Abstract
Cytokine responses can be regulated by a family of proteins termed suppressors of cytokine signaling (SOCS) which can inhibit the JAK/STAT pathway in a classical negative-feedback manner. While the SOCS are thought to target signaling intermediates for degradation, relatively little is known about how their turnover is regulated. Unlike other SOCS family members, we find that SOCS2 can enhance interleukin-2 (IL-2)- and IL-3-induced STAT phosphorylation following and potentiate proliferation in response to cytokine stimulation. As a clear mechanism for these effects, we demonstrate that expression of SOCS2 results in marked proteasome-dependent reduction of SOCS3 and SOCS1 protein expression. Furthermore, we provide evidence that this degradation is dependent on the presence of an intact SOCS box and that the loss of SOCS3 is enhanced by coexpression of elongin B/C. This suggests that SOCS2 can bind to SOCS3 and elongin B/C to form an E3 ligase complex resulting in the degradation of SOCS3. Therefore, SOCS2 can enhance cytokine responses by accelerating proteasome-dependent turnover of SOCS3, suggesting a mechanism for the gigantism observed in SOCS2 transgenic mice.
Cytokines such as interleukin-2 (IL-2) regulate the immune response via interaction with cell surface receptors on target cells. These receptors interact with cytoplasmic tyrosine kinases, specifically, members of the Janus kinase (JAK) family, which subsequently phosphorylate signal transducer and activator of transcription (STAT) proteins. Phosphorylation of STATs results in their dimerization and translocation to the nucleus and subsequent transcriptional activation of genes important for proliferation and differentiation (11). Inhibition of these signaling pathways is crucial for the control of the inflammatory response (16). The suppressors of cytokine signaling (SOCS/SSI/CIS) are thought to play a key role in this process and are upregulated by and inhibit the JAK/STAT pathway in a classic negative-feedback manner (7, 32, 39). Eight SOCS family proteins have been described, CIS (cytokine-inducible SH2 domain-containing protein) and SOCS1 to SOCS7 (10, 22, 29). These proteins are characterized by two common structural motifs, an SH2 domain and a C-terminal SOCS box. The SOCS box is thought to interact with elongin B/C, part of an E3 ubiquitin ligase complex that targets associated proteins for degradation through the ubiquitin pathway (27). As well as SOCS, a number of other protein subfamilies including the von Hippel-Lindau (VHL) tumor suppressor protein contain this SOCS box motif, indicating that it may have an important and conserved role (17). The SOCS box of VHL associates with an E3 ligase complex and induces the proteasomal degradation of hypoxia-inducible factor 1α (40). More recently, Asb, an adipocyte-specific ankyrin and SOCS box-containing protein, has been shown to interact with the adaptor protein APS (adapter protein with PH and SH2 domain) to enable recruitment of elongin B/C to the insulin receptor (43).
SOCS1 and SOCS3 are induced rapidly by a range of cytokines including prolactin (PRL), growth hormone (GH), leukemia inhibitory factor, IL-2, and IL-6 and act to negatively regulate the strength and duration of cytokine responsiveness (6, 45). SOCS1 and SOCS3 achieve this by binding to phosphorylated JAKs within the activation loop, thereby blocking kinase activity (44). SOCS1 is also thought to target JAK2 for proteasomal degradation by recruiting an elongin-containing Skp-cullin-F-box-like (SCF) E3 ligase complex to the phosphorylated JAK (41), a degradative mechanism similar to that reported for the TEL-JAK2 oncogene (translocation erythroblast transform leukemia-JAK2 fusion protein) (9, 23).
SOCS2 can inhibit GH and PRL signaling when expressed at very low levels. However, higher concentrations of SOCS2 restored responsiveness to these growth factors, perhaps by antagonizing the inhibitory effect of SOCS1 (5, 8, 34). This dual role of SOCS2 is reinforced by the observation that both knockout (31) and transgenic (12) SOCS2 mice displayed increased organ mass and excess weight gain and are therefore substantially (30%) larger than their wild-type littermates. These observations suggest that SOCS2 may have a dual suppressive and stimulating effect depending on its concentration in the cell (8, 12). Although the biochemical mechanism by which SOCS2 suppresses GH signaling remains unknown, the fact that SOCS2−/− mice are phenotypically similar to GH and IGF-1 transgenic mice supports the idea that SOCS2 may act to negatively regulate growth-promoting cytokines (18, 28, 34, 43).
Here, we investigate the ability of SOCS2 to enhance cytokine signaling and focus specifically on the effect of SOCS2 on the expression on other SOCS proteins. We show that SOCS2 can enhance IL-2- and IL-3-induced STAT phosphorylation and that while both SOCS2 and SOCS3 are induced following IL-2 treatment, SOCS3 was induced earlier and disappeared when SOCS2 was present. Expression of SOCS2 markedly reduced SOCS3 protein levels in response to cytokines, while SOCS3 mRNA remained unaffected. Therefore, SOCS2 can regulate SOCS3 protein expression in a proteasome-dependent manner and thus potentiate signaling by inducing degradation of other SOCS proteins.
MATERIALS AND METHODS
Antibodies.
Polyclonal rabbit anti-SOCS2 and -SOCS3 antibodies were supplied by Fusion Antibodies (Belfast, United Kingdom). FLAG M2 monoclonal antibody was purchased from Sigma Aldrich (Poole, United Kingdom). Anti-phosphotyrosine clone 4G10 was purchased from Upstate Biotechnology Inc. (Lake Placid, NY). Polyclonal rabbit anti-STAT5b was a generous gift of J. J. O'Shea (NIH, Bethesda, MD). Polyclonal anti-phospho-STAT3 was purchased from New England Biolabs (Hertfordshire, United Kingdom), and polyclonal anti-STAT3 was purchase from Santa Cruz Biotechnologies (Santa Cruz, CA). Monoclonal anti-phospho-extracellular signal-regulated kinase (ERK) and anti-ERK were purchased from Cell Signaling (Beverley, MA), and polyclonal anti-IL-2Rβ was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal anti-Myc and anti-His were purchased from Sigma Aldrich (Dorset, United Kingdom)
Quantitative real-time PCRs.
Quantitative PCR analysis was performed using the TaqMan Universal PCR Master Mix kit (Applied Biosystems, Foster City, CA). The probes were labeled with a reporter fluorescent dye, VIC, at the 5′ end and with a quencher fluorescent dye, 6-carboxytetramethylrhodamine, at the 3′ end. The resulting relative increase in reporter fluorescent dye emission was monitored in real time during PCR amplification using a sequence detection system (ABI Prism 7900 HT Sequence Detection; Applied Biosystems, Foster City, CA). Actin was used as an internal standard. The relative expression of target mRNA was computed from the target cycle threshold values and the actin cycle threshold value using the standard curve method (User Bulletin 2; Applied Biosystems, Foster City, CA). The following TaqMan forward (F) and reverse (R) primers and probes (P) were used (the nucleotide position for each open reading frame is given in parentheses: for SOCS2, (99)GGCGCGTCTGGCGA (F), (166)TAACAGTCATACTTCCCCAGTACCAT (R), and (118)CCCTGCGGGAGCTCGGTCAGA (P); for CIS, ATCTGCTGTGCATAGCCAAGAC (F), CGTAATGGAACCCCAATACCA (R), and AGCTCCCTTGTGTCCGTTTCCTGCC (P); for SOCS1, (448)GCGACTACCTGAGCTCCTTCC (F), (682)AACACGGCATCCCAGTTAATG (R), and (625)TCCAGATTTGACCGGCAGCGC (P); for SOCS3, (75)CAGCTCCAAGAGCGAGTACCA (F), (139)AGAAGCCGCTCTCCTGCAG (R), and (118)TGCGCACTGCGTTCACCACCA (P).
RT-PCR analysis.
RNA samples were extracted from 10 × 106 Ba/F3-SOCS2 cells using Stat60 reagent (Tel-Test Inc., Friendswood, Texas). Reverse transcription-PCR (RT-PCR) was carried out using the One Step RT-PCR system (QIAGEN, Crawley, United Kingdom). The forward and reverse primers used to amplify mouse cDNA are as follows: for CIS, (200)CTGGACTCTTAACTGCTTGTC (F) and (576)TAGGCAGCACCGAGTCAC (R); for SOCS1, (328)AACTGCTTTTTCGCCCTTAGC (F) and (390)AAAGTGCACGCGGATGCT (R); for SOCS2, (265)TCAGCTGGACCGACTAATCT (F) and (402)CAGGTGAACAGTCCCATTCC (R); for SOCS3, (77)GCTCCAAAAGCGAGTACCAG (F) and (286)GGATGCGTAGGTTCTTGGTC (R); for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), (58)CCACATCGCTCAGACACCAT (F) and (115)TGACCAGGCGCCCAAT (R); for Pim-1 (6536)CCCGAGCTATTGAAGTCTGA (F) and (6900)CTGTGCAGATGGATCTCAGA (R); for BCL-xL, (26)TGGTCGACTTTCTCTCCTAC (F) and (580)GAGATCCACAAAAGTGTCCC (R); for OSM, (99)GCTGCTCCAACTCTTCCTC (F) and (725)GACCCAGATTCTGCGGGTTC (R); for β-actin, (663)CATCACTATTGGCAACGAGC (F) and (1067)ACGCAGCTCAGTAACAGTCC (R).
