Abstract
We characterized and compared five geographically isolated hot springs with distinct red-layer communities in Yellowstone National Park. Individual red-layer communities were observed to thrive in temperatures ranging from 35 to 60°C and at pH 7 to 9. All communities were dominated by red filamentous bacteria and contained bacteriochlorophyll a (Bchl a), suggesting that they represented novel green nonsulfur (GNS) bacteria. The in vivo absorption spectra of individual sites were different, with two sites showing unusual Bchl a protein absorption bands beyond 900 nm. We prepared and analyzed 16S rRNA libraries from all of these sites by using a combination of general bacterial primers and new GNS-specific primers described here. These studies confirmed the presence of novel GNS-like bacteria in all five communities. All GNS-like clones were most similar to Roseiflexus castenholzii, a red filamentous bacterium from Japan that also contains only Bchl a. Phylogenies constructed by using GNS-like clones from Yellowstone red-layer communities suggest the presence of a moderately diverse new “red” cluster within the GNS lineage. Within this cluster, at least two well-supported subclusters emerged: YRL-A was most similar to Roseiflexus and YRL-B appeared to be novel, containing no known isolates. While these patterns showed some site specificity, they did not correlate with observed Bchl a spectrum differences or obvious features of the habitat.
The green nonsulfur (GNS) lineage has long been recognized as an evolutionarily and environmentally significant group of bacteria (12, 16). Representative GNS isolates include Chloroflexus, Chloronema, and Oscillochloris spp., which are all filamentous phototrophs that use bacteriochlorophyll (Bchl) c and Bchl a (17). However, the GNS lineage contains several members that defy the traditional “green” GNS type strain phenotype. These include chemotrophic genera, such as Herpetosiphon and Thermomicrobium (10, 16), as well as phototrophic Heliothrix and Roseiflexus, both of which use only Bchl a and appear orange and red, respectively (8, 20). Recently, several dozen uncultivated GNS-like sequences, all inferred chemotrophs, were redesignated as members of TM7, a proposed candidate division separate from the GNS (11, 13). Similar studies of Herpetosiphon isolates now suggest comparable novel clustering within this atypical GNS group (24).
In contrast with studies and information about chemotrophic GNS bacteria and GNS-like sequences, there is a relative paucity of comparable molecular diversity studies about phototrophic GNS organisms and sequences in natural environments. Improving our knowledge of diversity among traditional GNS should better elucidate the relationship between chemotrophs, “green” phototrophs, and “red-orange” phototrophs, a question with broad implications for better understanding the evolution of photosynthesis. In particular, the limited number of Bchl a-containing GNS representives in the database makes it difficult to assess whether these phototrophs represent a novel subgroup within the GNS lineage. A major goal of work in our laboratory is to improve understanding of molecular diversity in this lineage, with an emphasis on atypical, uncultured GNS phototrophs that have been difficult to isolate.
For nearly two decades, Heliothrix oregonensis has been the only known representative GNS that contains only Bchl a (18, 19, 20, 26). Originally cocultured from alkaline hot springs (pH 8.0, 40°C) in Oregon, Heliothrix filaments form orange surface communities. Filaments tolerate full sunlight, and the Bchl a maximally absorbs in vivo at 795 and 865 nm. Heliothrix-like communities or similar 16S rRNA sequences have been observed in comparable habitats outside of Oregon, including Octopus Spring, Yellowstone National Park (3, 17, 26). Recently, Hanada et al. (8) described Roseiflexus castenholzii, a novel red GNS bacterium containing only Bchl a from alkaline hot springs (pH 8.0, 40°C) in Japan.
Previously, we have reported our findings regarding Rabbit Creek Red (RCR), a novel red-layer bacterial community from Rabbit Creek Spouter, an alkaline hot spring (pH 8 to 8.2, 30 to 50°C) in Yellowstone National Park (3). Like Heliothrix, RCR was composed of photoheterotrophic filaments that contained Bchl a only. RCR was not amenable to isolation in culture, in contrast with Heliothrix. RCR filaments also grew deep within (5 to 10 mm between the surface) the laminated mat community, beneath layers dominated by cyanobacteria and Chloroflexus spp. Specific light wavelengths that penetrated to and were attenuated by the RCR layer were measured at 800 and 910 nm. Consistent with this finding, RCR Bchl a, in cell extracts, maximally absorbed at 807 and 913 nm, a result significantly different than that observed in vivo spectrum for Heliothrix Bchl a (795 and 865 nm) and Roseiflexus Bchl a (801 and 878 nm) (8, 18). Notably, only two other bacteria, Chromatium tepidum (6) and Roseospirillum parvum (7), both nonfilamentous purple proteobacteria, have Bchl a peaks of >900 nm. Whether RCR represents a new genus relative to Heliothrix will remain unknown because the Rabbit Creek Spouter source dried up before our recent molecular studies.
In this report, we have described and compared five red-layer microbial mat communities from thermal features in Yellowstone National Park. These communities constitute red-layer microbial observatory (RLMO) sites. RLMOs were geographically isolated and displayed different Bchl a absorption properties in vivo. To describe these communities, we characterized and analyzed 44 new GNS-like 16S RNA sequences and, in the process, developed new oligonucleotide primers specific for GNS phototrophs. Phylogenies generated with Yellowstone-derived red GNS sequences not only support the contention that red GNS sequences constitute a unique GNS subgroup but also suggest the presence of at least two distinct and heterogeneous clusters within this novel GNS subgroup.
MATERIALS AND METHODS
Habitat analyses and sample collection.
