Abstract
Reports of the biological multifunctional activity of various aminoacyl tRNA synthetases have recently accumulated in the literature. The primary function of these critical enzymes is to charge various tRNAs with their appropriate amino acids, thus producing the building blocks of protein synthesis. We have previously shown that lysyl tRNA synthetase (LysRS) associates with microphthalmia transcription factor (MITF) and regulates its activity by synthesis of Ap4A in mast cells. Here, we show for the first time that LysRS associates with another transcription factor, USF2, which unlike MITF, is ubiquitously expressed in eukaryotic cells. Using mast cells, we have found that USF2 is negatively regulated by Hint and Ap4A acts as a positive regulator of USF2 by a molecular mechanism similar to that described for MITF. Since USF2 plays a significant role in a variety of cellular functions, our finding suggests that LysRS and Ap4A may be involved in general regulation of gene transcription.
Aminoacyl tRNA synthetases (aaRSs) are essential proteins that are extremely conserved throughout the evolution from prokaryotes to eukaryotes. Each of the 20 different enzymes catalyzes the aminoacylation of the 20 specific tRNAs, thus providing the building blocks for protein synthesis. These proteins, however, have acquired various additional functions through evolution, which lead to specific aaRSs being involved in a broad repertoire of functions extending to several critical cellular activities, such as tRNA processing, RNA splicing, RNA trafficking, and transcriptional and translational regulation (25).
Among the aaRSs that posses multiple functions are TyrRS and TrpRS, which can also act as cytokines. Protease cleavage of TyrRS creates two distinct cytokines that induce angiogenesis and leukocyte recruitment (42), whereas alternative splicing of TrpRS transcripts produces antiangiogenic factor (29, 43). In our recent work, we provided evidence for the involvement of aaRS in transcriptional regulation by demonstrating the involvement of lysyl tRNA synthetase (LysRS) in the regulation of microphthalmia transcription factor (MITF) transcriptional activity (18). LysRS, as a part of the multiprotein complex with MITF and Hint, synthesizes Ap4A in close proximity to Hint. Ap4A binds to Hint, the suppressor of MITF's transcriptional activity, and this leads to the dissociation of Hint from MITF, thus allowing MITF to transactivate its target genes upon specific trigger.
LysRS is also known to synthesize Ap4A in mammals in a zinc-dependent chemical reaction (7, 13). Ap4A is a diadenosine polyphosphate, a naturally occurring group of molecules in which two adenosines are joined by three to six phosphate groups. These molecules are structural analogues of NTPs that are highly charged and have affinity to proteins (5). One such protein is Hint, which dissociates from MITF transcription factor on binding to an Ap4A molecule (18). An additional intracellular function proposed for Ap4A is a role in the induction of apoptosis, based on experiments showing that administration of Ap4A to a variety of cell lines causes cellular apoptosis (40).
Other basic helix-loop-helix leucine zipper (bHLH-Zip) transcription factors that belong to the same family of proteins as MITF are USF1 and USF2. These two transcription factors, unlike MITF, are ubiquitously expressed in eukaryotic cells where they are involved in a broad spectrum of biological activities (19, 28). For instance, both proteins are essential for embryonic development, since an embryonic lethal phenotype was observed with the double-null mouse mutants (34). Furthermore, USF2-null mice show a more severe phenotype than USF1-null mice, with severe growth defects, abnormalities in fertility, mammary gland malfunction, and an impaired transcriptional response to glucose in liver (12, 34, 39). Previously, we provided evidence for a direct connection between cell surface receptor-mediated USF2 nuclear translocation and cell viability (9). Another study demonstrated the involvement of USF in cell division as a negative regulator of cell proliferation by antagonizing the transforming function of Myc (24).
One of the ways to elucidate the precise functional regulation of USF2 activity could be performed by investigating its possible association with other molecules. A candidate protein for such an association with USF2 is Hint, which negatively regulates the transcriptional activity of MITF (18, 31). This ubiquitously expressed protein was recently determined to be a tumor suppressor gene (37) and is a member of the histidine triad (HIT) protein family, indicated by a conserved HIT motif sequence (23). This protein and other members of the HIT family are extremely conserved, ranging from prokaryotes to humans. In vitro assays show that HIT proteins have catalytic activity and are able to bind nucleotidyl substrates (6, 23), and Hint was shown to specifically interact with Ap4A by BIAcore analysis (18). Furthermore, the physical and genetic association of Hint with a part of the basal transcription factor TFIIH is conserved in yeast and humans, showing the functional significance of Hint (17). The possible interaction of Hint with USF2 would provide further insight into the molecular mechanisms underlying USF2 transcriptional activity.
In the present study, we show that USF2 is associated with Hint, which suppresses USF2 transcriptional activity. Furthermore, LysRS forms a multiprotein complex with USF2 and Hint and produces Ap4A, which dissociates Hint from USF2 and thus allows the transcription of USF2-responsive genes. This molecular mechanism of USF2 transcriptional activity closely follows the molecular mechanism of MITF activity. Here, we show the broadening of LysRS's function as a positive regulator of transcription factors via the synthesis of Ap4A in eukaryotic cells. This is a novel topic, which has implications for our understanding of a variety of cellular functions.
MATERIALS AND METHODS
Cell growth and permeabilization.
RBL and NIH 3T3 cells were maintained as previously described (31). RBL cells were permeabilized by cold shock (3), and cell viability was determined by trypan blue exclusion. Chinese hamster ovarian (CHO) cells that overexpress full-length LysRS (CKRS) and truncated LysRS (CKRS N̂) were kindly provided by M. Mirande (2). The Lys-101 CHO cell line (1) and Lys-101 stably transformed CHO cells with pSG/CKRS and pSG/CKRS N̂ (2) were maintained in at 34°C in RPMI 1640 medium supplemented with 8 mM l-glutamine, 2 mM nonessential amino acids, 100 units/ml penicillin, and 100 μg/ml streptomycin (Life Technologies, Inc.).
Plasmid construction.