Constructs.
SOCS2 and SOCS3 cDNAs were tagged with the FLAG epitope at their C termini by standard PCR-based methods. Each cDNA was amplified by PCR using a 5′ oligonucleotide containing an EcoRI site, an ATG codon, and a 3′ oligonucleotide containing a ClaI restriction site. The EcoRI/ClaI PCR fragment was subcloned between the EcoRI and ClaI sites of a modified pME18S vector in frame with the FLAG epitope. The pUHD plasmids expressing SOCS2 and SOCS3 were constructed by subcloning the human SOCS2 or SOCS3 FLAG cDNA. SOCS2 dSB, in which the consensus elongin B/C binding region was disrupted, contained the following mutations: L163A, C167Q, and I171Q. This construct was made using the QuikChange Multi site-directed mutagenesis kit (Stratagene, CA). Plasmids expressing elongin B and C were a kind gift from D. Hilton (WEHI, Melbourne, Australia).
Cells and transfections.
Peripheral blood mononuclear cells (PBMCs) were purified by Ficoll density gradient centrifugation from consenting healthy donors. Isolated PBMCs were plated in 100 ml of RPMI growth medium containing 10% fetal calf serum (FCS) in 175-cm2 flasks for 90 min to allow contaminating monocyte/macrophage to adhere. T lymphocytes were removed by pipetting RPMI growth medium over the base of the flask five times. The cells were activated for 72 h in 100 ml of 10% FCS RPMI growth medium (Gibco-BRL) supplemented with 2 mM glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, and 1.25 μg/ml phytohemagglutinin (PHA) (mitogenic stimulus that triggers synthesis of T lymphocytes) at 37°C in 5% CO2 and 100% humidity. Prior to stimulation with cytokine, the cells were washed and incubated for 12 h in 2% FCS RPMI medium at 37°C in 5% CO2 and 100% humidity.
The IL-3-dependent pro-B-cell line Ba/F3 cells expressing the TA transactivation repressor (Ba/F3TA) were maintained in RPMI medium supplemented with 10% FCS, 2 mM glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, 200 μg/ml puromycin, and 10 U/ml IL-3. SOCS2 and SOCS3 stable transfectants were generated by electroporation of Ba/F3TA (5 × 106 cells/ml) with pUHD10-3 containing the relevant SOCS using a Bio-Rad GenePulser (260 V, 960 mF) and selected using 1.25 μg/ml hygromycin. Cells were maintained in 4 μg/ml tetracycline and were removed from tetracycline 48 h prior to stimulation to allow gene expression. Transfections were performed in 293T cells grown to 60% confluence on p90 tissue culture plates in 5 ml of Dulbecco's modified Eagle's medium (Gibco-BRL) with 10% FCS using Fugene (Lewes, United Kingdom) transfection reagents according to the manufacturer's instructions. The 293T cells were transfected with 1 to 5 μg of either wild-type (WT) or SOCS box deletion plasmids in the presence of 1 μg of SOCS3 plasmid. After 24 h, the transfection medium was replaced with fresh medium. Cells were harvested 24 h later as outlined above.
Immunoprecipitation and immunoblotting.
Ba/F3 cells were maintained in RPMI medium supplemented with 5% FCS and 10 U/ml IL-3. Thereafter, the cells were collected, washed twice, and resuspended in RPMI medium containing 2% FCS. After 12 h of incubation, the cells were stimulated with 100 U/ml of IL-3 for different periods. Following treatment, cells were washed in 1× phosphate-buffered saline (PBS). If phosphorylation of the protein was being assessed, the 1× PBS was supplemented with Na3VO4 (0.2 mM) and lysed by adding a buffer composed of 0.875% (vol/vol) Brij 97, 50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM Na3V04, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride. After 15 min, cell lysates were clarified by centrifugation (13,400 × g), and the supernatants were subjected to immunoprecipitation. This was performed by exposure to either SOCS3 polyclonal antibody (1 μg) or STAT5b polyclonal antibody (1 μg) that was precoupled to protein A-Sepharose beads. After 1.5 h of incubation, the beads were subsequently collected by centrifugation and washed three times in the lysis buffer. The beads were then resuspended in Laemmli sample buffer containing 5% β-mercaptoethanol and boiled under reducing conditions for 5 min. For immunoblot analysis, the proteins were subjected to electrophoresis by 7.5% or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyscreen polyvinylidene difluoride transfer membranes. The membranes were blocked in PBS supplemented with 0.2% Tween 20 and either 3% bovine serum albumin or 5% Marvel milk and then incubated for 1 h (or overnight at 4°C) with a primary antibody (1:500 dilution of the anti-SOCS2 antibody, 1:1,000 dilution of anti-SOCS3, 1:1,000 dilution of anti-STAT3, 1:1,000 dilution of anti-ERK1/2, 1:1,000 dilution of anti-Myc, 1:1,000 dilution of anti-STAT5b, 1:5,000 dilution of the antiphosphotyrosine antibody, 1:2,000 dilution of anti-FLAG and phospho-STAT3 antibodies, and 1:3,000 dilution of anti-His). Thereafter, the membranes were washed three times for 15 min in PBS supplemented with 0.2% Tween and were subsequently incubated for 1 h with peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G (1:10,000) in PBS supplemented with 0.2% Tween 20 and 3% bovine serum albumin or 5% milk. The blots were extensively washed, and antibody binding was visualized by enhanced chemiluminescence (ECL; Amersham Biosciences, Chalfont, St. Giles, United Kingdom).
Interaction between SOCS2 and SOCS3.
For fusion protein pull-down experiments, 293T cells were transfected as described above. Cells were lysed in buffer containing 50 mM Tris-HCl, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM Na3V04, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride. Lysates were centrifuged at 13,400 × g for 10 minutes at 4°C, and supernatants were incubated with 3 μg of His-tagged SOCS2 fusion protein (Fusion Antibodies, Belfast, United Kingdom) which had been precoupled to 50 μl of 20% nickel-nitrilotriacetic acid beads (Promega, Madison, WI). Reaction mixtures were incubated for 2 to 4 h at 4°C in protein interaction buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole), and precipitates were washed with protein interaction buffer. Beads were resuspended in Laemmli sample buffer containing 5% β-mercaptoethanol and analyzed by SDS-PAGE as described above.
The endogenous association between SOCS2 and SOCS3 was analyzed in Ba/F3-SOCS2 cells grown in the presence and absence of tetracycline for 48 h. Cells were incubated overnight in 2% FCS and then stimulated with 100 U/ml IL-3. Proteasome inhibitors MG132 (0.5 μM) and LLnL (0.5 μM) were added 1 h before lysing. Cell lysates were immunoblotted with anti-FLAG or immunoprecipitated and immunoblotted with a SOCS3 antibody or anti-FLAG as described previously.
Trypan blue exclusion assay.
Ba/F3-SOCS2 cells were seeded at a density of 1 × 105 cells per ml and grown in RPMI medium containing 5% FCS and 10 U/ml IL-3 in the presence or absence of tetracycline (4 μg/ml). Every 12 h, 100-μl aliquots of cells were incubated at a 50:50 ratio with 0.1% trypan blue for 5 min, after which viable cells (able to exclude trypan blue) were counted using an hematocytometer (Invitrogen, Paisley, United Kingdom). Cell counts were carried out in triplicate.
MTT assay.
Ba/F3-SOCS2 cells were seeded at a density of 1 × 105 cells per ml and grown in a 96-well plate in RPMI medium supplemented with 5% FCS and 10 ng/ml IL-3 in the presence or absence of tetracycline (4 μg/ml). After 24 h and 48 h, 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide (MTT; 0.5 mg/ml) was added to a 100-μl cell aliquot and incubated at 37°C. After 2 h, the cell suspension was placed in a 1.5-ml Eppendorf tube, and the cells were pelleted by low-speed centrifugation (300 × g for 3 min). The supernatant was discarded using a Hamilton syringe, and 200 μl dimethyl sulfoxide was added to the cell pellet. Cells were mixed, placed in a 96-well plate, and incubated for a further 10 min at 37°C. The optical density of each sample was read at 570 nm using a microplate reader.
