in development (Fig. 4 and 6), and low-level expression of the more distal βwt-globin gene occurs beginning on day 12. Although there was a decline in βm-globin gene expression during adult erythropoiesis, the βm-globin gene was never completely silenced in spite of its location in the γ-globin gene domain. The elevated ɛ-globin gene expression during embryonic erythropoiesis suggests that location of the βm-globin gene 5′ to the Gγ-globin gene may give the ɛ-globin gene an expression advantage by downregulating γ-globin expression, especially in the absence of the βwt-globin gene. In day 12 fetal erythropoiesis, no γ-globin was observed in line 3 (Δβwt) mice, whereas in lines 1 and 2, γ-globin expression gradually declined, suggesting that the distal βwt-globin gene may exert a repressive effect on γ-globin expression.
A fetal γ-globin gene located proximally to the LCR is autonomously silenced during adult definitive erythropoiesis.
Five Aγm 5′ ɛ β-YAC transgenic mouse lines were established; each contained at least one intact copy of the β-YAC through the γ-globin genes (see Fig. S3 in the supplemental material). Three lines (1 to 3) carried at least one β-YAC extending from 5′HS3 through the β-globin gene, and two lines had deletions of the δ- and β-globin genes and downstream sequences (Δβ line 1 and Δβ line 2). Representative results of RNase protection assays are shown for line 3 (Fig. 7). ɛ- and γwt-globins were not expressed at any time during ontogeny. Aγm-Globin expression was initiated during primitive erythropoiesis and continued through fetal definitive erythropoiesis, as indicated by the presence of a 138-bp Aγm-globin exon 1 band. Aγm-Globin expression was not detected during adult definitive erythropoiesis in blood. β-Globin was expressed during fetal definitive erythropoiesis and continued throughout adulthood. Analysis of lines lacking the βwt-globin gene also demonstrated that ɛ-, Gγ-, and Aγwt-globins were not expressed at any stage (see Fig. S3 in the supplemental material, Δβ lines 1 and 2; data not shown). Interestingly, Aγm-globin expression was down-regulated even in the absence of the β-globin gene, suggesting that the 5.4-kb SspI fragment carrying the Aγm-globin gene contains a silencer region. In addition, these data demonstrate that γ-globin expression may be autonomously silenced, independently from competition with the β-globin gene for interaction with the LCR.
FIG. 7.
Aγm-Globin is silenced in adult Aγm 5′ ɛ β-YAC transgenic mice. RPAs of developmentally staged fetuses from Aγm 5′ ɛ β-YAC line 3. (A) Human β-like globin expression in blood. (B) Human and murine globin gene expression in hematopoietic tissues. Labeling is as for Fig. 2 and 4 with the addition of human Aγm-globin exon 1 (Aγm ex 1), 138 bp, and Gγ/Aγwt-globin exon 1 (Gγ/Aγwt ex 1), 118 bp. The Aγ-globin antisense probe hybridizes with exon 2 of the Aγwt-, Aγm-, these and Gγ-globin mRNAs; it also hybridizes with exon 1 of mRNAs. Thus, the exon 2 protected fragment is the sum of these three gene products, and the exon 1 protected fragments distinguish Aγm-globin mRNA from Gγ- and Aγwt-globin mRNAs.
Qualitative RT-PCR RFLP analysis was used to confirm whether γ-globin expression was exclusively Aγm-globin or whether it represented a mixture of Aγwt- and Aγm-globins. RNA derived from day 14 fetal liver was subjected to RT-PCR using primers encompassing the γ-globin gene 5′ UTR and exon 1 sequences. The mark in the Aγm-globin gene is a six-basepair deletion in the 5′ UTR of the Aγ-globin gene (41). The deletion removes a DdeI restriction enzyme site within the 5′ UTR. RT-PCR followed by digestion with DdeI confirmed that all of the γ-globin expressed in the Aγm 5′ ɛ β-YAC mice originated with the Aγm-globin gene (data not shown).
