Abstract
Signaling through the ErbB family of tyrosine kinase receptors in normal and cancer-derived cell lines contributes to cell growth and differentiation. In this work, we altered the levels of ErbB2 and ErbB3 receptors, individually and in combination, by using 6-finger and 12-finger synthetic zinc finger protein artificial transcription factors (ATFs) in an epidermoid squamous cell carcinoma line, A431. We successfully designed 12-finger ATFs capable of coregulating ErbB3 and ICAM-1 or ErbB2 and ErbB3. With ATFs, the effects of changes in ErbB2 and ErbB3 receptor levels were evaluated by using cell proliferation, cell migration, and cell signaling assays. Cell proliferation was increased when ErbB2 and ErbB3 were both overexpressed. Cell migration on collagen was decreased when ErbB2 was down-regulated, yet migration on laminin was significantly increased with ErbB3 overexpression. ErbB2 and ErbB3 overexpression also stimulated the phosphatidylinositol 3-kinase and mitogen-activated protein kinase pathways. Our ATF approach has elucidated differences in ErbB receptor-mediated proliferation, migration, and intracellular signaling that cannot be explained merely by the presence or absence of particular ErbB receptors and emphasizes the dynamic nature of the ErbB signaling system. The transcription factor approach developed here provides a gene-economical route to the regulation of multiple genes and may be important for complex gene therapies.
Signaling from ErbB tyrosine kinase receptors influences diverse aspects of a cell's biology that include growth, differentiation, migration, and apoptosis (29, 84). ErbB1 (EGFR/HER1) was the first member of the family identified. Based on homology to ErbB1, three additional family members, ErbB2 (HER2/p185), ErbB3 (HER3), and ErbB4 (HER4), were identified (41, 56, 60, 77). In normal development, binding of a growth factor ligand induces dimerization of ErbB receptors. Subsequently, the cytoplasmic tails are transphosphorylated. Each ErbB receptor has a unique pattern of phosphorylation sites that recruit various secondary signaling proteins (23, 51, 52, 55, 71). Ongoing research shows that the identity of the ligand bound, the amount of ligand, and the identities of dimers formed determine the activation of a particular intracellular signaling pathway such as the mitogen-activated protein kinase (MAPK), the stress-activated protein kinase, the protein kinase C, or the Akt pathway (53, 61, 72). The combination of at least 10 different ligands and 10 possible receptor dimers of the ErbB system form a signaling network essential for development (15, 34).
Various cancers, including those of the breast, head and neck, kidney, prostate, colon, pancreas, bladder, lung, and ovaries, are associated with overexpression of ErbB receptors (11, 59, 84). Research using breast cancer models has identified a dominant role for ErbB2 in tumor cell proliferation and metastasis (35, 64, 73, 74). ErbB2 is the preferred dimerization partner for all ErbB receptors, and dimers containing ErbB2 have higher ligand affinity and slower endocytosis rates compared to other dimers (6, 25, 29). Recent work has shown that down-regulation of ErbB3 inhibits proliferation of breast cancer cells to the same extent as inhibition of ErbB2 (33). Additional studies have established a role for ErbB3 and the ErbB2/ErbB3 heterodimer in the motility of cancer cells (1, 16, 30, 48, 75).
ErbB2-specific inhibition has been demonstrated by using a variety of recombinant protein-based strategies, nucleic acids, and small molecules (3, 5, 17, 18, 26, 57, 70, 83). Significantly, antibody therapies have proven efficacy in cancer treatment and small-molecule inhibitors of ErbB2 and ErbB1 are advancing through clinical trials (27). Specific inhibition of ErbB2 and ErbB3 at the level of transcription has been achieved with synthetic zinc finger protein (ZFP) artificial transcription factor (ATF) technology. This approach allows specific sequences to be targeted by using designed transcription factors (TFs) that are composed of zinc finger domains that are predefined to bind particular 3-bp sequences. For reviews of this technology, see the reports of Beerli and Barbas (7) and Blancafort et al. (10). E2C is a synthetic DNA-binding ZFP that recognizes an 18-bp binding site in the ErbB2 promoter, while E3 recognizes an 18-bp binding site in the ErbB3 promoter (9). When ZFPs E2C and E3 were fused to a repressor domain, KRAB, or to an activation domain, VP64, down- and up-regulation of receptor expression, respectively, provided the first examples of transcriptional control of endogenous gene expression (8). This ATF strategy allows both positive and negative regulation of gene transcription, in contrast to techniques using antibodies, small-molecule inhibitors, or small interfering RNA (siRNA) that act via posttranscriptional targeting.
While ATFs have been shown to provide targeted up- and down-regulation of gene expression, the delivery of transgenes in a therapeutic setting is limited, depending on the vector strategy used. For example, the capacity of retroviral vectors is limited to transgenes of less than 7 kb (44). Here we have studied the potential of linking independent TFs so that they can be expressed as a single gene cassette. Compared to the coexpression of two independent factors, this approach requires only a single promoter governing the fused TF and thus is more gene economic. This approach is predicted to facilitate the study of biological systems related to the coregulation of multiple genes.
To investigate the roles of ErbB2 and ErbB3 in driving cell proliferation and cell migration, a system using A431 cells and synthetic TFs was established. A431 cells were derived from an epidermoid squamous cell carcinoma and express ErbB1, ErbB2, and ErbB3 receptors (8, 24). In this work, we characterize a fusion of ATFs, E2C and E3, to create a novel TF that allows simultaneous regulation of ErbB2 and ErbB3 receptor expression (E2/3). To investigate the effects of changing ErbB receptor expression levels, ATFs with 6 (E2C or E3) and 12 zinc fingers (E2/3) were used to transcriptionally activate or repress ErbB2 and ErbB3 gene expression individually or in combination. Cell proliferation, cell migration, and intracellular signaling of transduced cells were evaluated. Dual regulation also allowed us to investigate whether synergistic or additive effects are a characteristic of ErbB expression in A431 biology.
MATERIALS AND METHODS
Cloning of 12-finger ZFPs.
The E2C and E3 six-finger ZFP constructs were previously defined (9). 31OPT was previously defined (46). The pMal-c2 vector was modified to include the following restriction sites and linker region (from 5′ to 3′): Sfi, XhoI, BsrFI, XmaI, 15L, BsrFI, XmaI, SpeI, and Sfi. The first ZFP was inserted by using the XhoI and 5′ XmaI sites. The second ZFP was inserted by using the 3′ XmaI and SpeI restriction sites. By using the flanking Sfi sites, the 12-finger cassette was ligated into a modified pMX vector for retroviral expression (45). The SS fragment codes for a single-chain Fab modified with stop codons that prevent expression.
Retroviral infection and flow cytometry analysis.
Infections were performed 48 and 63 h following transfection as described by Lund et al. (45). The antibodies used for staining were as follows: ErbB1, EGFR (R1) (5 μg/ml; Santa Cruz Biotechnology); ErbB2, FSP77 (2.5 μg/ml; N. H. Hynes laboratory); ErbB3, SGP1 (3 μg/ml; Lab Vision/NeoMarkers); ICAM-1, 31OPT (5 μg/ml; BD Pharmingen); control immunoglobulin G (IgG), mouse F(ab′)2 IgG1-UNLB control antibody (2.5 μg/ml; Southern Biotech). The secondary antibody used for all flow cytometry staining was 100 μl of 1:400-diluted, Cy-5 labeled, affinity-purified donkey F(ab′)2 anti-mouse IgG (Jackson ImmunoResearch).
Proliferation assay.
Cells were harvested from culture dishes after starvation overnight. Cells were plated in a 96-well tissue culture plate at a density of 1,500/well in 50 μl of medium. Cells were allowed to adhere for 1.5 h at 37°C, and then 50 μl of 20 ng/ml epidermal growth factor (EGF; Sigma) and/or 50 μl of 200 ng/ml heregulin-β (HRG) were added (R&D Systems). After addition of growth factors, cells were incubated for another 2.5 h at 37°C before adding 0.5 μCi per well (1 Ci = 37 GBq) of [3H]thymidine (ICN Radiochemicals) for the remaining 20 h of incubation. The cells were frozen at −80°C overnight and subsequently processed on a multichannel automated cell harvester (Cambridge Technology, Cambridge, MA) and counted in a liquid scintillation beta counter (Beckman Coulter). All experiments were performed in sextuplet, and the highest and lowest values were dropped from mean and standard deviation calculations.