Normal mouse Hint (381 bp) was inserted into pGEX3X vector (Amersham Biosciences, Uppsala, Sweden) and pcDNA 3.1. Mouse MITF (1,129 bp) was inserted into the pGEX-4T-3 vector (Stratagene, La Jolla, Calif.). Fidelity of the constructs was verified by direct sequencing. The luciferase reporter plasmid pSP72, containing the MITF binding region of the promoter and the first exon of the mMCP-6 gene (−191 to + 26), as well as a construct with a deleted MITF binding site (−151 to + 26), was generously provided by Y. Kitamura, Osaka, Japan. pCMV-USF2 and its reporter gene pU3ML, containing three USF2-responsive elements (E-box) (14), were kindly provided by M. Sawadogo (The University of Texas, Houston, Tex.). STAT3-C and the M67 pTATA tk-Luc reporter gene were kindly provided by J. E. Darnell (The Rockefeller University, New York, N.Y.). The PKA catalytic subunit beta (PKA-Cβ) luciferase reporter gene with a human promoter containing c-myc-responsive elements was kindly provided by R. Dalla-Favera (Columbia University, New York, N.Y.) (45).
In vitro GST pulldown assay.
Glutathione S-transferase (GST)-Hint fusion protein was expressed in protease-deficient Escherichia coli strain BL-21 and purified on glutathione-Sepharose beads (Amersham Biosciences). Pulldown assays were performed as described previously (21). The integrity and quantity of GST fusion proteins were confirmed by Gelcode Bluestain reagent (Pierce Biotechnology, Inc., Rockford, Ill.), and autoradiography detected the amount of retained radiolabeled 35S-labeled USF2.
Immunoprecipitation.
The immunoprecipitation of the specific proteins from RBL cells was carried out as previously described (21). The antibodies that were used for immunoprecipitation were anti-mouse USF2, anti-mouse MITF, anti-mouse Hint, and anti-human LysRS, which was kindly provided by L. Kleiman (Lady Davis Institute for Medical Research, Montreal, Canada).
Nucleotide assay.
This assay detects the relative amount of Ap4A present in extracts of mammalian cells. For each determination, cells from one well of a six-well plate were grown to about 80% confluence. The cell layer was washed with warm serum-free medium and was lysed with trichloroacetic acid. Extraction and measurement by luminometry of the nucleotides were performed as described previously (27).
PCR amplification.
Primers were used to amplify USF2 from mouse cDNA. The primers were designed by including a T7 promoter sequence and a ribosome binding site into the 5′ primers. The primers for USF2 were a sense 5′ (5′-CTAATACGACTCACTATAGGGAAGGAGATATACATATGGACATGCTG GACCCGGGTCTGGAT-3′) and anti-sense 3′ (5′-TTACTGCCGGGTACTCTCGCCCAC-3′).
Real-time quantitative PCR.
MITF-responsive genes were measured by using real-time quantitative PCR. Total RNA was extracted from RBL cells, and mRNA levels of various genes were quantified by SYBR-green incorporation (SYBR Green PCR Master Mix; Applied Biosystems, Foster City, Calif.). SYBR-green incorporation to double-strand DNA permits the direct detection of PCR product after each amplification cycle (ABI Prism 7000 sequence detection system; Applied Biosystems). The specificity of the amplification was controlled by electrophoresis. The genes whose mRNA levels were quantified by real-time PCR were Rat CXCR4, FcɛRI, TGF-β2, and actin genes.
Transient cotransfection and luciferase assay.
NIH 3T3 cells (2 × 105) were cotransfected by Transfast reagent (Promega Biosciences, San Luis Obispo, Calif.) with 0.5 μg of reporter pU3ML containing three E-boxes, 0.1 μg of pCMV-USF2, and 0.5 μg pcDNA-Hint and pcDNA alone as a nonspecific control. The cells were incubated in 24-well plates for 48 h. CHO cells (2 × 105) were transfected by Transfast reagent with 0.1 μg of various reporter genes: pSP72 for MITF, pU3ML for USF2, and PKA-Cβ for c-Myc. For STAT3, both the promoter M67 and constitutively active STAT3 (designated STAT3-C) were cotransfected. The luciferase activity was normalized to the total protein concentration. The ratio was expressed as relative luciferase activity.
RESULTS
Hint as a suppressor of LysRS-associated USF2 transcriptional activity.
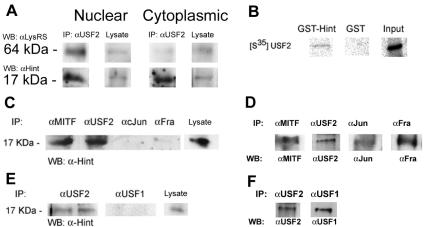
Regulation of transcriptional activity by LysRS was previously demonstrated with MITF (18). LysRS is associated with the MITF/Hint complex, and it is induced to synthesize Ap4A. The local accumulation of Ap4A causes the dissociation of MITF from its inhibitor, Hint, and thus transactivates MITF's target genes. To investigate whether LysRS is involved in the regulation of other transcription factors such as USF2, we first analyzed whether LysRS was in the same complex with USF2 in RBL cells. Nuclear and cytoplasmic extracts from 5 × 106 cells were immunoprecipitated with anti-USF2, subjected to polyacrylamide gel electrophoresis, and analyzed by Western blotting with anti-LysRS and anti-Hint antibodies. Figure 1A shows that LysRS immunoprecipitated together with USF2 only in the nuclear compartment and that Hint immunoprecipitated together with USF2 in the nuclear and cytoplasmic compartment of the cell. Furthermore, when nuclear and cytoplasmic extracts were immunoprecipitated with anti-Hint and subjected to Western blotting analysis with anti-USF2 and anti-LysRS, LysRS showed similar results (data not shown).
FIG. 1.
(A) Coimmunoprecipitation of USF2 with LysRS and Hint. RBL cells were subjected to nuclear and cytoplasmic extraction from 5 × 106 cells, and the lysates were incubated with anti-USF2 antibody. The resolved immunocomplexes and lysate input control (5%) were analyzed by Western blotting with anti-LysRS and anti-Hint antibodies. One representative of three is shown. (B) A pulldown assay of [35S]methionine-labeled USF2 by GST-Hint fusion protein. The GST-Hint protein bound to glutathione-Sepharose beads was incubated with [35S]methionine-labeled USF2 overnight at 4°C. One representative result of three is shown. (C) Coimmunoprecipitation of Hint with USF2, MITF, cJun, and Fra. RBL cell lysates were subjected to immunoprecipitation with anti-USF2, anti-MITF, anti-cJun, and anti-Fra antibodies. The resolved immunocomplexes and lysate input control (5%) were analyzed by Western blotting analysis with anti-Hint antibody. One representative result of three is shown. (D) Precipitation efficiency of anti-MITF, anti-USF2, anti-cJun, and anti-Fra antibodies. RBL lysates were immunoprecipitated with the antibodies indicated on the top, and the resolved immunocomplexes were subjected to Western blotting with the antibodies indicated on the bottom. (E) Coimmunoprecipitation of Hint with USF2 and USF1. RBL cell lysates were subjected to immunoprecipitation with anti-USF1 and anti-USF2 antibodies. The resolved immunocomplexes and lysate input control (5%) were analyzed by Western blotting analysis with anti-Hint antibody and are shown in duplicate. One representative result of three is shown. (F) Precipitation efficiency of anti-USF1 and anti-USF2 antibodies.
