Abstract
The Hey basic helix-loop-helix transcription factors are downstream effectors of Notch signaling in the cardiovascular system. Mice lacking Hey2 develop cardiac hypertrophy, often associated with congenital heart defects, whereas combined Hey1/Hey2 deficiency leads to severe vascular defects and embryonic lethality around embryonic day E9.5. The molecular basis of these disorders is poorly understood, however, since target genes of Hey transcription factors in the affected tissues remain elusive. To identify genes regulated by Hey factors we have generated a conditional Hey1 knockout mouse. This strain was used to generate paired Hey2- and Hey1/2-deficient embryonic stem cell lines. Comparison of these cell lines by microarray analysis identified GATA4 and GATA6 as differentially expressed genes. Loss of Hey1/2 leads to elevated GATA4/6 and ANF mRNA levels in embryoid bodies, while forced expression of Hey factors strongly represses expression of the GATA4 and GATA6 promoter in various cell lines. In addition, the promoter activity of the GATA4/6 target gene ANF was inhibited by Hey1, Hey2, and HeyL. Protein interaction and mutation analyses suggest that repression is due to direct binding of Hey proteins to GATA4 and GATA6, blocking their transcriptional activity. In Hey2-deficient fetal hearts we observed elevated mRNA levels of ANF and CARP. Expression of ANF and Hey2 is normally restricted to the trabecular and compact myocardial layer, respectively. Intriguingly, loss of Hey2 leads to ectopic ANF expression in the compact layer, suggesting a direct role for Hey2 in limiting ANF expression in this cardiac compartment.
The basic helix-loop-helix transcription factors Hey1, Hey2, and HeyL are primary mediators of the Delta-Notch signaling pathway, particularly in the cardiovascular system. Hey proteins, also known as Hesr, Hrt, Chf, Herp, and gridlock, have been shown to regulate a variety of cellular decisions in different processes such as cardiac development (for review see reference 9), angiogenesis (12), neurogenesis (43), gliogenesis (46), bone development (56), or the epithelial-to-mesenchymal transition (57). Besides their role in physiological developmental processes, Hey genes have also been implicated as possible tumor suppressors (20).
During heart development Hey1 and Hey2 show a rather unusual expression pattern. While Hey1 is expressed exclusively in the atria, Hey2 is restricted to the ventricles (29). We and others have shown that loss of Hey2 in mice leads to high postnatal lethality due to cardiac malformations and heart failure (7, 14, 26, 44). The hearts of newborn Hey2 knockout mice develop massive hypertrophy shortly after birth, and electron microscopy revealed structural defects in the myocardium of these mutants (14). In addition, there is a high incidence of ventricular septal defects, often accompanied by tricuspid atresia or dysplastic atrioventricular valves, atrial septal defects, and even pulmonary stenosis, depending on the genetic background (10).
Interestingly, Hey1 knockout mice are viable and do not show any overt defects (12). However, combined loss of Hey1 and Hey2 leads to embryonic lethality due to severe defects in blood vessel development and failure of arterial-venous differentiation. This phenocopies the cardiovascular defects seen in Notch1 and Notch1/Notch4 double-knockouts and the zebra fish gridlock mutant (12, 27, 59).
There is meanwhile profound knowledge on how expression of Hey genes is regulated by Notch signaling (33, 37), as well as by transforming growth factor beta/bone morphogenetic protein (23), but bona fide target genes of Hey proteins are still elusive. We have shown recently that the artery-specific markers ephrinB2, CD44, and neuropilin 1 are drastically downregulated in Hey1/Hey2 double-knockout mice (12). Nevertheless, it is not clear if these genes are direct targets of Hey1/2 or if their expression is lost due to the misregulation of another transcription factor. Henderson et al. (18) have shown a downregulation of vascular endothelial growth factor receptor 2 by Hey1 in endothelial cells, but again detailed promoter analysis is missing.
As is evident from our Hey double-knockout studies, Hey genes can act in a redundant manner, i.e., loss of Hey1 in endothelial cells can be compensated for by Hey2 and vice versa. Therefore, we have now established an in vitro system, based on embryonic stem cells, which eliminates this redundancy and allowed us to search for Hey1/Hey2-regulated genes by microarray analysis.
Here we show that Hey proteins repress expression of the zinc finger factors GATA4 and GATA6 and their target genes ANF and CARP. In addition, we found that in Hey2-deficient embryonic hearts there is ectopic ANF expression in the compact layer of the left ventricle. This represents an important step to elucidate the molecular basis of why Hey2-deficient mice develop fatal cardiac hypertrophy.
MATERIALS AND METHODS
Plasmids.
GATA4 reporter constructs are based on a 10.6-kbp KspI fragment that harbors the 5′-flanking region of murine GATA4, including half of the untranslated exon 1 from cosmid clone MPMGc121F03498Q (RZPD, Berlin, Germany), ligated into the SmaI site of the pGL3-basic vector (Promega). Constructs containing 0.5 kbp, 2.5 kbp, and 5 kbp of the GATA4 5′ region were obtained by religation after digestion with SacI, MluI, or NruI and PacI followed by a fill-in reaction.
To mutate the E-box sequence (CACGTG) −59 to −54 nucleotides upstream of putative exon 1 in the GATA4 promoter (NCBI GenBank accession number NT_039606.3, nucleotides 9602278 to 9602398, minus strand), pGL3-GATA4-10.6kb was linearized with PmlI, which cuts CACGTG, and an oligonucleotide linker (AGGGATCTGCTCGTCTAGAAAAAGAGC) was inserted. Two point mutations of the E-box were inserted into the pGL3-GATA4-0.5kb vector by PCR mutagenesis, changing CACGTG into TACATG. To generate a GATA6-luciferase reporter we cloned a 4.237-kb AatII fragment of bacterial artificial chromosome clone RZPDB737F092162D6 (RZPD, Berlin, Germany) into pGL3-basic. This fragment contains the 5′-flanking region of human GATA6, including the noncoding exons 1a/b, intron 1, and parts of exon 2 upstream of the translation start codon.
For mammalian expression of hemagglutinin (HA)-tagged GATA4, the complete coding sequence was amplified from adult murine heart cDNA and cloned into pcDNA3.1 containing an N-terminal hemagglutinin tag. Integrity and expression were verified by sequencing and immunofluorescence. To generate a FLAG-tagged full-length GATA6 expression vector we subcloned the GATA6 cDNA from pcDNA-GATA6 into a pCS2-FLAG vector.