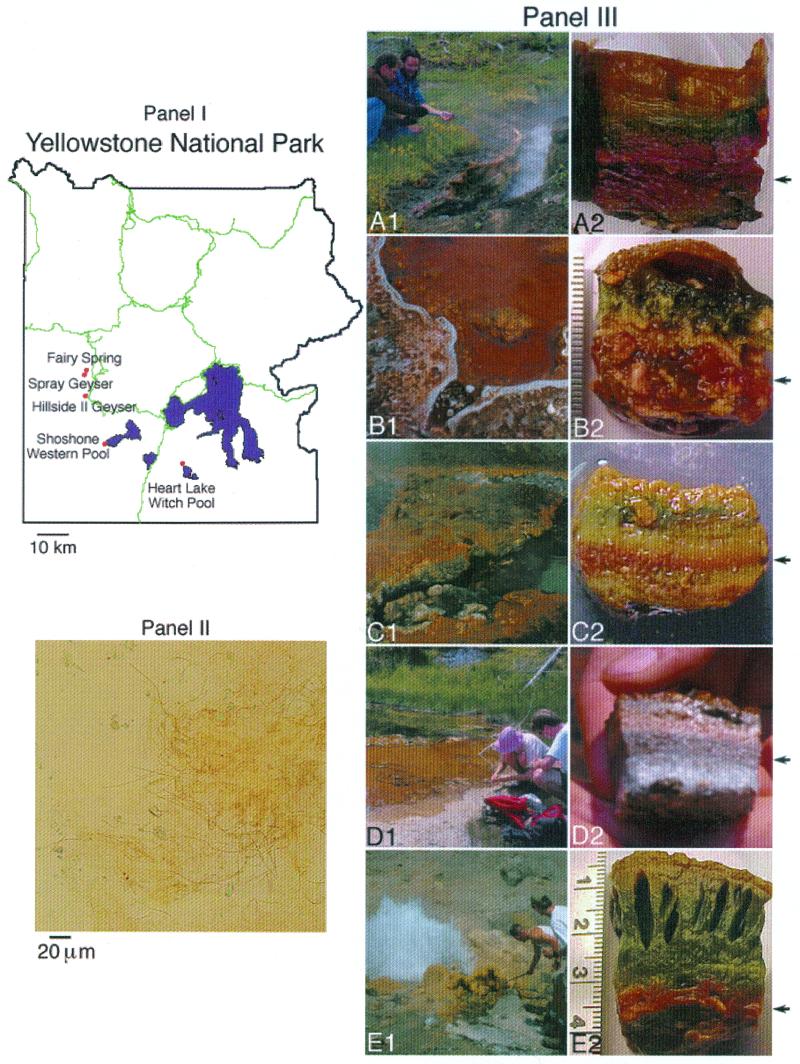
Global positioning system (GPS) coordinates for sites were obtained by using a hand-held GPS unit, Garmin model 12XL (Garmin International, Inc., Olathe, Kans.). GPS information was cross-referenced with the Yellowstone National Park Spatial Analysis Center laboratory and database to obtain accurate site nomenclature, if available (Ann Rodman and Steve Wheeler, unpublished data). The temperature within the red-layer community was recorded by using a Checktemp1 or Thermocouple Digital Thermometer (Atkins Technical, Inc., Gainesville, Fla.). The pH within the red-layer community was recorded by using ColorpHast pH paper (ranges 1 to 14 and 5 to 10; Gibbstown, N.J.). The cores of each mat (2-cm2 surface, variable thickness) were removed and refrigerated ca. 1 week. After returning from the field, we dissected the red layers from the rest of the mat. Half of each red-layer sample was used for pigment and microscopic analyses, and the other half was sectioned into ∼1.0-g pieces and stored at −70°C for DNA analyses. Microscopy was performed with an Olympus BX41 with a DP11 digital camera system (Olympus America, Inc., Melville, N.Y.). Site information was archived into a GIS database and merged with appropriate data sets from the Yellowstone Spatial Analysis Center. Relevant data from the data set was then used to generate the schematic map (Fig. 1, panel II) by using ArcView 3.2 (ESRI, Redlands, Calif.)
FIG. 1.
(Panel I) Schematic map of Yellowstone National Park indicating locations of research sites. (Panel II) Representative light micrograph of the Spray Geyser red layer. The bar represents 20 μm. (Panel III) Site photographs of Hillside II Spring mat (A1) and core (A2), Witch Pond mat (B1) and core (B2), Fairy Spring mat (C1) and core (C2), Shoshone/Western Pool mat (D1) and core (D2), and Spray Geyser mat (E1) and core (E2). The arrows at the right indicate the red layer of each core.
Pigment analysis.
In vivo absorption spectra of pigments were obtained from homogenized suspensions of red-layer mat sections in buffer as previously described (20, 21, 22). Methanol spectra were determined as previously described (20, 22). All spectra were recorded on a U-2000 UV/Vis spectrophotometer (Hitachi, Inc., Tokyo, Japan).
Genomic DNA extraction.
To target red filaments, we carefully dissected and rinsed all samples with buffer several times prior to DNA extraction. Filament-enriched preparations were subjected to extraction methods that were a modification of published methods with standard reagents (2, 9; Brian Hedlund, unpublished data). Samples were thawed in 10 to 20 ml of glucose-Tris-EDTA (GTE) buffer and teased apart with a watchmaker forceps. Filament masses were transferred through two to three GTE buffer rinses before being thoroughly ground in disposable nuclease-free mortar-pestle centrifuge tubes (MoBio Laboratory, Solana Beach, Calif.) with 300 μl of fresh GTE buffer. Rinsed filaments were examined microscopically for the presence of contaminating unicellular bacteria to crudely verify purity throughout the procedures. Homogenized samples were incubated with 1 μg of lysozyme and EDTA (0.1 M final concentration) for 30 min at 37°C. Sodium dodecyl sulfate (1% final concentration) was added prior to extractions with equal volumes of phenol-chloroform. DNA was precipated with sodium acetate (0.3 M final concentration) and 2 volumes of 95% ethanol. Pelleted DNA was resuspended in 50 to 100 μl of nuclease-free water to give a final concentration of ca. 100 ng/μl.
16S rRNA libraries.
Owing to potential site-specific variation in composition or chemistry (for example, pH or metals), all PCR amplifications of 16S rRNA genes were performed by using the suite of buffers in the MasterAmp PCR Optimization Kit (Epicentre Technologies, Madison, Wis.). Each 50-μl reaction contained ca. 100 ng of extracted DNA, 100 ng of each forward and reverse primer (described below), 3 U of ExTaq DNA Polymerase (TaKaRa Biomedicals/Takara Shuzo Co., Shiga, Japan), and buffer (one of twelve in the MasterAmp suite). Thermal cycling was carried out on a Perkin-Elmer DNA Thermal Cycler (original 1992 model) with 35 cycles as follows: 1 min of denaturation at 94°C, 1 min of annealing at 60°C, and 3 min of extension at 72°C (J. J. Gosink, unpublished data). All materials associated with PCR were handled in a plasmid-free space by using similarly dedicated reagents and pipetters to avoid contamination.