To determine the specificity of the USF2/Hint association, we carried out various biochemical analyses. First, the specific association of Hint with USF2 was determined by pulldown assay (Fig. 1B). Hint was expressed in bacteria as GST fusion protein, immobilized on glutathione-Sepharose beads, and was assayed for its ability to retain in vitro-translated USF2 labeled with [35S]methionine. No association between Hint and AP-1 proteins such as Jun and Fra was observed when RBL lysates were immunoprecipitated with anti-Jun and anti-Fra antibodies and were subjected to Western blot analysis using anti-Hint antibody (Fig. 1C). To show that the precipitation efficiency is similar among different antibodies, a Western blot for the corresponding immunoprecipitated proteins is shown in Fig. 1D. Since USF2 and USF1 are isoforms derived from different genes and work as heterodimers (11, 35, 36), we wanted to determine whether the association to Hint also occurred with USF1. Figure 1E shows that Hint associated with USF2 and not with USF1 when the precipitation efficiency in USF1 and USF2 was similar (Fig. 1F).
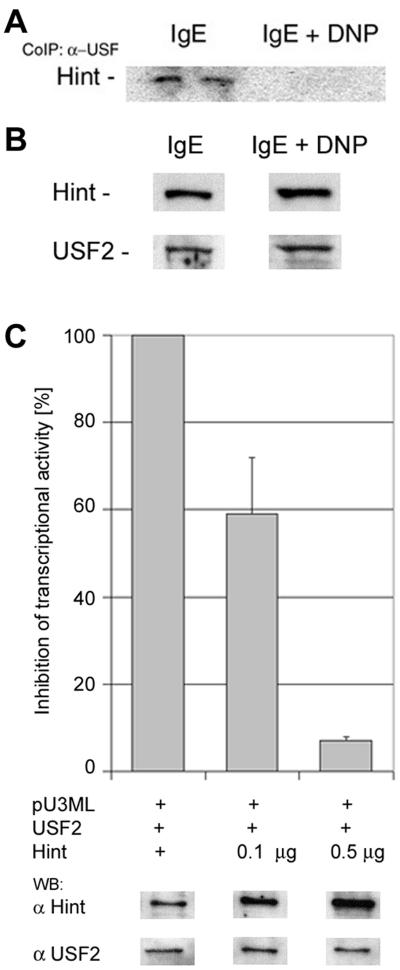
The physiological significance of the USF2 and Hint association was first determined by checking whether the signal from high-affinity immunoglobulin E (IgE) receptor FcɛRI led to any change in the protein-protein association, as was shown with MITF (18). Stimulation of RBL cells with IgE and 2,4-dinitrophenol (DNP) for 1 h dissociated Hint from USF2 (Fig. 2A). Both USF2 and Hint protein levels were unchanged with the stimulation of IgE and DNP (Fig. 2B). Next, the effect of Hint on the transcriptional activity of USF2 was assayed by using a luciferase reporter gene. Plasmids containing USF2, Hint, and the luciferase reporter gene with the promoter of USF2-responsive elements were cotransfected to NIH 3T3 cells. Increasing amounts of Hint showed a decrease in luciferase activity, suggesting that Hint serves as a suppressor of USF2 transcriptional activity (Fig. 2C).
FIG. 2.
Hint as an inhibitor of USF2 transcriptional activity. (A) RBL cells activated with IgE and DNP or IgE alone were lysed and subjected to immunoprecipitation with anti-USF2 antibody; the resolved immunocomplexes were analyzed by Western blotting with anti-Hint antibody. One representative result of three is shown. (B) The lysates from the IgE- and IgE-DNP-treated RBL cells were subjected to Western blot analysis for Hint and USF2 protein levels. One representative result of three is shown. (C) Hint-mediated inhibition of USF2 transcriptional activity in 3T3 NIH cells with 0.1 and 0.5 μg pcDNA-Hint. For each transfection, the total DNA concentration was constant by complementing with the empty vector pcDNA. The normalized value was expressed as percent luciferase activity. In addition, Western blot analysis for the levels of Hint and USF2 from the corresponding transfected 3T3 cells is shown.
Effects of Ap4A on USF2-Hint complex and on USF2 transcriptional activity.
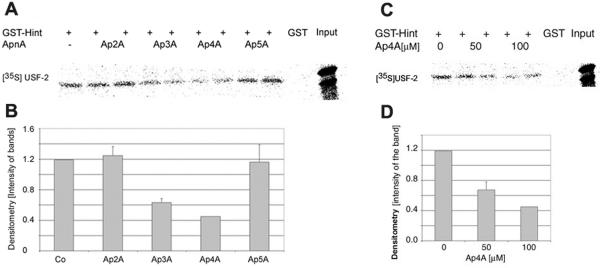
The effect of Ap4A on the USF2/Hint protein complex was first determined by a pulldown assay. GST-Hint fusion protein was incubated with [35S]methionine-labeled USF2 to form the USF2/Hint protein complex. Then various diadenosine oligophosphates (Ap2A, Ap3A, Ap4A, and Ap5A) at a concentration of 100 μM (each) were added to the USF2/Hint protein complex. Both Ap3A and Ap4A dissociated Hint from USF2, and Ap4A specifically dissociated Hint from USF2. However, Ap4A was more effective in dissociating Hint from USF2, as can be seen in Fig. 3A. These results were further analyzed by densitometry (Fig. 3B). Since Ap4A was more potent, we decided to concentrate on Ap4A. Moreover, Ap4A, which was found to be elevated upon mast cell stimulation (18), specifically reduced the association of USF2/Hint protein complex in a dose-dependent manner (Fig. 3C and D).
FIG. 3.