Mammalian expression plasmids pCS2p-Hey1, pCS2p-Hey2, and pCS2p-HeyL contain the full-length coding sequences either with or without N-terminal HA or FLAG tags as indicated. Vectors expressing enhanced green fluorescent protein (EGFP)-Hey1 fusion proteins were generated by cloning murine Hey1 PCR fragments encoding amino acids 1 to 299 (full length), 1 to 286, 1 to 176, 1 to 121, 1 to 93, 63 to 299, and 122 to 299 into pEGFPc2 or pEGFPc3 (Clontech). All primer sequences are available upon request.
Hey1-luciferase constructs have been described before (33). The 0.7-kb ANF- luciferase plasmid was provided by Mona Nemer (Montreal, Canada). pCS2-Fog2, pMT2-Fog1, and pcDNA-GATA6 expression plasmids are a kind gift of Stuart Orkin (Boston, MA) and pCI-FLAG-p300 was supplied by Werner Lutz (IMT, Marburg, Germany).
Generation of floxed Hey1 mice.
The targeting vector was constructed using the 129SVJ-derived genomic lambda clone SV3 (48). A 3.16-kb NotI/PvuII fragment containing the murine Hey1 promoter region and exon 1 (33) was cloned into pKSTKloxPneo, followed by a 0.76-kb PvuII fragment containing exons 2 to 4 that was inserted after the neo cassette. Intron 4 and part of exon 5 representing the short arm of homology were generated by PCR using primers EagIP1, GCATCGGCCGACTGCCTGCTTTGCTTTGTG, and NotIP2, ATCGCGGCCGCGTGTGGGTGATGTCCGAAG. The final vector contains a floxed PGK-neo cassette within intron 1 and an additional loxP site in intron 4. The integration sites were chosen not to affect splice sites.
R1 embryonic stem (ES) cells were electroporated and two positive clones (IIE8 and IVH10) could be identified by PCR and Southern blot analysis. The PGK-neo cassette was removed in IVH10 by transient transfection with pMC-Cre1 (15). Two clones (IVH10/2C7 and IVH10/2H9) that lost the neo cassette were used for blastocyst injection. Male chimeras with extensive ES cell contribution to the coat color were mated with C57BL/6 females for germ line transmission. For genotyping by PCR we used primers clik-atg, GCGGGATCCACATGAAGAGAGCTCACCCAG, Clikseq2, TGAGATCTTGCAGATGACTGTG, and Eagrev, ACAAAGCAAAGCAGGCAGTC. PCR products were 150 bp (floxed) and 260 bp (deleted). Primer sequences for Hey1-lacZ and Hey2-lacZ knockout mice have been described (12, 14).
Generation of Hey1fl/fl/Hey2−/− ES cells.
Hey1fl/fl/Hey2+/− mice were intercrossed and blastocysts were harvested at E3.5 as described (36). Blastocysts were cultured in ES medium on mitomycin-inactivated fibroblasts and forming ES cell colonies were further expanded and genotyped. Three Hey1fl/fl/Hey2−/− ES cell lines (A3, A5, and C1) with typical morphology were chosen for further experiments.
Cell culture and embryoid body formation.
HEK293, HEK293T, Cos7, and HeLa cells were cultured in Dulbecco's modified Eagle's medium (Gibco) with 10% fetal bovine serum. ES cells were maintained in ES medium containing Dulbecco's modified Eagle's medium with 10% ES cell qualified fetal bovine serum, 103 U/ml leukemia inhibitory factor, 50 mM β-mercaptoethanol, and minimal essential medium nonessential amino acids (Gibco) on mitomycin-inactivated fibroblasts. Before embryoid body formation ES cells were adapted to gelatin-coated culture dishes for three passages. On day 0 of induction ES cells were seeded on bacterial-grade petri dishes (Greiner) in ES medium lacking leukemia inhibitory factor and cultured for several days. Cells were fed every second day with fresh medium.
Tat-Cre treatment.
Recombinant Tat-Cre protein (HTNC) was produced as described (38). ES cells were treated with 1 μM HTNC for 4 h in ES medium. After TAT-Cre treatment cells were washed and seeded at low density (1,000 ES cells per 10-cm plate) onto feeder cells. Individual colonies were isolated and genotyped for Hey1 recombination.
RNA isolation, cDNA labeling, and microarray analysis.
Cells or embryoid bodies were washed in phosphate-buffered saline and total cellular RNA was isolated using Trizol reagent (Invitrogen) followed by DNase I treatment (RNeasy kit, QIAGEN); 30 μg total RNA was transcribed into cDNA using the CyScribe cDNA Post Labeling kit (Amersham Biosciences) with nonamer primers and modified amino-allyl-dUTP. After degradation of the RNA template the cDNA was labeled with Cy5 (samples) and Cy3 (reference). Microarrays containing 11.500 murine cDNA clones (Research Genetics) were generated as described using a GMS417 arrayer (Genetic MicroSystems, MWG Biotech, Germany) (3). Each hybridization probe was placed between two microarray slides (sandwich hybridization). After washing the arrays were scanned using a GMS 418 microarray scanner (Genetic MicroSystems, MWG Biotech, Germany) and analyzed with ImaGene 3.0 software (BioDiscovery Inc., Marina Del Rey, CA) as described (3).
RNA expression analysis.
Quantitative real-time RT-PCR was performed using a Bio-Rad iCycler (12). All primer sequences are available upon request. mRNA in situ hybridization was done essentially as described (28).
Luciferase assay.
HEK293, HEK293T, Cos7, HeLa, or ES cells were transiently transfected in 12-well plates using calcium phosphate coprecipitation or GeneJuice (Novagen) with the indicated plasmids; 36 to 48 h posttransfection cells were washed with phosphate-buffered saline and lysed. Luciferase activity was measured in a Berthold luminometer using the dual luciferase assay (Promega). Each experiment was repeated at least two times with each reaction measured in triplicate. Results are shown as mean values with standard deviations.
Coimmunoprecipitation.
HEK293T cells were transiently transfected with the indicated expression vectors for FLAG- or HA-tagged proteins. Coimmunoprecipitation was performed as described (54).
RESULTS
Generation of floxed Hey1 mice.