RESULTS
SOCS2 is highly expressed in T cells.
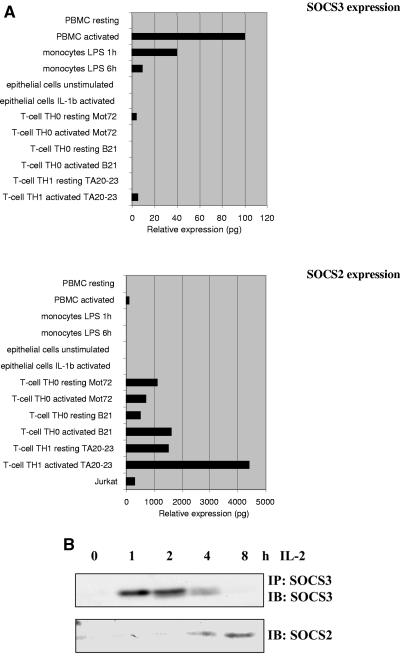
Although SOCS1 and SOCS3 are known to be expressed in many leukocyte lineages, the abundance of SOCS2 expression in the immune system has not been explored. To address this, a panel of cDNA libraries from various immune and nonimmune tissues was analyzed for expression of the SOCS mRNA. The copy number of individual genes within the cDNA libraries should not be altered, as they had not been amplified. SOCS3 was expressed in a number of libraries tested, appearing predominantly in cells of the monocyte lineage and PBMCs (Fig. 1A). SOCS3 expression was increased in some T-cell populations by activation with IL-2 ± anti-CD3 or PHA (data not shown). Expression of SOCS3 in activated PBMCs was confirmed by Northern blot analysis (data not shown). In contrast, expression of SOCS2 was more restricted to T cells and again particularly evident in activated T cells of both T-helper 1 (Th1) and T-helper 0 (Th0) lineages (Fig. 1A). Moreover, the relative levels of SOCS2 expression were over 3 logs higher in T-cell lineages than in other cells examined, suggesting that it may have an important function in regulating T-cell activation.
FIG. 1.
(A) Analysis of SOCS expression. A panel of cDNA libraries from different cell types were analyzed for SOCS expression by real-time PCR using specific primers for SOCS2 and SOCS3 as described in Materials and Methods. The abbreviations used in the figure are as follows: Th0, naive CD4+ T cells; B21 and T2023, Th1 clones; Mot72, CD4+ MEL14 naive T cells; LPS, lipopolysaccharide. (B) IL-2-induced expression of SOCS2 and SOCS3 proteins in PBMCs. PBMCs were PHA blasted for 72 h, incubated for 12 h in 2% FCS, and stimulated with 100 U/ml IL-2 for the indicated periods. Lysates were immunoblotted (IB) with anti-SOCS2 (top) or immunoprecipitated (IP) and immunoblotted with anti-SOCS3 (bottom).
SOCS2 and SOCS3 expression is reciprocally regulated in T cells.
In light of the real-time PCR data, SOCS protein expression in primary human PBMCs was analyzed. Treatment of PBMCs with PHA induced SOCS expression, but incubation in 2% FCS for 12 h reduced this to background levels. SOCS3 expression was observed 1 h following IL-2 stimulation and was greatly reduced at 4 h (Fig. 1B). In contrast, SOCS2 was induced at 4 h with a substantial increase in protein expression between 4 h and 8 h. A similar pattern of expression was observed in all samples examined, suggesting that it was not donor specific. These observations indicated that SOCS2 and SOCS3 were induced in T cells by IL-2 but with different kinetics, with SOCS3 being detectable much earlier and transiently and SOCS2 appearing later and for a more sustained period. Interestingly, SOCS3 expression was rapidly reduced to background levels following the appearance of SOCS2, suggesting that these proteins may be reciprocally regulated. These findings imply that SOCS3 plays an early role in regulating T-cell responses to IL-2 prior to SOCS2 induction and that SOCS2 may have a role later in the response.
SOCS2 potentiates cytokine-induced STAT5 phosphorylation.
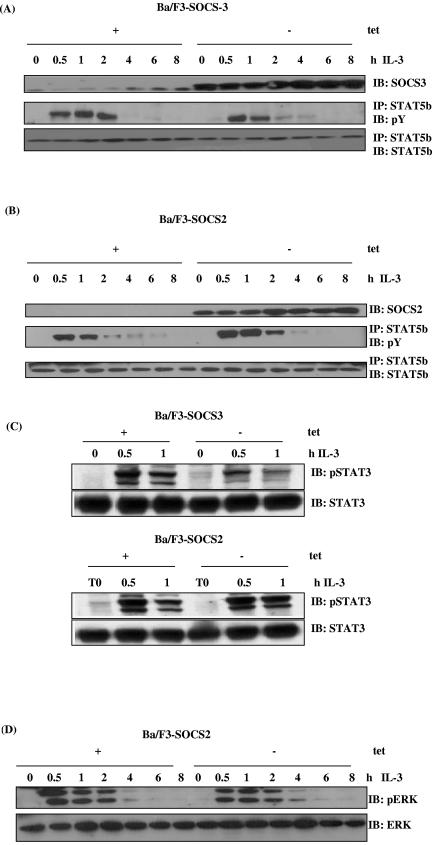
We next investigated whether SOCS2 could regulate IL-2- and IL-3-induced STAT5 activation. Ba/F3 cells expressing either SOCS2 or SOCS3 under the control of a tetracycline-inducible promoter were incubated overnight in 2% FCS followed by stimulation with IL-3 for the time course shown. Figure 2A (top) demonstrates the expression of SOCS3 when tetracycline was removed from the cells. To analyze whether STAT5b was phosphorylated upon stimulation with IL-3, lysates were immunoprecipitated with a STAT5b antibody and blotted with anti-phosphotyrosine. When cells were stimulated with IL-3, elevated levels of STAT5b tyrosine phosphorylation were observed between 0.5 h and 2 h (Fig. 2A, middle). Expression of SOCS3 decreased the level of STAT5 phosphorylation at 0.5 h and 1 h, which was almost undetectable 2 h following IL-3 treatment. In contrast, expression of SOCS2 increased and prolonged the level of STAT5 tyrosine phosphorylation following IL-3 treatment, with STAT5 phosphorylation being more pronounced 2 h following cytokine treatment (Fig. 2B, middle). The data suggest that SOCS2 could enhance STAT5 phosphorylation in response to IL-3. We therefore investigated whether other signaling responses, such as STAT3 phosphorylation, are potentiated in the presence of SOCS2 (Fig. 2C). To verify this, Ba/F3-SOCS2 and Ba/F3-SOCS3 cells were incubated as before and stimulated with IL-3 for the period shown. Again, expression of SOCS3 clearly inhibited phosphorylation of STAT3 at 0.5 h and 1 h (Fig. 2C, top), whereas expression of SOCS2 enabled the levels of phosphorylation to be maintained (Fig. 2C, bottom right).
FIG. 2.
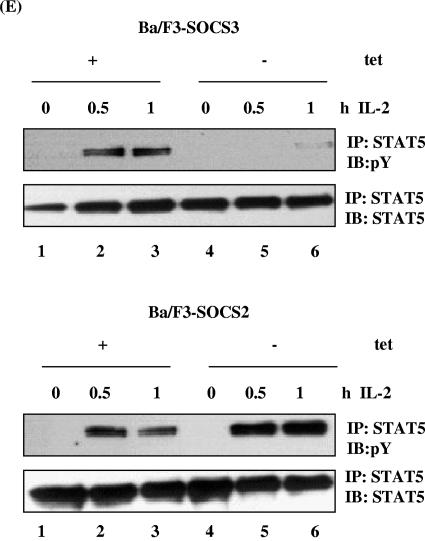
SOCS2 and SOCS3 have reciprocal effects on cytokine-induced STAT5 phosphorylation. Ba/F3 cells lacking SOCS3 (A) or SOCS2 (B) (with tetracycline [+tet]) or in which expression of SOCS3 (A) or SOCS2 (B) was induced by removal from tetracycline (−tet) were incubated for 12 h in 2% FCS before stimulation with IL-3 for the periods shown. Lysates (WCLs) were immunoblotted (IB) with anti-SOCS3 (A, top) or anti-SOCS2 (B, top). Alternatively, lysates were immunoprecipitated (IP) with anti-STAT5b and immunoblotted for anti-phosphotyrosine (A and B, middle). STAT5b levels were checked using a STAT5b antibody (A and B, bottom). (C) Ba/F3 cells treated as described above were stimulated with IL-3 as indicated, and lysates were immunoblotted with a phospho-STAT3 antibody (top). STAT3 levels were verified by immunoblotting with an anti-STAT3 antibody (bottom). (D) Lysates from Ba/F3-SOCS2 cells were treated as described above and immunoblotted with phospho-ERK monoclonal antibody, and protein levels were confirmed by immunoblotting with anti-ERK. (E) Ba/F3 cells which stably express the IL-2 receptor with SOCS2 (bottom) or SOCS3 (top) were stimulated with 100 U/ml IL-2 as indicated, and lysates were immunoprecipitated with anti-STAT5b and immunoblotted with antiphosphotyrosine. STAT5 levels were verified using an anti-STAT5b.