Ap4A mediates the dissociation of USF2/Hint complex. (A) The GST-Hint protein bound to glutathione-Sepharose beads was incubated with [35S]methionine-labeled USF2 overnight at 4°C. The USF2/Hint protein complex was exposed to Ap2A, Ap3A, Ap4A, and Ap5A compounds for 1 h at 37°C. One representative result of three is shown. (B) Densitometry analysis. The bands shown in panel A were subjected to densitometry analysis, and the results were normalized to GST control. (C) Dose-dependent dissociation of USF2/Hint protein complex by Ap4A. Increasing amounts of Ap4A were added to the USF2/Hint protein complex, and a pulldown assay was carried out. One representative result of three is shown. (D) Densitometry analysis. The bands shown in panel C were subjected to densitometry analysis, and the results were normalized to the GST control.
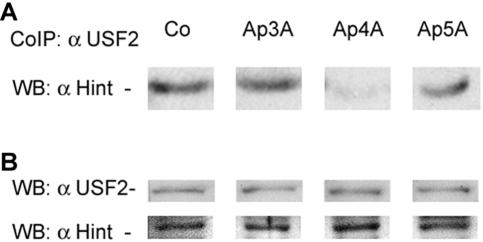
Next, RBL cells were exogenously introduced with different diadenosine polyphosphate compounds by the cell membrane permeabilization method, and cells were incubated at 37°C for 1 h. As can be seen in Fig. 4A, 100 μM Ap4A reduced immunoprecipitation of Hint, together with that of USF2. The protein levels of both Hint and USF2 remained unchanged upon the administration of various diadenosine polyphosphates (Fig. 4B).
FIG. 4.
The dissociation of USF2/Hint protein complex from Ap4A administered to RBL cells. (A) RBL cells were exogenously introduced with various ApnA compounds (concentration, 100 μM). The cells were lysed and subjected to immunoprecipitation with anti-USF2 antibody. The resolved immunocomplex was analyzed by Western blotting with anti-Hint antibody. One representative result of three is shown. (B) The RBL cell lysates with ApnA introduced were subjected to Western blot analysis to determine the protein levels of Hint and USF2.
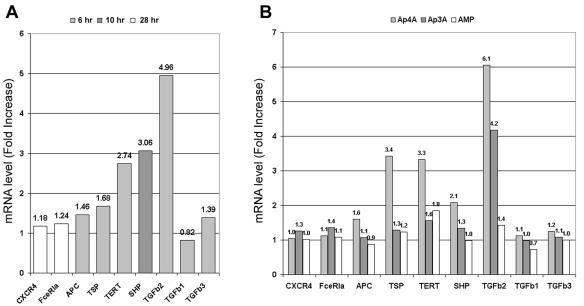
To determine the direct effect of Ap4A on USF2 transcriptional activity, the transcripts of USF2-responsive genes were measured at various time points after immunological triggering and after exogenous introduction of Ap3A, Ap4A, and AMP. Seven USF2-responsive genes—those for CXCR4, the alpha subunit of FcɛRI, adenomatous polyposis coli (APC) tumor suppressor, thrombospondin 1 (TSP-1), telomerase catalytic subunit (TERT), protein tyrosine phosphatase 1 (SHP), and transforming growth factor β2 (TGF-β2)—were tested. Of these target genes, only TERT, SHP, and TGF-β2 genes showed significant increase in their transcript levels upon 6 to 10 h of immunological triggering (Fig. 5A). The most profound elevation in the transcript level was observed with the TGF-β2 gene. The TGF-β2 gene is a specific target gene of USF2 (16), and it is involved in a broad range of cellular events such as cell growth, differentiation, and tissue morphogenesis (15, 26, 32); it is also considered a potent immunosuppressive cytokine. Next, the transcript levels of the above USF2 target genes in Ap4A-administered RBL cells were determined. The cells were incubated at 37°C for 12 h prior to RNA extraction. The introduction of Ap4A specifically increased the transcript level of the TGF-β2, TERT, and SHP genes. It should be noted that the transcript level of the TSP-1 gene was elevated due to the introduction of Ap4A, despite its nonsignificant elevation in the transcript level in immunologically activated cells (Fig. 5B). This indicates that immunologically induced USF2 activity on the transcription of TGF-β2, TERT, and SHP genes was mediated by Ap4A and that Ap4A alone might induce the TSP-1 gene. It is important to note that AMP, a by-product of Ap4A degradation, did not show any significant effect on the transcript levels of USF2-dependent genes, including TGF-β2. It is interesting to note that other TGF-β genes were not affected.
FIG. 5.
Immunological activation and Ap4A show positive effects on the transcript levels of USF2 target genes. (A) Expression level of USF2 target genes in IgE- and DNP-activated RBL cells. The RBL cells were incubated with 50-ng/ml IgE overnight, and then the cells were incubated 0, 6, 10, 20, and 28 h in the presence of DNP. The mRNA levels of CXCR4, FceRI, APC, TSP-1, TERT, SHP, TGFb2, TGFb1, and TGFb3 genes were measured and normalized to control. For each gene, the time point with the highest increase in the transcript level is shown. One representative result of three is shown. (B) Expression of the USF2 target genes in unstimulated RBL cells treated with 100 μM (each) Ap4A, Ap3A, and AMP. RNA from RBL cells was collected 12 h after the administration of Ap4A, Ap3A, and AMP. Values show the severalfold increase of the transcript levels normalized to that of the control. One representative result of three is shown.
Secondary function of LysRS as a regulator of transcription factors via Ap4A.
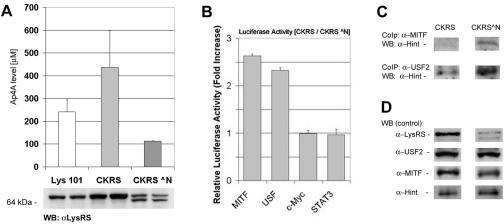
To determine a direct effect of LysRS on the transcriptional activity of USF2 and MITF, CHO cells that were stably transfected with either full-length or a truncated N terminus of LysRS were used. Lys-101 is a CHO cell line with a temperature-sensitive LysRS (1). These cells were further stably transfected with pSG/CKRS (full-length LysRS) and pSG/CKRS^N plasmids (LysRS with a truncated N terminus) (2). We have observed by Western blot analysis that LysRS is overexpressed by more than threefold in Lys-101 CHO cells that are stably transfected pSG/CKRS, compared to Lys-101 CHO cells (Fig. 6A). In Lys-101 CHO cells that are stably transfected with pSG/CKRS^N, there were two bands corresponding to the full-length LysRS and the N-terminus-truncated LysRS (Fig. 6A). The Ap4A levels were determined with Lys-101 CHO cells, Lys-101 CHO cells that were stably transfected with pSG/CKRS (CKRS), and pSG/CKRS^N (CKRS^N) plasmids. Ap4A levels were found to be more than twofold higher with CKRS than with CKRS^N cells (Fig. 6A).