To generate a conditional Hey1 null allele we used a targeting construct to introduce loxP sites into introns 1 and 4 of the Hey1 locus (Fig. 1). After confirming a homologous recombination event by PCR and Southern blotting, the neomycin resistance cassette was deleted by transient Cre recombinase expression in ES cells. Chimeric mice were generated from two independent lines by blastocyst injection and the floxed Hey1 locus was bred onto a C57BL/6 background for five generations. Hey1fl/fl offspring were obtained in a Mendelian ratio and appeared phenotypically normal and fertile. Embryonic fibroblasts derived from these animals efficiently recombined the floxed locus following Cre treatment, leading to deletion of exons 2 to 4 and a functionally null allele as verified by PCR and reverse transcription (RT)-PCR. (Fig. 1C and D).
FIG. 1.
Conditional inactivation of the Hey1 gene. (A) The targeting construct is shown above the genomic Hey1 locus containing five exons. The promoter and parts of exon 1 (3 kbp) were used as 5′ homology region while the 3′ homology consists of 1-kbp intron 4 and parts of exon 5. The PKG-neo cassette (Neo) is flanked by loxP sites (triangles) and an additional loxP site was inserted into intron 4. The herpes simplex virus thymidine kinase (TK) was used for negative selection. After homologous recombination the integrity of the Hey1 neo-flox allele was confirmed by Southern blot hybridization using probes P1, P2, and P3. B, BglII; E, Ecl136II; H, HindIII; *, stop codon. (B) Transient expression of Cre recombinase in ES cells yielded the final floxed Hey1 allele which contains loxP sites in introns 1 and 4. (C) Cre recombinase deletes the Hey1 gene. (D) Left: Genotyping of Tat-Cre recombinase-treated ES cells derived from conditional Hey1 knockout mice. As example PCR products of floxed/floxed (fl/fl), floxed/deleted (fl/del), and homozygous deleted ES cells are shown. Right: No Hey1 mRNA is detectable in Hey1del/del ES cells. RNA was isolated from Hey1fl/fl and Hey1del/del ES cells and, as a control, from Hey1-lacZ knockout and wild-type (WT) tail samples. RT-PCR shows the absence of Hey1 mRNA in Hey1del/del and Hey1-lacZ knockout cells. As a control expression of hypoxanthine phosphoribosyltransferase (Hprt) is shown below. Without reverse transcriptase (−RT), no products are obtained.
Establishment of Hey1fl/fl/Hey2−/− ES cells.
Mice carrying a conditional Hey1 allele were crossed with Hey2-lacZ mutants (14) to produce Hey1fl/fl/Hey2+/− mice. These were then intercrossed and blastocysts were harvested at embryonic day E3.5. Three independent Hey1fl/fl/Hey2−/− ES cell lines (A3, A5, and C1) were generated from these blastocysts.
To recombine, the floxed Hey1 locus cells were treated for 4 h with 1 μM of Cre recombinase fused to a human immunodeficiency virus Tat peptide and a nuclear localization signal. This fusion protein (HTNC) has been shown to cross cell membranes and to recombine with high efficiency in murine ES cells (38). In our hands HTNC treatment led to 19% homozygous (Hey1del/del) and 12% heterozygous deleted clones (Hey1fl/del). Two homozygous deleted clones, A3d6 and C1d14 (Hey1del/del/Hey2−/−), which represent Hey1/2 double-knockout cells, and two control Hey1fl/fl/Hey2−/− clones (A3f13 and C1f10), representing Hey2 “single” knockout cells were used for further experiments.
Microarray analysis of embryoid bodies with Hey1/2 loss.
To search for downstream target genes of Hey1/2 we used microarray technology and compared our Hey2 knockout with Hey1/2 double-knockout ES cell lines. These lines are ideally suited for this purpose as they were derived from the same parental cell line under identical conditions. Since our primary goal was to find Hey1/2 target genes particularly in the process of cardiovascular development, we decided to aggregate the ES cells and to differentiate these as embryoid bodies. It is well known that ES cells generate various cell types, including endothelial and smooth muscle cells or cardiomyocytes, under these conditions.
To identify the optimal time point for harvesting embryoid bodies, we tested the induction of various cardiovascular marker genes during EB formation by quantitative real-time PCR (qRT-PCR). After 9 days of culture, there was a strong and robust up-regulation of vascular endothelial growth factor recptor 2, Tie2, EfnB2, EphB4, Scl, platelet-derived growth factor β, and Tagln (SM22α) mRNAs, indicating the formation of endothelial cells and smooth muscle cells, which is in agreement with previous studies (55). In addition, PECAM staining showed the typical pattern of primitive endothelial tubes and around day 9 several of the embryoid bodies contained spontaneously contracting cells, a sign of cardiomyocyte formation. Therefore, RNA from 9-day-old embryoid bodies (lines A3f13, A3d6, C1f10, and C1d14) was prepared and labeled cDNA was hybridized to microarrays containing approximately 11,500 murine cDNAs.
In Hey1/2 double-knockout embryoid bodies 52 genes were up-regulated more than twofold with P values of < 0.05 compared to the corresponding Hey2 knockout lines, whereas 30 genes were down-regulated. The full microarray results will be published elsewhere. Among the regulated genes were GATA4 and GATA6, with 2.3- and 2.6-fold up-regulation, respectively (Fig. 2A). Both genes encode zinc finger transcription factors that are absolutely essential for heart development, as has been demonstrated by various mouse knockout models for a recent review see (41). In addition, in humans GATA4 mutations were found to cause congenital heart defects (13). Up-regulation of GATA4 in cardiomyocytes has been shown to lead to cardiac hypertrophy in mice (31). Hey2 knockout mice develop massive heart hypertrophy, so we speculated that this might in part be mediated through elevated GATA4 levels. We therefore examined the regulation of GATA4 and some of its target genes by Hey transcription factors in vitro and in vivo.
FIG. 2.
Loss of Hey1/2 up-regulates GATA4 and GATA6. Hey2 knockout (KO, gray bars) and Hey1/2 double-knockout (DKO, black bars) ES cells were differentiated into embryoid bodies and mRNA preparations were used for (A) microarray analysis and (C) quantitative real-time RT-PCR analysis. Loss of both Hey genes causes elevated GATA4 and GATA6 mRNA levels. (B) In undifferentiated ES cells GATA4 but not GATA6 gene expression is up-regulated in Hey1/2 double-knockout cells. For qRT-PCR analysis, expression levels were normalized to hypoxanthine phosphoribosyltransferase. (D) Expression of a 10.6-kb GATA4 luciferase reporter construct is significantly increased in Hey1/2-deficient undifferentiated ES cells. n.d., not determined
Loss of Hey1/2 leads to up-regulation of GATA factors and GATA target genes in ES cells.