γ- and β-globin gene expression was quantitated and compared to expression in wild-type β-YAC mice (Table 2). In all lines, mean γ-globin expression was approximately the same as that measured in wild-type β-YAC mice during primitive erythropoiesis, but expression varied widely between the mutant lines. Similarly, during fetal definitive erythropoiesis, γ-globin expression was variable between the different lines. The difference between the mutant and wild-type β-YAC lines in day 12 fetal liver erythropoiesis (40% versus 24%, respectively) is not statistically significant. γ-Globin expression was essentially the same in mutant and wild-type mice by day 14, and it was absent in adult definitive erythropoiesis. β-Globin was not detected during primitive erythropoiesis in either Aγm 5′ ɛ β-YAC mice or wild-type β-YAC mice. In day 12 and day 14 fetal liver, β-globin in the mutant line was 14% and 11% of the wild-type β-YAC level, respectively, reflecting delayed activation of β-globin gene transcription. During adult definitive erythropoiesis, mean β-globin expression was 61% of that measured in wild-type β-YAC mice. Analysis of the globin gene expression switching profile in blood confirmed that the switch from γ- to β-globin expression was delayed compared to results for wild-type β-YAC mice and occurred on approximately day 16 instead of on day 12 (data not shown).
TABLE 2.
Human globin mRNA levels per copy of transgene and copy of endogenous murine α- and ζ-globin in Aγm 5′ ɛ β-YAC transgenic mice and wild-type β-YAC control mice
| β-YAC line | % of murine α-plus ζ-globin mRNA (mean ± SD)
|
||||||
|---|---|---|---|---|---|---|---|
| ɛ-Globin mRNA during embryonic erythropoiesis; day 10 yolk sac | γ-Globin mRNA during embryonic erythropoiesis; day 10 yolk sac | γ-Globin mRNA during definitive erythropoiesis
|
β-Globin mRNA during definitive erythropoiesis
|
||||
| Day 12 liver | Day 14 liver | Day 12 liver | Day 14 liver | Adult blood | |||
| 1 | 0 | 104.5 ± 19.2 | 42.8 ± 21.0 | 0 | 7.4 ± 3.3 | 4.0 ± 1.5 | 24.5 ± 6.5 |
| 2 | 0 | 45.6 ± 1.8 | 52.0 ± 3.3 | 25.2 ± 8.7 | 0 | 10.3 ± 1.7 | 86.0 ± 15.4 |
| 3 | 0 | 72.6 ± 5.4 | 21.2 ± 4.0 | 14.7 ± 3.5 | 9.7 ± 1.2 | 9.1 ± 0.4 | 66.9 ± 19.6 |
| Δβ 1 | 0 | 31.0 ± 14.1 | 33.4 ± 14.8 | 0 | N/Ab | N/A | N/A |
| Δβ 2 | 0 | 80.0 ± 9.8 | 45.8 ± 19.9 | 20.5 ± 1.9 | N/A | N/A | N/A |
| Mean for lines 1-3 | 0 | 74.2 ± 29.5 | 38.7 ± 15.8 | 13.3 ± 12.7 | 5.7 ± 5.1 | 7.8 ± 3.4 | 59.2 ± 31.5 |
| Mean for Δβ lines 1, 2 | 55.5 ± 13.2 | 39.6 ± 0.7 | 10.3 ± 2.1 | N/A | N/A | N/A | |
| Mean for wt linesa | 5.3 ± 0.3 | 59.9 ± 2.4 | 24 ± 0.6 | 10.5 ± 1.0 | 41.2 ± 2.3 | 68.2 ± 6.2 | 97.4 ± 18.6 |
wt, wild-type β-YAC transgenic lines. Wild-type β-YAC controls, same as for Table 1.
N/A, not applicable.
These data demonstrate that the LCR-proximal Aγm-globin gene is preferentially expressed during development and that it represses downstream ɛ-, Gγ-, and Aγwt-globin transcription, perhaps by more effectively competing for interaction with the LCR during primitive and fetal definitive erythropoiesis. The Aγm-globin gene fragment contains an autonomous silencer element that functions in repression of Aγm-globin gene expression even in the absence of competition with the β-globin gene for LCR interaction during adult definitive erythropoiesis.
An embryonic globin gene inserted between two adult globin genes is not expressed.