Migration assay.
Cells were starved overnight in Dulbecco modified Eagle medium (DMEM)-0.5% fetal calf serum (FCS). Cells were trypsinized and washed once with phosphate-buffered saline (PBS), and their concentration was adjusted to 6 × 105/ml in assay medium (DMEM, 10 mM HEPES, 0.5% FCS). One hundred microliters of cell solution was added to the upper well of a 24-well Costar Transwell chamber (6.5 mm, 8-μm pore size). The undersides of the chambers were precoated with rat tail collagen or mouse Engelbreth-Holm-Swarm-derived laminin (Sigma) at 1 μg/ml and 0.25 μg/ml, respectively, in PBS overnight at 4°C and then washed twice with assay medium. Cells were allowed to migrate for 5.5 h at 37°C by using DMEM-5% FCS as a chemoattractant. For the inhibition studies, LY294002 (40 μM) and PD98059 (50 μM) (InvivoGen, San Diego, CA) were added to the cells 1 h prior to trypsinization and maintained in the medium of the migration assay. Before fixing and staining of the migrated cells with crystal violet (0.2 M boric acid, 0.05 M disodium tetraborate, 95% ethanol, PBS), cells that did not migrate were removed from the upper surface of the filters and cell migration was quantitated by counting and taking the sum of cells that migrated in four separate fields of at least three individual wells.
Western blot assay.
Cell lysates were collected in RIPA lysis buffer (100 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% deoxycholic acid, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) with freshly added 1 mM orthovanadate, 50 mM NaF, and protease inhibitor cocktail (Roche). Protein concentration was determined by the bicinchoninic acid assay (Pierce). Twenty micrograms of cell lysate was loaded in each lane, resolved by SDS-polyacrylamide gel electrophoresis (4 to 12% or 10%; Invitrogen), transferred to nitrocellulose membranes, and probed with specific antibodies. The antibodies used included ErbB3 C-17 (Santa Cruz Biotechnology), c-erbB-2 Ab-17 (Lab Vision, NeoMarkers), anti-β-actin (Sigma), Akt (Cell Signaling), phospho-Akt (Ser-473) (Cell Signaling), ERK1+2 (p42/44) (Cell Signaling), and phospho-ERK1+2 (Thr-202/Thr-204) (Cell Signaling). The secondary antibodies used were anti-mouse or anti-rabbit antibodies conjugated to horseradish peroxidase (HRP). Visualization of antibody binding was done by enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech).
RESULTS
Design and validation of a dual-specificity ATF strategy.
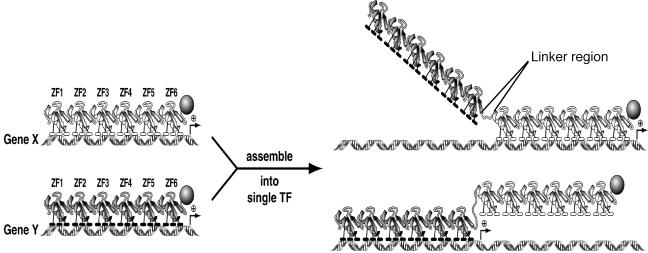
By using well-characterized ATFs, we designed a 12-finger ZFP that has two 6-finger ZFPs connected by a 15-amino-acid linker, 15L (TGGGGSGGGGTGEKP) (Fig. 1). The linker peptide was designed to be flexible and to have sufficient length to allow the linked six-finger ZFPs to function independently. To test the ability of a 12-finger ATF to regulate the expression of two independent target genes, a 12-finger ZFP was created by using two 6-finger ZFPs, E3 and 31OPT, that have specificity for regulatory elements in the ErbB3 and ICAM-1 genes, respectively (8, 9, 46). These two targets were selected because of their robust regulation by these ATFs and because they have not been shown to associate on the cell surface.
FIG. 1.
Illustration of the general assembly of 12-finger ZFP TFs. Two six-finger proteins that have binding sites independent of each other at two different promoters are linked by a 15-amino-acid linker (TGGGGSGGGGTGEKP). An effector domain can be attached at the N terminus (KRAB) or C terminus (VP64) of the ATF for repression or activation (shown here) of gene transcription, respectively.
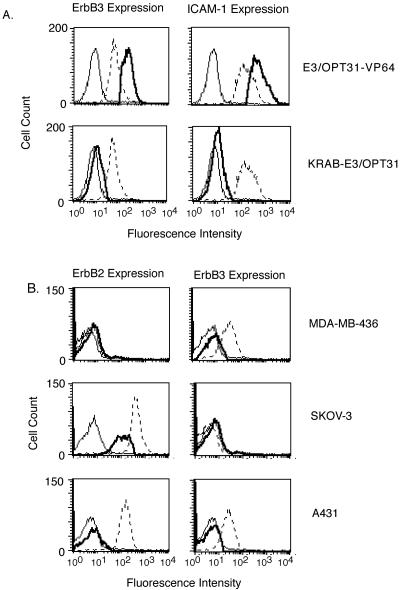
To inhibit gene transcription, the E3-15L-31OPT cassette was fused to an N-terminal repression domain, KRAB (Krüppel-associated box protein), and the 12-finger ATF was expressed from a retroviral vector in A431 cells (47). The flow cytometry analysis in Fig. 2A shows that A431 cells expressing the KRAB-E3-15L-31OPT protein exhibit a population of cells with reduced fluorescence intensity, a peak shifted to the left, compared with untransduced A431 cells. Reduced fluorescence indicates a decrease in the number of receptors that bound the fluorescently labeled receptor-specific antibody complex. The ability of a 12-finger ATF to increase gene expression was evaluated by using the dual-specificity 12-finger protein fused to the activation domain, VP64. A431 cells expressing E3-15L-31OPT-VP64 protein showed a peak with fluorescence intensity shifted to the right, indicating up-regulation of receptor levels compared with untransduced A431 cells (Fig. 2A). Expression of both 12-finger ATFs provided coregulated expression of ErbB3 and ICAM-1 receptors.
FIG. 2.
FACS analysis of 12-finger ATF-expressing cells. (A) Flow cytometry analysis of A431 cells transduced with ATF KRAB-E3-15L-31OPT or E3-15L-31OPT-VP64. (B) Flow cytometry analyses of MDA-MB-436, SKOV-3, and A431 cells transduced with ATF KRAB-E2C-15L-E3. Three days after transduction, cells were stained with ErbB3-, ICAM-1- or ErbB2-specific primary antibodies, followed by a Cy5-labeled secondary antibody. In all panels, the thin line represents the background fluorescence distribution of A431 cells stained with the IgG1 isotype control antibody, the dotted line is the fluorescence distribution for the level of ErbB2, ErbB3, or ICAM-1 expressed by untransduced A431 cells, and the bold line is the fluorescence distribution of A431 cells transduced with the ATF indicated.
Twelve-finger proteins were also assembled with a five-amino-acid TGEKP linker (5L) between E3 and 31OPT. Flow cytometry analysis of A431 cells expressing both the activation and repression versions of E3-5L-31OPT did not show a significant difference in the regulation of ErbB3 or ICAM-1 expression (data not shown). Based on the successful regulation achieved with ATFs linked with the 15-amino-acid linker, we hypothesize that the longer linker allows the six-finger ZFPs to function independently, whereas the five-amino-acid linker engages the proteins to make contiguous DNA contacts. Validation of this system encouraged us to examine the potential of this approach in a coregulatory strategy involving ErbB2 and ErbB3.
Regulation of ErbB2 and ErbB3 expression with a 12-finger ATF.
A 12-finger ATF containing E2C and E3 ZFPs was assembled by using the 15-amino-acid linker peptide and fused to the KRAB repressor domain (KRAB-E2/3). This ATF was expressed from a retroviral vector in three different cell lines: A431 (ErbB2+ ErbB3+); SKOV-3, an ovarian cancer cell line (ErbB2+ ErbB3−); and MDA-MB-436, a breast cancer cell line (ErbB2− ErbB3+). The effect of KRAB-E2/3 on expression of ErbB2 and ErbB3 in each of these cells lines is shown in Fig. 2B. ErbB2 expression was reduced in SKOV-3 cells, and ErbB3 expression was reduced in MDA-MB-436 cells. These studies further demonstrate that two ZFPs fused with a single effector domain can effectively regulate the expression of two different genes.