Endogenous GATA mRNA levels were measured in undifferentiated Hey2 knockout and Hey1/2 double-knockout ES cell lines A3f13 and A3d6 by quantitative real-time RT-PCR. There was a 3.1-fold up-regulation of GATA4 (P = 0.01) in cells lacking both Hey1 and Hey2 compared to those lacking only Hey2. The levels of GATA6 were equal in both lines (Fig. 2B). After embryoid body formation and culture for 9 days we detected elevated mRNA levels for both genes, GATA4 and GATA6 (3.5- and 5.3-fold, respectively), in Hey1/2 double-knockout embryoid bodies, confirming our initial microarray results (Fig. 2C). With C1f10 and C1d14 cells we found 3.4-fold higher GATA4 mRNA levels in undifferentiated Hey1/2 double-knockout ES cells while in embryoid bodies GATA4 and GATA6 were 4.9- and 8-fold up-regulated, respectively (data not shown). Furthermore, a luciferase reporter construct driven by a 10.6-kb GATA4 promoter revealed a similar regulation. Transfected Hey1/2 double-knockout cells exhibited a 2.5-fold higher luciferase activity compared to Hey2 knockout ES cells (Fig. 2D). Essentially the same results were obtained with cell lines C1f10 and C1d14 (data not shown).
Since GATA4 and GATA6 were up-regulated in Hey1/2-deficient embryoid bodies we tested whether this would also affect GATA target gene expression. Indeed, we found by qRT-PCR a 16- and 120-fold up-regulation of ANF mRNA in 9-day-old embryoid bodies of ES cell lines A3d6 and C1d14 (double-knockout), respectively, compared to the single-knockout A3f13 and C1f10 cells (P < 0.01). The expression of CARP, another GATA4/6-regulated gene, was elevated 16-fold in C1d14 cells (P < 0.05). In A3d6 we also found increased CARP levels, but this did not reach statistical significance (data not shown). On the other hand, comparison of embryoid bodies derived from A3f13 or C1f10 Hey2 knockout cells with those from wild-type WW6 ES cells did not reveal any significant differences in ANF, GATA4, and GATA6 mRNA expression.
Hey1, Hey2, and HeyL repress the GATA4 and GATA6 promoter in vitro.
Since loss of Hey factors leads to increased GATA4 and GATA6 expression, we next asked if overexpression of Hey proteins would in turn decrease their levels. The 10.6-kb GATA4 luciferase reporter construct was transfected into HEK293 cells together with increasing amounts of Hey1, Hey2, or HeyL expression vector. Compared to empty control vector, all three Hey factors strongly repressed the activity of the GATA4 promoter in a dose-dependent manner (Fig. 3A), with Hey1 being the strongest repressor. Similar results were obtained in repeat experiments with HEK293T, Cos7, and HeLa cells.
FIG. 3.
Hey factors downregulate the GATA4 promoter. (A) Dose-dependent repression of GATA4; 1 μg of the 10.6-kb GATA4 luciferase reporter was coexpressed in 293 cells with empty pCS2 plasmid or with increasing amounts (0.1, 0.25, 0.5, 0.75, and 1.0 μg) of pCS2-Hey1, -Hey2, or -HeyL expression vector. Inset: Equal protein expression levels of Hey1, Hey2, and HeyL in luciferase experiments were confirmed by Western blotting (B) Deletion analysis of the mouse GATA4 promoter. Hey factors are able to repress the minimal 0.5-kb promoter. (C) Mutation of the E-box CACGTG by introducing a 25-bp oligonucleotide in the 10.6-kb GATA4 promoter almost abolishes its basal activity. However, Hey1 can still repress this reporter construct. (D) Mutation analysis of the 0.5-kb GATA4 promoter. Destruction of the E-box by two point mutations does not prevent repression of the GATA4 promoter by Hey factors, indicating that Hey proteins act in a manner independent of interaction with their proposed DNA binding site. For all experiments, basal luciferase activity with empty pCS2 plasmid was given a value of 100%.
Next we cotransfected HEK293 cells with a 4.2-kb GATA6 luciferase reporter and Hey1, Hey2, or HeyL expression plasmid. Similar to the results seen with the GATA4 promoter, all Hey factors efficiently repressed the GATA6 promoter by more than 95% (data not shown).
To map elements that mediate the GATA4 repression we analyzed shortened GATA4-luciferase constructs. As depicted in Fig. 3B, we observed comparable repression for the 10.6-kb, 5-kb, 2.5-kb, and even the 0.5-kb GATA4 promoters. Hey1 and Hey2 almost completely abolished GATA4 promoter activity (95 to 98% and 80 to 94%, respectively) and HeyL still achieved repression rates of 57 to 95%.
We have shown previously that murine Hey1 and Hey2 proteins preferentially bind to the E-box sequence CACGTG in vitro (11). The identical element was also proposed as the optimal binding site for Xenopus Hey (40). Within 11 kb of the GATA4 promoter and 5′ untranslated region this element can be found only once at −59 to −54 nucleotides upstream of the noncoding exon 1. To test if Hey proteins exert repression via this element, we disrupted the CACGTG box in the 10.6-kb GATA4 luciferase promoter by separating the two half-sites with a 25-bp oligonucleotide. This mutation reduced basal promoter activity in HEK293 cells to less than 10% compared to the wild-type construct. Hey1 especially still exhibited some repression activity, however (Fig. 3C).
To generate a less severe mutation without physically extending the promoter in this presumably critical region, we introduced two point mutations changing CACGTG into TACATG in the 0.5-kb GATA4 luciferase promoter. This should prevent any binding of Hey factors to the mutated element. The point mutations decreased the basal activity only by about 25%, but Hey proteins could still repress the 0.5-kb GATA4 promoter carrying this mutated E-box, albeit to a somewhat lesser extent (Fig. 3D). These results suggest that Hey proteins do not effect repression through direct binding to their cognate target sequence, but may rather act through a different mechanism, e.g., by sequestering other activators or by indirect repression through other DNA-binding factors.
Basic helix-loop-helix domain of Hey1 is essential for repression activity.
