Abstract
When cultured in 20% O2, human cytotrophoblasts fuse to form the syncytiotrophoblast with marked induction of hCYP19 (aromatase) gene expression. When cultured in 2% O2, cytotrophoblast fusion and induced hCYP19 expression are prevented. These effects of hypoxia are mediated by increased expression of mammalian achaete/scute homologue-2 (Mash-2), which increases levels of upstream stimulatory factors 1 and 2 (USF1/2) and their binding as heterodimers to E-boxes surrounding the hCYP19 promoter. In studies to define mechanisms for O2 regulation of syncytiotrophoblast differentiation, we found that hypoxia and overexpression of Mash-2 markedly increased cyclin B1 levels in cultured trophoblasts and the proportion of cells at the G2/M transition. Unlike USF proteins, USF1/2 mRNA levels are unaffected by O2 tension. To determine whether increased O2 might enhance proteasomal degradation of USF1/2, human trophoblasts were cultured in 2% or 20% O2 with or without proteasome inhibitors. In cells cultured in 20% O2, proteasome inhibitors increased USF1/2 protein levels and blocked spontaneous induction of hCYP19 expression, cell fusion, and differentiation. Like hypoxia, inhibitory effects of proteasome inhibitors on hCYP19 expression were mediated by increased binding of USF1/2 to the E-boxes. In human trophoblast cells cultured in 20% O2, increased polyubiquitylation of USF1/2 proteins was observed. Thus, early in gestation when the placenta is relatively hypoxic, increased USF1/2 may block trophoblast differentiation and hCYP19 gene expression. In the second trimester, increased O2 tension promotes proteasomal degradation of USF1/2, resulting in syncytiotrophoblast differentiation and induction of hCYP19 expression.
In addition to its primary role as an organ of nutrient and gas exchange for the fetus, the placenta produces a number of steroid, peptide, and polypeptide hormones that are important for maintenance of pregnancy and fetal growth and development. The basic unit of nutrient and gas exchange and of hormone production by the human placenta is the floating chorionic villous, comprised of an outer multinuclear cell layer, the syncytiotrophoblast, which is bathed in maternal blood and overlies an inner core comprised of cytotrophoblast and stromal components. Growth of the placenta is driven by replication of cytotrophoblasts. As the cytotrophoblasts mature within the floating chorionic villi, they stop dividing and fuse to form the terminally differentiated syncytiotrophoblast layer (25, 36).
The human placenta has an extraordinary capacity to synthesize estrogens by aromatization of C19/androgen precursors secreted by the fetal adrenals (39). The aromatization reaction is catalyzed by an enzyme complex comprised of two polypeptides, NADPH-cytochrome P450 reductase and a unique form of cytochrome P450, aromatase P450 (P450arom, product of the CYP19 gene) (45). In most vertebrates, CYP19 expression is restricted to the gonads and to discrete nuclei of the brain; however, in humans, it also is expressed in the syncytiotrophoblast layer of the placenta, stromal cells of adipose tissue, bone, fetal liver, and in vascular smooth muscle and endothelial cells (23, 40). Expression of the human CYP19 (hCYP19) gene in various estrogen-producing tissues is driven by tissue-specific promoters upstream of tissue-specific alternative first exons, which encode the 5′ untranslated regions of hCYP19 mRNA transcripts. These alternative first exons, located from ∼110 bp to ∼100,000 bp upstream of the hCYP19 translation initiation site in exon II, are alternatively spliced onto a common site just upstream of the translation start site so that the protein synthesized by each of these tissues is identical (31). In placenta, the majority of the hCYP19 mRNA transcripts contain 5′ untranslated sequences encoded by exon I.1, which lies ∼95,000 bp upstream of the start site of translation in exon II (23). Studies using transgenic mice suggest that as little as 246 bp of hCYP19 exon I.1 5′ flanking DNA and 103 bp of exon I.1 are sufficient to direct reporter gene expression exclusively to the placenta, specifically to the labyrinthine and trophoblast giant cell layers (22, 24). Since mouse placenta does not express aromatase, these findings indicate that placental transcription factors that mediate hCYP19 gene expression are conserved between mouse and human, while the genetic response elements that bind these factors are not.
Estrogen synthesis by human placenta is increased markedly after the ninth week of gestation (9) in association with cytotrophoblast invasion, remodeling, and enlargement of the uterine arterioles. This invasion results in a pronounced increase in blood flow to the intervillous space and concomitant increase in O2 availability to cells of the floating chorionic villi (18). When cytotrophoblasts isolated from midgestation human placenta are cultured in a 20% O2 environment, they spontaneously fuse to form the syncytiotrophoblast; this occurs in concert with a marked induction of hCYP19 gene expression and of aromatase activity (19). By contrast, when cytotrophoblasts are cultured in a 2% O2 (hypoxic) environment (similar to the placental O2 tension during the first trimester of gestation), they manifest increased rates of DNA synthesis and fail to fuse to form the syncytiotrophoblast, and hCYP19 expression remains undetectable (19). Thus, by use of this culture system, we are able to mimic morphological and biochemical events associated with placental differentiation.
In previous studies to define the mechanisms for O2 regulation of trophoblast differentiation, we identified a basic-helix-loop-helix (bHLH) transcription factor, Mash-2 (mammalian achaete/scute homologue-2), that manifested elevated levels of expression in freshly isolated human cytotrophoblasts, declined with syncytiotrophoblast differentiation in a 20% O2 environment, and was maintained at elevated levels by hypoxia (19). Overexpression of Mash-2 in cultured human trophoblast cells maintained in 20% O2 caused marked inhibition of hCYP19 expression and blocked syncytiotrophoblast differentiation (19).
In more recent studies, we observed that Mash-2 is primarily a cytoplasmic protein that does not bind to the hCYP19 gene directly; rather, it acts to increase expression of the bHLH-zipper transcription factors, upstream stimulatory factor 1 (USF1) and USF2, which bind as heterodimers to E-box sequences within the 5′ flanking region and in placenta-specific exon I.1 of the hCYP19 gene to inhibit hCYP19I.1 promoter activity (20). In contrast to Mash-2, USF proteins are localized to the nucleus. Nuclear levels of USF1 and USF2 were elevated in freshly isolated cytotrophoblasts and in trophoblasts cultured in 2% O2 and declined with syncytiotrophoblast differentiation in 20% O2. Importantly, overexpression of USF1 in human trophoblasts cultured in 20% O2 blocked syncytiotrophoblast differentiation and induction of hCYP19 gene expression (20). Surprisingly, unlike USF proteins, which were markedly induced by hypoxia and by Mash-2 overexpression, USF1 (19) and USF2 (B. Jiang and C. R. Mendelson, unpublished observations) mRNA levels were unaffected. These findings suggest that hypoxia-induced expression of USF1/2 proteins is due to changes in mRNA translatability and/or protein stability. Thus, increased protein levels of USF1/2 mediate the inhibitory effects of hypoxia and of Mash-2 on syncytiotrophoblast differentiation and hCYP19 gene expression.
Our goal in the present study was to define the mechanisms whereby O2 tension alters USF protein expression and differentiation of human trophoblast cells. We observed that proteasome inhibitors blocked the O2-mediated decline in USF1/2 protein levels, increased their binding to E boxes surrounding hCYP19 promoter I.1, and prevented syncytiotrophoblast differentiation and induction of hCYP19 gene expression. These findings together with the observation that increased O2 tension promoted USF polyubiquitylation, suggest that increased degradation of USFs via the proteasome pathway plays a key role in O2-mediated trophoblast differentiation and induction of hCYP19 gene expression during human placental development. The finding that hypoxia and Mash-2 overexpression with associated increases in USFs markedly induced cyclin B1 expression and increased the proportion of cultured trophoblast cells at the G2/M boundary further suggests that increased USF1/2 expression may promote cytotrophoblast proliferation at the expense of differentiation.
MATERIALS AND METHODS
Primary culture of human trophoblast cells.