Selective down- and up-regulation of ErbB2 and ErbB3.
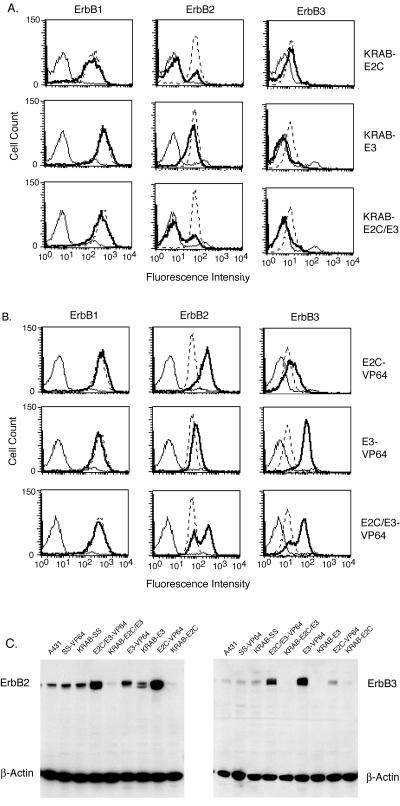
Retroviral transduction of A431 cells was used to integrate 6-finger or 12-finger ATF DNA cassettes for stable expression of ATF protein. The expression levels of ErbB1, -2, and -3 were analyzed by flow cytometry 5 days after retroviral transduction. Six different populations of ATF-expressing A431 cells were characterized by study of their expression of cell surface ErbB1, ErbB2, and ErbB3 receptors as shown in Fig. 3A and B. The fluorescence-activated cell sorter (FACS) profiles represent the ErbB protein expression profiles of the transduced population of cells which were used in the subsequent assays described. When A431 cells were transduced to express KRAB-E2C, 79% of the cells transduced showed a 10-fold reduction in ErbB2 expression and no change in ErbB1 or ErbB3 receptor expression. Expression of ErbB3 was completely inhibited in 82% of the A431 cells transduced with KRAB-E3. Transduction of KRAB-E2/3 resulted in 68% of the cells showing down-regulation of both ErbB2 and ErbB3, with no change in ErbB1 expression. Transduction of the activation-associated TFs showed a 10-fold increase in ErbB2 expression in the 66% of the transduced A431 cells transduced and no change in ErbB1 or ErbB3 expression levels. Expression of ErbB3 was increased 10-fold in 92% of cells transduced to express E3-VP64, accompanied by a slight increase in ErbB2 expression (8%) and no change in ErbB1 expression. When E2/3-VP64 was delivered by transduction, 61% of the cells had increased levels of both ErbB2 and ErbB3, with up-regulation of ErbB3 achieved in a greater number of cells than ErbB2 up-regulation. Controls were also evaluated that included a stuffer fragment of DNA fused to either the KRAB or the VP64 effector domain and a 12-finger protein with two ZFPs that do not have binding sites in the ErbB promoters. These controls demonstrated that either the KRAB or the VP64 domain by itself or in the context of an irrelevant ATF does not change the level of endogenous ErbB2 or ErbB3 expression (data not shown). To confirm the ErbB receptor regulation shown by FACS analysis (Fig. 3A and B), Western blot analyses of cell lysates derived from ATF-expressing cells were performed by using anti-ErbB2 and anti-ErbB3 antibodies (Fig. 3C). ErbB2 and ErbB3 protein level changes, as measured by blotting, corresponded to the levels of ErbB receptor expression observed by flow cytometry analysis.
FIG. 3.
FACS analysis of 6-finger and 12-finger ATF-expressing A431 cells. (A and B) Flow cytometry analysis shows A431 cells transduced with ZFPs fused to an N-terminal KRAB repression domain (A) or to a C-terminal VP64 activation domain (B). Three days after transduction, cells were stained with ErbB1-, ErbB2-, and ErbB3-specific primary antibodies, followed by a Cy5-labeled secondary antibody. In all panels, the thin line represents the fluorescence distribution of cells stained with the IgG1 isotype control antibody, the dotted line is the fluorescence distribution for the level of ErbB2 or ErbB3 expressed by untransduced A431 cells, and the bold line is the fluorescence distribution of A431 transduced with the ATF indicated. (C) Western blot analysis of 6-finger and 12-finger ATF-expressing A431 cells. Cell lysates were separated on 4 to 12% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. After incubation with ErbB2-, ErbB3-, and β-actin-specific primary antibodies, followed by HRP-conjugated anti-mouse antibodies, protein-antibody complexes were detected by using the ECL system. SS refers to the control stuffer DNA fragment.
Up-regulation of ErbB3 increased cell proliferation.
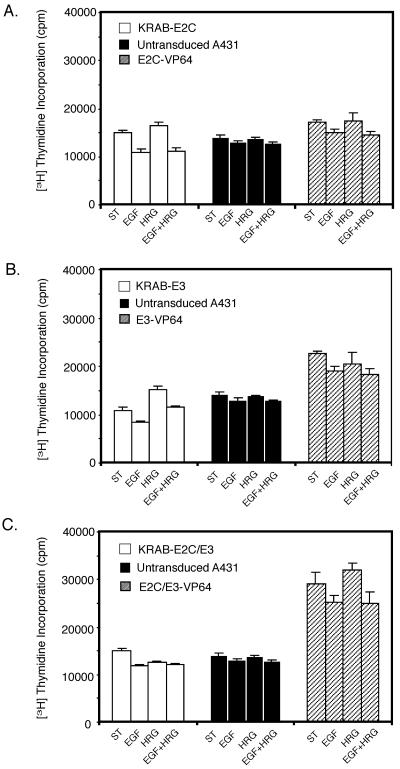
Proliferation of ATF-expressing A431 cells in the absence or presence of exogenous growth factors was monitored by using [3H]thymidine incorporation assays (Fig. 4). EGF, with specificity for ErbB1, and HRG, which binds to ErbB3 or ErbB4, were used to evaluate ligand-induced proliferation of A431 cells (36). A431 cells showed no significant change in proliferation with addition of EGF or HRG, which is consistent with studies with squamous cell carcinoma cell lines (76). However, when expression of ErbB2 and/or ErbB3 was altered by using ATFs, the proliferation profiles in the absence and presence of growth factors were altered. When ErbB2 expression was down-regulated, a 27% decrease in proliferation was observed upon addition of EGF. Down-regulation of ErbB3 decreased proliferation 23% in the absence of growth factors. Addition of EGF decreased the basal proliferation another 24%, while addition of HRG increased basal proliferation by 40%. Addition of both EGF and HRG did not change proliferation from basal levels. Simultaneous down-regulation of ErbB2 and ErbB3 (E2/3) resulted in a proliferation profile similar to that of untransduced A431 cells, although basal proliferation was increased. Overall, up-regulation of ErbB2 and/or ErbB3 increased basal proliferation. Up-regulation of ErbB2 increased the proliferation of A431 cells 22%. Up-regulation of ErbB3 increased basal proliferation 59%, and dual up-regulation of E2/3 increased basal proliferation of A431 cells 106%.
FIG. 4.
Proliferation of A431 cells expressing ATFs. After overnight incubation of cells in assay medium, cells were plated in 96-well plates. Cells were left untreated or incubated with 10 ng/ml EGF, 100 ng/ml HRG, or 10 ng/ml EGF plus 100 ng/ml HRG for 4 h before addition of [3H]thymidine for an additional 20 h. Data are expressed as the mean counts per minute in quadruplicate samples of each cell population. Error bars represent the average standard deviation. (A) From left to right, proliferation of A431 cells with down-regulated ErbB2 expression (KRAB-E2C), wild-type A431 cells, and A431 cells with up-regulated ErbB2 expression (E2C-VP64). (B) From left to right, proliferation of A431 cells with down-regulated ErbB3 expression (KRAB-E3), untransduced A431 cells, and A431 cells with up-regulated ErbB3 expression (E3-VP64). (C) From left to right, proliferation of A431 cells with down-regulated ErbB2 and ErbB3 expression (KRAB-E2C/E3), untransduced A431 cells, and A431 cells with up-regulated ErbB2 and ErbB3 expression (E2C/E3-VP64).