Similarly, we tested the effect of SOCS2 expression on cytokine-induced ERK phosphorylation. Lysates from Ba/F3-SOCS2 cells were directly immunoblotted with anti-phospho-ERK. SOCS2 had little effect on the phosphorylation of ERK in response to IL-3 (Fig. 2D), suggesting that the target of SOCS2 may be limited to the JAK/STAT pathway.
Signaling in response to IL-2 was also investigated in Ba/F3 cells that stably express the IL-2 receptor (Fig. 2E). In the absence of SOCS3, levels of phosphorylation increased at 0.5 h and were still observed 1 h following IL-2 treatment. Again, SOCS3 expression strongly inhibited STAT5 tyrosine phosphorylation following IL-2 stimulation (Fig. 2E, top), whereas expression of SOCS2 prevented this inhibition (Fig. 2E, bottom). We also observed an increase in IL-2 receptor β-chain phosphorylation in the presence of SOCS2 (data not shown). Overall, these data suggest that SOCS2 can potentiate IL-2- and IL-3-mediated responses (5, 8).
SOCS2 augments cytokine-induced proliferation.
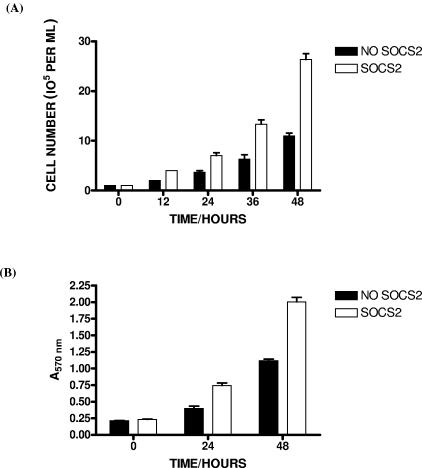
These findings raised the possibility that SOCS2 expression may also affect proliferation in response to these cytokines. To examine this, Ba/F3-SOCS2 cells were seeded at 1 × 105 cells per ml in the presence and absence of tetracycline and cultured in RPMI 1640 medium supplemented with 5% FCS and 10 U/ml of IL-3. Samples were taken every 12 h over 48 h, and numbers of viable cells were determined using the trypan blue exclusion assay. At 48 h, cells expressing SOCS2 proliferated at double the rate of controls (Fig. 3A). Results were taken in triplicate and are representative of three separate experiments. This result demonstrates that SOCS2 can enhance cytokine-induced proliferation following IL-3 stimulation. To confirm this observation, proliferation was also examined in Ba/F3-SOCS2 cells using the MTT assay. Again, cells expressing SOCS2 proliferated at almost double the rate of the control cells when they were cytokine stimulated (Fig. 3B). These results confirm that SOCS2 can enhance IL-3-induced proliferation, in contrast to other members of the SOCS family, and support the observation that SOCS2 can promote lymphokine signaling and proliferative responses as has been reported for GH and PRL (8, 33).
FIG. 3.
Effect of SOCS2 on cell proliferation. (A) Ba/F3-SOCS2 cells seeded at 1 × 105 cells/ml were cultured with tetracycline (no SOCS2, black box) or without tetracycline (SOCS2, white box). Viability was determined at 12-h intervals by trypan blue exclusion assay. (B) Ba/F-SOCS2 cells were seeded at 1 × 105 cells/ml and grown in the presence or absence of 4 μg/ml tetracycline (bars are as described above). At 24 h and 48 h, 10 μl of MTT (0.5 mg/ml) was added to 100 μl of cell culture and incubated at 37°C for 2 h. The cells were centrifuged (300 × g for 3 min), and the supernatant was removed. Dimethyl sulfoxide (200 μl) was added and incubated for 10 min at 37°C. Plates were read at an optical density at 570 nm using a microplate reader.
SOCS2 drives SOCS3 degradation by a proteasome-dependent mechanism.
Given that SOCS2 and SOCS3 are induced under similar conditions in T cells, but with different kinetics, we can assume that SOCS3 functions prior to SOCS2 in the cytokine response. Since these proteins are reciprocally regulated in T cells, we investigated whether SOCS2 may enhance signaling by regulating SOCS3 expression.
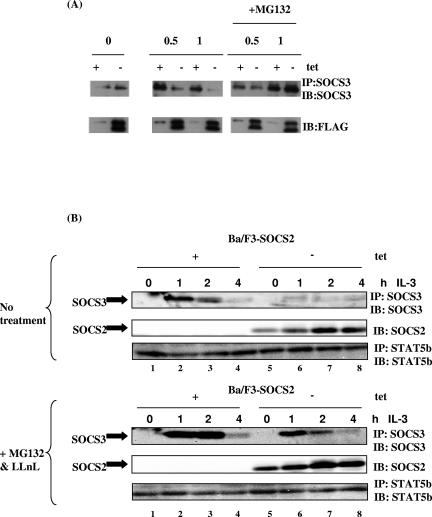
To investigate the effects of SOCS2 on endogenous SOCS3, Ba/F3-SOCS2 cells were used. In these cells, endogenous SOCS3 is rapidly induced upon IL-3 stimulation. Ba/F3-SOCS2 cells were cultured for 48 h in the presence or absence of tetracycline before being incubated for 12 h in RPMI medium supplemented with 5% FCS. The cells were then stimulated for 0.5 h and 1 h with 100 U/ml IL-3 (Fig. 4A). To examine endogenous levels of SOCS3, the lysates were immunoprecipitated with anti-SOCS3 and analyzed by Western blot, while SOCS2 protein levels were determined from whole-cell lysates (WCLs). Figure 4A (bottom) shows the expression of SOCS2 following tetracycline withdrawal. SOCS3 was induced 0.5 h and still expressed 1 h after IL-3 stimulation in the absence of SOCS2 (Fig. 4A, top, lanes 3 and 5), and SOCS3 protein levels were markedly reduced in the presence of SOCS2 (lanes 4 and 6). The addition of the proteasome inhibitor MG132 restored SOCS3 protein levels (Fig. 4A, lanes 8 and 10). In Fig. 4B, a similar experiment was performed over a longer time course. In this case, cells were stimulated for 1 h, 2 h, and 4 h in the presence (Fig. 4B, bottom) or absence (top) of the proteasome inhibitors MG132 and LLnL. Again, in the absence of SOCS2, SOCS3 was induced at 1 h following IL-3 stimulation (Fig. 4B, lane 2). This was sustained until 2 h (Fig. 4B, lane 3) and returned to background levels at 4 h (lane 4). In contrast, when SOCS2 protein was expressed (Fig. 4B, lanes 5 to 8), the levels of endogenous SOCS3 were markedly decreased, being almost undetectable. This implies that the presence of SOCS2 causes the downregulation of endogenous SOCS3. To further investigate the turnover of SOCS3 by SOCS2, cells were also treated with the proteasome inhibitors for 30 min prior to lysing (Fig. 4B, bottom). The lysates were immunoprecipitated and immunoblotted as described above. The addition of MG132 and LLnL increased SOCS3 levels, such that expression was still detectable after 4 h, suggesting that the transient expression was due to rapid degradation of SOCS3 by the proteasome. In the presence of SOCS2 and proteasome inhibitors, SOCS3 protein expression was rescued (Fig. 4B, bottom, lanes 5 to 8). These results indicated that SOCS2 accelerates the degradation of SOCS3 in a proteasome-dependent manner.
FIG. 4.
Effects of SOCS2 on SOCS3 protein expression. (A) Ba/F3 cells in which SOCS2 expression was controlled by the removal of tetracycline were treated as described in the text. Cells were stimulated with 100 U/ml IL-3 for the indicated times. Lysates were immunoprecipitated (IP) and immunoblotted (IB) with a SOCS3 antibody (top). Alternatively, lysates were subjected to immunoblotting with M2-FLAG antibody (bottom). (B) Ba/F3-SOCS2 cells were stimulated as described above but treated with the proteasome inhibitors MG132 (0.5 μM) and LLnL (0.5 μM). Lysates were immunoprecipitated and immunoblotted with anti-SOCS3 (top). WCLs were immunoblotted with SOCS2 antibody.
Expression of SOCS2 enhanced SOCS mRNA induced by cytokines.