Table 1 summarizes the amplification strategy used to prepare each site library. For the Hillside II Spring, the first red-layer community analyzed, the broad-specificity primer 1492FPL was combined with the Bacterium-specific 8FPL (23). For subsequent sites, we designed more specific primers, 77FGNS and 953RRED (see “primer design,” below). Forward primer 77FGNS was predicted to amplify all known phototrophic GNS 16S rRNA sequences. Reverse primer 953RRED was predicted to amplify sequences that correspond to Bchl a-only GNS 16S rRNA sequences. 1492FPL and 77FGNS were applied to Witch Pond, generating a phototrophic GNS library. 77FGNS and 953RRED were applied to Fairy, Spray, and Shoshone red-layer samples, generating red GNS libraries in each case.
TABLE 1.
Summary of red layer communities
| Site, basin | GPS coordinates | Yr | Temp (°C) | pH | Bchl a absorbance values in vivo (nm) | 16S library (generating primers)a | GNS-like clones (accession no.) |
|---|---|---|---|---|---|---|---|
| Hillside II, Upper Basin | 44°28′56″N, 110°51′98″W | 1998 | 47–50 | 8.7 | 804, 882 | General bacterial (F, 8FPL; R, 1492FPL)d | |
| 1999 | 53 | 8.5 | 802, 888 | Hs2-1 (AF421761), Hs2-3 (AF421747), Hs2-8 (AF421762), Hs2-10 (AF421759), Hs2-26 (AF421746), Hs2-35 (AF421760), Hs2-38 (AF421748), Hs2-44 (AF421751), Hs2-48 (AF421750), Hs2-53 (AF421756) | |||
| 2000 | 45 | 7.5 | 801, 880 | ||||
| Witch Pond, Heart Lake | 44°17′49″N, 110°30′53″W | 1999 | 47–50 | 8.7 | 801, 889 | Phototrophic GNS (F, 77GNS; R, 1492FPL) | Witch-1 (AF421752), Witch-5 (AF421749), Witch-7 (AF421755), Witch-8 (AF421757), Witch-10 (AF421758), Witch-11 (AF421753), Witch-14 (AF421754) |
| Fairy Spring, Lower Basin | 44°32′53″N, 110°51′68″W | 1999 | 48–53 | 8 | 803, 900 | Red GNS (F, 77GNS; R, 953RRED) | Fairy-3 (AF421744), Fairy-9 (AF421743), Fairy-27 (AF421742), Fairy-14 (AF421737). Fairy-32 (AF421740), Fairy-37 (AF421741), Fairy-3b (AF421739) |
| 2000 | 53 | 8 | 805, 911 | ||||
| Western Pool, Shoshone Basin | 44°21′070″N, 110°48′342″W | 2000 | 41 | 8 | 801, 879 | Red GNS (F, 77GNS; R, 953RRED) | Shosh-3 (AF421733), Shosh-4 (AF421736), Shosh-11 (AF421729), Shosh-16 (AF421732), Shosh-17 (AF421738), Shosh-24 (AF421730), Shosh-26 (AF421734), Shosh-28 (AF421745), Shosh-29 (AF421731), Shosh-38 (AF421735) |
| Spray Geyser, Middle Basin | 44°31′93″N, 110°52′41″W | 1999 | 38.8 | 8.5 | 801, 905 | Red GNS (F, 77GNS; R, 953RRED) | |
| 2000 | 35 | 8.5 | 805, 907 | Spray-3 (AF421728), Spray-6 (AF421719), Spray-7 (AF421720), Spray-8 (AF421726), Spray-10 (AF421721), Spray-19 (AF421725), Spray-20 (AF421722), Spray-21 (AF421724), Spray-24 (AF421723) | |||
| Rabbit Creek Redb | 30–50 | 8–8.2 | 807, 913 | ||||
| Roseiflexusc | 50 | 7.5–8.0 | 801, 878 |
Amplified products were ligated into the vector, pCR2.1-TOPO (Invitrogen/Life Sciences, Carlsbad, Calif.) and transformed into chemically competent One Shot Escherichia coli TOP10 cells (Invitrogen/Life Sciences). Libraries for each site were prepared and screened by using a restriction fragment length polymorphism specified by two insert-flanking EcoRI sites. Insert-bearing clones were stored in 50:50 65% glycerol-Luria-Bertani broth at −70°C until further analysis (2).
Nucleotide sequence analysis.
Nucleotide sequence analysis was performed in our laboratory on a Li-Cor 4200 Gene ReadIR Single Dye system (Li-Cor Inc., Lincoln, Nebr.). As recommended by Li-Cor, all plasmid templates were prepared by using the Promega Wizard Midi-Prep Kit (Promega, Madison, Wis.). Templates were combined with fluorescently labeled IRD800 primers (Li-Cor, Inc.), and dideoxynucleotide chain termination methods were carried out by using the SequiTherm Excel II DNA Sequencing Kit-LC (Epicentre Technologies). Li-Cor-recommended reactions were modified for our thermal cycler to include an initial denaturation step (2 min at 92°C), followed by 30 cycles of 30 s of denaturation at 92°C, 15 s of annealing at 50°C, and 30 s of extension at 70°C. For GNS-like clones, the nucleotide sequence was determined for the entire insert by using a combination of vector-specific universal primers and internal custom primers. For non-GNS sequences, partial nucleotide sequences were determined with only universal primers. In all cases, nucleotide sequence information was edited and assembled by using Base ImagIR 4.0 Image Analysis Software (Li-Cor, Inc.).
Phylogenetic analyses.
All sequences were compared to the National Center for Biotechnology Information (NCBI) database by using the Basic Local Alignment Search Tool (BLAST) (1) to make preliminary phylogenetic inferences about relatedness. In the case of the Hillside II Spring General Bacterial Library, cloned sequences fell into one of two categories: GNS-like (10 of 16 clones) or non-GNS-like (6 of 16 clones). In the case of all other site libraries, generated by using GNS-specific primers, cloned sequences were GNS-like. Subsequent phylogenetic analyses were performed by using only GNS-like sequences. GNS-like sequences were also subjected to Check_Chimera on the Ribosomal Database Project website (14).
For phylogenetic analyses of GNS-like sequences, we compiled a data set that consisted of all GNS sequences on the NCBI Taxonomy Database that were 900 bases or longer. Additionally, two other sequences, eubacterial sp. (M62775) and eubacterial sp.-Os4 (M62774) (27) were added due to strong BLAST similarity to all our GNS-like sequences. This combined data set was compiled in the sequence management program SeqPup (D. G. Gilbert [SeqPup, version 0.9]).