Midtrimester human placental tissues were obtained in accordance with the Donors Anatomical Gift Act of the State of Texas after obtaining consent in writing. In all cases, consent forms and protocols were approved by the Institutional Review Board of the University of Texas Southwestern Medical Center at Dallas. A placental primary culture system (26) was modified for isolation and culture of cytotrophoblasts from midgestation human placenta (21). Briefly, the placental tissues were washed with Hanks' balanced salt solution, pH 7.4 (GIBCO, Grand Island, NY), and then finely minced and digested with 0.125% trypsin in Hanks' balanced salt solution at 37°C for 20 min. This procedure was repeated three times. At the end of each digestion step, the supernatant was collected, layered over 10 ml of serum, and then briefly centrifuged. The pellet was suspended in Dulbecco's modified Eagle's medium (DMEM; GIBCO, Grand Island, NY), filtered, and layered over a Percoll gradient (70% to 5%). The gradients were centrifuged at 1,200 × g for 20 min at room temperature, and cells in the middle layer (density, 1.045 to 1.062 g/ml) were collected, washed, and counted. The cells were then resuspended in DMEM supplemented with 10% fetal bovine serum (FBS) and 1.2% antibiotic/antimycotic solution (GIBCO, Grand Island, NY) and plated at a density of 2 × 106 cells per dish in 35-mm culture dishes or 15 × 106 cells per dish in 100-mm dishes. They were then incubated at 37°C in a humidified atmosphere of 95% air-5% CO2 (20% O2) or placed in a modular incubator chamber (Billups-Rothenberg, Inc., Del Mar, CA) in an atmosphere containing 2% O2, 93% N2, and 5% CO2. Cells were cultured overnight, and the medium was then changed to DMEM containing 2% FBS. Cells that were cultured either in 20% or 2% O2 were harvested at ambient O2 tension. To ensure that the findings obtained using cells cultured at 2% O2 were not altered by harvesting in room air, a parallel experiment was performed in which cells cultured in 2% O2 were harvested in 2% O2 using a glove box.
siRNA transfection.
One day before transfection, JEG3 cells were trypsinized, counted, and plated in 60-mm dishes in growth medium without antibiotics to achieve 30 to 50% confluence at the time of transfection. Small interfering RNA (siRNA)-Oligofectamine complexes were prepared as follows: 10 μl of 20 μM USF1 or USF2 siRNA or USF1 plus USF2 siRNA oligonucleotide (Ambion, Inc., Austin, TX) was added to 175 μl of Opti-MEM I “reduced serum medium” (Invitrogen, Carlsbad, CA) and incubated for 5 to 10 min. Three microliters of Oligofectamine (Invitrogen, Carlsbad, CA) was combined with 12 μl of Opti-MEM for addition to each well of a six-well plate. The diluted oligonucleotides were combined with the diluted Oligofectamine (total volume, 200 μl), mixed gently, and incubated for 15 to 20 min at room temperature to allow the siRNA-Oligofectamine complexes to form. The growth medium was removed, and the cells were washed once with medium without serum. Medium without serum (800 μl) was added to each well containing cells, followed by addition of siRNA-Oligofectamine complexes (200 μl). The cells were then maintained at 37°C in a CO2 incubator for 4 h. Growth medium containing three times the normal concentration of serum (500 μl) was then added to each dish without removing the transfection mixture. After culture for 48 h, the transfected cells were scraped from the dishes, passed through a 21-gauge needle for cell disruption, and centrifuged at 10,000 × g. Total cell lysates (20 μg of protein) were analyzed for USF1 and USF2 protein by immunoblotting or placed in RNA lysis solution (Ambion, Inc., Austin, TX) for RNA isolation and analysis of hCYP19 mRNA or 28S rRNA by Northern blotting.
Infection of trophoblast cells with recombinant adenoviruses.
Freshly isolated cytotrophoblasts plated at a density of 2 × 106 cells per 35-mm dish in DMEM containing 10% FBS were infected with recombinant adenoviruses containing a fusion gene comprised of 501 bp of hCYP19 exon I.1 5′ flanking sequence linked to hGH (human growth hormone gene) as reporter at a multiplicity of infection (MOI) of 0.5 and cultured in a 20% O2 environment in the absence (dimethyl sulfoxide [DMSO] vehicle) or presence of the proteasome inhibitors, LLnL (N-acetyl-l-leucyl-l-leucyl-l-norleucinal) (Sigma, St. Louis, MO) and MG-115 (Calbiochem, La Jolla, CA). After an overnight incubation, the medium was removed and replaced with fresh DMEM containing 2% FBS, with or without the proteasome inhibitors. The cells were incubated for up to 72 h; medium was collected and replaced daily and analyzed for hGH by radioimmunoassay (Nichols Institute Diagnostics, San Juan Capistrano, CA).
In other experiments, recombinant adenoviruses containing CMV-Mash-2 (where CMV is the cytomegalovirus promoter) or CMV-β-galactosidase (CMV-β-gal, as a control), constructed as described previously (19), were used to infect freshly isolated cytotrophoblasts at an MOI of 5.0 and cultured in a 2% or 20% O2 environment. The infected cells were analyzed for stage of the cell cycle or for expression of cyclin B1 by immunofluorescence, as described below.
Cell cycle analysis.
Human trophoblasts cultured for 72 h in a 2% or 20% O2 environment or in a 20% O2 environment after infection with recombinant adenoviruses expressing Mash-2 or β-Gal, were trypsinized and fixed in 70% ethanol overnight at 4°C. After fixation, the cells were washed in phosphate-buffered saline (PBS) and incubated with RNase (50 μg/ml; Roche, Indianapolis, IN) for 1 h and stained with propidium iodide (25 μg/ml; Sigma-Aldrich, St. Louis, MO) for 15 min at 4°C. The stained cells were analyzed by FACScan using CellQUEST software (Becton Dickinson, Franklin, NJ).
Immunofluorescence staining.
Freshly isolated cytotrophoblasts were plated on glass coverslips and infected with recombinant adenoviruses containing CMV-Mash-2 (MOI, 5) or the same amount of CMV-β-gal overnight (as described above) and then cultured for 3 days in DMEM containing 2% FBS in an atmosphere of 20% O2. Cells were fixed in methanol (100%) at −20°C for 6 min and then incubated with antibody to cyclin B1 (5 μg/ml) (Pharmingen, San Diego, CA) in PBS containing 0.5% bovine serum albumin (Sigma, St. Louis, Mo.) for 45 min. Goat anti-rabbit immunoglobulin G (IgG)-fluorescein conjugate (Molecular Probes, Eugene, OR) was used as secondary antibody. Slides were examined by immunofluorescence microscopy using a B-2 filter for fluorescein isothiocyanate (Nikon, Kanagawa, Japan).
Morphological analysis.
Cytotrophoblasts were cultured on glass coverslips in DMEM containing 2% FBS in a 20% O2 environment in the absence or presence of proteasome inhibitors, MG-115 or MG-132 (Calbiochem, La Jolla, CA). After cells were cultured for 16 h in 20% O2, medium was removed, half of the dishes were rinsed with PBS, and the cells were fixed in 75% ethanol. Fresh medium without proteasome inhibitors was added to the remaining dishes, which were cultured for an additional 16 h and then rinsed and fixed in 75% ethanol. Hematoxylin and eosin Y were used to stain nuclei and cytoplasm, respectively. Morphology was analyzed by light microscopy.
Coimmunoprecipitation and immunoblot analysis.