Up-regulation of ErbB2 or ErbB3 increased cell migration on laminin.
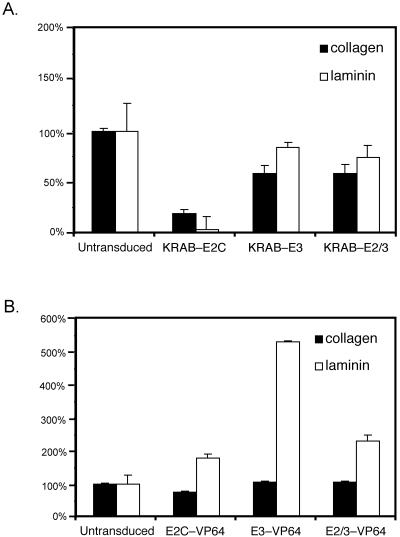
Growth factor receptor-mediated signaling has been shown to coordinate with integrin-mediated signaling (12, 42). Immunoprecipitation studies have shown an association between ErbB1 and ErbB2 receptors with integrins α6β4 and α6β1 in carcinoma cell lines overexpressing ErbB2 (22). Based on studies with keratinocytes that showed an effect of ErbB2 expression on integrin-mediated migration, we evaluated the haptotactic migration of ATF-expressing A431 cells on two major components of the extracellular matrix, collagen and laminin (31). The data are shown in Fig. 5. ErbB2 down-regulation significantly reduced A431 migration on collagen (by 80%) and laminin (by 96%). Down-regulation of ErbB3 and dual down-regulation of E2/3 both resulted in a 40% decrease in migration on collagen and a 16% or 26% decrease in migration on laminin, respectively. When ErbB2 was up-regulated, migration on collagen was inhibited 25% and migration on laminin increased 180%. When ErbB3 alone or both ErbB2 and ErbB3 were up-regulated, no difference in migration on collagen was observed, yet migration on laminin was significantly increased by 530% and 230%, respectively. Additional migration assays were done to investigate the increased migration on laminin observed with ErbB overexpression. Migration assays with E2C-VP64, E3-VP64, E2/3-VP64, SS-VP64, and untransduced A431 were repeated with and without the presence of the phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002 and the MAPK inhibitor PD98059. The results of these assays were complete inhibition of migration in all of the samples in the presence of LY294002 and no change in migration in the presence of PD98059 (data not shown). Thus, migration of A431 cells on laminin is PI3K dependent and not MAPK dependent.
FIG. 5.
Migration of ATF-expressing A431 cells on collagen and laminin. Cell migration assays were performed in Transwell chambers precoated with 1 μg/ml collagen or 0.25 μg/ml laminin. Cells were starved overnight in assay medium prior to plating in assay medium in the top chamber. Bottom chambers contained medium with 5% FCS. Cells were incubated at 37°C for 5.5 h. Migration was quantified by counting cells that had migrated through the filters. The bar graphs illustrate the percent cell migration relative to that of wild-type A431 cells (100%) determined from the average of triplicate assays for each sample. Error bars represent the average standard deviation. (A) Effect of ErbB down-regulation on migration. From left to right, untransduced A431 cells, A431 cells with down-regulated ErbB2 (KRAB-E2C), A431 cells with down-regulated ErbB3 (KRAB-E3), and A431 cells with both ErbB2 and ErbB3 down-regulated (KRAB-E2/3). (B) Effect of ErbB up-regulation on migration. From left to right, untransduced A431 cells, A431 cells with up-regulated ErbB2 (E2C-VP64), A431 cells with up-regulated ErbB3 (E3-VP64), and A431 cells with both ErbB2 and ErbB3 up-regulated (E2/3-VP64).
In summary, a decrease in migration was associated with down-regulation of ErbB3 and especially with down-regulation of ErbB2, where migration was almost completely abolished (Table 1). In contrast, a laminin-specific increase in migration was observed when ErbB2 or ErbB3 was overexpressed, and migration on laminin requires signaling through the PI3K pathway. The most dramatic increase in migration was associated with ErbB3 up-regulation alone. The role of ErbB2 in migration confirms results of previous studies (4, 20, 48, 65), while to the best of our knowledge, this is the first study to point to a role for ErbB3 in laminin-specific migration.
TABLE 1.
Summary of data from ErbB2 and ErbB3 regulation in A431 cells using ATFsa
| Regulation | % Proliferation | % Migration
|
Signalingd | |
|---|---|---|---|---|
| COLb | LINc | |||
| Down | ||||
| ErbB2 (KRAB-E2C) | 106 | 20 | 4 | Akt, NC; ERK1/2, ↓ |
| ErbB3 (KRAB-E3) | 77 | 60 | 84 | Akt, ↓; ERK1/2, ↓ |
| ErbB2 + ErbB3 (KRAB-E2C-15L-E3) | 106 | 60 | 74 | Akt, ↓; MAPK, ↓ |
| Up | ||||
| ErbB2 (E2C-VP64) | 122 | 75 | 180 | Akt, ↓; ERK1/2, NC |
| ErbB3 (E3-VP64) | 159 | 105 | 530 | Akt, ↑; ERK1/2, ↑ |
| ErbB2 + ErbB3 (E2C-15L-E3-VP64) | 206 | 106 | 230 | Akt, NC; ERK1/2, ↓ |
All values were calculated by using the value of untreated A431 cells as 100% of the effect.
COL, collagen.
LIN, laminin.
Signaling was evaluated in the absence of growth factors. NC, no change.
Effect of ErbB2 and ErbB3 regulation on Akt and ERK1/2 phosphorylation.
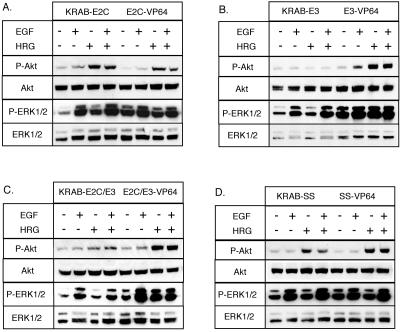
The PI3K and MAPK signaling pathways are associated with ErbB receptor signaling (79, 80). Akt is a signaling intermediate of the PI3K pathway, and ERK1 and -2 are signaling intermediates of the MAPK pathway (50, 58). We monitored the effects of ErbB2 and ErbB3 expression on the PI3K and MAPK signaling pathways by evaluating the phosphorylation levels of these intermediates. Addition of EGF to untransduced A431 cells (data not shown) or to effector domain control cells, KRAB-SS or SS-VP64, increased the phosphorylation of ERK1/2, and addition of HRG increased the phosphorylation of Akt (Fig. 6D). Figure 6A shows the signaling profile of A431 cells with altered ErbB2 expression in response to the presence and absence of EGF and HRG. When ErbB2 expression was reduced, there was a slight increase in the phosphorylation of Akt in the presence of HRG. ErbB2 down-regulation also resulted in a decrease in ERK1/2 phosphorylation in the absence of added growth factors and increasing levels of phosphorylated ERK1/2 with HRG, EGF, and EGF plus HRG, respectively. For ErbB2 overexpression, the levels of phosphorylated Akt were lower compared with control cells, while levels of phosphorylated ERK1/2 were increased when EGF was present (Fig. 6A). Figure 6B shows the signaling profile of A431 cells with decreased and increased ErbB3 expression. As expected with down-regulated ErbB3 expression, Akt signaling was greatly diminished. Strong ERK1/2 activation was observed, except in the absence of growth factors or in the presence of HRG alone. When ErbB3 was up-regulated, induction of phosphorylated Akt was observed in the presence of HRG and EGF, unlike in the other ErbB samples. Phosphorylation of ERK1/2 also showed less induction from basal levels with addition of EGF or HRG. When ErbB3 expression was altered, either up or down relative to untransduced A431 cells, there was a striking decrease in unphosphorylated ERK1/2 levels. Effects on the phosphorylation of Akt and ERK1/2 in A431 cells with increased or decreased expression of both ErbB2 and ErbB3 is shown in Fig. 6C. When ErbB2 and ErbB3 expression was down-regulated, Akt and ERK1/2 phosphorylation was reduced. However, in the presence of EGF, the phosphorylation of ERK1/2 increased. When both receptors were up-regulated, there was no difference in Akt phosphorylation from control samples. ERK1/2 phosphorylation was decreased in the absence of growth factors, and induction of phosphorylation was strongest with addition of EGF alone. Figure 6D shows the similar signaling profiles of the KRAB-SS and SS-VP64 control transduced cells, indicating that these controls did not affect signaling. Overall, the signaling data demonstrate that altering ErbB3 expression has a greater effect on the PI3K and MAPK signaling pathways than changing ErbB2 expression. Consistent with the data collected in the migration studies, signaling observed in cells with both ErbB2 and ErbB3 affected showed different dominant contributions from ErbB2 versus ErbB3. For example, when both ErbB2 and ErbB3 were down-regulated, the signaling profile paralleled the signaling profile of cells with only ErbB3 down-regulated. In contrast, when both ErbB2 and ErbB3 were up-regulated, the resulting signaling profile paralleled that of cells with up-regulated ErbB2. These results emphasize the utility of evaluating the effects of changing ErbB2 and ErbB3 at the same time and illustrate the sensitivity of the ErbB signaling system to changes in the ratio of ErbB receptor expression.