Compiled sequences were preliminarily aligned by using CLUSTAL W (J. Thompson, T. Gibson, and D. Higgins [CLUSTAL W, version 1.7]) with identity matrix settings that assigned equal weight to all nucleotides. The alignment was trimmed to the length of our shortest GNS-like sequences (corresponding to bases 101 to 1025 of E. coli). Any NCBI-derived sequences lacking information in this region were removed from the analysis at this time.
Preliminary phylogenetic trees were generated by parsimony methods with a 2:1 transversion-transition correction matrix with PAUP 4.0b8 (25). Thermomicrobium roseum was used as the outgroup in our final tree because, in our preliminary tree, it was the closest relative to the ingroup. Of a total of 1,033 bases, 586 were usable characters. The trees were then tested for robustness with bootstrap resampling methods. Based on the preliminary tree, all red-layer GNS-like sequences fell into one of two large subgroups. Further phylogenetic analyses were performed on the subgroup that included all red-layer GNS-like sequences. This final data set was refined by hand based on the secondary structure of T. roseum (R. Gutell et al. [http://www.rna.icmb.utexas.edu/]), and trees were generated by using the parsimony and bootstrap methods described above. There were 353 usable characters out of 980 total characters in the final analysis. The topology of the single most parsimonious tree was selected from multiple most parsimonious trees on the basis of likelihood scores. The range of the log likelihood scores of most parsimonious trees was 8500 to 8506.
Primer design.
Two novel GNS-specific primers were designed to facilitate amplification of specific 16S rRNA genes in this study: 77FGNS was predicted to amplify known red and green phototrophic GNS sequences, and 953RRED was predicted to amplify only red GNS sequences. To design the forward primer, 77FGNS, we compiled a data set that consisted of all known or inferred phototrophic GNS sequences in GenBank (December 1999), all Herpetosiphon and Thermomicrobium sequences, and one to two representative genera from all major bacterial lineages (Brian Hedlund, unpublished data). Data were transferred to SeqPup and aligned by using CLUSTAL W as described for the phylogenetic analyses above. Alignments were inspected for obvious distinct regions that could serve as primers. Specifically, we located regions that were conserved for all phototrophic GNS sequences but variable for non-GNS and chemotrophic-GNS sequences. Candidate sequences were analyzed by using Check Probe (version 2.1r) on the RDP website (14). Forward primer 77FGNS (5′-GTGGCGMACGGCTGACC-3′; Tm = 50°C, %GC = 73) was predicted to amplify all known phototrophic GNS bacterial 16S rRNA sequences, including those of Chloroflexus. To design the reverse primer, 953RRED, we used a strategy similar to that described for 77FGNS. In this case, the data set considered included Roseiflexus and all Hillside II-derived GNS-like sequences but did not contain Heliothrix because no full-length 16S rRNA clones of this isolate have been described. Reverse primer 953RRED (5′-GACGGNCCCTCGNAGGC-3′; Tm = 52°C, %GC = 76%) was predicted to amplify sequences that correspond to Bchl a-only GNS16S rRNA sequences.
Nucleotide sequence access numbers.
The nucleotide sequence accession numbers for GNS-like clones reported in this study are listed in Table 1.
Website and database information.
All data in this publication and related materials can be found at the RLMO website (www.wou.edu/las/natsci_math/biology/boomer/ALLRESEARCH/allresearch.html).
RESULTS
Description of red-layer microbial habitats.
For this comparative study of red-layer microbial communities in Yellowstone National Park, we located, surveyed, and described five distinct thermal features that contain filamentous bacteria which resemble RCR and Roseiflexus. Mat communities at all sites contained several different-colored layers, and red layers were located beneath upper green, orange, and/or flesh-colored laminations. Based on similar, well-characterized layers, upper layers were likely supported by cyanobacteria and green GNS bacteria (3, 4). Our five red-layer community sites included the following locations: (i) Hillside II Springs (Upper Geyser Basin), (ii) Witch Pond (Witch Creek/Heart Lake Basin), (iii) Fairy Springs (Fairy Group, Lower Basin), (iv) Shoshone/Western Pool (Shoshone Basin, Western Group), and (v) Spray Geyser (Imperial Group, Middle Basin). These five sites are between 2 and 45 km from each other, with Witch Pond and Shoshone being separated from the rest by the Continental Divide. Each of these red-layer mat sites is described and summarized in Fig. 1 and Table 1.
Hillside II Springs.
Hillside II Springs is located on the southern boundary of the Upper Geyser Basin. This feature has been a consistently active thermal source (80 to 90°C, pH 7.5 to 8.5) that continuously splashes adjacent rocks, supporting a variably thick microbial mat. Temperature in the red layer of this mat ranged from 45 to 53°C, and the pH was 7.5 to 8.5. 16S rRNA analysis was performed only with the 1999 red-layer sample. Adjacent Hillside I Spring contains virtually identical red-layer communities.
Witch Pond.
Witch Pond is located in the remote Witch Creek Thermal Basin near Heart Lake. Witch Pond was one of many still thermal pools along Witch Creek that harbored submerged mats. Temperature in the red layer of this mat ranged from 47 to 50°C, and the pH was 8.7. Surveys of adjacent Heart Lake Thermal Basin, the Fissure Group, and the Upper Group demonstrated no mat communities with visible red layers.
Fairy Geyser.
Fairy Geyser, located off-trail in the Lower Geyser Basin, has been a constantly active spouter that erupts from a series of vents, supporting an extensive splash mat of various thickness. The temperature in the red layer of this mat ranged from 48 to 53°C, and the pH was 8.0. 16S rRNA analysis was performed only with the 1999 red-layer sample.
Shoshone/Western Pool.
Western Pool is located in the remote and pristine Shoshone Basin. Western Pool was a still pool that harbored a submerged mat. Temperature in the maroon-brown layer of this mat was 41°C, and the pH was 8.0. The Western Pool mat lacked distinct green upper layers, unlike anything we have seen before in our survey of potential red-layer communities. Western Pool was the only obvious feature we found in the Shoshone vicinity that contained a visible red-layer community.
Spray Geyser.