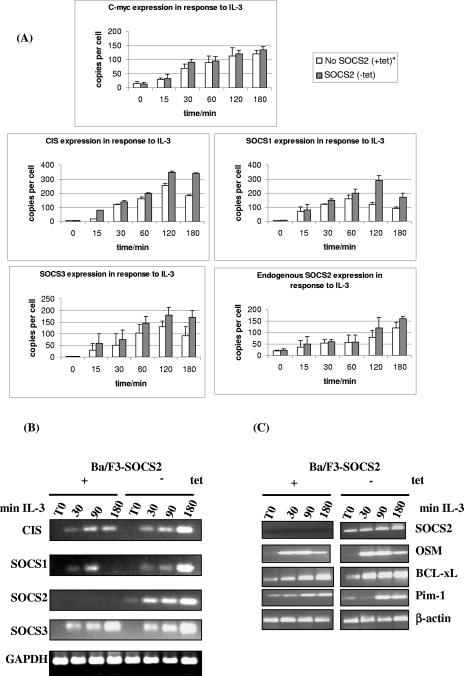
If SOCS2 accelerates the turnover of SOCS3, the prediction would be that there would be increased activity in the signaling systems leading to more SOCS expression. To investigate this, real-time PCR probes were designed to measure levels of mRNA for the SOCS genes and c-myc as a control in Ba/F3-SOCS2 cells (Fig. 5A). Cells were stimulated with IL-3 as described above, and mRNA was extracted and analyzed using real-time PCR. The patterns of mRNA induction in the absence of SOCS2 mimicked the protein expression discussed above. CIS, SOCS1, and SOCS3 mRNAs were induced rapidly following stimulation and declined slightly at 180 min. However, in the presence of SOCS2, a stronger signal was observed for all three SOCS family members, consistent with the enhanced STAT activation detected. SOCS2 was detected using two different sets of real-time PCR probes, one set which detected both endogenous and exogenous SOCS2 and another where the forward primer was located upstream of the coding region and was therefore only able to detect endogenous SOCS2. Expression of exogenous SOCS2 had little or no effect on the endogenous levels of the message. When primers specific for c-myc were used, in the presence of SOCS2, levels of c-myc mRNA were enhanced and prolonged, again consistent with enhanced cytokine responses.
FIG. 5.
SOCS2 enhances mRNA levels of CIS, SOCS1, and SOCS3. (A) Ba/F3-SOCS2 cells were incubated overnight in 2% FCS and stimulated with 100 U/ml IL-3 as shown. mRNA levels were analyzed by real-time PCR using specific primers for CIS, SOCS1, SOCS2, SOCS3, and c-myc. Data are expressed as relative mRNA levels. (B) RT-PCR was performed with 1 μg of total RNA using primer pairs specific for CIS, SOCS1, SOCS2, SOCS3, and GAPDH. The PCR products were resolved by 2% agarose gel electrophoresis and visualized with ethidium bromide staining. (C) RT-PCR was performed with 1 μg of total RNA using primer pairs specific for Pim-1, OSM, BCL-xL, SOCS2, and β-actin. The PCR products were resolved by 2% agarose gel electrophoresis and visualized with ethidium bromide staining. tet, tetracycline.
To substantiate this observation, SOCS mRNA levels were analyzed by RT-PCR using 1 μg of total RNA (Fig. 5B). Again, the presence of SOCS2 did not diminish the cytokine-induced mRNA levels of CIS, SOCS1, and SOCS3. SOCS2 did not affect the level of SOCS mRNA at 30 or 60 min but maintained expression of CIS, SOCS1, and SOCS3 at 180 min (Fig. 5B). We further examined other IL-3-responsive genes including Pim1, OSM, and BclxL and did not observe markedly enhanced expression of these genes in two separate experiments (Fig. 5C). The enhanced mRNA expression observed in the presence of SOCS2 supports the idea that SOCS2 can boost signaling and sustain the activation of some STAT5-dependent genes.
SOCS2 causes degradation of SOCS3 in a SOCS box-dependent manner.
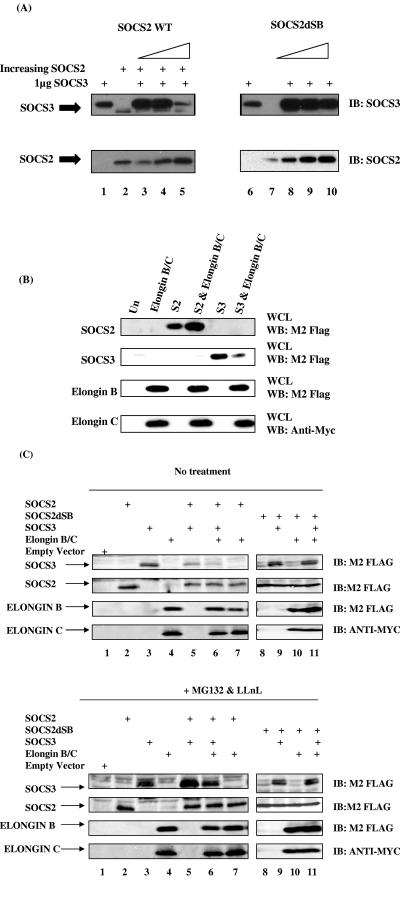
Taken together, the data suggest that SOCS2 can restore JAK/STAT signaling via degradation of SOCS3. We therefore wished to determine the mechanism by which SOCS2 could affect SOCS3 protein levels. Since the SOCS box motif is thought to interact with an E3 ligase complex and target associated proteins for degradation (15, 27, 45), a mutant SOCS2 construct was made in which the elongin B/C binding motif was deleted (SOCS2 dSB). We coexpressed SOCS3 with the SOCS2 wild type or mutant in 293T cells. When SOCS2 and SOCS3 were coexpressed, high levels of SOCS2 targeted SOCS3 for degradation (Fig. 6A, top, lane 5) with 5 μg of SOCS2 plasmid almost completely blocking detectable SOCS3 expression. However, transfection of up to 5 μg of the SOCS2 dSB mutation did not have this effect (Fig. 6A, top, lane 10), suggesting that SOCS2 targets SOCS3 for degradation in a SOCS box-dependent manner.
FIG. 6.
SOCS2 causes degradation of SOCS3 via its SOCS box. (A) SOCS2 wild type (WT) or SOCS2 mutant in which the elongin C binding region was mutated (SOCS2 dSB) was coexpressed with SOCS3 as indicated. Lysates were immunoblotted (IB) with anti-SOCS3 (top) or anti-SOCS2 (bottom). The first lane of each panel represents 1 μg of SOCS3 expressed alone, while the second lane represents 1 μg of SOCS2 expressed alone. Thereafter, the lanes represent increasing amounts of SOCS2 expressed with 1 μg of SOCS3. (B) Five micrograms of FLAG-tagged SOCS2 and 2 μg of SOCS3, elongin B, and Myc-tagged elongin C cDNAs were transfected into 293T cells. Lysates were immunoblotted with anti-FLAG or anti-Myc, respectively. (C) Five micrograms of SOCS2 or SOCS2 dSB mutant and 2 μg SOCS3 and elongin B/C were transfected into 293T cells. Cells were treated with (bottom panels) or without (top panels) the proteasome inhibitors MG132 (0.5 μM) and LLnL (0.5 μM). WB, Western blot. Un, untransfected.
The SOCS box region has been shown to bind to an E3 ligase complex containing elongin B/C, Cullin5, and Rbx2 (26). We therefore wished to determine whether this complex mediated SOCS3 degradation. Interestingly, when SOCS3 was expressed with elongin B/C, there was a significant reduction in SOCS3 protein levels compared to when SOCS3 was expressed alone (Fig. 6B, last lane). In contrast, when SOCS2 was expressed with elongin B/C, the level of SOCS2 protein detected was elevated (Fig. 6B). This suggests that elongin B/C stabilized SOCS2, whereas SOCS3 was less stable in the presence of the elongins. This would hold true if SOCS2 could bind elongin B/C and activate an E3 ligase complex that targets SOCS3 for proteasomal degradation. To investigate this possibility, SOCS2 or SOCS2 dSB, SOCS3, and elongin B/C were coexpressed in 293T cells (Fig. 6C). SOCS2, SOCS3, and elongin B were all FLAG tagged, while elongin C was Myc tagged. Again, when SOCS3 was coexpressed with SOCS2, we observed a reduction in SOCS3 levels (Fig. 6C, top, lane 5). Moreover, when SOCS3 was coexpressed with both SOCS2 and elongin B/C, SOCS3 protein levels were much more markedly reduced (Fig. 6C, top, lane 6) compared to when SOCS3 was expressed alone (lane 3) or with SOCS2 (lane 5). This implied that expression of SOCS2 resulted in the loss of SOCS3 protein and that this effect was enhanced further in the presence of elongin B/C. This reduction in SOCS3 levels was not observed when SOCS3 was expressed with SOCS2 dSB (Fig 6C, top, lane 9 versus lane 11), further reinforcing the hypothesis that the loss of SOCS3 is dependent on an intact SOCS box. When SOCS2 was expressed with elongin B/C, SOCS2 protein levels were elevated (Fig. 6C, top, lanes 6 and 7), strengthening the idea that elongin B/C stabilized SOCS2. To verify that this degradation was proteasome dependent, the cells were treated with the proteasome inhibitors MG132 and LLnL. Figure 6C (bottom) shows that the presence of these inhibitors rescued SOCS3 expression even in the presence of SOCS2 (lane 5) and when expressed with both SOCS2 and elongin B/C (lane 6). These data suggested that SOCS2 targets SOCS3 for proteasomal degradation.