Nuclear and cytosolic fractionation and immunoblotting procedures have been described in detail previously (20). For coimmunoprecipitation, nuclear and cytosolic fractions were isolated from trophoblast cells that had been cultured in 2% O2 or 20% O2 for 16 h in the absence (DMSO) or presence of the proteasome inhibitors LLnL, MG-115, or MG-132. For immunoprecipitation and immunoblot analysis, cytosolic (500 μg) and nuclear proteins (150 μg) were precleared by incubation with nonimmune rabbit IgG (0.25 μg/ml) for 30 min at 4°C and then incubated with 1 μg of USF1 or USF2 rabbit polyclonal antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for 1 h at 4°C, followed by addition of 20 μl of protein A agarose beads (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The mixture was then incubated on a rotating mixer at 4°C overnight. The protein A-agarose beads were washed with RIPA buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) four times, and bound proteins were eluted using 40 μl of 1× SDS sample buffer. The eluates (40 μl) were electrophoresed on 12% SDS-polyacrylamide gels and electrophoretically transferred to nitrocellulose membranes (Midwest Scientific, Valley Park, MO), which were incubated with mouse monoclonal antiubiquitin antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Sheep anti-mouse IgG horseradish peroxidase-linked F(ab′)2 fragment (Amersham Pharmacia Biotech Inc., Piscataway, NJ) was used as a secondary antibody. For standard immunoblot analysis of USF1/2, nuclear and cytosolic proteins (20 μg) were electrophoresed on 12% SDS-polyacrylamide gels and electrophoretically transferred to nitrocellulose membranes. Membranes were blocked using 5% milk and incubated with antibodies to either human USF1 or human USF2 (0.5 μg/ml). Donkey anti-rabbit IgG horseradish peroxidase-linked F(ab′)2 fragment (Amersham Pharmacia Biotech Inc., Piscataway, NJ) was used as a secondary antibody. Proteins assessed by coimmunoprecipitation and immunoblotting were detected using an ECL kit (Amersham Pharmacia Biotech Inc., Piscataway, NJ) following the manufacturer's protocol.
Northern blot analysis.
Total RNA was extracted using an RNA purification kit according to the manufacturer's protocol (Ambion Inc., Austin, TX) from JEG3 cells that were transfected with either USF1 siRNA, USF2 siRNA, or USF1 siRNA plus USF2 siRNA and incubated for 48 h. Aliquots (20 μg) of total RNA were size fractionated on a 7.4% formaldehyde-0.9% agarose gel, transferred to Zeta-Probe blotting membrane (Bio-Rad Laboratories, Inc. Hercules, CA), and hybridized for 2 h at 65°C to radiolabeled cDNA probes for hCYP19 mRNA (19). The cDNAs were 32P-labeled using a Prime-It RmT random primer labeling kit (Stratagene, La Jolla, Calif.). After samples were washed, relative levels of mRNA were assessed by autoradiography. The Northern blots were stripped and reprobed using a radiolabeled 28S cDNA (American Type Culture Collection, Manassas, VA) to assess loading and transfer of RNA.
Tritiated water assay of aromatase activity in placental cells.
Freshly isolated cytotrophoblast cells were cultured in a 20% O2 environment in the absence or presence of proteasome inhibitors (LLnL and MG-115) for 16 h. Aromatase activity was assayed using a tritiated water assay as described previously (1). 1β-[3H]androstenedione (NEN Life Science Products, Boston, MA) was added to the culture medium during the last 1 h of incubation. The medium was then removed and placed in ice-cold 30% (wt/vol) trichloroacetic acid. The incorporation of tritium from 1β-[3H]androstenedione into water was assayed in aqueous scintillation fluid after extraction with 4 volumes of chloroform and 1 volume of dextran-charcoal suspension. The adherent cells were analyzed for protein (29). Aromatase activity is expressed as picomoles of 1β-[3H]androstenedione metabolized to estrogen/min/mg protein.
EMSA.
Nuclear proteins isolated from trophoblasts cultured in a 20% O2 environment in the absence or presence of proteasome inhibitors (LLnL or MG-132) for 16 h were incubated with double-stranded oligonucleotides (GIBCO, Carlsbad, CA; Integrated DNA Technologies, Inc., Coralville, IA) containing three E-box sequences (underlined) within placenta-specific exon I.1 and its 5′ flanking region (20) (E1 at −325 bp, 5′-ACTCCCATGACACTTGCTGAGGTCTT-3′; E2 at −58 bp, 5′-TTTGTTCAATCACATGCTTCAGTCAT-3′; E3 at +26 bp, 5′-GAGGGCTGAACACGTGGAGGCAAACA-3′) (GenBank accession no. M30795). Binding activity was analyzed by electrophoretic mobility shift assay (EMSA) as described in detail previously (20). Briefly, the nuclear proteins were incubated for 1 h at 4°C in binding buffer in the absence or presence of IgG (1 μg) for human USF1 (hUSF1) or hUSF2 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Radiolabeled E1, E2, or E3 oligonucleotides were added to the reaction mixture, and the incubation was continued for another 30 min at room temperature, before separation on 5% native polyacrylamide gels and visualization by autoradiography.
RESULTS
Endogenous USFs negatively regulate hCYP19 gene expression.
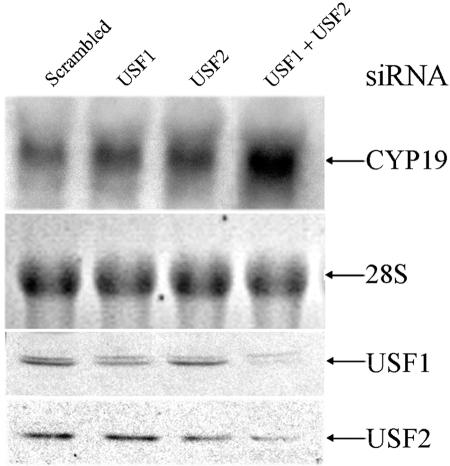
Previously, we observed that USF1 overexpression markedly inhibited hCYP19 gene expression in human trophoblasts cultured in a 20% O2 environment (20). In the present study, siRNA technology was used to determine the effects of silencing USF1 and USF2 expression on expression of the endogenous hCYP19 gene in JEG3 choriocarcinoma cells. As shown in the top two panels of Fig. 1A, transfection of JEG3 cells with either USF1 or USF2 siRNA modestly reduced the respective endogenous USF1 and USF2 protein levels and failed to alter hCYP19 gene expression. On the other hand, in cells cotransfected with siRNAs for USF1 and USF2, a far more pronounced decrease in endogenous USF1 and USF2 protein levels was observed in association with markedly increased levels of endogenous hCYP19 mRNA. These findings provide important evidence for the inhibitory role of endogenous USF1/2 on hCYP19 expression in cultured trophoblast cells. We suggest that the more pronounced inhibitory effect on USF1 and USF2 protein levels observed when their respective siRNAs were cotransfected may be due, in part, to the fact that newly synthesized USF1 and USF2 proteins preferentially form heterodimers (11), which may be more resistant to proteasomal degradation. Thus, when either USF1 or USF2 protein is reduced by transfection of a single siRNA, the remaining protein will form heterodimers that are relatively stable. On the other hand, when both USF1 and USF2 proteins are reduced by cotransfection of both siRNAs, the remaining proteins are more susceptible to proteasomal degradation because of their reduced capacity to form heterodimers. Importantly, the findings obtained indicate that USF1/2 protein and CYP19 mRNA levels are reciprocally regulated.
FIG. 1.
Endogenous USF1 and USF2 inhibit hCYP19 gene expression in JEG3 cells. JEG3 cells were transfected with USF1 siRNA, USF2 siRNA, USF1 siRNA plus USF2 siRNA or with scrambled siRNA, as a control, and cultured for an additional 48 h. hCYP19 mRNA levels were analyzed by Northern blotting using a full-length 32P-labeled hCYP19 cDNA as probe. The blot was reprobed for 28S rRNA as a control for RNA loading and transfer. USF1 and USF2 protein levels in lysates from the same cells were analyzed by immunoblotting (two lower panels).
Hypoxia and Mash-2 overexpression increase cell cycle progression and cyclin B1 expression.