Located in the Middle Geyser Basin, Spray Geyser is the closest feature in this study to the former Rabbit Creek Spouter (4 km). The thermal source and splash mats at Spray Geyser have changed dramatically over the last 2 years. Temperature and pH measurements within the red layer of this mat, taken during cooler eruption intervals, were 35 to 38.8°C and pH 8.5, respectively. 16S rRNA analysis was performed only with the 2000 red-layer sample.
Pigment analysis and microscopy.
After collection in the field, all samples described above were dissected, examined microscopically, and subjected to pigment analysis. In all cases, microscopy revealed complex communities dominated by reddish orange filaments, the diameters of which comprised a broad range (0.8 to 1.5 μm); a representative micrograph of the Spray Geyser red-layer community is shown (Fig. 1, panel II). Methanol-extracted pigment from all red-layer samples maximally absorbed light at 769 to 771 nm, a finding consistent with Bchl a spectra (data not shown). In vivo absorption maxima of the pigment-protein complexes in cell extracts from each red layer varied and are summarized in Table 1. Notably, all spectra lacked major absorption maxima in the 735- to 750-nm range, typical of Bchl c, and thus were unlike Chloroflexus spectra. All in vivo spectra had one absorption band between 800 and 807 nm and a second variable band. Based on the position of the second band, red-layer communities fell into one of two distinct groups: (i) those with the second band between 879 to 889 nm, typified by the red-layer communities at Hillside II, Witch Pond, and Shoshone/Western Pool, or (ii) those with the second band at 900 nm or greater, typified by the red-layer communities at Spray and Fairy.
16S rRNA library design and analysis.
When we began this study, we knew little about GNS diversity in terms of red-orange organisms. Heliothrix and two Heliothrix-like sequences from Octopus Spring (27) were the most similar candidates in GenBank for designing primers given our suspicion that red-layer bacteria were related. Thus, we began this work by analyzing members of a General Bacterial 16S rRNA library from Hillside II Springs (1999 sample). Based on BLAST similarity analyses, members of the Hillside II Springs general bacterial library included six non-GNS sequences (data submitted to GenBank [not shown]; these sequences and BLAST information are available via our RLMO website) and ten GNS-like sequences. All GNS-like sequences were most similar to Roseiflexus. Specific information about these clones and this library are shown in Table 1.
The next site library was prepared from Witch Pond by using 77GNS, a new forward primer predicted to recognize all phototrophic GNS bacteria (77FGNS) and Hillside GNS-like sequences. We combined 77GNS with the broad-specificity reverse primer, 1492RPL (23), to generate a phototrophic GNS library. Analyzed members from this library contained only GNS-like sequences, and all were most similar to Roseiflexus. Specific information about these clones and this library is shown in Table 1.
We next designed a specific reverse primer, 953R, that was predicted to amplify Bchl a-only-containing GNS 16S rRNA. This primer was combined with the GNS-specific forward primer 77FGNS and was used to generate red GNS libraries from the red-layer communities at Spray, Shoshone, and Fairy. All libraries contained only GNS-like sequences, and all were most similar to Roseiflexus. Specific information about these clones and libraries is shown in Table 1.
Check_Chimera.
All GNS-like sequences and Roseiflexus were submitted to the RDP Program, Check_Chimera (14). Full-length clones and Roseiflexus were not predicted to have any chimeric regions. However, all clones derived with 77FGNS appeared to contain one chimeric region in their first 110 bases (this region corresponded to positions 100 to 210 in E. coli). Notably, when Roseiflexus was trimmed to the size of our non-full-length clones and subjected to Check_Chimera analysis, it was also predicted to contain the same chimeric region as our shortened clones.
Phylogenetic analysis and comparison of red-layer GNS isolates.
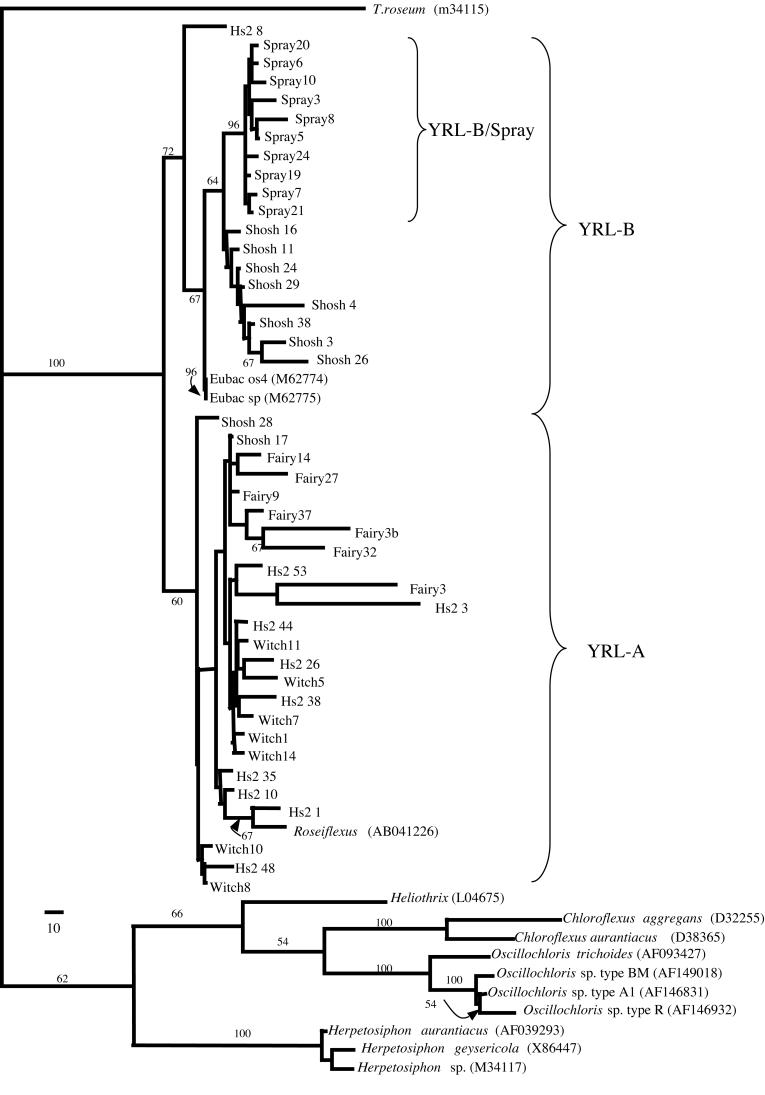
All of the following data refer to the phylogenetic tree shown in Fig. 2. All GNS-like clones from this study, Roseiflexus, and two previously described isolates from Octopus Spring (27) formed a distinct cluster relative to all other GNS sequences examined in this analysis. This “red” cluster was supported with a bootstrap value of 100%. Within this cluster, two distinct subclusters, YRL-A and YRL-B, were supported with bootstrap values of 60 and 72%, respectively.