FIG. 6.
Phosphorylation of Akt or ERK1/2 in ATF-expressing A431 cells. Transduced cells were incubated overnight in assay medium before addition of the following growth factor(s): 10 ng/ml EGF, 100 ng/ml HRG, or 10 ng/ml EGF plus 100 ng/ml HRG. Cells without addition of growth factors were used as a control. After incubation with growth factors, cells were lysed, separated on 10% SDS-polyacrylamide gels, and transferred to nitrocellulose membranes. After incubation with phosphorylated-Akt- and phosphorylated-ERK1/2-specific primary antibodies, followed by HRP-conjugated anti-mouse antibodies, protein-antibody complexes were detected by using the ECL system. Detection of unphosphorylated Akt and ERK1/2 served as controls for changes in phosphorylation. (A) Western blot analysis of cell lysates with ErbB2 down-regulated (KRAB-E2C) and ErbB2 up-regulated (E2C-VP64). (B) Western blot analysis of cell lysates with ErbB3 down-regulated (KRAB-E3) and ErbB3 up-regulated (E3-VP64). (C) Western blot analysis of cell lysates with ErbB2 and ErbB3 down-regulated (KRAB-E2C/E3) and cell lysates with ErbB2 and ErbB3 up-regulated (E2C/E3-VP64). (D) Western blot analysis of cell lysates following transduction with KRAB-SS and SS-VP64 control vectors.
DISCUSSION
In this study, dual regulatory ATFs were prepared by linking well-defined TFs that target two independent genes for transcriptional regulation. This approach was validated with the construction and testing of ATFs that target both the ErbB3 and ICAM-1, or the ErbB2 and ErbB3, cell surface receptors. The bispecific ATFs carried either an activation (VP64) or a repression (KRAB) domain in order to overexpress, or repress, the two different genes by using the same TF. While the repression and overexpression that we produced could have been achieved by using combinations of several siRNAs or cDNAs, these methods are limited. For example, in various tumor cell lines, alternatively spliced variants of oncogenic proteins have been observed (21, 37, 66, 67). Alternatively spliced ErbB2 proteins have been shown to mediate effects that differ from those of the full-length receptor protein (2, 62, 82). Even if several ErbB2 cDNAs were used for overexpression, it would be difficult to mimic the effects of a single ATF. Similarly, when inhibiting gene expression by using RNA interference, multiple regions of the mRNA are generally targeted by using multiple siRNAs to ensure successful target down-regulation; this increases the potential for undesirable off-target effects (13, 39). Zinc finger TFs allow either up-regulation of all splice variants or efficient down-regulation through targeting of a single sequence. In this study, bispecific ErbB2 and ErbB3 regulators were used to investigate the effects of singular or coordinate regulation of these genes.
ErbB2 and ErbB3 are relevant cancer targets based on the successful use of ErbB2 blocking antibodies in the treatment of breast cancer patients and based on studies that associate ErbB3 with a role in cancer progression (3, 33, 75, 78). Recent studies suggest that the role of ErbB2 in proliferation is a result of signaling through an ErbB2/ErbB3 heterodimer (33). In this study, we made the observation that increased expression of ErbB3 was associated with increased proliferation. More interestingly, we found that simultaneous overexpression of ErbB2 and ErbB3 produced a greater increase in proliferation than overexpression of ErbB3 in cells that already have ErbB2 expressed on the cell surface. Differences in ErbB receptor trafficking with overexpression have been observed to alter ErbB2/ErbB3 dimerization patterns (81). Consistent with the role for ErbB3 in increased proliferation, down-regulation of ErbB3 expression produced a decrease in basal proliferation. When both ErbB2 and ErbB3 were down-regulated, the basal proliferation of cells was largely unaffected. Down-regulation of ErbB2 alone, or ErbB2 and ErbB3 simultaneously, resulted in levels of proliferation that were comparable to untransduced A431 cell proliferation. This result is consistent with siRNA inhibition of ErbB1 in A431 cells and studies of proliferation in other squamous cell lines that show that the ErbB1 receptor plays a key role in maintaining cell proliferation (28, 49, 63). With growth factor stimulation, the most dynamic changes in proliferation were observed with down-regulation of ErbB3. When ErbB3 was down-regulated, cells showed a proliferative response to the combined stimulation of EGF and HRG that was greater than that observed for EGF alone. Other samples did not show a response to the combination of growth factors that was greater than the stimulation observed with the addition of either single factor. Although ErbB3 is considered the main receptor for HRG on A431 cells in the absence of detectable levels of ErbB4 expression (68), even with ErbB3 down-regulation, proliferation was stimulated in response to HRG.
Advanced stages of cancer are characterized by the metastasis of the primary tumor to secondary sites in the body. A key step in this process is the migration of cancer cells. We evaluated changes in migration as a result of changes in ErbB receptor expression. Collagen and laminin are central proteins in the extracellular matrix of the epidermis and have been associated with cancer cell motility (32, 54). Previous studies have demonstrated the role of ErbB2 in cell motility by using an intrabody for the down-regulation of ErbB2 expression (48, 65). Consistent with these studies, we observed that down-regulation of ErbB2 significantly inhibited migration on both collagen and laminin. As also shown by Kawahara et al., decreased MAPK activity, as a result of ErbB down-regulation, correlated with decreased migration (38, 43). Surprisingly, simultaneous down-regulation of ErbB2 and ErbB3 did not inhibit migration to the extent observed for down-regulation of ErbB2 alone. These results were unexpected yet are consistent with the differences in signaling we observed (Fig. 6). The specific role of ErbB3 in migration has not, to our knowledge, been investigated. When ErbB3 was down-regulated, migration on collagen and laminin decreased. However, with ErbB3 overexpression, significant increases in migration on laminin were observed. Migration on laminin, even at basal levels, was PI3K dependent, as determined by using a PI3K inhibitor. The increase in migration observed with ErbB3 overexpression is consistent with efficient coupling of ErbB3 signaling with the PI3K signaling pathway (23) and may represent increased ErbB2/3 heterodimer formation and signaling through PI3K. A similar disparity in migration profiles between KRAB-E2C and KRAB-E2/3 and between E3-VP64 and E2/3-VP64 was noted. In these samples, migratory changes were greater in the cells that were modulated in their expression of a single receptor. Therefore, by using the bispecific TFs, we were able to determine that changes in the expression of one receptor were not independent of changes in other ErbB receptor populations. Although studies have examined the effect of stimulated signaling through ErbB1 versus ErbB3, or through activation of particular integrins on migration (31, 38, 48, 65), bispecific ATFs provide a new set of tools for further study of the molecular details of ErbB receptor expression and the motility of cancer cells.