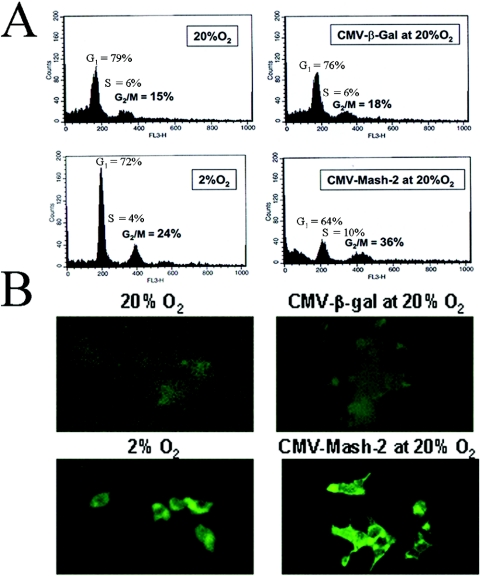
Hypoxia and Mash-2 overexpression increase USF1/2 protein levels (20) and enhance DNA synthesis and proliferation of cultured trophoblast cells (19). As mentioned above, USF was found to bind to and activate transcription of the cyclin B1 gene in HeLa cells (6). Since cyclin B1 is essential for the G2/M transition and progression through the cell cycle, in the present study, we analyzed the effects of O2 tension and of Mash-2 overexpression on cell cycle progression and on cyclin B1 expression. As can be seen in Fig. 2A, the proportion of trophoblast cells in G2/M was considerably increased when cells were cultured in 2% versus 20% O2 and when cells were infected with recombinant adenovirus expressing Mash-2 compared to control adenovirus expressing β-Gal. Effects of hypoxia and of Mash-2 overexpression on cell cycle progression were associated with a marked induction in the levels of cyclin B1 protein in the cells (Fig. 2B). These findings suggest that the actions of hypoxia and of increased Mash-2 and USF1/2 expression to stimulate trophoblast proliferation and inhibit syncytiotrophoblast differentiation may be mediated, in part, by induction of cyclin B1.
FIG. 2.
Hypoxia and Mash-2 overexpression increase cell cycle progression and cyclin B1 expression in human trophoblast cells. Human trophoblast cells in primary culture were maintained in either a 2% or 20% O2 environment for 3 days. Parallel dishes of cells were infected with recombinant adenoviruses containing CMV-Mash-2 or CMV-β-Gal and cultured in 20% O2 for 3 days. (A) Cells were fixed in 70% ethanol and stained with propidium iodide for cell cycle analysis by FACScan. (B) In the same experiment, cells were plated onto coverslips and analyzed for immunoreactive cyclin B1 levels by immunofluorescence.
USF1 and USF2 protein levels are increased by proteasome inhibitors in cells cultured in a 20% O2 environment.
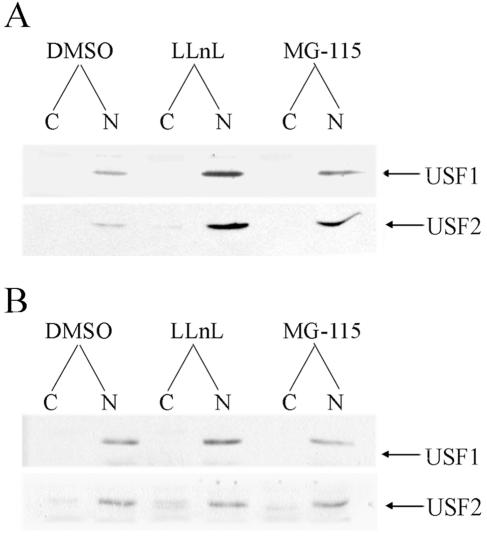
Previously, we observed that although USF1/2 protein levels declined with syncytiotrophoblast differentiation in 20% O2 and were markedly induced by hypoxia and by Mash-2 overexpression, USF1 (20) and USF2 (B. Jiang and C. R. Mendelson, unpublished observations) mRNA levels were relatively unaffected. This suggested that the hypoxia- and Mash-2-induced expression of USF1/2 proteins was not due to altered mRNA transcription but, rather, to changes in mRNA translatability and/or protein stability. To test the hypothesis that O2-mediated induction of trophoblast differentiation and hCYP19 gene expression is mediated by increased degradation of USF1 and USF2 protein levels via the proteasome degradation pathway, we analyzed the effects of two proteasome inhibitors, LLnL and MG-115 on cytoplasmic and nuclear levels of USF1 and USF2 proteins in human trophoblast cells cultured in a 20% (Fig. 3A) or 2% (Fig. 3B) O2 environment. As can be seen, the proteasome inhibitors caused a marked increase in USF1 and USF2 protein levels in trophoblasts cultured in a 20% O2 environment (Fig. 3A). By contrast, these agents had no effect on USF protein levels in cells cultured in a hypoxic environment (Fig. 3B). These findings suggest that proteasomal degradation of USF1/2 is induced when trophoblasts are cultured under conditions of increased O2 tension. It should be noted that although it appears that USF1/2 protein levels were unaffected by O2 tension, the immunoblot of trophoblasts cultured in 20% O2 was exposed to X-ray film for considerably (about five times) longer than the immunoblot of cells cultured in 2% O2.
FIG. 3.
Proteasome inhibitors increase USF1 and USF2 protein levels in cultured trophoblasts. Nuclear (N) and cytoplasmic (C) fractions were prepared from human trophoblasts cultured for 16 h in a 20% O2 (A) or 2% O2 (B) environment in the absence or presence of proteasome inhibitors, LLnL (20 μM) and MG-115 (10 μM). Twenty micrograms of cytoplasmic or nuclear proteins was analyzed for USF1 and USF2 by immunoblotting. It should be noted that the exposure time of the autoradiogram of USF1/2 in cells cultured in the 20% O2 environment was considerably longer than that of the cells cultured in 2% O2 so that the effects of proteasome inhibitors could be assessed. However, the overall levels of USF1/2 protein in the immunoblot shown in panel A were substantially lower than those shown in panel B.
Proteasome inhibitors prevent induction of endogenous aromatase activity and hCYP19 gene expression and reduce hCYP19 promoter activity in human trophoblasts cultured in a 20% O2 environment.
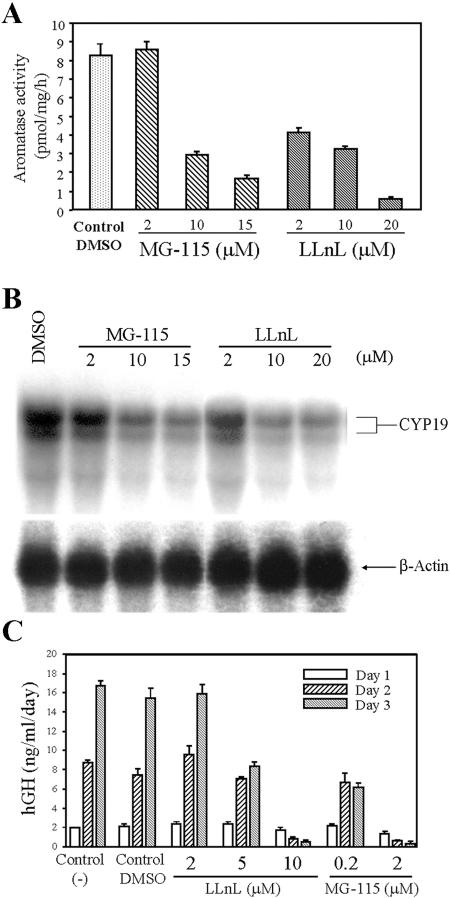
We previously reported that increased USF1/2 protein levels mediated hypoxia inhibition of aromatase activity, hCYP19 mRNA, and hCYP19 promoter I.1 activity (20). To analyze the effects of proteasome inhibitors on aromatase activity and hCYP19 gene expression, human trophoblast cells were cultured for 16 h in a 20% O2 environment in the absence or presence of increasing concentrations of MG-115 and LLnL. MG-115 and LLnL caused a dose-dependent inhibition of aromatase activity (Fig. 4A) and of hCYP19 mRNA levels (Fig. 4B) compared to DMSO controls. The inhibition of aromatase activity and of hCYP19 mRNA levels was associated with the effects of these inhibitors to increase USF1 and USF2 protein levels.
FIG. 4.