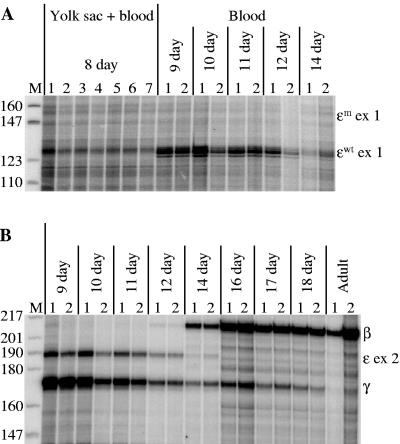
Three ɛm 3′ δ β-YAC lines were obtained (see Fig. S4 in the supplemental material). Analysis of ɛ-globin gene expression revealed that only wild-type ɛ-globin was expressed during primitive erythropoiesis; no signal was observed from the ɛm-globin gene at any stage of development (Fig. 8A). These data were confirmed by semiquantitative RT-PCR RFLP analysis as described in Materials and Methods (Fig. 9). ɛwt-Globin synthesis detected in day 12 and 14 fetal liver is due to the presence of primitive erythrocytes from earlier yolk sac erythropoiesis in the fetal circulation (31). Although transcription was not observed, the integrity of the inserted ɛm-globin gene was confirmed by Southern blot hybridization and PCR analyses (see Fig. S4 in the supplemental material; also data not shown). Expression of the gene was confirmed by RPA of erythroid cells containing an ɛm-globin plasmid (data not shown). The insertion of an ɛm-globin gene between the δ- and β-globin genes had no effect on either the temporal or spatial pattern of globin gene expression within the locus (Fig. 8B). More recently, a derivative of the ɛm 3′ δ β-YAC construct was synthesized that lacks the normally located ɛwt-globin gene. Transgenic lines produced from this construct also did not display ɛm-globin gene expression at any stage of development as measured by semiquantitative RT-PCR (data not shown).
FIG. 8.
ɛm-Globin is not expressed in ɛm 3′ δ β-YAC transgenic mice. RPAs of developmentally staged fetuses from ɛm 3′ δ β-YAC lines. Labeling is as for Fig. 2, 4, and 7. (A) Wild-type ɛ-globin mRNA is detected as a 127-bp exon 1 protected fragment, whereas ɛm-globin mRNA is detected as a 152-bp exon 1 protected fragment. (B) The temporal pattern of globin transgene expression.
FIG. 9.
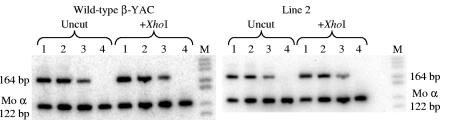
Semiquantitative RT-PCR RFLP analysis of ɛ-globin gene expression in ɛm 3′ δ β-YAC transgenic mice. RT-PCRs were performed for wild-type β-YAC and three ɛm 3′ δ β-YAC mouse lines. Total mRNAs from 10 day embryonic yolk sac (lanes 1), 12-day fetal liver (lanes 2), 14-day fetal liver (lanes 3), and adult blood (lanes 4) were analyzed using primers for the 5′ UTR and exon 1/exon 2 boundary of the ɛ-globin gene (see Materials and Methods). The full-length ɛwt-globin product is 164 bp; the ɛm-globin product is 172 bp. The wild-type ɛ-globin product is not digested with XhoI, whereas the ɛm-globin product is digested and generates 155-bp and 17-bp fragments. Left panel, wild-type 213-kb β-YAC transgenic mouse line 4 (wild-type β-YAC) (40); right panel, ɛm 3′ δ β-YAC mouse line 2 (Line 2). Results were the same for ɛm 3′ δ β-YAC transgenic mouse lines 1 and 3.
DISCUSSION
Many mammalian loci show a correlation between spatial arrangement of genes in the DNA sequence (gene order) and the temporal order of expression during development. Such an arrangement may facilitate cis regulatory mechanisms, allowing fine-tuning of gene activation and repression circuitry and maintenance of the delicate balance between these developmentally controlled processes. Examples include the human β-globin locus, the homeobox (hox) gene loci of Drosophila and mammals, and mammalian α-fetoprotein and albumin genes (5, 20, 29).