The signal transduction data from our study confirmed the differences observed between one-gene versus two-gene regulation. For example, cells that overexpressed ErbB3 showed differences in migration on laminin compared to cells that overexpressed both ErbB2 and ErbB3; differences were also evident in basal MAPK signaling and GF-induced signaling (Fig. 6). Overexpression of ErbB3 in cells stimulated with EGF activated both the PI3K and MAPK pathways, whereas EGF stimulation of cells that overexpressed both proteins stimulated only MAPK signaling. These types of differences emphasize that ErbB receptor signaling is based not on absolute levels of receptor expression but rather on the ratio of ErbB receptors expressed and the dynamic nature of homodimeric and heterodimeric interactions.
The ability to regulate two receptors at a time was a result of novel TF design and provided insight into the dynamics of the ErbB receptor signaling network in A431 cells. Our data highlighted the synergistic relationship between ErbB2 and ErbB3 in cell proliferation, identified a role for ErbB3 in laminin-mediated migration, and provided signaling data that showed that ErbB receptors respond to dimer-inducing growth factors differently, depending on the ratio of ErbB receptors expressed. Characterization of the ErbB receptor network in additional cell lines will refine the model of ErbB receptor interactions in both normal and cancer cells and will further our understanding of the contribution of these receptors to the initiation, progress, and metastasis of various cancers. The application of bispecific ATF technology also has the potential to provide insight into other signaling pathways since current zinc finger technology allows any gene to be targeted (7, 10). In addition, the application of bispecific ATFs with dual regulatory abilities is promising for cancer therapy, as evidenced by multiple studies that show the efficacy of drug combinations in chemotherapy (14, 19, 40, 69).
Acknowledgments
We thank Karin Effertz and Dave Segal for early contributions to this study and Brian Eliceiri for critical reading of the manuscript. We thank N. H. Hynes for the FSP77 antibody.
This work was supported by NIH grant R01CA086258. C.V.L. is a Skaggs Predoctoral Fellow.
REFERENCES
- 1.Adelsman, M. A., J. B. McCarthy, and Y. Shimizu. 1999. Stimulation of beta1-integrin function by epidermal growth factor and heregulin-beta has distinct requirements for erbB2 but a similar dependence on phosphoinositide 3-OH kinase. Mol. Biol. Cell 10:2861-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aigner, A., H. Juhl, C. Malerczyk, A. Tkybusch, C. C. Benz, and F. Czubayko. 2001. Expression of a truncated 100 kDa HER2 splice variant acts as an endogenous inhibitor of tumour cell proliferation. Oncogene 20:2101-2111. [DOI] [PubMed] [Google Scholar]
- 3.Albanell, J., J. Codony, A. Rovira, B. Mellado, and P. Gascon. 2003. Mechanism of action of anti-HER2 monoclonal antibodies: scientific update on trastuzumab and 2C4. Adv. Exp. Med. Biol. 532:253-268. [DOI] [PubMed] [Google Scholar]
- 4.Baeckstrom, D., P. J. Lu, and J. Taylor-Papadimitriou. 2000. Activation of the alpha2beta1 integrin prevents c-erbB2-induced scattering and apoptosis of human mammary epithelial cells in collagen. Oncogene 19:4592-4603. [DOI] [PubMed] [Google Scholar]
- 5.Barbacci, E. G., L. R. Pustilnik, A. M. Rossi, E. Emerson, P. E. Miller, B. P. Boscoe, E. D. Cox, K. K. Iwata, J. P. Jani, K. Provoncha, J. C. Kath, Z. Liu, and J. D. Moyer. 2003. The biological and biochemical effects of CP-654577, a selective erbB2 kinase inhibitor, on human breast cancer cells. Cancer Res. 63:4450-4459. [PubMed] [Google Scholar]
- 6.Baulida, J., M. H. Kraus, M. Alimandi, P. P. Di Fiore, and G. Carpenter. 1996. All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. J. Biol. Chem. 271:5251-5257. [DOI] [PubMed] [Google Scholar]
- 7.Beerli, R. R., and C. F. Barbas III. 2002. Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 20:135-141. [DOI] [PubMed] [Google Scholar]
- 8.Beerli, R. R., B. Dreier, and C. F. Barbas III. 2000. Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl. Acad. Sci. USA 97:1495-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beerli, R. R., D. J. Segal, B. Dreier, and C. F. Barbas III. 1998. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. USA 95:14628-14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blancafort, P., D. J. Segal, and C. F. Barbas III. 2004. Designing transcription factor architectures for drug discovery. Mol. Pharmacol. 66:1361-1371. [DOI] [PubMed] [Google Scholar]
- 11.Blume-Jensen, P., and T. Hunter. 2001. Oncogenic kinase signalling. Nature 411:355-365. [DOI] [PubMed] [Google Scholar]
- 12.Cabodi, S., L. Moro, E. Bergatto, E. Boeri Erba, P. Di Stefano, E. Turco, G. Tarone, and P. Defilippi. 2004. Integrin regulation of epidermal growth factor (EGF) receptor and of EGF-dependent responses. Biochem. Soc. Trans. 32:438-442. [DOI] [PubMed] [Google Scholar]
- 13.Campbell, T. N., and F. Y. Choy. 2005. RNA interference: past, present and future. Curr. Issues Mol. Biol. 7:1-6. [PubMed] [Google Scholar]
- 14.Caponigro, F., R. Formato, M. Caraglia, N. Normanno, and R. V. Iaffaioli. 2005. Monoclonal antibodies targeting epidermal growth factor receptor and vascular endothelial growth factor with a focus on head and neck tumors. Curr. Opin. Oncol. 17:212-217. [DOI] [PubMed] [Google Scholar]
- 15.Casalini, P., M. V. Iorio, E. Galmozzi, and S. Menard. 2004. Role of HER receptors family in development and differentiation. J. Cell. Physiol. 200:343-350. [DOI] [PubMed] [Google Scholar]
- 16.Chausovsky, A., H. Waterman, M. Elbaum, Y. Yarden, B. Geiger, and A. D. Bershadsky. 2000. Molecular requirements for the effect of neuregulin on cell spreading, motility and colony organization. Oncogene 19:878-888. [DOI] [PubMed] [Google Scholar]
- 17.Chen, C. H., G. A. Chernis, V. Q. Hoang, and R. Landgraf. 2003. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc. Natl. Acad. Sci. USA 100:9226-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang, S. Y., R. W. Burli, C. C. Benz, L. Gawron, G. K. Scott, P. B. Dervan, and T. A. Beerman. 2000. Targeting the ets binding site of the HER2/neu promoter with pyrrole-imidazole polyamides. J. Biol. Chem. 275:24246-24254. [DOI] [PubMed] [Google Scholar]
- 19.Chu, I., K. Blackwell, S. Chen, and J. Slingerland. 2005. The dual ErbB1/ErbB2 inhibitor, lapatinib (GW572016), cooperates with tamoxifen to inhibit both cell proliferation- and estrogen-dependent gene expression in antiestrogen-resistant breast cancer. Cancer Res. 65:18-25. [PubMed] [Google Scholar]
- 20.D'Souza, B., F. Berdichevsky, N. Kyprianou, and J. Taylor-Papadimitriou. 1993. Collagen-induced morphogenesis and expression of the alpha 2-integrin subunit is inhibited in c-erbB2-transfected human mammary epithelial cells. Oncogene 8:1797-1806. [PubMed] [Google Scholar]
- 21.Eicheler, W., D. Zips, A. Dorfler, R. Grenman, and M. Baumann. 2002. Splicing mutations in TP53 in human squamous cell carcinoma lines influence immunohistochemical detection. J. Histochem. Cytochem. 50:197-204. [DOI] [PubMed] [Google Scholar]
- 22.Falcioni, R., A. Antonini, P. Nistico, S. Di Stefano, M. Crescenzi, P. G. Natali, and A. Sacchi. 1997. Alpha 6 beta 4 and alpha 6 beta 1 integrins associate with ErbB-2 in human carcinoma cell lines. Exp. Cell Res. 236:76-85. [DOI] [PubMed] [Google Scholar]