Aromatase activity, CYP19 mRNA levels, and CYP19 promoter activity are decreased in a dose-dependent manner by proteasome inhibitors. (A) Freshly isolated cytotrophoblasts were cultured in a 20% O2 environment for 16 h in the absence (DMSO vehicle alone) or presence of proteasome inhibitors, LLnL and MG-115, in various concentrations. Aromatase activity was analyzed by measuring the incorporation of tritium from 1β-[3H]androstenedione into water. (B) Aliquots (20 μg) of total RNA obtained from trophoblasts cultured for 16 h in a 20% O2 environment in the absence (DMSO) or presence of proteasome inhibitors (LLnL and MG-115) were analyzed for P450arom mRNA transcripts by Northern blotting using a 32P-labeled human P450arom cDNA probe. The blot was reprobed for β-actin mRNA as a control for RNA loading and transfer and to assess the effects of the proteasome inhibitors on levels of a constitutively regulated transcript. (C) Freshly isolated trophoblast cells maintained in a 20% O2 environment were infected overnight with recombinant adenovirus containing a hCYP19I.1-501-hGH fusion gene (MOI, 0.5). The next morning, the medium was removed and the cells were incubated in the absence (Control and Control DMSO) or presence of proteasome inhibitors in a 20% O2 environment. Media were collected and changed daily for a 72-h period. Shown is the accumulation of hGH secreted into the medium during the first 24 h (Day 1), between 24 h and 48 h (Day 2), and between 48 h and 72 h (Day 3) of incubation. Data are the means ± standard errors of the means of triplicate values from two independent experiments.
To analyze effects of the proteasome inhibitors on activity of the hCYP19 gene placenta-specific promoter (promoter I.1), human trophoblasts in primary culture were infected with recombinant adenoviruses containing a fusion gene comprised of 501 bp of hCYP19 exon I.1 5′ flanking DNA linked to hGH, as reporter. The transfected cells were then incubated for up to 3 days in the absence or presence of the proteasome inhibitors LLnL and MG-115 in increasing concentrations; hGH secreted into the medium over each 24-h period was assayed as an index of hCYP19 promoter I.1 activity. As previously observed, when trophoblasts were cultured in control medium (21) or in medium containing DMSO, hCYP19 promoter I.1 activity was relatively low on day 1 and increased markedly with time in culture in association with syncytiotrophoblast differentiation (Fig. 4C). Little or no effect of the proteasome inhibitors was observed after 1 day of culture when promoter I.1 activity was relatively low. With syncytiotrophoblast differentiation on days 2 and 3, both proteasome inhibitors caused a marked and dose-dependent inhibition of hCYP19 promoter I.1 activity (Fig. 4C). This was associated with their effects to increase USF1/2 protein levels.
Peptide aldehyde proteasome inhibitors, such as MG-115 and MG132, also inhibit lysosomal and calcium activated proteases (32). To verify specificity for the proteasome, we also tested the effects of the epoxyketone containing natural product epoxomicin, which has been found to specifically target the 26S proteasome (33). We observed that epoxomicin at a concentration as low as 200 nM caused a pronounced increase in USF1 and USF2 protein levels and markedly inhibited hCYP19 mRNA levels in human trophoblast cells cultured in 20% O2 (data not shown). These findings indicate that O2-mediated degradation of USF1/2 likely occurs via the 26S proteasome pathway.
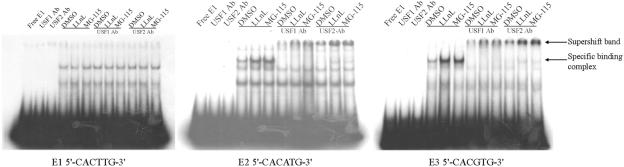
Proteasome inhibitors increase USF1 and USF2 binding to E-boxes in DNA within hCYP19 exon I.1 and its 5′ flanking region.
We previously identified three potential E-box binding sites for USF1/2 upstream and within placenta-specific hCYP19 exon I.1 (E1, −325 bp; E2, −58 bp; E3, +26 bp) (20). By EMSA, we found that binding activity for all three E-boxes was increased in nuclear extracts of freshly isolated cytotrophoblasts compared to syncytiotrophoblast, but that only the E2 and E3 boxes had the capacity to bind USF1/2 (20). In the present study, EMSA was used to analyze the effects of proteasome inhibitors on USF binding activity in trophoblast nuclear extracts for the E2 and E3 boxes; nuclear protein binding to the E1 box was analyzed as a control. As can be seen in Fig. 5, binding activity for the E2 and E3 boxes of nuclear proteins isolated from human trophoblasts cultured in 20% O2 for 16 h in the presence of the two proteasome inhibitors was increased markedly compared to extracts from cells incubated with the DMSO vehicle. The finding that the specific binding complexes were completely supershifted by either USF1 or USF2 antibody indicates that the proteasome inhibitors increased binding of USF1 and USF2 proteins as a heterodimer to E2 and E3 boxes (Fig. 5). By contrast, no effect of the proteasome inhibitors was observed for nuclear protein binding to the E1 box. These findings suggest that like hypoxia, proteasome inhibitors inhibit hCYP19 gene expression by increasing USF1/2 binding to the E2 and E3 boxes.
FIG. 5.
Binding of USF1 and USF2 to the E2 and E3 boxes in the hCYP19 promoter and first exon of hCYP19 gene is increased by proteasome inhibitors. Nuclear extracts from human cytotrophoblasts cultured in a 20% O2 environment for 16 h in the absence or presence of proteasome inhibitors were incubated with 32P-labeled oligonucleotides containing the E1, E2, and E3 boxes in the absence or presence of antibodies specific for USF1 and USF2 and analyzed by EMSA.
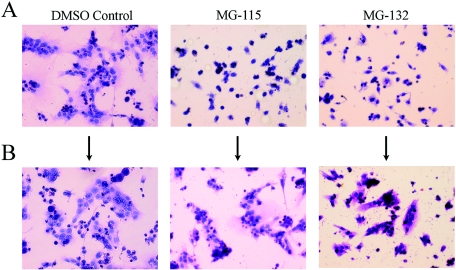
Proteasome inhibitors prevent trophoblast cell fusion and syncytiotrophoblast differentiation.
Previously, we observed that hypoxia (19) and USF1 overexpression (20) inhibited trophoblast cell fusion and differentiation. To assess the effects of proteasome inhibitors on trophoblast cell fusion and differentiation, human trophoblast cells were cultured in a 20% O2 environment for 16 h in the absence or presence of MG-115 and MG-132. As shown in Fig. 6A, cell fusion to form syncytiotrophoblast was readily apparent in cells incubated with the DMSO vehicle alone. By contrast, MG-115 (10 μM) and MG-132 (2 μM) had marked effects to inhibit cell fusion and syncytiotrophoblast differentiation (Fig. 6A). We previously found that the inhibitory effects of hypoxia on trophoblast cell fusion and differentiation were reversible (19). To ensure that the inhibitory effects of proteasome inhibitors on trophoblast differentiation were not due to toxicity, parallel dishes of cells incubated with MG-115 and MG-132 for 16 h were rinsed and cultured for an additional 16 h in control medium. As shown in Fig. 6B, within 16 h of removal of the proteasome inhibitors, trophoblast cell fusion and differentiation were evident.
FIG. 6.
Proteasome inhibitors prevent fusion and differentiation of cultured human trophoblasts. Freshly isolated cytotrophoblasts were cultured in a 20% O2 atmosphere in the absence or presence of proteasome inhibitors for 16 h (A). After 16 h, the medium in some dishes was removed; fresh medium without proteasome inhibitors was then added, and the cells were cultured for an additional 16 h (B). Shown are light micrographs of hematoxylin- and eosin Y-stained cells (magnification, ×200).
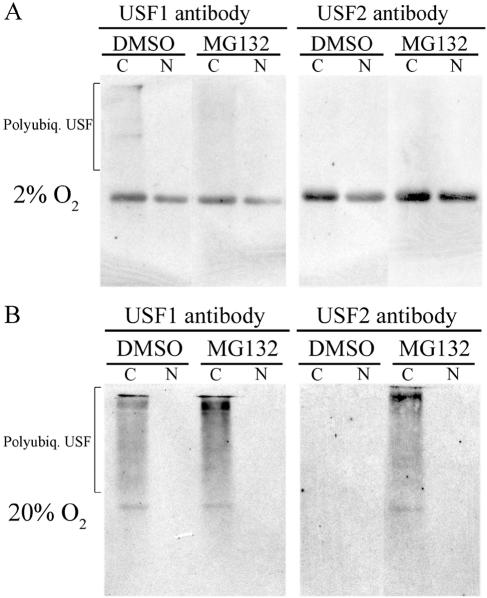
O2-induced degradation of USF1 and USF2 proteins is associated with increased polyubiquitylation.