The question of the role of gene order in developmental regulation has been previously addressed with various approaches. We demonstrated that when two globin genes from the same developmental stage were arranged tandemly and linked to the LCR, the gene more proximal to the LCR was expressed preferentially throughout development, indicating that proximity to the LCR was a determinant of globin gene expression, perhaps due to the increased frequency of interaction of the proximal gene with the LCR (39). One such arrangement occurs naturally within the human β-globin locus. The Gγ- and Aγ-globin coding sequences are nearly identical, but the Gγ-globin gene is expressed at higher level than the Aγ-globin gene early in fetal erythropoiesis; the ratio inverts as development proceeds (22). When the γ- and β-globin genes were linked tandemly to the LCR, γ-globin was expressed at a higher level than β-globin during fetal definitive erythropoiesis, and the ratio was reversed during adult definitive erythropoiesis (39). This result occurred regardless of the order of the γ- and β-globin genes with respect to the LCR, indicating that the trans-acting environment was the primary determinant of globin gene expression, overcoming other motifs, including proximity to the LCR. Although these experiments suggest that proximity to the LCR plays a role in correct gene expression, the distances between the LCR and linked genes were artificially shortened, leaving open the question as to whether gene order was important.
Another experimental system utilized two cosmid constructs encompassing portions of the human β-globin locus that were ligated together just prior to producing transgenic mice (10). One set of mouse lines contained a locus in which a βm-globin gene was placed upstream of the δ-globin gene; in another set of lines the ɛ-globin gene was replaced with the βm-globin gene. In the lines where the βm-globin gene replaced the ɛ-globin gene, βm-globin was expressed throughout development; βwt-globin comprised only a small percentage of expression in adult tissues. In addition, γ-globin gene expression was silenced. In the lines where the βm-globin gene was located near the δ-globin gene, βm-globin accounted for 75% of total β-globin synthesis and βwt-globin for 25%. In a third set of experiments, the genes of the human β-globin locus were inverted with respect to the LCR in β-YAC transgenic mice (44). In this instance the β-globin gene was proximal, and it was constitutively expressed. The ɛ-globin gene was most distal; it was not expressed at all, and γ-globin expression was restricted to primitive erythropoiesis. These data suggested that any gene located near the LCR will be strongly expressed throughout ontogeny.
In this study, we used complete, intact locus constructs containing all potential regulatory elements and all the wild-type genes of the locus in their normal spatial configuration. The 213-kb human β-globin locus YAC contains 187 kb of the locus, and mice generated with this YAC have been shown to display correct tissue-specific and temporal expression patterns (36, 38). A marked β-globin gene was placed upstream of either the ɛ-globin gene or the Gγ-globin gene. A marked Aγ-globin gene was placed upstream of the ɛ-globin gene. A marked ɛ-globin gene was placed between the δ- and β-globin genes. Several conclusions can be drawn from the expression patterns we observed.
Placement of the marked β-globin gene upstream to the ɛ-globin gene resulted in predominantly βm-globin expression during yolk sac erythropoiesis. γ-Globin expression in embryonic cells was decreased by 50% compared to that for the wild-type β-YAC γ-globin gene control, and there was no detectable ɛ-globin gene expression. Only βm-globin gene expression could be detected during definitive erythropoiesis in fetal liver and adult erythroid cells. Similar results were observed in a previous study in which a βm-globin gene replaced the ɛ-globin gene (10). The most likely explanation for these results is preferential interaction of the βm-globin gene with the LCR due to its proximity to this regulatory element. The absence of downstream βwt-globin gene expression can be attributed to competition. It is of interest that although there was γ-globin gene expression in the embryonic cells, ɛ-globin gene expression was absent. This finding suggests that structural (or distance) constraints exist that determine (or influence) the interaction between the LCR and the downstream globin genes. Perhaps the distance between the βm- and ɛ-globin genes is too short and prohibits the interaction of the LCR with the nearby downstream ɛ-globin gene. Similarly, Dillon et al. (10) showed that γ-globin was expressed during embryonic erythropoiesis when the βm-globin gene replaced the ɛ-globin gene, but expression was completely suppressed in the fetal liver. Thus, the distance between the ɛ-globin gene (or the βm-globin gene in this case) and the γ-globin gene is sufficient to allow interaction of either of these genes with the LCR.