FIG. 2.
Maximum parsimony tree of Yellowstone red-layer GNS-like clones. Representative GNS organisms are indicated in italics, with the GenBank accession number in parentheses. Red-layer clones are signified by site and clone number (abbreviations: Hillside II [Hs2], Witch Pond [Witch], and Shoshone/Western Pool [Shosh]). Three significant clusters are shown in brackets and are labeled YRL-A, YRL-B, and YRL-B/Spray. The bar represents 10 nucleotide changes. In this analysis, there were 980 total characters, and 353 were usable for parsimony analysis. A total of 100 bootstrap replicates were performed, and the bootstrap values are indicated (those that were <50% are not shown).
The YRL-A subcluster included Roseiflexus, all clones from Witch Pond, all clones from Fairy, 9 of 10 Hillside clones, and 2 of 10 Shoshone clones. All internal topologies within this cluster had bootstraps of <50% and are not shown.
The second subcluster, YRL-B, included the two previously described clones from Octopus Spring (27), all clones from Spray Geyser, 8 of 10 Shoshone clones, and 1 of 10 Hillside clones. In contrast to YRL-A, the internal topologies within YLR-B were moderately supported. All Spray and Shoshone clones formed a group within this subcluster that was supported with a 64% bootstrap. Within this group, all Spray clones formed a subgroup that was supported with a 96% bootstrap. The two clones from Octopus Spring also formed a distinct group within YRL-B that was supported with a 96% bootstrap.
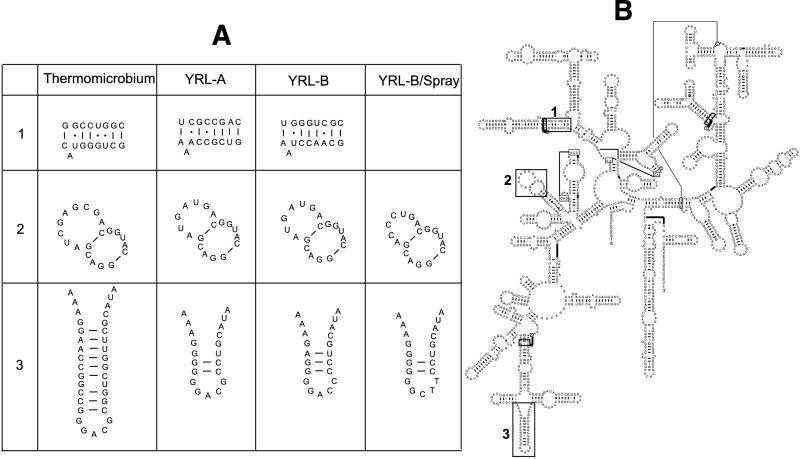
Structure comparison.
Given the putative phylogeny of GNS-like clones, we analyzed our alignments in conjunction with 16S rRNA structures for E. coli and T. roseum (Gutell et al. [http://www.rna.icmb.utexas.edu/]). From our GNS clones, we used consensus data for each of the three well-supported clusters observed in this study: YRL-A, YRL-B, and YRL-B/Spray. Based on these analyses, we noted three distinct regions that corresponded to observed phylogenetic clustering (Fig. 3). The first region forms a stem portion that corresponds to positions 588 to 596 and positions 644 to 651 in E. coli (region 1, Fig. 3). This region contains cluster-specific mutations that reflect differences between YRL-A and YRL-B. In this region, there were 9 of 17 base changes between YRL-A and YRL-B. The second region is a stem-loop region that corresponds to positions 179 to 218 in E. coli (region 2, Fig. 3). In this region, T. roseum contains a large stem-loop structure determined by a unique insertion. GNS-like clones are predicted to have a small stem-loop structure and contain cluster-specific mutations that reflect differences between YRL-A, YRL-B, and YRL-B/Spray. The third region is a complex loop, and the corresponding structure in E. coli (positions 440 to 490) is larger than that for both T. roseum and the predicted structures for our GNS-like sequences (region 3, Fig. 3). While this loop is conserved among YRL-A and YRL-B, it contains changes that are specific to YRL-B/Spray. Of 26 bases in this loop, five bases are specific to Spray clones only.
FIG. 3.
(A) Secondary structure prediction of Yellowstone red-layer GNS-like clones. Alignments were analyzed in conjunction with the secondary structure of T. roseum (B). In this analysis, three areas were found that corresponded to significant phylogenetic clustering (see Fig. 2): YRL-A, YRL-B, and YRL-B/Spray. These three areas are boxed in panel B and labeled 1, 2, and 3. In panel A, these areas are enlarged for T. roseum, and the corresponding predicted areas for the three clusters are shown.
DISCUSSION
Our comparative survey of five red-layer communities in Yellowstone National Park suggests the presence of a diverse and distinct group of uncultured GNS-like bacteria, the closest known isolate of which is R. castenholzii, a red filamentous Bchl a-only-containing bacterium from a similar hot springs in Japan. This hypothesis was supported by16S rRNA phylogenetic studies, in vivo Bchl a absorption spectra comparisons, and morphologic assessment of each mat red-layer sample.
Within this new red GNS cluster, two distinct and well-supported phylogenetic subclusters emerge: YRL-A was most similar to Roseiflexus, and YRL-B was most similar to two unclassified 16S rRNA sequences originally retrieved from Octopus Spring, Yellowstone National Park (27). A potential third cluster that contains all Spray Geyser GNS-like clones represents a well-supported subgroup within YRL-B. While our analyses were based on a somewhat limited character set (353 usable characters out of 980 aligned positions), we believe that our observed trees represent supported hypotheses given the following: (i) the observed bootstrap values we generated by using maximum parsimony analysis (Fig. 2); (ii) virtually identical results were generated when the aligned data sets were analyzed by maximum-likelihood analysis with bootstrapping (data not shown); and (iii) subclustering was observed to correlate with variation at several 16S rRNA structural elements (Fig. 3).