The finding that proteasome inhibitors increased USF1/2 levels and binding to hCYP19 promoter I.1 in human trophoblasts cultured in 20% O2 suggests that increased O2 tension promotes USF1/2 polyubiquitylation and degradation via the 26S proteasome pathway. To determine the effects of O2 tension on USF1/2 polyubiquitylation, human trophoblasts were incubated for 16 h in 2% or 20% O2 environments in the absence or presence of MG-132 (10 μM). Nuclear and cytoplasmic fractions were prepared, and USF1 or USF2 antibodies were used to immunoprecipitate the respective proteins, which were then resolved by SDS-polyacrylamide gel electrophoresis and electroblotted to nitrocellulose membranes that were then analyzed for ubiquitylated forms of USF1/2 by immunoblotting using an antiubiquitin antibody. As can be seen, cytoplasmic levels of polyubiquitylated USF1 and USF2 were greatly increased in trophoblasts cultured in 20% (Fig. 7B), compared to 2% O2 (Fig. 7A). In trophoblast cells cultured in 20% O2, the levels of polyubiquitylated USF1/2 were further increased by treatment with MG-132. These findings suggest that increased O2 tension enhances USF1 and USF2 protein degradation by increasing their polyubiquitylation.
FIG. 7.
Polyubiquitylation of USF1 and USF2 is increased in human trophoblasts treated with MG-132 and cultured in 20% O2. Cytoplasmic (C) and nuclear (N) proteins isolated from human trophoblast cells cultured in the absence or presence of MG-132 in either 2% O2 (A) or 20% (B) O2 environments for 16 h were subjected to immunoprecipitation using rabbit polyclonal antisera for hUSF1 or hUSF2. The immunoprecipitated proteins were separated using SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and analyzed for ubiquitylated forms of USF1/2 by immunoblotting using a mouse monoclonal antibody to ubiquitin.
DISCUSSION
Hypoxia has been reported to stimulate cytotrophoblast proliferation (10), impair cell fusion (3) and differentiation into invasive cells (14), decrease placental polypeptide hormone production (16), and mimic the placental defect associated with preeclampsia (12, 14). Using trophoblast cells from midgestation human placenta in primary culture, we previously observed that differentiation of cytotrophoblasts to syncytiotrophoblast in a 20% O2 environment was associated with a marked induction of aromatase activity and of hCYP19 gene expression (21). By contrast, when the cells were cultured in a hypoxic (2% O2) environment, syncytiotrophoblast differentiation and induction of hCYP19 gene expression were prevented (19). These effects of hypoxia were associated with increased nuclear levels of the bHLH zipper transcription factors USF1 and USF2 and their binding as heterodimers to two E-boxes within hCYP19 exon I.1 and its 5′ flanking region (20). The findings that endogenous USF1/2 protein levels declined with syncytiotrophoblast differentiation and that overexpression of USF1 inhibited syncytiotrophoblast differentiation, hCYP19 promoter activity, and endogenous hCYP19 gene expression (20) suggest that USFs play a role to prevent trophoblast differentiation and inhibit hCYP19 gene expression. Interestingly, we observed that, whereas hypoxia increased USF1 (20) and USF2 (B. Jiang and C. R. Mendelson, unpublished observations) protein levels, mRNA levels were unaffected.
The goal of this study was to further define the mechanisms whereby USFs sense changes in O2 tension and mediate hypoxia inhibition of trophoblast differentiation and CYP19 gene expression. siRNA technology was applied to JEG3 cells to assess the inhibitory role of endogenous USFs on hCYP19 gene expression. JEG3 is a choriocarcinoma cell line that is cytotrophoblast-like. In this regard, these cells are highly proliferative and manifest low levels of hCYP19 gene expression (21). Interestingly, JEG3 cells have increased expression levels of USF1/2, compared to primary cultures of human trophoblast cells (B. Jiang and C. R. Mendelson, unpublished observations). We observed that cotransfection of JEG3 cells with siRNAs corresponding to both USF1 and USF2 caused a pronounced decrease in nuclear levels of USF1 and USF2 proteins and a marked induction of hCYP19 mRNA levels. These findings indicate that increased expression of endogenous USFs in the choriocarcinoma cells suppresses hCYP19 gene expression, which is a manifestation of differentiative function.
Based on our previous finding that changes in O2 tension altered USF protein levels but had no effect on USF1/2 mRNA, we hypothesized that O2 regulation of trophoblast differentiation and hCYP19 gene expression may be mediated by alterations in USF1 and USF2 protein degradation via the proteasome pathway. To test this hypothesis, we analyzed the effects of proteasome inhibitors on human trophoblast cells in primary culture. We found that these agents increased USF1/2 protein levels and binding to the E-boxes surrounding hCYP19 promoter I.1. This was associated with inhibitory effects of the proteasome inhibitors on aromatase activity, hCYP19 gene expression, hCYP19 promoter activity, and trophoblast cell fusion and differentiation. Furthermore, we observed increased levels of polyubiquitylated USF1 and USF2 in cytoplasm of human trophoblast cells cultured in 20% O2 in the presence of proteasome inhibitors, compared to cells cultured in a 2% O2-containing environment. We propose that with increased blood flow through the placental spiral arteries after the ninth week of human gestation with an associated increase in local O2 tension, USF1/2 levels decline because of increased proteasomal degradation. This, in turn, results in enhanced syncytiotrophoblast differentiation with increased biosynthesis of steroid and polypeptide hormones.
A number of bHLH transcription factors containing PAS (Per-aryl hydrocarbon receptor nuclear translocator [ARNT]-Sim) domains have been found to be hypoxia inducible and mediate enhanced expression of target genes, including those encoding glycolytic enzymes, glucose transporters, and growth factors that induce erythropoiesis and angiogenesis (4, 7, 38). These bHLH-PAS domain transcription factors include hypoxia inducible factor 1α (HIF-1α) (38), human endothelial PAS domain protein 1 (EPAS-1/HIF-2α) (8), and their common obligate heterodimeric partner ARNT/HIF-1β (46). Protein levels of HIF-1α (38) and EPAS-1 (47) decline under normoxic conditions because of increased proteasomal degradation triggered by posttranslational hydroxylation of a conserved proline residue in its oxygen-dependent degradation domain. This, in turn, promotes recruitment of von Hippel-Lindau tumor-suppressor protein (VHL), the recognition component of an E3 ubiquitin ligase, leading to polyubiquitylation and targeting of HIF-1α and EPAS-1 to the 26S proteasome. Increased expression of HIF-1α (5), VHL, and EPAS-1 (13) was observed in human trophoblast villous explants cultured under hypoxic conditions; levels of these proteins declined when explants were cultured in 20% O2. Although we have observed that EPAS-1 levels decline in human trophoblast cells during culture in 20% O2 and are induced by hypoxia and Mash-2 overexpression (20), we were unable to detect EPAS-1 binding to the E2 and E3 boxes surrounding hCYP19 promoter I.1 (20). This is likely due to the fact that HIF transcription factors bind as heterodimers with ARNT to DNA sequences termed “hypoxia response elements” that have a GTG common to USF binding sites as the 3′ half-site (which binds ARNT) and a unique 5′ half-site (43). Only ARNT homodimers have the capacity to bind to the palindromic E-box core sequences (CACGTG) (42, 43) that bind USF1/2 heterodimers. Furthermore, HIF-1α protein levels are low in our primary human trophoblast cultures and appear to be unaffected by changes in O2 tension or proteasome inhibitors (B. Jiang and C. R. Mendelson, unpublished observations). Using coimmunoprecipitation, we were unable to detect a direct interaction between USF1/2 and VHL, although in the same coimmunoprecipitation analysis, HIF-1α in HeLa cells incubated with CoCl2 interacted strongly with VHL (B. Jiang and C. R. Mendelson, unpublished observations). Thus, it is likely that USF1 and USF2 interact with another E3 ligase(s) to mediate their proteasomal degradation.