To map the critical protein domain of Hey1 that is responsible for repression we generated different deletion constructs of Hey1 that were amino-terminally fused to enhanced green fluorescent protein. This allowed us to visualize the localization of the fusion protein in transfected cells. All deletion mutants, even those lacking the basic domain, which contains a presumed nuclear localization signal, efficiently entered the nucleus. Upon cotransfection with the 10.6-kb GATA4 luciferase reporter plasmid the basic helix-loop-helix domain was found to be critical for the repression activity of Hey1 in HEK293 and HeLa cells (Fig. 4). Interestingly, the highly conserved YRPW motif at the carboxy terminus was dispensable for repression in this case.
FIG. 4.
Basic helix-loop-helix (bHLH) domain of Hey1 mediates repression activity. (Left) Schematic drawing of Hey1 deletion mutants. All mutants contained an EGFP protein at their N terminus to visualize expression and localization. b, basic domain; HLH, helix-loop-helix domain; O, orange domain; Y, carboxy-terminal YRPW-TEIGAF motif. (Right) Full-length (1 to 299) Hey1 strongly represses the 10.6-kb GATA4 promoter. Lack of the basic domain significantly reduces repression activity and lack of the basic helix-loop-helix domain completely abolishes repression of GATA4-luciferase.
Hey1, Hey2, and HeyL inhibit ANF expression in vitro.
Since Hey proteins can repress GATA4 and GATA6 expression, we asked if the expression of GATA target genes would also be affected by Hey1, Hey2, and HeyL. It has been shown before that GATA4 and GATA6 can activate the ANF promoter (for review, see reference 34). A 0.7-kb ANF promoter-luciferase reporter was strongly up-regulated upon cotransfection with GATA4 and GATA6 expression plasmids in HEK293 cells. When Hey1, Hey2, or HeyL was added, a more than 90% inhibition of GATA4/6-driven ANF promoter activity was observed, even below noninduced levels (Fig. 5A).
FIG. 5.
Hey proteins downregulate the ANF promoter in vitro and they interact with GATA proteins. (A) GATA4/6-driven ANF expression is repressed by Hey1, Hey2, and HeyL. HEK293 cells were transfected with the ANF-luciferase reporter together with GATA4 and GATA6 and the different Hey1, Hey2, and HeyL expression plasmids. Basal activity with empty pCS2 vector was set to 100%. GATA4 and -6 activate the ANF promoter, which is completely blocked by Hey proteins (B) In HEK293 cells, basal ANF promoter activity is repressed by Hey1, Hey2, and to a lesser extent by HeyL. (C) Hey proteins interact with GATA4 in mammalian cells. HEK293T cells were cotransfected with HA-GATA4 and either FLAG-Hey1, -Hey2, -HeyL, or empty pCS2-FLAG expression vector. After 40 h cells were lysed and protein expression was verified by Western blotting (left). Lysates were incubated with an anti-HA antibody linked to CNBr-Sepharose to pull down HA-GATA4 and interacting proteins. After washing and sodium dodecyl sulfate-polyacrylamide gel electrophoresis immunoprecipitates were probed with anti-FLAG antibody to identify interacting FLAG-tagged Hey proteins. This interaction is specific, as no FLAG-Hey1 could be detected in the absence of HA-GATA4. (D) Hey proteins interact with GATA6. FLAG-GATA6 was pulled down from lysates of cotransfected cells with an anti-FLAG antibody bound to protein A-Sepharose. Protein interaction with HA-tagged Hey proteins is detected with an anti-HA antibody.
These results may be explained by two different mechanism of repression. First, Hey proteins may directly repress the ANF promoter. Indeed overexpression of Hey1, Hey2, and HeyL in HEK293 cells inhibited ANF-luciferase levels by 92%, 78%, and 66%, respectively (Fig. 5B), implying that Hey basic helix-loop-helix factors may directly inhibit the ANF promoter.
A second mechanism of repression could be that Hey proteins interact with GATA proteins and thereby inhibit GATA-driven gene activation. By coimmunoprecipitation of FLAG-tagged Hey and HA-tagged GATA4 proteins we could indeed show that Hey1, Hey2, and HeyL directly interact with GATA4 in mammalian cells (Fig. 5C). In addition, we could also demonstrate a direct protein-protein interaction between HA-tagged Hey proteins and FLAG-tagged GATA6 (Fig. 5D). While this work was in progress, Elagib et al. showed that Hey1 can bind directly to the amino-terminal zinc fingers of GATA1 and inhibit its function in erythropoiesis (8). In addition Kathiriya et al. demonstrated a Hey1-GATA4 protein interaction (24). Repression of GATA4-driven gene expression might by caused by a displacement of the cognate GATA4 transcriptional cofactors Friend-of-GATA Fog1 or Fog2/Zfpm2 by Hey proteins, since Fog proteins also interact with the amino-terminal zinc fingers of GATA (51, 53). This appears unlikely, however, as overexpression of Fog1 or Fog2 in HEK293 and Cos7 cells did not significantly alter the repression of GATA4-driven ANF expression by Hey factors (data not shown).
Repression activity of Hey proteins is independent of histone deacetylase activity and p300.
It has been shown before that Hey proteins can interact with the corepressors mSin3 and SIRT1, which possess histone deacetylase activity (22, 50). To determine if Hey factors mediate repression of the GATA4 and ANF promoter by recruiting histone deacetylases we treated HEK293 cells with trichostatin A, a potent histone deacetylase inhibitor. However, trichostatin A treatment in doses of 0.1, 0.5, and 1.0 μM could not prevent the repression capacity of Hey1, Hey2, and HeyL. This suggests that Hey transcription factors act in a histone deacetylase-independent way on the GATA4 and ANF promoter.
On the other hand, it is well known that GATA4 function is critically dependent on binding of the transcriptional coregulator p300, a protein with acetyltransferase activity (6). We therefore asked if Hey proteins may compete with p300 for binding to GATA4. To analyze a potential interaction we overexpressed p300 together with Hey1 and different GATA4- and ANF-luciferase reporters in transfected cells. However, this did not prevent the repression activity of the Hey proteins. Thus, our data suggest that repression of the GATA4 and ANF promoters by Hey transcription factors is not mediated either by recruitment of histone deacetylase activity or by interfering with the binding of the transcriptional activator p300.
GATA4 target genes ANF and CARP are overexpressed in Hey2-deficient hearts.