Another conclusion from this study is that it is not so much the placement of a gene within the locus that determines globin gene switching but rather the presence of silencing sequences and their interactions with repressor complexes. Because the β-globin gene lacks developmentally controlled silencing elements in its promoter, switching is abolished when that gene is located at the beginning of the locus. Conversely, when an Aγm-globin gene was placed at the same location upstream of the ɛ-globin gene, the Aγm-globin gene was turned off during definitive erythropoiesis and the switch from γ- to β-globin gene expression occurred. These results provide support for the concept that to a major degree, globin gene switching is controlled negatively by the presence of silencing elements in the embryonic and fetal globin gene promoters and the interaction of these elements with the prevailing trans-acting environment. For the γ-globin genes, the silencing mechanism is leaky and can be overcome by several physiological manipulations, acquired pathological conditions, and mutations in vivo in humans. Also of note here is the fact that the level of βm-globin gene expression in the adult stage of development decreased to about 50% of that in the embryonic stage. As will be discussed later, this finding may reflect a change (in chromatin modification?) of the embryonic gene chromatin domain in which the βm-globin gene is located.
The placement of the βm-globin gene upstream of the Gγ-globin gene resulted in phenotypes that could have been predicted on the basis of the hypothesis that gene order and proximity to the LCR determine globin gene expression. Thus, in the embryonic cells there was ɛ-, βm-, and γ-globin gene expression, but there was no expression of the distal wild-type β-globin gene. ɛ-Globin expression switched off at the onset of fetal erythropoiesis; γ-globin gene expression subsequently switched off later. The distal βwt-globin gene was expressed at 1/10 the level of the upstream βm-globin gene during fetal or adult definitive erythropoiesis. All of these observations were expected based on the hypothesis. Significantly, however, instead of the expected increase of β-globin gene expression as development proceeded, a consistent decrease of βm-globin gene expression was observed, so that total β-globin gene expression (βwt+βm) in the adult cells was about 25% of that in the embryonic cells. As mentioned earlier, a decrease of βm-globin expression to 50% of that in embryonic cells was noted in adult erythropoiesis when the βm-globin gene was located upstream of the ɛ-globin gene. How can these unexpected declines in βm- and βwt-globin gene expression be explained? Changes in the transcriptional environment cannot be the reason, because the environment of the adult erythroid cells is expected to be more conducive to the interaction between the LCR and the β-globin gene promoter. Perhaps, as suggested, the whole architecture of the β-globin locus chromatin changes during development, so that an initial embryonic chromatin domain is succeeded by a fetal and finally an adult domain (17). Although this change in the chromatin domain still remains hypothetical, it could provide an explanation for our observations. For example, chromatin modification in the embryonic domain might affect the efficiency of looping of the LCR and decrease its interaction with the βm-globin gene placed in that domain. Chromatin modification studies may provide further insight into this question.
The control of γ-globin gene expression during development has been the subject of considerable discussion. Initial studies by two laboratories suggested that competition between the γ- and β-globin genes for interaction with the LCR, coupled with the preferential interaction of the β-globin gene with the LCR during the adult stage of development, are responsible for the silencing of the γ-globin gene in adult erythropoiesis (3, 6, 12). When the γ-globin gene was linked to a μLCR cassette, it was not silenced in the adult stage (13); it was, however, silenced in a cosmid containing the Gγ- to β-globin gene region with these genes in their normal chromosomal location (11). On the other hand, studies with transgenic mice carrying a mini-LCR linked to a γ-globin gene suggested that silencing could be autonomous, since several (but not all) of these transgenics did not express γ-globin in adult cells (9). The results of our study in which the Aγm-globin gene was placed upstream of the ɛ-globin gene provide support for the possibility of autonomous silencing, since the γ-globin gene, even that close to the LCR, was turned off in adult erythropoiesis. This experiment also provided information on the relative roles of gene order and transcriptional environment in determining globin gene expression, because only the upstream Aγm-globin gene was expressed in these transgenic mice; the distal γ-globin genes remained silent. Since the downstream γ-globin genes remained totally silent during the embryonic/fetal stage of development, it is unlikely that a fetal environment alone is sufficient for γ-globin gene expression. The results are best explained by assuming that proximity to the LCR is the major determinant of the order of γ-globin gene expression during development.