FIG. 6.
Overexpression of LysRS via elevated amounts of Ap4A increases the transcriptional activity of USF2 and MITF. (A) Ap4A levels were determined with Lys-101 CHO, CKRS, and CKRS N̂ cells. The measurement of Ap4A was determined luminometrically as described in Materials and Methods. The bars show means ± standard error for three experiments. In addition, LysRS protein levels in these cell lines are shown at the bottom of the graph. From each cell line, 30 μg of total protein was analyzed by Western blotting with anti-LysRS antibody. Duplicates are shown for each cell line. (B) USF2, MITF, c-Myc, and STAT3 transcriptional activity in Lys-101 CHO cells overexpressing LysRS. CKRS and CKRS^N cells were transfected with a luciferase reporter gene containing the USF2-, MITF-, and c-Myc-responsive elements and a PKA-Cβ reporter gene with STAT3-C. Normalized values are relative severalfold increases in luciferase activity in CKRS cells compared to those in CKRS^N cells. Values are means ± standard error for three experiments. (C) Coimmunoprecipitation of either USF2 or MITF with Hint in cells overexpressing LysRS. CKRS and CKRS^N cells were lysed and incubated with either anti-USF2 or anti-MITF antibody. The resolved immunocomplexes were analyzed by Western blotting with anti-Hint antibody. One representative result of three is shown. (D) The lysates of CKRS and CKRS^N cells were subjected to Western blotting with anti-LysRS, anti-Hint, anti-MITF, and anti-USF2 to show the protein levels in various CHO cells. One representative result of three is shown.
Since the CHO cells that overexpressed full-length LysRS and CHO cells that overexpressed the truncated form of LysRS showed the greatest difference in the levels of Ap4A, we continued to use only these two cell lines and excluded the use of Lys-101 CHO cells. Furthermore, it has been reported that the expression LysRS in Lys-101 CHO cells is expressed only at 34°C and not at 40.1°C (1). Therefore, at 40.1°C the expression of the endogenous LysRS goes down and allows only the expression of the stably transfected vectors. However, we observed that at both 34°C and 40.1°C these cells expressed similar levels of LysRS (data not shown). The cells that overexpressed full-length LysRS and the truncated form of LysRS showed similar results at both 34°C and 40.1°C (data not shown). Furthermore, Ap4A levels were found to be similar in these cells at both 34°C and 40.1°C (data not shown). Thus, all experiments were carried out at 34°C.
Next, the transcriptional activity of USF2 and MITF in the cells that overexpress LysRS was determined by transfecting them with the luciferase reporter gene consisting of either USF2- or MITF-responsive elements. Lys-101 CHO cells that overexpress full-length LysRS significantly showed higher transcriptional activity of both USF2 and MITF (Fig. 6B). The transcriptional activity of other transcription factors such as c-Myc and STAT3 was tested and showed no change in transcriptional activity in Lys-101 CHO cells that overexpressed the truncated LysRS, compared to Lys-101 CHO cells that overexpressed full-length LysRS. Western blot analysis of these CHO cell lysates verified the presence of MITF, USF2, c-Myc, and STAT-3 (data not shown).
To determine whether there was any differences in the USF/Hint or MITF/Hint protein complexes, Lys-101 CHO cells stably transformed with pSG/CKRS and pSG/CKRS^N cells were subjected to coimmunoprecipitation analysis. Lys-101 CHO cells that overexpressed LysRS, thus having higher Ap4A levels, showed a decrease in the USF2/Hint and MITF/Hint protein complexes (Fig. 6C). This set of experiments clearly showed that LysRS was an inducer of USF2 (as well as MITF) transcriptional activity via the synthesis of Ap4A. As we have previously shown with MITF, it can be logically postulated that the accumulated Ap4A binds to Hint (18) and liberates USF2 to transactivate the target genes of USF2.
DISCUSSION
LysRS has been known for 3 decades to be the main cellular source of Ap4A from both prokaryotes and eukaryotes (30, 46, 47). In our previous and present studies, we have revealed the biological significance of LysRS as an enzyme that participates in intracellular signaling. By using a genetically altered CHO cell line, we demonstrated the involvement of LysRS in activating transcription factors (Fig. 6). CKRS cells showed twofold increases in the production of Ap4A and elevation in MITF and USF2 transcriptional activities. This specific molecular mechanism in which LysRS serves as a regulator of transcription factors via the production of Ap4A was first described for MITF in our recent publication (18). Here, we strengthen our previously proposed model for the regulation of MITF transcriptional activity with our current study of the regulation of USF2 transcriptional activity via the synthesis of Ap4A.
In contrast to MITF, which is expressed in specific cell types, USF2 is a ubiquitously expressed transcription factor, which has been shown to regulate many genes in various cell types by binding to the E-box element in the promoter. For instance, in mesangial cells USF2 specifically mediates the glucose-induced TSP-1 and TSP-1-dependent TGF-β activity (44). In nontumorigenic mammary epithelial cells, USF2 regulates the transcript levels of insulin-like growth factor 2 receptor, but interestingly it loses its ability to transactivate insulin-like growth factor 2 receptor in breast cancer cells (38). Furthermore, extensive studies were carried out on the regulation of USF2 expression in interleukin 3 activated mast cells, where an induction of USF2 protein synthesis was observed via increased translational efficiency (22, 48). We previously utilized cell permeable peptides as inhibitors of the activity of the USF2 proteins, to demonstrate its possible role in mast cell survival (9). Here, we demonstrate for the first time the association of LysRS and Ap4A with the molecular mechanism that regulates USF2 activity. Since USF2 plays a significant role in variety of cellular functions, it suggests that perhaps LysRS and Ap4A are involved in general regulation of transcriptional activity.