Since Hey proteins inhibit GATA4 and GATA6, loss of Hey genes may lead to increased GATA4/6 activity and this may in turn result in enhanced expression of GATA4/6 targets such as atrial natriuretic factor (ANF). One has to keep in mind, however, that there is redundancy among Hey genes and Hey1 can compensate for the loss of Hey2 in places where both are coexpressed (12). Therefore, we concentrated our analysis on the developing heart, where Hey1 is expressed in the atria and Hey2 in the ventricles, while the third family member, HeyL, is not expressed in either place. GATA4 and GATA6 show uniform expression in the fetal heart.
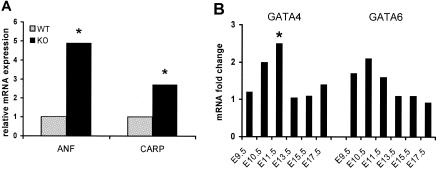
Using qRT-PCR we detected no significant changes of GATA4, GATA6, and ANF mRNA levels in Hey1-deficient atria and ventricles, indicating that GATA4/6 activity may be kept at correct levels by other pathways not affected by Hey1 loss. However, in Hey2 knockout hearts at stage E17.5 (6 knockouts and 10 controls) we found 4.9-fold higher (P = 0.01) ANF levels in the ventricles (Fig. 6A). Interestingly, even at stage E15.5 we observed 5.2-fold higher (P < 0.01) ANF levels in Hey2 knockout ventricles, whereas in earlier stages (E9.5 to E13.5) the amount of ANF mRNA was not significantly increased. In addition, mRNA levels of cardiac ankyrin repeat protein (CARP), another target gene of GATA4 and GATA6, were also up-regulated 2.7-fold (P < 0.05) in the ventricles of Hey2-deficient mice at stage E17.5. GATA4 and GATA6 mRNA levels were slightly elevated in Hey2 knockout hearts in all stages analyzed, but this only reached statistical significance at E11.5 for GATA4 (2.5-fold, P = 0.016) (Fig. 6B). In the atria of Hey2 knockout hearts ANF, CARP, GATA4, and GATA6 levels were unchanged, consistent with the ventricular restriction of Hey2 expression. These data suggest that Hey2 limits the levels of GATA4/6-driven genes to a certain level in the heart, while loss of Hey2 function leads to increased ANF and CARP levels.
FIG. 6.
Expression levels of ANF, CARP, GATA4, and GATA6 mRNAs in Hey2−/− hearts. Endogenous mRNA levels of GATA4, GATA6, and the GATA target genes ANF and CARP were measured by qRT-PCR in ventricles of wild-type and Hey2−/− fetal hearts and normalized to expression of the housekeeping gene hypoxanthine phosphoribosyltransferase (Hprt). (A) Elevated mRNA levels of GATA4/6 target genes ANF and CARP in Hey2-deficient hearts at embryonic stage E17.5. (B) Relative changes of GATA4 and GATA6 mRNAs in Hey2 knockout hearts versus wild-type controls. GATA4 and -6 are up-regulated in Hey2−/− hearts, but only at E11.5 does this reach statistical significance for GATA4 (*, P < 0.05).
Ectopic expression of ANF in the compact layer of Hey2 knockout hearts.
To determine which of the cardiac cells produce elevated amounts of ANF and CARP in embryonic Hey2 knockout hearts we employed RNA in situ hybridization on whole hearts and on paraffin sections. Interestingly, Hey2 expression is limited to the compact myocardial layer in E17.5 mouse embryo hearts, a fact not mentioned in previous expression pattern analyses (5, 29). This restriction to the compact layer can already be seen at earlier stages, e.g., at E12.5 (Fig. 7D). In control embryos CARP, GATA4, and GATA6 showed uniform expression throughout the myocardium, whereas ANF was detected in the atria and only the trabecular layer of both ventricles. Thus, in wild-type ventricles ANF and Hey2 expression domains are completely separate, with Hey2 in the compact and ANF in the trabecular layer (Fig. 7A and B). In Hey2 knockout hearts, however, there was a strong ANF expression detectable in the compact layer of the anterior and posterior wall of the left ventricle (Fig. 7E to H). Thus, loss of Hey2 leads to ectopic expression of the GATA4 and GATA6 target gene ANF in the compact ventricular layer.
FIG. 7.
Expression of Hey2, ANF, and GATA4 in wild-type and Hey2 knockout hearts. Transverse sections show Hey2 expression in the compact layer of the ventricular myocardium (A and D), whereas ANF has a complementary expression pattern with strong expression in both atria and the trabecular layer of both ventricles of control hearts (B). (E) In Hey2 knockout hearts there is ectopic expression of ANF in the compact myocardium of the left ventricle (arrow). GATA4 expression is found ubiquitously in the myocardium of both wild-type and Hey2−/− (C and F). (G and H) Whole-mount in situ hybridization of wild-type (left) and Hey2−/− (right) hearts. Anterior (G) and posterior (H) views show ectopic ANF expression (arrowheads) in the compact compartment of the left ventricular wall. All hearts are derived from embryos at stage E17.5 except for (D), which is E12.5. a, atrium; v, ventricle; tra, trabecular layer; com, compact layer.
DISCUSSION
The Notch signaling pathway regulates many aspects of cardiovascular development (1) and our recent data showed that Hey genes are the primary mediators during these processes (9). However, there is very little insight into the molecular mechanism of how Hey proteins regulate their target genes. Our study extends prior evidence that Hey proteins act as transcriptional repressors. It appears that this effect is at least to some extent not mediated via classical E box binding, but through interaction with other transcription factors. We could show that GATA4, GATA6, ANF, and CARP are up-regulated after loss of Hey factors in vitro and in vivo. In addition, the elevated levels of GATA target genes that we have found in Hey2-deficient hearts can explain some of why these mice develop fatal cardiac hypertrophy.
Genetic redundancy can pose severe problems in the search for target genes of a transcription factor family. Our analysis of Hey1/2 double-knockout mice revealed a strong redundancy between these genes, at least in endothelial cells (12). The comparison of Hey1+/+ with Hey1−/− cells on a Hey2−/− background eliminates such potential compensatory mechanisms. This was achieved through the use of conditional Hey1 knockout mice. These mice will be a valuable tool in the future when combined with tissue-specific or inducible Cre transgenes in adult animals. For the current study, however, we employed the floxed Hey1 allele to establish Hey1fl/fl/Hey2−/− ES cell lines that were in turn used to create paired cell lines representing the Hey2 single-knockout situation and Hey1/2 double deficiency. These lines are ideally suited for comparative microarray expression analysis since they are derived from the same parental cells under identical conditions, ruling out variability between independently generated lines.