- 23.Fedi, P., J. H. Pierce, P. P. di Fiore, and M. H. Kraus. 1994. Efficient coupling with phosphatidylinositol 3-kinase, but not phospholipase C gamma or GTPase-activating protein, distinguishes ErbB-3 signaling from that of other ErbB/EGFR family members. Mol. Cell. Biol. 14:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giard, D. J., S. A. Aaronson, G. J. Todaro, P. Arnstein, J. H. Kersey, H. Dosik, and W. P. Parks. 1973. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 51:1417-1423. [DOI] [PubMed] [Google Scholar]
- 25.Graus-Porta, D., R. R. Beerli, J. M. Daly, and N. E. Hynes. 1997. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 16:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graus-Porta, D., R. R. Beerli, and N. E. Hynes. 1995. Single-chain antibody-mediated intracellular retention of ErbB-2 impairs Neu differentiation factor and epidermal growth factor signaling. Mol. Cell. Biol. 15:1182-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross, M. E., R. L. Shazer, and D. B. Agus. 2004. Targeting the HER-kinase axis in cancer. Semin. Oncol. 31:9-20. [DOI] [PubMed] [Google Scholar]
- 28.Hansen, L. A., R. L. Woodson II, S. Holbus, K. Strain, Y. C. Lo, and S. H. Yuspa. 2000. The epidermal growth factor receptor is required to maintain the proliferative population in the basal compartment of epidermal tumors. Cancer Res. 60:3328-3332. [PubMed] [Google Scholar]
- 29.Harari, D., and Y. Yarden. 2000. Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene 19:6102-6114. [DOI] [PubMed] [Google Scholar]
- 30.Hijazi, M. M., E. W. Thompson, C. Tang, P. Coopman, J. A. Torri, D. Yang, S. C. Mueller, and R. Lupu. 2000. Heregulin regulates the actin cytoskeleton and promotes invasive properties in breast cancer cell lines. Int. J. Oncol. 17:629-641. [DOI] [PubMed] [Google Scholar]
- 31.Hintermann, E., M. Bilban, A. Sharabi, and V. Quaranta. 2001. Inhibitory role of alpha 6 beta 4-associated erbB-2 and phosphoinositide 3-kinase in keratinocyte haptotactic migration dependent on alpha 3 beta 1 integrin. J. Cell Biol. 153:465-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hintermann, E., and V. Quaranta. 2004. Epithelial cell motility on laminin-5: regulation by matrix assembly, proteolysis, integrins and erbB receptors. Matrix Biol. 23:75-85. [DOI] [PubMed] [Google Scholar]
- 33.Holbro, T., R. R. Beerli, F. Maurer, M. Koziczak, C. F. Barbas III, and N. E. Hynes. 2003. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. USA 100:8933-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holbro, T., and N. E. Hynes. 2004. ErbB receptors: directing key signaling networks throughout life. Annu. Rev. Pharmacol. Toxicol. 44:195-217. [DOI] [PubMed] [Google Scholar]
- 35.Hudelist, G., C. F. Singer, M. Manavi, K. Pischinger, E. Kubista, and K. Czerwenka. 2003. Co-expression of ErbB-family members in human breast cancer: Her-2/neu is the preferred dimerization candidate in nodal-positive tumors. Breast Cancer Res. Treat. 80:353-361. [DOI] [PubMed] [Google Scholar]
- 36.Jones, J. T., R. W. Akita, and M. X. Sliwkowski. 1999. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett. 447:227-231. [DOI] [PubMed] [Google Scholar]
- 37.Kalnina, Z., P. Zayakin, K. Silina, and A. Line. 2005. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer 42:342-357. [DOI] [PubMed] [Google Scholar]
- 38.Kawahara, E., N. Nakada, T. Hikichi, J. Kobayashi, and I. Nakanishi. 2002. EGF and beta1 integrin convergently regulate migration of A431 carcinoma cell through MAP kinase activation. Exp. Cell Res. 272:84-91. [DOI] [PubMed] [Google Scholar]
- 39.Kim, V. N. 2003. RNA interference in functional genomics and medicine. J. Korean Med. Sci. 18:309-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klos, K. S., X. Zhou, S. Lee, L. Zhang, W. Yang, Y. Nagata, and D. Yu. 2003. Combined trastuzumab and paclitaxel treatment better inhibits ErbB-2-mediated angiogenesis in breast carcinoma through a more effective inhibition of Akt than either treatment alone. Cancer 98:1377-1385. [DOI] [PubMed] [Google Scholar]
- 41.Kraus, M. H., W. Issing, T. Miki, N. C. Popescu, and S. A. Aaronson. 1989. Isolation and characterization of ERBB3, a third member of the ERBB/epidermal growth factor receptor family: evidence for overexpression in a subset of human mammary tumors. Proc. Natl. Acad. Sci. USA 86:9193-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, J. W., and R. Juliano. 2004. Mitogenic signal transduction by integrin- and growth factor receptor-mediated pathways. Mol. Cell 17:188-202. [PubMed] [Google Scholar]
- 43.Lindberg, L. E., S. Hedjazifar, and D. Baeckstrom. 2002. c-erbB2-induced disruption of matrix adhesion and morphogenesis reveals a novel role for protein kinase B as a negative regulator of α2β1 integrin function. Mol. Biol. Cell 13:2894-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorens, J. B., C. Sousa, M. K. Bennett, S. M. Molineaux, and D. G. Payan. 2001. The use of retroviruses as pharmaceutical tools for target discovery and validation in the field of functional genomics. Curr. Opin. Biotechnol. 12:613-621. [DOI] [PubMed] [Google Scholar]
- 45.Lund, C. V., P. Blancafort, M. Popkov, and C. F. Barbas III. 2004. Promoter-targeted phage display selections with preassembled synthetic zinc finger libraries for endogenous gene regulation. J. Mol. Biol. 340:599-613. [DOI] [PubMed] [Google Scholar]
- 46.Magnenat, L., P. Blancafort, and C. F. Barbas III. 2004. In vivo selection of combinatorial libraries and designed affinity maturation of polydactyl zinc finger transcription factors for ICAM-1 provides new insights into gene regulation. J. Mol. Biol. 341:635-649. [DOI] [PubMed] [Google Scholar]
- 47.Margolin, J. F., J. R. Friedman, W. K. Meyer, H. Vissing, H. J. Thiesen, and F. J. Rauscher III. 1994. Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. USA 91:4509-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marone, R., D. Hess, D. Dankort, W. J. Muller, N. E. Hynes, and A. Badache. 2004. Memo mediates ErbB2-driven cell motility. Nat. Cell Biol. 6:515-522. [DOI] [PubMed] [Google Scholar]
- 49.Nagy, P., D. J. Arndt-Jovin, and T. M. Jovin. 2003. Small interfering RNAs suppress the expression of endogenous and GFP-fused epidermal growth factor receptor (erbB1) and induce apoptosis in erbB1-overexpressing cells. Exp. Cell Res. 285:39-49. [DOI] [PubMed] [Google Scholar]
- 50.Nicholson, K. M., and N. G. Anderson. 2002. The protein kinase B/Akt signalling pathway in human malignancy. Cell. Signal. 14:381-395. [DOI] [PubMed] [Google Scholar]
- 51.Olayioye, M. A. 2001. Update on HER-2 as a target for cancer therapy: intracellular signaling pathways of ErbB2/HER-2 and family members. Breast Cancer Res. 3:385-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olayioye, M. A., D. Graus-Porta, R. R. Beerli, J. Rohrer, B. Gay, and N. E. Hynes. 1998. ErbB-1 and ErbB-2 acquire distinct signaling properties dependent upon their dimerization partner. Mol. Cell. Biol. 18:5042-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olayioye, M. A., R. M. Neve, H. A. Lane, and N. E. Hynes. 2000. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 19:3159-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Toole, E. A. 2001. Extracellular matrix and keratinocyte migration. Clin. Exp. Dermatol. 26:525-530. [DOI] [PubMed] [Google Scholar]