It is interesting to note that the induction of USF2 transcriptional activity via Ap4A in a mast cell line affects only certain USF2 target genes such as TGF-β2. Thus, it could be assumed that Ap4A mediates the regulation of a limited number of USF2 target genes. This is despite the fact that USF2 is broadly expressed in many cell types, has multiple target genes, and is involved in variety of physiological functions.
TGF-β2 belongs to the TGF protein family, which is one of the seven major families of growth factors that control numerous intracellular processes. This growth factor controls the differentiation, proliferation, and activation states of lymphocytes, macrophages, and dendritic cells and thus plays a critical role in the mechanism of tolerance autoimmunity and in anti-inflammatory processes. Furthermore, TGF-β is an immunoregulatory molecule, which serves initially as an immune response inducer and then acts as an immunosuppressive agent (20). Additionally, TGF-β knockout mice develop inflammation in various tissues (33). Therefore, it is intriguing to observe that among the USF2 target genes, only TGF-β2 shows a significant response to activated mast cells and ectopically introduced Ap4A, a positive regulator of USF2 transcription factor that is elevated upon immunological triggering in mast cells.
It is interesting to note that only specific USF2 target genes are affected (TGF-β2, SHP, TERT, and TSP-1 genes) in a manner similar in both IgE-Ag-triggered and Ap4A-administered mast cells. Mast cells express numerous cell surface molecules and undergo a wide variety of activations. Thus, we assume that the activation of these specific USF2 target genes is specific to IgE-Ag stimulation via Ap4A, whereas other USF2 target genes that were not affected by IgE-Ag and Ap4A are affected by other stimulation and signal transduction. Further studies are needed to determine the specific signal transduction and pathways, which lead to Ap4A synthesis and its role in the USF2 transcriptional activity.
As mentioned above, the TSP-1-dependent TGF-β activity in mesangial cells (44) might be explained by the fact that both these genes are USF2-responsive genes and are elevated upon Ap4A administration, as was shown in the present study. Thus, it is possible that in mesangial cells under specific stimulus, USF2 transcriptional activity increases via Ap4A and thus activates the TSP-1 and TGF-β2 genes. It is interesting to note that in both immunologically triggered and Ap4A-administered mast cells, the increase in transcription levels of the TGF-β2 gene is higher than in the TSP-1 gene (Fig. 5). This observation strengthens the possibility that this unique molecular mechanism plays a more general role in different cell types.
It can be seen that Ap3A, a molecule closely related to Ap4A, has a similar effect on the USF2/Hint protein complex (Fig. 3A) and on USF2-mediated TGF-β2 transcription (Fig. 5B). We have previously observed that exogenously introduced Ap3A to mast cells caused elevation of a specific MITF target gene (c-kit) (18). Furthermore, it was reported that LysRS can synthesize Ap3A when the level of ADP is higher than that of ATP, despite the fact that Ap4A synthesis is the more predominant reaction (4). The effects of Ap3A and Ap4A are controversial in physiological and cellular functions. Some studies show that Ap3A and Ap4A have similar antiapoptotic effects on neutrophils when administered together with granulocyte-macrophage colony-stimulating factor (10). Other studies demonstrate an elevation of Ap3A levels in differentiating HL60 cells, compared to an elevation of Ap4A levels in apoptotic HL60 cells (41). Therefore, further studies are required to evaluate the difference in the roles, if any, played by Ap3A and Ap4A in mast cell signaling and in other mammalian cells.
This study and our previous study (18) both demonstrate that immunological stimulation of RBL leads to elevated levels of Ap4A by LysRS, which is in complex with either USF2 or MITF, enhancing the transcriptional activity of USF2 and MITF. LysRS is involved in protein synthesis; thus, one of the challenging questions raised is what might be the possible mechanism leading to this alternate function of LysRS. In vitro studies show that phosphorylation of aaRS does not effect the aminoacylation reaction but increases up to sixfold in Ap4A production (8). It is possible that phosphorylation of LysRS leads to its association with either USF2 or MITF. In addition, it will be interesting to determine the fraction of LysRS that is bound to either USF2 or MITF. Further studies are needed to understand the mechanism of this receptor induced LysRS activation.
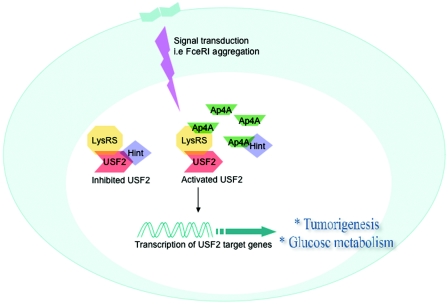
The two bHLH-zip transcription factors, USF2 and MITF, which are activated in immunologically triggered RBL cells, are regulated by an unusual regulatory mechanism. These two transcription factors are suppressed in resting RBL cells by Hint, which prevents them from trans-activating their responsive genes. Interestingly, LysRS, when it is associated with USF2 and MITF, acquires an unconventional function of synthesizing Ap4A in immunologically triggered RBL cells. The local accumulation of Ap4A leads to its binding to Hint, possibly causing conformational changes, and thus dissociating the transcription factors USF2 and MITF from their inhibitor (Fig. 7). Thus, deciphering the involvement of Ap4A in USF2 transcriptional activity may provide new insights into the mechanisms by which transcription factors of the bHLH-zip family are regulated.
FIG. 7.
Proposed model for the general molecular mechanism of USF2 transcriptional activity by LysRS and Ap4A. Following a specific stimulus, LysRS produces Ap4A in close proximity to the multiprotein complex LysRS/USF2/Hint. Ap4A then binds to Hint and liberates USF2, thus allowing USF2 to transactivate its target genes that are involved in physiological processes such as carcinogenesis and diabetes.
Acknowledgments
We thank G. Kay for figure and manuscript preparation. We thank M. Mirande for the Lys-101 CHO cell line and Lys-101 CHO cell lines stably transfected with full-length LysRS and truncated LysRS.
This work was supported by the United States-Israel Binational Science Foundation (grant 2003009; E.R.), the Israeli Academy of Science (grant 144/04; E.R.), and the German-Israeli Foundation for Scientific Research and Development (grant I-726-10.2/2002; E.R.).