GATA factor genes are regulated by Hey genes.
Among the genes that are differentially regulated between embryoid bodies from single- and double-knockout cells were the zinc finger transcription factors GATA4 and GATA6, which are known as key cardiac regulators. They have been shown to activate transcription of numerous cardiac genes, such as the ANF, BNP, CARP, myosin light chain, and myosin heavy chain genes (41). GATA4 up-regulation was already seen in two independent Hey1/2 double-knockout ES cell lines compared to their Hey2−/− control lines. These data are functionally rather intriguing as it has been shown that transgenic GATA4 overexpression in cardiomyocytes leads to cardiac hypertrophy (31). Hey2-deficient hearts also develop massive cardiac hypertrophy shortly after birth suggesting that this hypertrophy in Hey2 knockout mice may at least in part be caused by elevated levels of GATA factors.
Despite the great interest in GATA4 as one of the best studied cardiac regulators, very little is known about factors that drive GATA4 expression in vivo. Only for the zebrafish GATA4 gene have T-box binding sites been identified as essential for cardiac activity (17). In our transfection studies, all Hey proteins strongly repressed the GATA4 promoter. The basic domain of Hey1 appears to be most important for this effect since deletion of the amino-terminus with or without the helix-loop-helix domain almost abolishes repression activity. Most surprisingly, the evolutionarily conserved carboxy terminus may not be important in this context. This agrees with previous findings by Iso et al. (22), who also mapped the critical region for repression by Hey2-GAL4 fusion proteins to the basic helix-loop-helix domain.
In previous studies we and others could show that Hey proteins can bind to DNA at the E-box sequence CACGTG (11, 21, 40). This element is found only once in the 10.6-kb GATA4 promoter, but its mutation only partially reduced Hey repression activity. Our results imply that Hey proteins, at least to some extent, repress GATA4/6 transcription indirectly through protein-protein interaction with another transcription factor. Indeed, it has been shown before that Hey proteins can repress transcription by interfering with heterodimer formation of transcription factors such as MyoD/E47 and ARNT/EPAS1 (5, 49).
Effects of Hey proteins on the ANF promoter.
Given the clear repression of the GATA4 promoter by Hey proteins and the elevated expression of GATA genes after Hey1/2 deletion in our embryoid bodies, we speculated that a similar effect might be seen for GATA target genes. We indeed found a strong induction of ANF in Hey1/2 double-knockout embryoid bodies. ANF is a key regulator of electrolyte and body fluid homoeostasis, and it is expressed from the earliest stage of heart development in both atria and ventricles. Around birth, expression is switched off in the ventricles, whereas atrial expression remains high throughout adulthood (2, 58). In response to chronic volume or pressure overload, ANF expression is reactivated (42) and represents a molecular hallmark of cardiac hypertrophy. Therefore, understanding the regulation of ANF expression should provide valuable insights into the pathophysiology of cardiac hypertrophy and high blood pressure, two very common human diseases.
The transcription factors GATA4, GATA6, Nkx2.5, Tbx5, MEF2, and serum response factor are well described activators of the ANF promoter (for a summary see reference 39), while Tbx2 and HOP are known to be repressors (4, 16, 47). Our data add the Hey factors to the group of ANF repressors. Hey proteins may repress the ANF promoter activity indirectly through inhibition of GATA gene expression, but there seems to be an even stronger and direct effect on GATA4/6-driven transcriptional activation. Furthermore, we found a down-regulation of the ANF-luciferase reporter in HEK293 cells even in the absence of exogenous GATA factors. The latter is in contrast to the results of Kathiriya et al., who found no direct ANF repression by Hey factors in HeLa cells (24). This obvious discrepancy may be due to differences in cellular context, since we could just barely detect any ANF-luciferase activity in these cells. The ANF promoter even contains potential binding sites for Hey factors, but we cannot rule out that the ANF promoter is driven by GATA4/6 only in 293 cells, where forced Hey expression interferes with GATA-dependent activation.
An indirect mode of repression through complex formation with unrelated activating factors appears rather unusual for classical basic helix-loop-helix factors such as Hey proteins. Nevertheless, it has been shown very recently that Hey proteins, after induction through c-Jun, inhibit GATA1 and GATA2 transcriptional activity through binding to one of its zinc fingers in K562 cells (8). This GATA1-Hey1 complex could still bind to a minimal GATA-dependent promoter lacking any E-box sequences. While this manuscript was in preparation, Kim et al. (25) provided evidence that the transcription factor jumonji can also repress ANF expression. Again this is apparently not mediated by direct DNA binding, but by protein-protein interaction with GATA4 and Nkx2.5. These factors can still bind to the ANF promoter but are devoid of transcriptional activity when jumonji is bound. These observations parallel very nicely the way in which Hey proteins seem to act on the ANF promoter.
The nature of the alteration in transcriptional properties of GATA factors remains unclear, however. Fog proteins are critical coregulators that also interact with the amino terminal zinc finger of GATA factors. In the developing heart Fog2 expression overlaps with GATA4,5,6 and Fog2−/− embryos die between E12.5 and E15.5 with complex cardiac defects (52). It has been reported that Fog2 can either activate or repress GATA4 function, depending on cell type and p300 levels (19, 32). It is conceivable that the Hey and Fog proteins compete for GATA binding. Our data do not support this model, however, as forced expression of Fog did not alter the repression of GATA-driven ANF expression by Hey factors. In addition, we also ruled out the possibility that Hey proteins interfere with the transcriptional coactivator p300.
Consistent with our data, Elagib et al. provided evidence that Hey1 acts in a Fog-independent manner on GATA1 (8). Another mode of action for Hey proteins would be through histone modification. It was shown that Hey proteins can recruit corepressors such as N-Cor or the histone deacetylase complex mSin3 (22). These proteins can modify the chromatin structure and decrease the expression of the affected genomic regions. However, we found an unaltered repression capacity of Hey factors when we blocked histone deacetylase activity with trichostatin A, leaving open the mode of action of Hey proteins in this case.
Hey2 effects in vivo.