As mentioned earlier, some evidence suggests that the β-globin locus may be divided into developmental stage-specific chromatin subdomains that have a role in determining gene expression (14, 17, 23). Our data show that an open adult chromatin subdomain does not activate ɛm-globin gene expression when this gene is placed in the adult chromatin subdomain, between the δ- and β-globin genes. More-recent data from our laboratory support this notion, because an ɛm-globin gene located between the δ- and β-globin genes was not expressed even in the absence of the wild-type ɛ-globin gene (unpublished data).
Three major conclusions may be drawn from our data. First, any gene located near the LCR will be strongly expressed throughout ontogeny, unless some gene-specific silencing mechanism exists. Second, the Aγ-globin gene is autonomously silenced. Third, both gene order and gene-proximal cis regulatory elements are important for correct developmental expression of the globin genes. Gene order, per se, may not be a direct determinant of gene expression, but ultimately an inherent property of spatial gene order is the maintenance of the necessary distance between the LCR and the globin genes to avoid aberrant gene expression. Thus, gene order constrains distance and LCR proximity. To some extent, alteration of normal LCR proximity supersedes the dominance of the trans-acting environment, particularly when the gene does not have a mechanism of autonomous silencing.
There may be a selective advantage for clustering genes in a specific order. First, physical organization of the genes in the order they are expressed during development might facilitate correct temporal expression of these genes, as is evident for the β-globin and hox gene loci (20, 29). Second, genes may be linked in an ordered array to assist in the functional interaction of products of polymorphic alleles or to facilitate sequence exchange. Order is essential for somatic recombination of immunoglobulin genes, T-cell receptors, and protocadherins (45). Close proximity of similarly imprinted genes may assist in establishing epigenetically regulated domains, such as occurs in the coordinated regulation of the Igf-2 and H19 genes by the CTCF transcription factor (33). Analysis of the chromosomal location and expression patterns of 105 different metabolic pathways in 5 different species demonstrated that 30% to 98% of genes involved in different metabolic pathways were clustered (24). The expression patterns of housekeeping genes and highly expressed genes subject them to selective pressure for maintaining active genes within the genome due to their essential functions within cells. Therefore, these genes should be coregulated and evolutionarily conserved. One cis-acting motif by which this is accomplished may be the formation and maintenance of active gene clusters or gene order in multigenic loci on an evolutionary time scale. These chromosomal regulatory landscapes comprise domains in which multiple genes share one or more common regulatory motifs.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK53510 (NIDDK), HL67336 (NHLBI), and DK61804 (NIDDK) and a Lila and Madison Self Faculty Scholar award to K.R.P. and by a University of Kansas Medical Center Biomedical Research Training Scholarship awarded to S.H.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ausubel, F. M. 1992. Current protocols in molecular biology. John Wiley, New York, N.Y.
- 2.Baralle, F. E., C. C. Shoulders, and N. J. Proudfoot. 1980. The primary structure of the human ɛ-globin gene. Cell 21:621-626. [DOI] [PubMed] [Google Scholar]
- 3.Behringer, R. R., T. M. Ryan, R. D. Palmiter, R. L. Brinster, and T. M. Townes. 1990. Human γ- to β-globin gene switching in transgenic mice. Genes Dev. 4:380-389. [DOI] [PubMed] [Google Scholar]
- 4.Bulger, M., J. H. von Doorninck, N. Saitoh, A. Telling, C. Farrell, M. A. Bender, G. Felsenfeld, R. Axel, and M. Groudine. 1999. Conservation of sequence and structure flanking the mouse and human β-globin loci: the β-globin genes are embedded within an array of odorant receptor genes. Proc. Natl. Acad. Sci. USA 96:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, H., J. O. Egan, and J. F. Chiu. 1997. Regulation and activities of α-fetoprotein. Crit. Rev. Eukaryot. Gene Expr. 7:11-41. [DOI] [PubMed] [Google Scholar]