SOCS2 interacts with SOCS3.
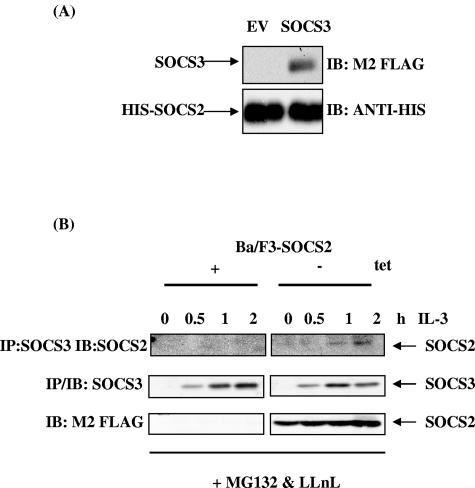
Recently, SOCS2 has been shown to bind elongin B/C (13), suggesting that it can form an E3 ligase complex. We have shown that SOCS2 can target SOCS3 for degradation via the proteasome and that this is dependent on an intact elongin B/C binding site (Fig. 6C). In order for SOCS3 to be degraded by SOCS2, we assumed that SOCS2 and SOCS3 can associate. To examine this, we analyzed the interaction between SOCS2 and SOCS3 in a fusion protein pull-down assay. As shown in Fig. 7A, His-tagged SOCS2 fusion protein bound SOCS3 in vitro. To determine if this interaction could occur endogenously, Ba/F3 SOCS2 cells were grown in the presence and absence of tetracycline for 48 h. The cells were incubated in 2% FCS overnight and stimulated with IL-3 as shown. Cell lysates were immunoprecipitated with anti-SOCS3 and immunoblotted for the presence of SOCS2. When SOCS2 is expressed with endogenous SOCS3, 1 h and 2 h after IL-3 stimulation, both proteins coprecipitate when proteasome activity is blocked, suggesting that an endogenous association between SOCS2 and SOCS3 can occur (Fig. 7B), which suggests that SOCS3 degradation is being accelerated by SOCS2. Although the nature of this interaction is yet to be determined, the findings demonstrate that SOCS2 can form a complex with SOCS3 and suggest a mechanism which enables SOCS3 to be brought into contact with an E3 ligase complex that ultimately results in its turnover by the proteasome.
FIG. 7.
SOCS2 and SOCS3 interaction. (A) His-tagged SOCS2 fusion protein (3 μg) was precoupled to 50 μl of 20% nickel-nitrilotriacetic acid beads in protein interaction buffer for 2 h at 4°C. The beads were collected by centrifugation, and lysates from 293T cells transiently transfected with 2 μg empty vector (Ev) or FLAG-tagged SOCS3 were added. Reaction mixtures were incubated for 2 to 4 h at 4°C in protein interaction buffer, washed, and analyzed by SDS-PAGE as described in the text. Membranes were immunoblotted (IB) with anti-FLAG (top) to detect SOCS3 and reprobed with anti-His to detect His-tagged SOCS2 fusion protein (bottom). (B) Ba/F3-SOCS2 cells were grown in the presence and absence of tetracycline for 48 h. Cells were incubated overnight in 2% FCS and stimulated with 100 U/ml IL-3 as shown. Proteasome inhibitors MG132 (0.5 μM) and LLnL (0.5 μM) were added 1 h before lysing. Cell lysates were immunoprecipitated (IP) with SOCS3 antibody and immunoblotted with anti-FLAG (top) or anti-SOCS3 (middle). SOCS2 expression is shown in the bottom panel.
DISCUSSION
A clear picture has emerged in recent years showing that many members of the SOCS family have an important role in inhibition of the JAK/STAT pathway. The mechanisms by which CIS, SOCS1, and SOCS3 function to block cytokine signaling have been well documented (19). All three have been shown to be induced rapidly and act in a feedback loop to inhibit JAK/STAT activity by binding the JAKs, the receptor, or both (6, 32, 39). However, although SOCS2 can inhibit GH and PRL responses, it can also potentiate the signals induced by many cytokines. In this study, we found that, in contrast to other members of this family, SOCS2 appears to enhance cytokine signaling rather than suppress it.
To date, the mechanism of action of SOCS2 is unclear. One previous report suggested that SOCS2 may have a dual role (12) since low concentrations of SOCS2 inhibited GH action and higher concentrations of SOCS2 enhanced GH signaling. Accordingly, both mice overexpressing SOCS2 and mice lacking SOCS2 displayed a gigantism phenotype (31). This implies that SOCS2 can have a positive and negative role in GH signaling. It is therefore tempting to speculate that in other systems, SOCS2 acts as an accelerator rather than an inhibitor of cytokine signaling. Moreover, SOCS2-null mice show improved responses to growth hormone, indicating an enhanced effect on cytokine responses. However, higher levels of SOCS2 also block GH responses, suggesting that antagonism of other SOCS may occur in these cells. Our observations support the theory that expression of SOCS2 can enhance cytokine responses, most likely by driving degradation of other SOCS proteins.
We have demonstrated that both SOCS2 and SOCS3 are cytokine-induced genes in human PBMCs but that the proteins appear with different kinetics. SOCS3 protein was detected rapidly (30 min following stimulation), while SOCS2 did not appear until later. We also provide evidence that while SOCS3 inhibited both IL-2- and IL-3-induced tyrosine phosphorylation and proliferation, SOCS2 was able to enhance signaling. Since SOCS3 protein expression was maintained in the presence of proteasome inhibitors and mRNA levels of SOCS3 were not reduced by SOCS2, it was evident that SOCS2 at least partially enhanced signaling by reducing SOCS3 protein levels via a proteasome-dependent mechanism. These observations are further supported by the finding that IL-3-induced gene expression was also enhanced when SOCS2 was highly expressed. It is possible that SOCS2 blocks the expression of other SOCS family members, particularly since all SOCS proteins compete for binding to the same E3 ligases. Indeed, our preliminary data suggest that SOCS2 may also reduce SOCS1 expression (data not shown).
It is intriguing that cytokine-induced SOCS2 is expressed much later in peripheral blood T cells. This suggests that SOCS2 is not involved in the feedback loop that inhibits cytokine signaling but may limit this inhibition. Moreover, the finding that SOCS2 is still expressed 24 h after IL-2 treatment suggests that it may be important to potentiate the proliferation of these rapidly dividing cells, perhaps by keeping the expression of other SOCS at reduced levels. This is also true of IL-3-induced SOCS2 in Ba/F3 cells (J. A. Johnston, unpublished observations).
When expressed at high levels, SOCS2 has been demonstrated to antagonize the inhibitory effects of SOCS1 on PRL and GH signaling (5, 8, 33) by an unknown mechanism. CIS levels are also lower in the presence of SOCS2, although the effects are not as marked as for SOCS3 (data not shown), and CIS is also induced later than SOCS1 or SOCS3 in response to cytokine stimulation (3). Clearly, we have observed SOCS3 degradation in the presence of overexpressed SOCS2, but since both proteins are induced by many ligands and appear to be reciprocally regulated, the SOCS2-induced degradation of SOCS3 would presumably function under normal physiological conditions. This would provide a mechanism for eliminating SOCS3 and the other SOCS family members and thus resensitize cells for further cytokine-mediated responses.
The mechanism of action of SOCS2 remains unclear, although several theories have been suggested. One theory holds that high concentrations of SOCS2 may overcome the effect of endogenous SOCS3 on GH signaling (8). SOCS3, unlike SOCS2, contains a kinase-inhibitory region at its N terminus which is thought to directly inhibit JAK activity (36). It has been suggested that SOCS2 may compete with SOCS3 for binding to the GH receptor, and the lack of a kinase-inhibitory region on SOCS2 may result in the continuation of JAK activity and enhanced signaling (12). However, recently, Greenhalgh et al. reported that SOCS2 binds to the GH receptor at Y487 and Y595, which are not classic immunoreceptor tyrosine-based inhibitory motifs, suggesting that SOCS3 will not compete to bind these sites (13). Furthermore, the SH2 domains of SOCS2 and SOCS3 differ significantly and are therefore likely to interact with different phosphotyrosine sequences on different target molecules.