On the other hand, a role for VHL in placental differentiation is likely, since mouse embryos that are homozygous null for the vhl gene die at embryonic day 9.5 (E9.5) to E10.5 because of a defect in placental vasculogenesis (15). Furthermore, Arnt−/− embryos also die by E10.5 because of a failure of labyrinthine trophoblast development and placental vascularization (27). The Arnt−/− placentas also manifest marked decreases in diploid spongiotrophoblasts and increased numbers of giant cells (2). Interestingly, ARNT was found to be required for differentiation of trophoblast stem cells into spongiotrophoblasts under hypoxia conditions (2). On the other hand, Hif-1α−/− embryos die by E11 because of neural tube defects, lack of cephalic vascularization, and increased cell death and cardiovascular malformations (17, 37), while Epas-1−/− mice die after E12.5 because of presumed defects in catecholamine homeostasis. Neither Hif-1α- nor Epas-1-deficient embryos manifest defects in placental development or vascularization. Thus, it is likely that ARNT acts either as a homodimer (42) or as a heterodimer with another bHLH transcription factor to promote labyrinthine trophoblast development. The finding that embryonic lethality occurs in mice that are homozygous for targeted deletion in the usf2 gene and either homozygous or heterozygous for a mutation in the usf1 gene suggests that USF proteins are essential for embryonic development (41). However, their role in placental development remains unknown since the cause of embryonic death in these mutant mice was not reported.
The mechanism(s) whereby increased expression levels of USF1/2 prevent syncytiotrophoblast differentiation has not been defined. In previous studies, we found that hypoxia and overexpression of Mash-2 markedly increased DNA synthesis in cultured trophoblast cells (19). In the present study, we found that this was associated with a pronounced increase in the number of cells at the G2/M transition and induction in cytoplasmic levels of cyclin B1, a protein that plays a critical role in control of the G2/M transition and progression through the somatic cell cycle (see reference 35 for a review). As mentioned above, Mash-2 overexpression markedly enhances USF1/2 nuclear protein levels in human trophoblast cells (20). Interestingly, it has been found that USF activates expression of the gene for cyclin B1 in HeLa cells (6). Furthermore, it was observed that USF DNA-binding activity was enhanced in a G2-dependent manner (6). Thus, it appears that increased levels of USF1/2 in cytotrophoblasts cultured in a hypoxic environment may promote cell proliferation and block cellular pathways leading to differentiation.
The mechanisms whereby USFs inhibit hCYP19 gene expression have not been determined. USFs are known to serve both as transcriptional activators (11, 34) and as repressors of a number of target genes, including the ABCA1 transporter (48), Xenopus MyoD (30), and the rabbit CYP19A1 gene (44). USF1 was found to inhibit transcription of the rabbit CYP19A1 gene by competing with ARNT for binding to the promoter (44). In the case of Xenopus MyoD, USF was found to block autoactivation of the promoter by competing for binding with myogenic activating transcription factors (30). We postulate that under hypoxic conditions in human trophoblast cells, increased nuclear levels of USF1/2 may compete with transcriptional activators for binding to the E2 and E3 boxes. USF1/2 binding, in turn, may facilitate the recruitment of transcriptional corepressors leading to a closing of chromatin structure and silencing of hCYP19 expression. We further suggest that with the developmental increase in placental vascularization and increased O2 availability to trophoblast cells, the inhibitory USFs are degraded, which may allow placenta-specific transcription factors to bind to the E boxes and promote placenta-specific expression. A potential candidate is the bHLH-leucine zipper transcription factor, TFEB-A, which is highly and selectively expressed in placenta (28). Interestingly, like hCYP19, TFEB has numerous tissue-specific first exons suggesting its control via alternative promoters (28). In recent studies, we found that a fusion gene comprised of 246 bp of hCYP19 exon I.1 5′ flanking DNA and 103 bp of exon I.1, containing the E2 and E3 boxes, was expressed in a placenta-specific and developmentally regulated manner in transgenic mice (24). Studies are in progress to define the transcription factors bound to the E2 and E3 boxes under hypoxic and normoxic conditions and their roles in developmental and placental cell-specific regulation of hCYP19 promoter I.1 activity.
Acknowledgments
We thank Jo Smith and Vickey Chau for preparation of cytotrophoblast cells and Margaret Hinshelwood and George DeMartino for their advice and for helpful discussion.
This work was funded by Public Health Service grant 5-R01 DK031206 from the National Institute of Diabetes and Digestive and Kidney Diseases (C.R.M.).
REFERENCES
- 1.Ackerman, G. E., M. E. Smith, C. R. Mendelson, P. C. MacDonald, and E. R. Simpson. 1981. Aromatization of androstenedione by human adipose tissue stromal cells in monolayer culture. J. Clin. Endocrinol. Metab. 53:412-417. [DOI] [PubMed] [Google Scholar]
- 2.Adelman, D. M., M. Gertsenstein, A. Nagy, M. C. Simon, and E. Maltepe. 2000. Placental cell fates are regulated in vivo by HIF-mediated hypoxia responses. Genes Dev. 14:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alsat, E., P. Wyplosz, A. Malassine, J. Guibourdenche, D. Porquet, C. Nessmann, and D. Evain-Brion. 1996. Hypoxia impairs cell fusion and differentiation process in human cytotrophoblast, in vitro. J. Cell. Physiol. 168:346-353. [DOI] [PubMed] [Google Scholar]
- 4.Bruick, R. K., and S. L. McKnight. 2002. Transcription. Oxygen sensing gets a second wind. Science 295:807-808. [DOI] [PubMed] [Google Scholar]
- 5.Caniggia, I., H. Mostachfi, J. Winter, M. Gassmann, S. J. Lye, M. Kuliszewski, and M. Post. 2000. Hypoxia-inducible factor-1 mediates the biological effects of oxygen on human trophoblast differentiation through TGFβ(3). J. Clin. Investig. 105:577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cogswell, J. P., M. M. Godlevski, M. Bonham, J. Bisi, and L. Babiss. 1995. Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 gene promoter. Mol. Cell. Biol. 15:2782-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ema, M., K. Hirota, J. Mimura, H. Abe, J. Yodoi, K. Sogawa, L. Poellinger, and Y. Fujii-Kuriyama. 1999. Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 18:1905-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ema, M., S. Taya, N. Yokotani, K. Sogawa, Y. Matsuda, and Y. Fujii-Kuriyama. 1997. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. USA 94:4273-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett, R. B., and P. C. MacDonald. 1979. Endocrinology of the placenta. Annu. Rev. Med. 30:473-488. [DOI] [PubMed] [Google Scholar]
- 10.Fox, H. 1970. Effect of hypoxia on trophoblast in organ culture. A morphologic and autoradiographic study. Am. J. Obstet. Gynecol. 107:1058-1064. [DOI] [PubMed] [Google Scholar]
- 11.Gao, E., Y. Wang, J. L. Alcorn, and C. R. Mendelson. 2003. Transcription factor USF2 is developmentally regulated in fetal lung and acts together with USF1 to induce SP-A gene expression. Am. J. Physiol. 284:L1027-L1036. [Google Scholar]
- 12.Genbacev, O., R. Joslin, C. H. Damsky, B. M. Polliotti, and S. J. Fisher. 1996. Hypoxia alters early gestation human cytotrophoblast differentiation/invasion in vitro and models the placental defects that occur in preeclampsia. J. Clin. Investig. 97:540-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genbacev, O., A. Krtolica, W. Kaelin, and S. J. Fisher. 2001. Human cytotrophoblast expression of the von Hippel-Lindau protein is downregulated during uterine invasion in situ and upregulated by hypoxia in vitro. Dev. Biol. 233:526-536. [DOI] [PubMed] [Google Scholar]