REFERENCES
- 1.Adair, G. M., and J. H. Carver. 1979. Unstable, non-mutational expression of resistance to the thymidine analogue, trifluorothymidine in CHO cells. Mutat. Res. 60:207-213. [DOI] [PubMed] [Google Scholar]
- 2.Agou, F., S. Quevillon, P. Kerjan, M. T. Latreille, and M. Mirande. 1996. Functional replacement of hamster lysyl-tRNA synthetase by the yeast enzyme requires cognate amino acid sequences for proper tRNA recognition. Biochemistry 35:15322-15331. [DOI] [PubMed] [Google Scholar]
- 3.Berger, N. A., and E. S. Johnson. 1976. DNA synthesis in permeabilized mouse L cells. Biochim. Biophys. Acta 425:1-17. [DOI] [PubMed] [Google Scholar]
- 4.Blanquet, S., P. Plateau, and A. Brevet. 1983. The role of zinc in 5′,5′-diadenosine tetraphosphate production by aminoacyl-transfer RNA synthetases. Mol. Cell. Biochem. 52:3-11. [DOI] [PubMed] [Google Scholar]
- 5.Brenner, C., H. Cadiou, H. L. Vieira, N. Zamzami, I. Marzo, Z. Xie, B. Leber, D. Andrews, H. Duclohier, J. C. Reed, and G. Kroemer. 2000. Bcl-2 and Bax regulate the channel activity of the mitochondrial adenine nucleotide translocator. Oncogene 19:329-336. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, C., P. Garrison, J. Gilmour, D. Peisach, D. Ringe, G. A. Petsko, and J. M. Lowenstein. 1997. Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins. Nat. Struct. Biol. 4:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brevet, A., P. Plateau, B. Cirakoglu, J. P. Pailliez, and S. Blanquet. 1982. Zinc-dependent synthesis of 5′,5′-diadenosine tetraphosphate by sheep liver lysyl- and phenylalanyl-tRNA synthetases. J. Biol. Chem. 257:14613-14615. [PubMed] [Google Scholar]
- 8.Dang, C. V., and J. A. Traugh. 1989. Phosphorylation of threonyl- and seryl-tRNA synthetase by cAMP-dependent protein kinase. A possible role in the regulation of P1, P4-bis(5′-adenosyl)-tetraphosphate (Ap4A) synthesis. J. Biol. Chem. 264:5861-5865. [PubMed] [Google Scholar]
- 9.Frenkel, S., G. Kay, H. Nechushtan, and E. Razin. 1998. Nuclear translocation of upstream stimulating factor 2 (USF2) in activated mast cells: a possible role in their survival. J. Immunol. 161:2881-2887. [PubMed] [Google Scholar]
- 10.Gasmi, L., A. G. McLennan, and S. W. Edwards. 1996. The diadenosine polyphosphates Ap3A and Ap4A and adenosine triphosphate interact with granulocyte-macrophage colony-stimulating factor to delay neutrophil apoptosis: implications for neutrophil:platelet interactions during inflammation. Blood 87:3442-3449. [PubMed] [Google Scholar]
- 11.Gregor, P. D., M. Sawadogo, and R. G. Roeder. 1990. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 4:1730-1740. [DOI] [PubMed] [Google Scholar]
- 12.Hadsell, D. L., S. Bonnette, J. George, D. Torres, Y. Klementidis, S. Gao, P. M. Haney, J. Summy-Long, M. S. Soloff, A. F. Parlow, M. Sirito, and M. Sawadogo. 2003. Diminished milk synthesis in upstream stimulatory factor 2 null mice is associated with decreased circulating oxytocin and decreased mammary gland expression of eukaryotic initiation factors 4E and 4G. Mol. Endocrinol. 17:2251-2267. [DOI] [PubMed] [Google Scholar]
- 13.Hilderman, R. H., and B. J. Ortwerth. 1987. A preferential role for lysyl-tRNA4 in the synthesis of diadenosine 5′,5‴-P1,P4-tetraphosphate by an arginyl-tRNA synthetase-lysyl-tRNA synthetase complex from rat liver. Biochemistry 26:1586-1591. [DOI] [PubMed] [Google Scholar]
- 14.Ismail, P. M., T. Lu, and M. Sawadogo. 1999. Loss of USF transcriptional activity in breast cancer cell lines. Oncogene 18:5582-5591. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, D. L., and A. Rizzino. 1999. Growth regulatory factors and carcinogenesis: the roles played by transforming growth factor beta, its receptors and signaling pathways. Anticancer Res. 19:4791-4807. [PubMed] [Google Scholar]
- 16.Kingsley-Kallesen, M., T. A. Luster, and A. Rizzino. 2001. Transcriptional regulation of the transforming growth factor-β2 gene in glioblastoma cells. In Vitro Cell. Dev. Biol. Anim. 37:684-690. [DOI] [PubMed] [Google Scholar]
- 17.Korsisaari, N., and T. P. Makela. 2000. Interactions of Cdk7 and Kin28 with Hint/PKCI-1 and Hnt1 histidine triad proteins. J. Biol. Chem. 275:34837-34840. [DOI] [PubMed] [Google Scholar]
- 18.Lee, Y. N., H. Nechushtan, N. Figov, and E. Razin. 2004. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FɛRI-activated mast cells. Immunity 20:145-151. [DOI] [PubMed] [Google Scholar]
- 19.Lemaitre, J. M., R. S. Buckle, and M. Mechali. 1996. c-Myc in the control of cell proliferation and embryonic development. Adv. Cancer Res. 70:95-144. [DOI] [PubMed] [Google Scholar]
- 20.Letterio, J. J., and A. B. Roberts. 1996. Transforming growth factor-β1-deficient mice: identification of isoform-specific activities in vivo. J. Leukoc. Biol. 59:769-774. [DOI] [PubMed] [Google Scholar]
- 21.Levy, C., H. Nechushtan, and E. Razin. 2002. A new role for the STAT3 inhibitor, PIAS3: a repressor of microphthalmia transcription factor. J. Biol. Chem. 277:1962-1966. [DOI] [PubMed] [Google Scholar]
- 22.Lewin, I., J. Jacob-Hirsch, Z. C. Zang, V. Kupershtein, Z. Szallasi, J. Rivera, and E. Razin. 1996. Aggregation of the FcɛRI in mast cells induces the synthesis of Fos-interacting protein and increases its DNA binding-activity: the dependence on protein kinase C-β. J. Biol. Chem. 271:1514-1519. [DOI] [PubMed] [Google Scholar]
- 23.Lima, C. D., M. G. Klein, and W. A. Hendrickson. 1997. Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science 278:286-290. [DOI] [PubMed] [Google Scholar]