We were surprised to find that, in the context of this new data set, H. oregonensis still remained clustered with traditional “green” GNS organisms. This finding contradicts one previously published hypothesis that Yellowstone “red” GNS organisms may represent new species of Heliothrix (3). Notably, this hypothesis was based on structural and metabolic comparisons and was made prior to the isolation of Roseiflexus (8).
The phylogenetic position of clones within YRL-A and -B bore no relationship to the in vivo absorption spectra of the samples. The long-wavelength (>900-nm) absorption bands were observed in samples from both groups. Nevertheless, Heliothrix was not observed to cluster within either of these groups, and its in vivo absorption maxima for Bchl a were at shorter wavelengths than all of the others. Taken together, our data do not suggest any correlation between 16S phylogenetic clustering and in vivo Bchl a spectral phenotype. This observation is consistent with phylogenies generated from representative purple nonsulfur bacteria that also contain only Bchl a (7).
We observed site-specific phylogenetic clustering at Spray Geyser (YRL-B only), Fairy Geyser (YRL-A only), and Witch Pond (YRL-A only). These data suggest several possibilities: simple genetic drift, geographical isolation, and/or distinct selection factors present at specific sites. Potential selection factors include temperature, pH, the presence of specific nutrients or minerals, and the presence of other microorganisms. Recently, temperature selection was found to correlate with 16S rRNA phylogeny among related Synechococcus spp. in thermal environments (15). Given variable red-layer temperatures, particularly at intermittent or splash mats, we found it inappropriate to attempt to draw relationships between this habitat feature and observed phylogenetic clusters. Only a long-term study of temperature variation and population dynamics could address this hypothesis. We also found it inappropriate to sort our data based on pH for the same reasons. More precise nutrient and mineral analyses offer a potential future direction for our habitat survey work, although the presence of microenvironments within mats would make this difficult to assess.
Hillside and Shoshone clones were observed in both clusters, suggesting that these sites contain mixed populations of red GNS-like bacteria in the same community or in microenvironments within the same macroscopic layer. Observed data from other sites do not preclude the possibility that these sites also contain mixed populations. Whether these putative populations reside in specific niches within the red layer remains to be seen. Given the highly distinct laminations observed in alkaline hot spring communities, we believe that depth-related selection offers a more testable hypothesis as it relates to observed red GNS clustering. Position in a mat community is determined by a variety of specific selective factors, including light attenuation and the presence or absence of specific nutrients, minerals, and oxygen. This has long been known to be the case for genus- or lineage-specific layers (4). However, in our hypothesized model a given red layer would, in fact, be composed of two or more populations of GNS-like bacteria. Similar depth-determined distributions have been observed in open ocean bacterial communites (5). We are currently attempting to address this question through refined red-layer sectioning and the application of cluster-specific probes in fluorescent in situ hybridization (FISH) studies.
The 16S rRNA structure studies described here serve three important functions. First, they offer regions for cluster-specific probes that are being designed by our lab for FISH analyses. Second, they provide structural motifs that can be correlated with the three observed clusters or subclusters in this study. Third, the fact that they are found in multiple clones suggests that they are not random Taq errors generated during initial PCR amplification or DNA sequencing reactions.
The presence of chimeras can be an important artifact in phylogenetic analyses such as this. Check_Chimera suggested that all of our non-full-length clones contained a chimeric fragment but, for the following reasons, we believe that these sequences are not truly chimeric. First, it is unlikely that all of these clones would have recombined at the same position and with the same parents given the diversity of the population sampled. Second, no chimeric fragments were detected among our full-length clones or Roseiflexus. Third, when we analyzed truncated Roseiflexus, purposely trimmed to the exact size of our non-full-length sequences, this shortened sequence was predicted to contain the same chimeric fragment as our short clones. Given that Roseiflexus was derived from a clonal population, there should have been no potential for artificial recombination. The fact that both full-length and non-full-length Roseiflexus, when analyzed by using Check_Chimera, give conflicting results with this program is problematic and may reflect, again, the limited representation of phototrophic GNS sequences in this and other databases.
In this study, we have described a novel and diverse group of GNS-like clones from five distinct red-layer communities in Yellowstone National Park. Phylogenetically supported clustering patterns did not correlate with variation in the in vivo Bchl a absorption spectra for each community. The diversity of pigmentation patterns and phylogenies suggest the presence of a large number of diverse populations of red GNS bacteria, although additional hybridization studies are needed to explicitly link the molecular sequences described here with specific organisms in the mat community. Nevertheless, we believe it is significant that 10 of 16 Hillside II clones from this general bacterial library were Roseiflexus-like given that we physically and visibly enriched for red filaments prior to DNA isolation. Furthermore, all Witch Pond clones from this phototrophic GNS library were also Roseiflexus-like, despite the fact that the selective forward primer used to generate this library was predicted to amplify both red and green GNS phototrophs. Taken together with observed pigment signatures, these data suggest that enriched filaments were more likely novel red GNS and not cohabiting Chloroflexus. The use of highly specific primers to amplify 100% Roseiflexus-like clones Spray, Shoshone, and Fairy does not directly support a relationship between observed red filaments and retrieved red GNS-like sequences in these communities. However, concerns that this primer could be too specific were diminished by the observed diversity of distinct red GNS clusters. The red GNS-specific primers described in this study represent probes that we are currently applying to FISH analyses, the goal being to directly link observed filaments with retrieved GNS-like sequences.
Two other lines of evidence support that these phylogenies represent a true red line of descent within the GNS. First, all of these sequences are most similar to R. castenholzii, a Bchl a-containing red filamentous GNS bacterium found in comparable habitats in Japan. Second, GNS phototrophs are the only known filamentous bacteria that contain only Bchl a. While the clones described here are most similar to Roseiflexus, they are not the same, whether in terms of 16S rRNA sequence or in vivo Bchl a spectra. Our efforts to culture organisms from Spray, Fairy, and Hillside II by the methods described for Roseiflexus (8) failed. We are continuing to address these issues in our ongoing survey of Yellowstone thermal features for RLMOs and red GNS diversity.