- 14.Genbacev, O., Y. Zhou, J. W. Ludlow, and S. J. Fisher. 1997. Regulation of human placental development by oxygen tension. Science 277:1669-1672. [DOI] [PubMed] [Google Scholar]
- 15.Gnarra, J. R., J. M. Ward, F. D. Porter, J. R. Wagner, D. E. Devor, A. Grinberg, M. R. Emmert-Buck, H. Westphal, R. D. Klausner, and W. M. Linehan. 1997. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc. Natl. Acad. Sci. USA 94:9102-9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huot, R. I., J. M. Foidart, and K. Stromberg. 1979. Effects of culture conditions on the synthesis of human chorionic gonadotropin by placental organ cultures. In Vitro 15:497-502. [DOI] [PubMed] [Google Scholar]
- 17.Iyer, N. V., L. E. Kotch, F. Agani, S. W. Leung, E. Laughner, R. H. Wenger, M. Gassmann, J. D. Gearhart, A. M. Lawler, A. Y. Yu, and G. L. Semenza. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jauniaux, E., A. L. Watson, J. Hempstock, Y. P. Bao, J. N. Skepper, and G. J. Burton. 2000. Onset of maternal arterial blood flow and placental oxidative stress. A possible factor in human early pregnancy failure. Am. J. Pathol. 157:2111-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, B., A. Kamat, and C. R. Mendelson. 2000. Hypoxia prevents induction of aromatase expression in human trophoblast cells in culture: potential inhibitory role of the hypoxia-inducible transcription factor Mash-2 (mammalian achaete-scute homologous protein-2). Mol. Endocrinol. 14:1661-1673. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, B., and C. R. Mendelson. 2003. USF1 and USF2 mediate inhibition of human trophoblast differentiation and CYP19 gene expression by Mash-2 and hypoxia. Mol. Cell. Biol. 23:6117-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamat, A., J. L. Alcorn, C. Kunczt, and C. R. Mendelson. 1998. Characterization of the regulatory regions of the human aromatase (P450arom) gene involved in placenta-specific expression. Mol. Endocrinol. 12:1764-1777. [DOI] [PubMed] [Google Scholar]
- 22.Kamat, A., K. H. Graves, M. E. Smith, J. A. Richardson, and C. R. Mendelson. 1999. A 500-bp region, approximately 40 kb upstream of the human CYP19 (aromatase) gene, mediates placenta-specific expression in transgenic mice. Proc. Natl. Acad. Sci. USA 96:4575-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamat, A., M. M. Hinshelwood, B. A. Murry, and C. R. Mendelson. 2002. Mechanisms in tissue-specific regulation of estrogen biosynthesis in humans. Trends Endocrinol. Metab. 13:122-128. [DOI] [PubMed] [Google Scholar]
- 24.Kamat, A., M. E. Smith, J. M. Shelton, J. A. Richardson, and C. R. Mendelson. 2005. Genomic regions that mediate placental cell-specific and developmental regulation of human CYP19 (aromatase) gene expression in transgenic mice. Endocrinology 146:2481-2488. [DOI] [PubMed] [Google Scholar]
- 25.Kaufmann, P. 1982. Development and differentiation of the human placental villous tree. Bibl. Anat. 22:29-39. [PubMed] [Google Scholar]
- 26.Kliman, H. J., J. E. Nestler, E. Sermasi, J. M. Sanger, and J. F. Strauss III. 1986. Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology 118:1567-1582. [DOI] [PubMed] [Google Scholar]
- 27.Kozak, K. R., B. Abbott, and O. Hankinson. 1997. ARNT-deficient mice and placental differentiation. Dev. Biol. 191:297-305. [DOI] [PubMed] [Google Scholar]
- 28.Kuiper, R. P., M. Schepens, J. Thijssen, E. F. Schoenmakers, and A. G. van Kessel. 2004. Regulation of the MiTF/TFE bHLH-LZ transcription factors through restricted spatial expression and alternative splicing of functional domains. Nucleic Acids Res. 32:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 30.Lun, Y., M. Sawadogo, and M. Perry. 1997. Autoactivation of Xenopus MyoD transcription and its inhibition by USF. Cell Growth Differ. 8:275-282. [PubMed] [Google Scholar]
- 31.Means, G. D., M. S. Mahendroo, C. J. Corbin, J. M. Mathis, F. E. Powell, C. R. Mendelson, and E. R. Simpson. 1989. Structural analysis of the gene encoding human aromatase cytochrome P-450, the enzyme responsible for estrogen biosynthesis. J. Biol. Chem. 264:19385-19391. [PubMed] [Google Scholar]
- 32.Mellgren, R. L. 1997. Specificities of cell permeant peptidyl inhibitors for the proteinase activities of μ-calpain and the 20 S proteasome. J. Biol. Chem. 272:29899-29903. [DOI] [PubMed] [Google Scholar]
- 33.Meng, L., R. Mohan, B. H. Kwok, M. Elofsson, N. Sin, and C. M. Crews. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 96:10403-10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowak, M., A. Helleboid-Chapman, H. Jakel, G. Martin, D. Duran-Sandoval, B. Staels, E. M. Rubin, L. A. Pennacchio, M. R. Taskinen, J. Fruchart-Najib, and J. C. Fruchart. 2005. Insulin-mediated down-regulation of apolipoprotein A5 gene expression through the phosphatidylinositol 3-kinase pathway: role of upstream stimulatory factor. Mol. Cell. Biol. 25:1537-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porter, L. A., and D. J. Donoghue. 2003. Cyclin B1 and CDK1: nuclear localization and upstream regulators. Prog. Cell Cycle Res. 5:335-347. [PubMed] [Google Scholar]
- 36.Ringler, G. E., and J. F. Strauss III. 1990. In vitro systems for the study of human placental endocrine function. Endocr. Rev. 11:105-123. [DOI] [PubMed] [Google Scholar]
- 37.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semenza, G. L. 2001. HIF-1, O2, and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell 107:1-3. [DOI] [PubMed] [Google Scholar]
- 39.Simpson, E. R., and P. C. MacDonald. 1981. Endocrine physiology of the placenta. Annu. Rev. Physiol. 43:163-188. [DOI] [PubMed] [Google Scholar]
- 40.Simpson, E. R., Y. Zhao, V. R. Agarwal, M. D. Michael, S. E. Bulun, M. M. Hinshelwood, S. Graham-Lorence, T. Sun, C. R. Fisher, K. Qin, and C. R. Mendelson. 1997. Aromatase expression in health and disease. Rec. Prog. Horm. Res. 52:185-213. [PubMed] [Google Scholar]
- 41.Sirito, M., Q. Lin, J. M. Deng, R. R. Behringer, and M. Sawadogo. 1998. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc. Natl. Acad. Sci. USA 95:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sogawa, K., R. Nakano, A. Kobayashi, Y. Kikuchi, N. Ohe, N. Matsushita, and Y. Fujii-Kuriyama. 1995. Possible function of Ah receptor nuclear translocator (Arnt) homodimer in transcriptional regulation. Proc. Natl. Acad. Sci. USA 92:1936-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanson, H. I., W. K. Chan, and C. A. Bradfield. 1995. DNA binding specificities and pairing rules of the Ah Receptor, ARNT, and SIM proteins. J. Biol. Chem. 270:26292-26302. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi, Y., K. Nakayama, S. Itoh, Y. Fujii-Kuriyama, and T. Kamataki. 1997. Inhibition of the transcription of CYP1A1 gene by the upstream stimulatory factor 1 in rabbits. Competitive binding of USF1 with AhR · Arnt complex. J. Biol. Chem. 272:30025-30031. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, E. A., Jr., and P. K. Siiteri. 1974. The involvement of human placental microsomal cytochrome P-450 in aromatization. J. Biol. Chem. 249:5373-5378. [PubMed] [Google Scholar]
- 46.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiesener, M. S., H. Turley, W. E. Allen, C. Willam, K. U. Eckardt, K. L. Talks, S. M. Wood, K. C. Gatter, A. L. Harris, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 1998. Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1α. Blood 92:2260-2268. [PubMed] [Google Scholar]
- 48.Yang, X. P., L. A. Freeman, C. L. Knapper, M. J. Amar, A. Remaley, H. B. Brewer, Jr., and S. Santamarina-Fojo. 2002. The E-box motif in the proximal ABCA1 promoter mediates transcriptional repression of the ABCA1 gene. J Lipid Res. 43:297-306. [PubMed] [Google Scholar]