- 6.Choi, O. R., and J. D. Engel. 1988. Developmental regulation of β-globin gene switching. Cell 55:17-26. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 8.DeLaat, W., and F. G. Grosveld. 2003. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 11:447-459. [DOI] [PubMed] [Google Scholar]
- 9.Dillon, N., and F. Grosveld. 1991. Human γ-globin genes silenced independently of other genes in the β-globin locus. Nature 350:252-254. [DOI] [PubMed] [Google Scholar]
- 10.Dillon, N., T. Trimborn, J. Strouboulis, P. Fraser, and F. Grosveld. 1997. The effect of distance on long-range chromatin interactions. Mol. Cell 1:131-139. [DOI] [PubMed] [Google Scholar]
- 11.Enver, T., M. Brice, J. Karlinsey, G. Stamatoyannopoulos, and T. Papayannopoulou. 1991. Developmental regulation of fetal to adult globin gene switching in human fetal erythroid x mouse erythroleukemia cell hybrids. Dev. Biol. 148:129-137. [DOI] [PubMed] [Google Scholar]
- 12.Enver, T., N. Raich, A. J. Ebens, T. Papayannopoulou, F. Costantini, and G. Stamatoyannopoulos. 1990. Developmental regulation of human fetal-to-adult globin gene switching in transgenic mice. Nature 344:309-313. [DOI] [PubMed] [Google Scholar]
- 13.Enver, T., A. J. Ebens, W. C. Forrester, and G. Stamatoyannopoulos. 1989. The human β-globin locus activation region alters the developmental fate of a human fetal globin gene in transgenic mice. Proc. Natl. Acad. Sci. USA 86:7033-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furukawa, T., P. A. Navas, B. M. Josephson, K. R. Peterson, T. Papayannopoulou, and G. Stamatoyannopoulos. 1995. Coexpression of ɛ, Gγ and Aγ globin mRNA in embryonic red blood cells from a single copy β-YAC transgenic mouse. Blood Cells Mol. Dis. 21:168-178. [DOI] [PubMed] [Google Scholar]
- 16.Gnirke, A., C. Huxley, K. Peterson, and M. V. Olson. 1993. Microinjection of intact 200- to 500-Kb fragments of YAC DNA into mammalian cells. Genomics 15:659-667. [DOI] [PubMed] [Google Scholar]
- 17.Gribnau, J., K. Diderich, S. Pruzina, R. Calzolari, and P. Fraser. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell 5:377-386. [DOI] [PubMed] [Google Scholar]
- 18.Hanscombe, O., D. Whyatt, P. Fraser, N. Yannoutsos, D. Greaves, N. Dillon, and F. Grosveld. 1991. Importance of globin gene order for correct developmental expression. Genes Dev. 5:1387-1394. [DOI] [PubMed] [Google Scholar]
- 19.Hardison, R., J. L. Slightom, D. L. Gumucio, M. Goodman, N. Stojanovic, and W. Miller. 1997. Locus control regions of mammalian β-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene 205:73-94. [DOI] [PubMed] [Google Scholar]
- 20.Harju, S., K. J. McQueen, and K. R. Peterson. 2002. Chromatin structure and control of β-like globin gene switching. Exp. Biol. Med. 227:683-700. [DOI] [PubMed] [Google Scholar]
- 21.Harju, S., and K. R. Peterson. 2001. Sensitive ribonuclease protection assay employing glycogen as a carrier and a single inactivation/precipitation step. BioTechniques 30:1198-1204. [DOI] [PubMed] [Google Scholar]
- 22.Huisman, T. H., H. Harris, and M. Gravely. 1977. The chemical heterogeneity of the fetal hemoglobin in normal newborn infants and in adults. Mol. Cell Biochem. 17:45-55. [DOI] [PubMed] [Google Scholar]
- 23.Im, H., J. A. Grass, H. M. Christensen, A. Perkins, and E. H. Bresnick. 2002. Histone deacetylase-dependent establishment and maintenance of broad low-level histone acetylation within a tissue-specific chromatin domain. Biochemistry 41:15152-15160. [DOI] [PubMed] [Google Scholar]
- 24.Lee, J. M., and E. L. Sonnhammer. 2003. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 13:875-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Q., C. Clegg, K. Peterson, S. Shaw, N. Raich, and G. Stamatoyannopoulos. 1997. Binary transgenic mouse model for studying the trans control of globin gene switching: evidence that GATA-1 is an in vivo repressor of human ɛ gene expression. Proc. Natl. Acad. Sci. USA 94:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Q., S. Harju, and K. R. Peterson. 1999. Locus control regions: coming of age at a decade plus. Trends Genet. 15:403-408. [DOI] [PubMed] [Google Scholar]