- 24.Luo, X., and M. Sawadogo. 1996. Antiproliferative properties of the USF family of helix-loop-helix transcription factors. Proc. Natl. Acad. Sci. USA 93:1308-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinis, S. A., P. Plateau, J. Cavarelli, and C. Florentz. 1999. Aminoacyl-tRNA synthetases: a family of expanding functions. Mittelwihr, France, October 10-15, 1999. EMBO J. 18:4591-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massague, J. 2000. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 1:169-178. [DOI] [PubMed] [Google Scholar]
- 27.Murphy, G. A., D. Halliday, and A. G. McLennan. 2000. The Fhit tumor suppressor protein regulates the intracellular concentration of diadenosine triphosphate but not diadenosine tetraphosphate. Cancer Res. 60:2342-2344. [PubMed] [Google Scholar]
- 28.Nechushtan, H., and E. Razin. 1998. Deciphering the early-response transcription factor networks in mast cells. Immunol. Today 19:441-444. [DOI] [PubMed] [Google Scholar]
- 29.Otani, A., B. M. Slike, M. I. Dorrell, J. Hood, K. Kinder, K. L. Ewalt, D. Cheresh, P. Schimmel, and M. Friedlander. 2002. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc. Natl. Acad. Sci. USA 99:178-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randerath, K., C. M. Janeway, M. L. Stephenson, and P. C. Zamecnik. 1966. Isolation and characterization of dinucleoside tetra- and tri-phosphates formed in the presence of lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 24:98-105. [DOI] [PubMed] [Google Scholar]
- 31.Razin, E., Z. C. Zhang, H. Nechushtan, S. Frenkel, Y. N. Lee, R. Arudchandran, and J. Rivera. 1999. Suppression of microphthalmia transcriptional activity by its association with protein kinase C-interacting protein 1 in mast cells. J. Biol. Chem. 274:34272-34276. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, A. B., and M. B. Sporn. 1993. Physiological actions and clinical applications of transforming growth factor-beta (TGF-beta). Growth Factors 8:1-9. [DOI] [PubMed] [Google Scholar]
- 33.Shull, M. M., I. Ormsby, A. B. Kier, S. Pawlowski, R. J. Diebold, M. Yin, R. Allen, C. Sidman, G. Proetzel, D. Calvin, et al. 1992. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature 359:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirito, M., Q. Lin, J. M. Deng, R. R. Behringer, and M. Sawadogo. 1998. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc. Natl. Acad. Sci. USA 95:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirito, M., Q. Lin, T. Maity, and M. Sawadogo. 1994. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 22:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sirito, M., S. Walker, Q. Lin, M. T. Koslowski, W. H. Klein, and M. Sawadogo. 1992. Member of the USF family of helix-loop-helix proteins bind DNA as homo- as well as heterodimers. Gene Expr. 2:231-240. [PMC free article] [PubMed] [Google Scholar]
- 37.Su, T., M. Suzui, L. Wang, C. S. Lin, W. Q. Xing, and I. B. Weinstein. 2003. Deletion of histidine triad nucleotide-binding protein 1/PKC-interacting protein in mice enhances cell growth and carcinogenesis. Proc. Natl. Acad. Sci. USA 100:7824-7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szentirmay, M. N., H. X. Yang, S. A. Pawar, C. Vinson, and M. Sawadogo. 2003. The IGF2 receptor is a USF2-specific target in nontumorigenic mammary epithelial cells but not in breast cancer cells. J. Biol. Chem. 278:37231-37240. [DOI] [PubMed] [Google Scholar]
- 39.Vallet, V. S., M. Casado, A. A. Henrion, D. Bucchini, M. Raymondjean, A. Kahn, and S. Vaulont. 1998. Differential roles of upstream stimulatory factors 1 and 2 in the transcriptional response of liver genes to glucose. J. Biol. Chem. 273:20175-20179. [DOI] [PubMed] [Google Scholar]
- 40.Vartanian, A., I. Alexandrov, I. Prudowski, A. McLennan, and L. Kisselev. 1999. Ap4A induces apoptosis in human cultured cells. FEBS Lett. 456:175-180. [DOI] [PubMed] [Google Scholar]
- 41.Vartanian, A., I. Prudovsky, H. Suzuki, I. Dal Pra, and L. Kisselev. 1997. Opposite effects of cell differentiation and apoptosis on Ap3A/Ap4A ratio in human cell cultures. FEBS Lett. 415:160-162. [DOI] [PubMed] [Google Scholar]
- 42.Wakasugi, K., and P. Schimmel. 1999. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 284:147-151. [DOI] [PubMed] [Google Scholar]
- 43.Wakasugi, K., B. M. Slike, J. Hood, A. Otani, K. L. Ewalt, M. Friedlander, D. A. Cheresh, and P. Schimmel. 2002. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. USA 99:173-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, S., J. Skorczewski, X. Feng, L. Mei, and J. E. Murphy-Ullrich. 2004. Glucose up-regulates thrombospondin 1 gene transcription and transforming growth factor-beta activity through antagonism of cGMP-dependent protein kinase repression via upstream stimulatory factor 2. J. Biol. Chem. 279:34311-34322. [DOI] [PubMed] [Google Scholar]
- 45.Wu, K. J., M. Mattioli, H. C. Morse III, and R. Dalla-Favera. 2002. c-MYC activates protein kinase A (PKA) by direct transcriptional activation of the PKA catalytic subunit beta (PKA-Cβ) gene. Oncogene 21:7872-7882. [DOI] [PubMed] [Google Scholar]
- 46.Zamecnik, P. C. 1969. An historical account of protein synthesis, with current overtones—a personalized view. Cold Spring Harb. Symp. Quant. Biol. 34:1-16. [DOI] [PubMed] [Google Scholar]
- 47.Zamecnik, P. C., M. L. Stephenson, C. M. Janeway, and K. Randerath. 1966. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 24:91-97. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Z. C., H. Nechushtan, J. Jacob-Hirsch, D. Avni, O. Meyuhas, and E. Razin. 1998. Growth-dependent and PKC-mediated translational regulation of the upstream stimulating factor-2 (USF2) mRNA in hematopoietic cells. Oncogene 16:763-769. [DOI] [PubMed] [Google Scholar]