- 55.Penington, D. J., I. Bryant, and D. J. Riese II. 2002. Constitutively active ErbB4 and ErbB2 mutants exhibit distinct biological activities. Cell Growth Differ. 13:247-256. [PubMed] [Google Scholar]
- 56.Plowman, G. D., J. M. Culouscou, G. S. Whitney, J. M. Green, G. W. Carlton, L. Foy, M. G. Neubauer, and M. Shoyab. 1993. Ligand-specific activation of HER4/p180erbB4, a fourth member of the epidermal growth factor receptor family. Proc. Natl. Acad. Sci. USA 90:1746-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rabindran, S. K., C. M. Discafani, E. C. Rosfjord, M. Baxter, M. B. Floyd, J. Golas, W. A. Hallett, B. D. Johnson, R. Nilakantan, E. Overbeek, M. F. Reich, R. Shen, X. Shi, H. R. Tsou, Y. F. Wang, and A. Wissner. 2004. Antitumor activity of HKI-272, an orally active, irreversible inhibitor of the HER-2 tyrosine kinase. Cancer Res. 64:3958-3965. [DOI] [PubMed] [Google Scholar]
- 58.Roux, P. P., and J. Blenis. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68:320-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salomon, D. S., R. Brandt, F. Ciardiello, and N. Normanno. 1995. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit. Rev. Oncol. Hematol. 19:183-232. [DOI] [PubMed] [Google Scholar]
- 60.Schechter, A. L., D. F. Stern, L. Vaidyanathan, S. J. Decker, J. A. Drebin, M. I. Greene, and R. A. Weinberg. 1984. The neu oncogene: an erb-B-related gene encoding a 185,000-Mr tumour antigen. Nature 312:513-516. [DOI] [PubMed] [Google Scholar]
- 61.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 62.Scott, G. K., R. Robles, J. W. Park, P. A. Montgomery, J. Daniel, W. E. Holmes, J. Lee, G. A. Keller, W. L. Li, B. M. Fendly, et al. 1993. A truncated intracellular HER2/neu receptor produced by alternative RNA processing affects growth of human carcinoma cells. Mol. Cell. Biol. 13:2247-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sibilia, M., A. Fleischmann, A. Behrens, L. Stingl, J. Carroll, F. M. Watt, J. Schlessinger, and E. F. Wagner. 2000. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell 102:211-220. [DOI] [PubMed] [Google Scholar]
- 64.Slamon, D. J., G. M. Clark, S. G. Wong, W. J. Levin, A. Ullrich, and W. L. McGuire. 1987. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235:177-182. [DOI] [PubMed] [Google Scholar]
- 65.Spencer, K. S., D. Graus-Porta, J. Leng, N. E. Hynes, and R. L. Klemke. 2000. ErbB2 is necessary for induction of carcinoma cell invasion by ErbB family receptor tyrosine kinases. J. Cell Biol. 148:385-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Staalesen, V., J. Falck, S. Geisler, J. Bartkova, A. L. Borresen-Dale, J. Lukas, J. R. Lillehaug, J. Bartek, and P. E. Lonning. 2004. Alternative splicing and mutation status of CHEK2 in stage III breast cancer. Oncogene 23:8535-8544. [DOI] [PubMed] [Google Scholar]
- 67.Sternberg, L. R., J. C. Byrd, G. C. Hansson, K. F. Liu, and R. S. Bresalier. 2004. Alternative splicing of the human MUC2 gene. Arch. Biochem. Biophys. 421:21-33. [DOI] [PubMed] [Google Scholar]
- 68.Stoll, S. W., S. Kansra, S. Peshick, D. W. Fry, W. R. Leopold, J. F. Wiesen, M. Sibilia, T. Zhang, Z. Werb, R. Derynck, E. F. Wagner, and J. T. Elder. 2001. Differential utilization and localization of ErbB receptor tyrosine kinases in skin compared to normal and malignant keratinocytes. Neoplasia 3:339-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sudbo, J., and A. Reith. 2005. The evolution of predictive oncology and molecular-based therapy for oral cancer prevention. Int. J. Cancer 115:339-345. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki, T., B. Anderegg, T. Ohkawa, A. Irie, O. Engebraaten, M. Halks-Miller, P. S. Holm, D. T. Curiel, M. Kashani-Sabet, and K. J. Scanlon. 2000. Adenovirus-mediated ribozyme targeting of HER-2/neu inhibits in vivo growth of breast cancer cells. Gene Ther. 7:241-248. [DOI] [PubMed] [Google Scholar]
- 71.Sweeney, C., and K. L. Carraway III. 2000. Ligand discrimination by ErbB receptors: differential signaling through differential phosphorylation site usage. Oncogene 19:5568-5573. [DOI] [PubMed] [Google Scholar]
- 72.Sweeney, C., D. Fambrough, C. Huard, A. J. Diamonti, E. S. Lander, L. C. Cantley, and K. L. Carraway III. 2001. Growth factor-specific signaling pathway stimulation and gene expression mediated by ErbB receptors. J. Biol. Chem. 276:22685-22698. [DOI] [PubMed] [Google Scholar]
- 73.Thor, A. D., S. Liu, S. Edgerton, D. Moore II, K. M. Kasowitz, C. C. Benz, D. F. Stern, and M. P. DiGiovanna. 2000. Activation (tyrosine phosphorylation) of ErbB-2 (HER-2/neu): a study of incidence and correlation with outcome in breast cancer. J. Clin. Oncol. 18:3230-3239. [DOI] [PubMed] [Google Scholar]
- 74.Tovey, S. M., C. J. Witton, J. M. Bartlett, P. D. Stanton, J. R. Reeves, and T. G. Cooke. 2004. Outcome and human epidermal growth factor receptor (HER) 1-4 status in invasive breast carcinomas with proliferation indices evaluated by bromodeoxyuridine labelling. Breast Cancer Res. 6:R246-R251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsai, M. S., L. A. Shamon-Taylor, I. Mehmi, C. K. Tang, and R. Lupu. 2003. Blockage of heregulin expression inhibits tumorigenicity and metastasis of breast cancer. Oncogene 22:761-768. [DOI] [PubMed] [Google Scholar]
- 76.Tsang, D. K., and D. L. Crowe. 1999. The mitogen activated protein kinase pathway is required for proliferation but not invasion of human squamous cell carcinoma lines. Int. J. Oncol. 15:519-523. [PubMed] [Google Scholar]
- 77.Ullrich, A., L. Coussens, J. S. Hayflick, T. J. Dull, A. Gray, A. W. Tam, J. Lee, Y. Yarden, T. A. Libermann, J. Schlessinger, et al. 1984. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature 309:418-425. [DOI] [PubMed] [Google Scholar]
- 78.van der Horst, E. H., M. Murgia, M. Treder, and A. Ullrich. 2005. Anti-HER-3 MAbs inhibit HER-3-mediated signaling in breast cancer cell lines resistant to anti-HER-2 antibodies. Int. J. Cancer 115:519-527. [DOI] [PubMed] [Google Scholar]
- 79.Vijapurkar, U., M. S. Kim, and J. G. Koland. 2003. Roles of mitogen-activated protein kinase and phosphoinositide 3′-kinase in ErbB2/ErbB3 coreceptor-mediated heregulin signaling. Exp. Cell Res. 284:291-302. [DOI] [PubMed] [Google Scholar]
- 80.Walker, F., A. Kato, L. J. Gonez, M. L. Hibbs, N. Pouliot, A. Levitzki, and A. W. Burgess. 1998. Activation of the Ras/mitogen-activated protein kinase pathway by kinase-defective epidermal growth factor receptors results in cell survival but not proliferation. Mol. Cell. Biol. 18:7192-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wiley, H. S. 2003. Trafficking of the ErbB receptors and its influence on signaling. Exp. Cell Res. 284:78-88. [DOI] [PubMed] [Google Scholar]
- 82.Xia, W., L. H. Liu, P. Ho, and N. L. Spector. 2004. Truncated ErbB2 receptor (p95ErbB2) is regulated by heregulin through heterodimer formation with ErbB3 yet remains sensitive to the dual EGFR/ErbB2 kinase inhibitor GW572016. Oncogene 23:646-653. [DOI] [PubMed] [Google Scholar]
- 83.Yang, G., K. Q. Cai, J. A. Thompson-Lanza, R. C. Bast, Jr., and J. Liu. 2004. Inhibition of breast and ovarian tumor growth through multiple signaling pathways by using retrovirus-mediated small interfering RNA against Her-2/neu gene expression. J. Biol. Chem. 279:4339-4345. [DOI] [PubMed] [Google Scholar]
- 84.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2:127-137. [DOI] [PubMed] [Google Scholar]