Acknowledgments
We greatly appreciate the assistance and support of Yellowstone National Park, in particular Ann Deutch (former Research Permit Facilitator and Director of the Center for Resources), Ann Rodman, and Steve Wheeler (Yellowstone National Park Spatial Analysis Center Laboratory). We thank Brian Hedlund, John Gosink, and Russ Herwig for technical assistance and advice regarding DNA extraction, 16S rRNA isolation and analysis, and primer design. We gratefully acknowledge Dick Castenholz for recommendations about red-layer community sites based on his extensive experience in Yellowstone. We also acknowledge Joel Klappenbach for insightful comments about the manuscript and phylogenetic analyses. Many 16S rRNA clones from Hillside II and Fairy Springs were initially screened in the context of an advanced course in molecular biology. Students who partipated in this research-based curriculum included Ramon Larios, Chris Coverdill, Kevin Larson, Michelle Hase, Nathan Fitzpatrick, Jeremy Taylor, Daniel Lodge (then an undergraduate), Peter Williams, Nicole Mullins, Norm McIntosh, Jeannine Earnest, Sarah Crosky, and Jim Erdman. All samples were collected during field research trips with Western Oregon University senior-level biology majors. Students who participated in Yellowstone survey and sampling included Ben Stern, Robin Leitch, Daniel Russell, Joseph Hayes, Kody Phillis, Chris Coverdill, Kevin Larson, Rex Addis, Michelle Hase, Nicole Mullins, Eric Stroup, Mandy Ziglinski, Jessica Cameron, and Jeannine Earnest. Two former colleagues, Brian Hedlund and Chandell Terwilliger, participated in coleadership on two undergraduate research trips to Yellowstone.
A large portion of this work was supported by an NSF Microbial Observatories/Research at Undergraduate Institute Grant (NSF-MO/RUI 0074452). The Li-Cor DNA sequencer was acquired via an NSF Improved Laboratory Instrumentation Grant (NSF-ILI DUE-9851322). All research trips to Yellowstone were supported by annual Western Oregon University Foundation Grants.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 3.Boomer, S. M., B. K. Pierson, R. Austinhirst, and R. W. Castenholz. 2000. Characterization of novel bacteriochlorophyll-a-containing red filaments from alkaline hot springs in Yellowstone National Park. Arch. Microbiol. 174:152–161. [DOI] [PubMed] [Google Scholar]
- 4.Castenholz, R. W. 1984. Microbial mats: stromatolites. Alan R. Liss, Inc., New York, N.Y.
- 5.Field, K. G., D. Gordon, T. Wright, M. Rappe, E. Urback, K. Vergin, and S. J. Giovannoni. 1997. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl. Environ. Microbiol. 63:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia, D., P. Parot, A. Vermeglio, and M. T. Madigan. 1986. The light-harvesting complexes of a thermophilic purple bacterium Chromatium tepidum. Biochim. Biophys. Acta 850:390–395. [Google Scholar]
- 7.Glaeser, J., and J. Overmann. 1999. Selective enrichment and characterization of Roseospirillum parvum, gen. nov. and sp. nov., a new purple nonsulfur bacterium with unusual light absorption properties. Arch. Microbiol. 171:405–416. [DOI] [PubMed] [Google Scholar]
- 8.Hanada, S., S. Takaichi, K. Matsuura, and K. Nakamura. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium which lacks chlorosomes. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 9.Holben, W. E. 1997. Isolation and purification of bacterial community DNA from environmental samples, p.431–436. In C. J. Hurst, M. J. Knudsen, L. D. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 10.Holt, J. G., and R. A. Lewin. 1968. Herpetosiphon aurantiacus gen. et sp. n., a new filamentous gliding organism. J. Bacteriol. 95:2407–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67:411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, S. R., and R. W. Castenholz. 2000. Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 66:4222–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oyaizu, H., B. A. Debrunner-Vossbrinck, L. Mandelco, J. A. Studier, and C. R. Woese. 1987. The green non-sulfur bacteria: a deep branching in the eubacterial line of descent. Syst. Appl. Microbiol. 9:47–53. [DOI] [PubMed] [Google Scholar]
- 17.Pierson, B. K., and R. W. Castenholz. 1992. The family Chloroflexacae, p.3754–3774. In A. Balows, H. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, Berlin, Germany.
- 18.Pierson, B. K., S. J. Giovannoni, and R. W. Castenholz. 1984. Physiological ecology of a gliding bacterium containing bacteriochlorophyll a. Appl. Environ. Microbiol. 47:576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierson, B. K., S. J. Giovannoni, D. A. Stahl, and R. W. Castenholz. 1985. Heliothrix oregonensis, gen. nov., sp. nov., a phototrophic filamentous gliding bacterium containing bacteriochlorophyll a. Arch. Microbiol. 142:164–167. [DOI] [PubMed] [Google Scholar]
- 20.Pierson, B. K., A. Oesterle, and G. L. Murphy. 1987. Pigments, light penetration, and photosynthetic activity in the multi-layered microbial mats of Great Sippewissett Salt Marsh, Massachusetts. FEMS Microbiol. Ecol. 45:365–376. [Google Scholar]
- 21.Pierson, B. K., V. M. Sands, and J. L. Frederick. 1990. Spectral irradiance and distribution of pigments in a highly layered marine microbial mat. Appl. Environ. Microbiol. 56:2327–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierson, B. K., D. Valdez, M. Larsen, E. Morgan, and E. E. Mack. 1994. Chloroflexus-like organisms from marine and hypersaline environments: distribution and diversity. Photosynthesis Res. 41:35–52. [DOI] [PubMed] [Google Scholar]
- 23.Reysenbach, A. L., G. S. Wickham, and N. R. Pace. 1994. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 60:2113–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sly, L. I., M. Taghavi, and M. Fegan. 1998. Phylogenetic heterogeneity within the genus Herpetosiphon: transfer of the marine species Herpetosiphon cohaerens, Herpetosiphon nigricans, and Herpetosiphon persicus to the genus Lewinella gen. nov. in the Flexibacter-Bacteroides-Cytophaga phylum. Int. J. Syst. Bacteriol. 48:731–737. [DOI] [PubMed] [Google Scholar]
- 25.Swofford, D. L. 2000. PAUP 4.0b8. Sinauer Associates Inc., Sunderland, Mass.
- 26.Weller, R., M. M. Bateson, B. K. Heimbuch, E. D. Kopczynski, and D. M. Ward. 1992. Uncultivated cyanobacteria, Chloroflexus-like inhabitants, and spirochete-like inhabitants of a hot spring microbial mat. Appl. Environ. Microbiol. 58:3964–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weller, R., J. W. Weller, and D. M. Ward. 1991. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl. Environ. Microbiol. 57:1146–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]