- 27.Li, Q., K. R. Peterson, X. Fang, and G. Stamatoyannopoulos. 2002. Locus control regions. Blood 100:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Q., and J. A. Stamatoyannopoulos. 1994. Position independence and proper developmental control of γ-globin gene expression require both a 5′ locus control region and a downstream sequence element. Mol. Cell. Biol. 14:6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGinnis, W., and R. Krumlauf. 1992. Homeobox genes and axial patterning. Cell 68:283-302. [DOI] [PubMed] [Google Scholar]
- 30.Navas, P. A., K. R. Peterson, Q. Li, M. McArthur, and G. Stamatoyannopoulos. 2001. The 5′HS4 core element of the human β-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis. J. Mol. Biol. 312:17-26. [DOI] [PubMed] [Google Scholar]
- 31.Navas, P. A., K. R. Peterson, Q. Li, E. Skarpidi, A. Rohde, S. E. Shaw, C. H. Clegg, H. Asano, and G. Stamatoyannopoulos. 1998. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol. Cell. Biol. 18:4188-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Omori, A., O. Tanabe, J. D. Engel, A. Fukamizu, and K. Tanimoto. 2005. Adult stage γ-globin silencing is mediated by a promoter direct repeat element. Mol. Cell. Biol. 25:3443-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pant, V., S. Kurukuti, E. Pugacheva, S. Shamsuddin, P. Mariano, R. Renkawitz, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 24:3497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patrinos, G. P., M. de Krom, E. de Boer, A. Langeveld, A. M. Imam, J. Stouboulis, W. de Laat, and F. G. Grosveld. 2004. Multiple interactions between regulatory regions are required to stabilize an active chromatin domain. Genes Dev. 18:1495-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson, K. R. 2003. Transgenic mice carrying yeast artificial chromosomes. Exp. Rev. Mol. Med. 5:1-25. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, K. R., C. H. Clegg, C. Huxley, B. M. Josephson, H. S. Haugen, T. Furukawa, and G. Stamatoyannopoulos. 1993. Transgenic mice containing a 248-Kb yeast artificial chromosome carrying the human β-globin locus display proper developmental control of human globin genes. Proc. Natl. Acad. Sci. USA 90:7593-7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson, K. R., C. H. Clegg, Q. Li, and G. Stamatoyannopoulos. 1997. Production of transgenic mice with yeast artificial chromosomes. Trends Genet. 13:61-66. [DOI] [PubMed] [Google Scholar]
- 38.Peterson, K. R., Q. Li, C. H. Clegg, T. Furukawa, P. A. Navas, E. J. Norton, T. G. Kimbrough, and G. Stamatoyannopoulos. 1995. Use of yeast artificial chromosomes (YACs) in studies of mammalian development: production of β-globin locus YAC mice carrying human globin developmental mutants. Proc. Natl. Acad. Sci. USA 92:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson, K. R., and G. Stamatoyannopoulos. 1993. Role of gene order in developmental control of human γ- and β-globin gene expression. Mol. Cell. Biol. 13:4836-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson, K. R., P. A. Navas, Q. Li, and G. Stamatoyannopoulos. 1998. LCR-dependent gene expression in β-globin YAC transgenics: detailed structural studies validate functional analysis even in the presence of fragmented YACs. Hum. Mol. Genet. 7:2079-2088. [DOI] [PubMed] [Google Scholar]
- 41.Rixon, M. W., and R. E. Gelinas. 1988. A fetal globin gene mutation in Aγ nondeletion hereditary persistence of fetal hemoglobin increases promoter strength in a nonerythroid cell. Mol. Cell. Biol. 8:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamatoyannopoulos, G. 2001. The molecular basis of blood diseases, 3rd ed. W. B. Saunders, Philadelphia, Pa.
- 43.Stamatoyannopoulos, G., B. Josephson, J. W. Zhang, and Q. Li. 1993. Developmental regulation of human γ-globin genes in transgenic mice. Mol. Cell. Biol. 13:7636-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanimoto, K., Q. Liu, J. Bungert, and J. D. Engel. 1999. Effects of altered gene order or orientation of the locus control region on human β-globin gene expression in mice. Nature 398:344-348. [DOI] [PubMed] [Google Scholar]
- 45.Wu, Q., and T. Maniatis. 1999. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97:779-790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.