Since our data imply that in the presence of SOCS2, SOCS3 protein expression is downregulated, a more likely theory is that SOCS2 may compete with SOCS3 for binding to the elongin B/C complex. This interaction could occur via the BC box region of the SOCS box. Other SOCS box-containing proteins, including VHL (21), Muf1 (24), elongin A (1), and SOCS1 (15), are thought to be stabilized by this interaction. It has also been shown that tyrosine phosphorylation in the SOCS box of SOCS3 (14) and serine/threonine phosphorylation of SOCS1 (2) disrupt elongin binding. Therefore, SOCS2 may bind to or compete with SOCS3 for elongin B/C, resulting in reduced SOCS3 protein stability.
Another possibility is that SOCS2 may form part of an E3 ligase complex similar to that of VHL. The C-terminal domain of the VHL protein is homologous to the SOCS box and interacts with the elongin B/C complex which in turn binds the Cullin family member Cul2 and RING finger protein Rbx1 to form an E3 ligase complex (25-27). Under normoxic conditions, hypoxia-inducible factor 1α binds to the VHL E3 ligase complex, resulting in ubiquitination and proteasomal degradation (20, 21). SOCS1 has been proposed to form part of an E3 ligase complex containing Cul5 and Rbx1 (24) to target associated proteins for degradation. More recently, it has been suggested that SOCS1 lacks a Cul5 binding site within the SOCS box (26); however, it is plausible that other Cullin proteins may interact with SOCS1.
As shown in Fig. 7, SOCS2 and SOCS3 can associate both in vitro and in vivo, although the precise nature of this interaction remains unknown. However, this study brings us a step closer to understanding when the interaction between SOCS2 and SOCS3 can occur. Our data imply that both proteins associate upon cytokine stimulation when proteasome activity is blocked. It is yet to be determined whether phosphorylation plays a role in regulating the SOCS2-SOCS3 association. Since we have observed association of these proteins, it is plausible to suggest that SOCS2 may act as a linker which brings an E3 ligase complex into close proximity with SOCS3. This may be a potential mechanism by which SOCS2 could result in the loss of SOCS3 protein.
SOCS1 has been reported to regulate the half-life of VAV (4) and the insulin receptor substrates IRS1 and IRS2 (35). Also, SOCS1 inhibits the kinase activity of JAK2 (41) and the TEL-JAK2 oncogene (9) in a phosphorylation-dependent manner by inducing SOCS box-dependent proteasomal degradation. SOCS1 therefore targets these substrates to the proteasome for degradation. Also, SOCS1 and SOCS3 promoted polyubiquitination and degradation of focal adhesion kinase in a SOCS box-dependent manner which inhibited focal adhesion kinase-dependent signaling events (30). Likewise, the SOCS box has been implicated in the inhibition of granulocyte colony-stimulating factor signaling, suggesting a role for proteasomal degradation mediated via SOCS1 and SOCS3 in downregulating granulocyte colony-stimulating factor responses (42). This evidence suggests that the SOCS box is involved in the proteasomal targeting of specific substrates, including perhaps other SOCS. Therefore, SOCS2 may act as part of an E3 ligase and target SOCS3, and perhaps other SOCS, for ubiquitination and degradation via the 26S proteasome.
Despite these observations, questions remain concerning how these findings relate to immune homeostasis and disease. SOCS3 deficiency leading to sustaining IL-6-induced STAT3 activation is thought to contribute to inflammatory diseases such as rheumatoid arthritis, Crohn's disease, and inflammatory bowel disease (38). In comparison, sustained SOCS3 expression in T cells skews differentiation towards a Th2 response which may lead to Th2-related allergy (37). It will be interesting to establish if altered SOCS3 expression in these instances is due to defects in the ability of SOCS2 to regulate SOCS3 expression.
Acknowledgments
Blood packs were kindly donated by the Northern Ireland Blood Transfusion Service. This work has been sponsored by the Wellcome Trust (grant no. 070304/2/03/2) and the BBSRC (grant no. 81/C17863).
We thank G. Clarke for his continuing support and James Burrows, Massimo Gadina, and Karim Dib for their helpful comments on the manuscript.
REFERENCES
- 1.Aso, T., D. Haque, R. J. Barstead, R. C. Conaway, and J. W. Conaway. 1996. The inducible elongin A elongation activation domain: structure, function and interaction with the elongin BC complex. EMBO J. 15:5557-5566. [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, X. P., J. A. Losman, S. Cowan, E. Donahue, S. Fay, B. Q. Vuong, M. C. Nawijn, D. Capece, V. L. Cohan, and P. Rothman. 2002. Pim serine/threonine kinases regulate the stability of Socs-1 protein. Proc. Natl. Acad. Sci. USA 99:2175-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohney, S. J., D. Sanden, N. A. Cacalano, A. Yoshimura, A. Mui, T. S. Migone, and J. A. Johnston. 1999. SOCS-3 is tyrosine phosphorylated in response to interleukin-2 and suppresses STAT5 phosphorylation and lymphocyte proliferation. Mol. Cell. Biol. 19:4980-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Sepulveda, P., S. Ilangumaran, and R. Rottapel. 2000. Suppressor of cytokine signaling-1 inhibits VAV function through protein degradation. J. Biol. Chem. 275:14005-14008. [DOI] [PubMed] [Google Scholar]
- 5.Dif, F., E. Saunier, B. Demeneix, P. A. Kelly, and M. Edery. 2001. Cytokine-inducible SH2-containing protein suppresses PRL signaling by binding the PRL receptor. Endocrinology 142:5286-5293. [DOI] [PubMed] [Google Scholar]
- 6.Elliott, J., and J. A. Johnston. 2004. SOCS: role in inflammation, allergy and homeostasis. Trends Immunol. 25:434-440. [DOI] [PubMed] [Google Scholar]
- 7.Endo, T. A., M. Masuhara, M. Yokouchi, R. Suzuki, H. Sakamoto, K. Mitsui, A. Matsumoto, S. Tanimura, M. Ohtsubo, H. Misawa, T. Miyazaki, N. Leonor, T. Taniguchi, T. Fujita, Y. Kanakura, S. Komiya, and A. Yoshimura. 1997. A new protein containing an SH2 domain that inhibits JAK kinases. Nature 387:921-924. [DOI] [PubMed] [Google Scholar]
- 8.Favre, H., A. Benhamou, J. Finidori, P. A. Kelly, and M. Edery. 1999. Dual effects of suppressor of cytokine signaling (SOCS-2) on growth hormone signal transduction. FEBS Lett. 453:63-66. [DOI] [PubMed] [Google Scholar]
- 9.Frantsve, J., J. Schwaller, D. W. Sternberg, J. Kutok, and D. G. Gilliland. 2001. Socs-1 inhibits TEL-JAK2-mediated transformation of hematopoietic cells through inhibition of JAK2 kinase activity and induction of proteasome-mediated degradation. Mol. Cell. Biol. 21:3547-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujimoto, M., and T. Naka. 2003. Regulation of cytokine signaling by SOCS family molecules. Trends Immunol. 24:659-666. [DOI] [PubMed] [Google Scholar]
- 11.Gadina, M., D. Hilton, J. A. Johnston, A. Morinobu, A. Lighvani, Y. J. Zhou, R. Visconti, and J. J. O'Shea. 2001. Signaling by type I and II cytokine receptors: ten years after. Curr. Opin. Immunol. 13:363-373. [DOI] [PubMed] [Google Scholar]
- 12.Greenhalgh, C. J., P. Bertolino, S. L. Asa, D. Metcalf, J. E. Corbin, T. E. Adams, H. W. Davey, N. A. Nicola, D. J. Hilton, and W. S. Alexander. 2002. Growth enhancement in suppressor of cytokine signaling 2 (SOCS-2)-deficient mice is dependent on signal transducer and activator of transcription 5b (STAT5b). Mol. Endocrinol. 16:1394-1406. [DOI] [PubMed] [Google Scholar]
- 13.Greenhalgh, C. J., E. Rico-Bautista, M. Lorentzon, A. L. Thaus, P. O. Morgan, T. A. Willson, P. Zervoudakis, D. Metcalf, I. Street, N. A. Nicola, A. D. Nash, L. J. Fabri, G. Norstedt, C. Ohlsson, A. Flores-Morales, W. S. Alexander, and D. J. Hilton. 2005. SOCS2 negatively regulates growth hormone action in vitro and in vivo. J. Clin. Investig. 115:397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haan, S., P. Ferguson, U. Sommer, M. Hiremath, D. W. McVicar, P. C. Heinrich, J. A. Johnston, and N. A. Cacalano. 2003. Tyrosine phosphorylation disrupts elongin interaction and accelerates SOCS3 degradation. J. Biol. Chem. 278:31972-31979. [DOI] [PubMed] [Google Scholar]
- 15.Hanada, T., T. Yoshida, I. Kinjyo, S. Minoguchi, H. Yasukawa, S. Kato, H. Mimata, Y. Nomura, Y. Seki, M. Kubo, and A. Yoshimura. 2001. A mutant form of JAB/SOCS1 augments the cytokine-induced JAK/STAT pathway by accelerating degradation of wild-type JAB/CIS family proteins through the SOCS-box. J. Biol. Chem. 276:40746-40754. [DOI] [PubMed] [Google Scholar]
- 16.Hibbert, L., and J. A. Johnston. 2001. Cytokine signalling and disease. Expert Opin. Ther. Targets 5:641-653. [DOI] [PubMed] [Google Scholar]
- 17.Hilton, D. J., R. T. Richardson, W. S. Alexander, E. M. Viney, T. A. Willson, N. S. Sprigg, R. Starr, S. E. Nicholson, D. Metcalf, and N. A. Nicola. 1998. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc. Natl. Acad. Sci. USA 95:114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoeflich, A., M. Wu, S. Mohan, J. Foll, R. Wanke, T. Froehlich, G. J. Arnold, H. Lahm, H. J. Kolb, and E. Wolf. 1999. Overexpression of insulin-like growth factor-binding protein-2 in transgenic mice reduces postnatal body weight gain. Endocrinology 140:5488-5496. [DOI] [PubMed] [Google Scholar]
- 19.Ihle, J. N. 1995. Cytokine receptor signalling. Nature 377:591-594. [DOI] [PubMed] [Google Scholar]
- 20.Ivan, M. and W. G. Kaelin, Jr. 2001. The von Hippel-Lindau tumor suppressor protein. Curr. Opin. Genet. Dev. 11:27-34. [DOI] [PubMed] [Google Scholar]
- 21.Iwai, K., K. Yamanaka, T. Kamura, N. Minato, R. C. Conaway, J. W. Conaway, R. D. Klausner, and A. Pause. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 96:12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, J. A. 2003. Are SOCS suppressors, regulators, and degraders? J. Leukoc. Biol. 75:743-748. [DOI] [PubMed] [Google Scholar]
- 23.Kamizono, S., T. Hanada, H. Yasukawa, S. Minoguchi, R. Kato, M. Minoguchi, K. Hattori, S. Hatakeyama, M. Yada, S. Morita, T. Kitamura, H. Kato, K. Nakayama, and A. Yoshimura. 2001. The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J. Biol. Chem. 276:12530-12538. [DOI] [PubMed] [Google Scholar]
- 24.Kamura, T., D. Burian, Q. Yan, S. L. Schmidt, W. S. Lane, E. Querido, P. E. Branton, A. Shilatifard, R. C. Conaway, and J. W. Conaway. 2001. Muf1, a novel elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276:29748-29753. [DOI] [PubMed] [Google Scholar]
- 25.Kamura, T., M. N. Conrad, Q. Yan, R. C. Conaway, and J. W. Conaway. 1999. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamura, T., K. Maenaka, S. Kotoshiba, M. Matsumoto, D. Kohda, R. C. Conaway, J. W. Conaway, and K. I. Nakayama. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18:3055-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamura, T., S. Sato, D. Haque, L. Liu, W. G. Kaelin, Jr., R. C. Conaway, and J. W. Conaway. 1998. The elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopchick, J. J., L. L. Bellush, and K. T. Coschigano. 1999. Transgenic models of growth hormone action. Annu. Rev. Nutr. 19:437-461. [DOI] [PubMed] [Google Scholar]
- 29.Kubo, M., T. Hanada, and A. Yoshimura. 2003. Suppressors of cytokine signaling and immunity. Nat. Immunol. 4:1169-1176. [DOI] [PubMed] [Google Scholar]
- 30.Liu, E., J. F. Cote, and K. Vuori. 2003. Negative regulation of FAK signaling by SOCS proteins. EMBO J. 22:5036-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metcalf, D., C. J. Greenhalgh, E. Viney, T. A. Willson, R. Starr, N. A. Nicola, D. J. Hilton, and W. S. Alexander. 2000. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature 405:1069-1073. [DOI] [PubMed] [Google Scholar]
- 32.Naka, T., M. Narazaki, M. Hirata, T. Matsumoto, S. Minamoto, A. Aono, N. Nishimoto, T. Kajita, T. Taga, K. Yoshizaki, S. Akira, and T. Kishimoto. 1997. Structure and function of a new STAT-induced STAT inhibitor. Nature 387:924-929. [DOI] [PubMed] [Google Scholar]
- 33.Pezet, A., H. Favre, P. A. Kelly, and M. Edery. 1999. Inhibition and restoration of prolactin signal transduction by suppressors of cytokine signaling. J. Biol. Chem. 274:24497-24502. [DOI] [PubMed] [Google Scholar]
- 34.Quaife, C. J., L. S. Mathews, C. A. Pinkert, R. E. Hammer, R. L. Brinster, and R. D. Palmiter. 1989. Histopathology associated with elevated levels of growth hormone and insulin-like growth factor I in transgenic mice. Endocrinology 124:40-48. [DOI] [PubMed] [Google Scholar]
- 35.Rui, L., M. Yuan, D. Frantz, S. Shoelson, and M. F. White. 2002. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J. Biol. Chem. 277:42394-42398. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki, A., H. Yasukawa, A. Suzuki, S. Kamizono, T. Syoda, I. Kinjyo, M. Sasaki, J. A. Johnston, and A. Yoshimura. 1999. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells 4:339-351. [DOI] [PubMed] [Google Scholar]
- 37.Seki, Y., H. Inoue, N. Nagata, K. Hayashi, S. Fukuyama, K. Matsumoto, O. Komine, S. Hamano, K. Himeno, K. Inagaki-Ohara, N. Cacalano, A. O'Garra, T. Oshida, H. Saito, J. A. Johnston, A. Yoshimura, and M. Kubo. 2003. SOCS-3 regulates onset and maintenance of T(H)2-mediated allergic responses. Nat. Med. 9:1047-1054. [DOI] [PubMed] [Google Scholar]
- 38.Shouda, T., T. Yoshida, T. Hanada, T. Wakioka, M. Oishi, K. Miyoshi, S. Komiya, K. Kosai, Y. Hanakawa, K. Hashimoto, K. Nagata, and A. Yoshimura. 2001. Induction of the cytokine signal regulator SOCS3/CIS3 as a therapeutic strategy for treating inflammatory arthritis. J. Clin. Investig. 108:1781-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Starr, R., T. A. Willson, E. M. Viney, L. J. Murray, J. R. Rayner, B. J. Jenkins, T. J. Gonda, W. S. Alexander, D. Metcalf, N. A. Nicola, and D. J. Hilton. 1997. A family of cytokine-inducible inhibitors of signalling. Nature 387:917-921. [DOI] [PubMed] [Google Scholar]
- 40.Tanimoto, K., Y. Makino, T. Pereira, and L. Poellinger. 2000. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 19:4298-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ungureanu, D., P. Saharinen, I. Junttila, D. J. Hilton, and O. Silvennoinen. 2002. Regulation of Jak2 through the ubiquitin-proteasome pathway involves phosphorylation of Jak2 on Y1007 and interaction with SOCS-1. Mol. Cell. Biol. 22:3316-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Geijn, G. J., J. Gits, and I. P. Touw. 2004. Distinct activities of suppressor of cytokine signaling (SOCS) proteins and involvement of the SOCS box in controlling G-CSF signaling. J. Leukoc. Biol. 76:237-244. [DOI] [PubMed] [Google Scholar]
- 43.Wilcox, A., K. D. Katsanakis, F. Bheda, and T. S. Pillay. 2004. Asb6, an adipocyte-specific ankyrin and SOCS box protein, interacts with APS to enable recruitment of elongins B and C to the insulin receptor signaling complex. J. Biol. Chem. 279:38881-38888. [DOI] [PubMed] [Google Scholar]
- 44.Yasukawa, H., A. Sasaki, and A. Yoshimura. 2000. Negative regulation of cytokine signaling pathways. Annu. Rev. Immunol. 18:143-164. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, J. G., A. Farley, S. E. Nicholson, T. A. Willson, L. M. Zugaro, R. J. Simpson, R. L. Moritz, D. Cary, R. Richardson, G. Hausmann, B. J. Kile, S. B. Kent, W. S. Alexander, D. Metcalf, D. J. Hilton, N. A. Nicola, and M. Baca. 1999. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. USA 96:2071-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]