Our finding that ANF is ectopically expressed in the compact myocardial layer of Hey2 knockout mice further extends our in vitro data that Hey2 is a repressor of ANF expression. It is interesting that in the embryonic heart ANF and Hey2 are expressed in different compartments of the ventricles, with Hey2 in the compact and ANF in the trabecular layer. Around birth, expression of both genes is switched off in the ventricle. Therefore it is tempting to speculate that a physiological function of Hey2 is to prevent ANF activation in the compact layer. Although the changes in GATA4 and GATA6 mRNA levels in Hey1- or Hey2-deficient hearts did not reach statistical significance in most of the embryonic stages analyzed, it is interesting that GATA4 and -6 were up-regulated about twofold in Hey2-deficient hearts around E11.5. This misregulation might be involved in the development of ventricular septal defects, as septation of the ventricles begins during this time. The elevated ANF mRNA levels we found in older Hey2 knockout hearts cannot be explained by increased amounts of GATA4/6, however, the loss of functional repression of GATA factors on the protein level should suffice to explain the elevated ventricular ANF expression in Hey2 mutants.
Our findings of increased GATA4 and GATA6 levels in embryoid bodies are very intriguing not only for cardiac but also for blood vessel development. Hey1/2-deficient mice die due to severe defects in blood vessel formation and the specification of arteries. GATA6 is known to be expressed in arterial smooth muscle cells (35) and it regulates genes for the synthetic function of vascular smooth muscle cells (30). Hey2/CHF1 has also been shown to regulate vascular smooth muscle cell proliferation and migration (45) and it is tempting to speculate that GATA factors are misregulated in the developing vasculature of Hey1/2 double-knockout embryos. We indeed found an up-regulation of GATA4 and GATA6 in the yolk sacs of these mutants, but this effect did not fully reach statistical significance (A. Fischer and M. Gessler, unpublished data).
Based on our current findings, it should be quite revealing to generate and analyze conditionally Hey-overexpressing mice in the future. Forced Hey2 expression in the entire myocardium may repress GATA-dependent genes in the trabecular layer of the developing heart or generally at later stages of life, when the endogenous Hey2 locus has long been silenced. It will also be interesting to test if temporary Hey induction slows down the development of cardiac hypertrophy in mouse model systems.
Acknowledgments
We thank Anja Winkler, Sabrina Schrauth, Susanne Spahr, and Vanessa Cook for excellent technical assistance and Alexandra Klaus for maintaining the mouse colonies. We are in debt to Birgit Zirn, Michael Krause, Birgit Samans, and Martin Eilers for their help with the microarray experiments. We gratefully acknowledge Mona Nemer, Stuart Orkin, and Werner Lutz for providing probes.
This work was supported by grants from the Deutsche Forschungsgemeinschaft to C.E. (DFG En 280/6-1) and M.G. (DFG Ge539-9) and the DFG Graduiertenkolleg 1048 (Organogenesis).
REFERENCES
- 1.Alva, J. A., and M. L. Iruela-Arispe. 2004. Notch signaling in vascular morphogenesis. Curr. Opin. Hematol. 11:278-283. [DOI] [PubMed] [Google Scholar]
- 2.Argentin, S., A. Ardati, S. Tremblay, I. Lihrmann, L. Robitaille, J. Drouin, and M. Nemer. 1994. Developmental stage-specific regulation of atrial natriuretic factor gene transcription in cardiac cells. Mol. Cell. Biol. 14:777-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berwanger, B., O. Hartmann, E. Bergmann, S. Bernard, D. Nielsen, M. Krause, A. Kartal, D. Flynn, R. Wiedemeyer, M. Schwab, H. Schafer, H. Christiansen, and M. Eilers. 2002. Loss of a FYN-regulated differentiation and growth arrest pathway in advanced stage neuroblastoma. Cancer Cell 2:377-386. [DOI] [PubMed] [Google Scholar]
- 4.Chen, F., H. Kook, R. Milewski, A. D. Gitler, M. M. Lu, J. Li, R. Nazarian, R. Schnepp, K. Jen, C. Biben, G. Runke, J. P. Mackay, J. Novotny, R. J. Schwartz, R. P. Harvey, M. C. Mullins, and J. A. Epstein. 2002. Hop is an unusual homeobox gene that modulates cardiac development. Cell 110:713-723. [DOI] [PubMed] [Google Scholar]
- 5.Chin, M. T., K. Maemura, S. Fukumoto, M. K. Jain, M. D. Layne, M. Watanabe, C. M. Hsieh, and M. E. Lee. 2000. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J. Biol. Chem. 275:6381-6387. [DOI] [PubMed] [Google Scholar]
- 6.Dai, Y. S., and B. E. Markham. 2001. p300 Functions as a coactivator of transcription factor GATA-4. J. Biol. Chem. 276:37178-37185. [DOI] [PubMed] [Google Scholar]
- 7.Donovan, J., A. Kordylewska, Y. N. Jan, and M. F. Utset. 2002. Tetralogy of fallot and other congenital heart defects in Hey2 mutant mice. Curr. Biol. 12:1605-1610. [DOI] [PubMed] [Google Scholar]
- 8.Elagib, K. E., M. Xiao, I. M. Hussaini, L. L. Delehanty, L. A. Palmer, F. K. Racke, M. J. Birrer, G. Shanmugasundaram, M. A. McDevitt, and A. N. Goldfarb. 2004. Jun blockade of erythropoiesis: role for repression of GATA-1 by HERP2. Mol. Cell. Biol. 24:7779-7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer, A., and M. Gessler. 2003. Hey genes in cardiovascular development. Trends Cardiovasc. Med. 13:221-226. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, A., B. Klamt, N. Schumacher, C. Glaeser, I. Hansmann, H. Fenge, and M. Gessler. 2004. Phenotypic variability in Hey2-/- mice and absence of HEY2 mutations in patients with congenital heart defects or Alagille syndrome. Mamm Genome 15:711-716. [DOI] [PubMed] [Google Scholar]
- 11.Fischer, A., C. Leimeister, C. Winkler, N. Schumacher, B. Klamt, H. Elmasri, C. Steidl, M. Maier, K. P. Knobeloch, K. Amann, A. Helisch, M. Sendtner, and M. Gessler. 2002. Hey bHLH factors in cardiovascular development. Cold Spring Harb. Symp. Quant. Biol. 67:63-70. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, A., N. Schumacher, M. Maier, M. Sendtner, and M. Gessler. 2004. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 18:901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garg, V., I. S. Kathiriya, R. Barnes, M. K. Schluterman, I. N. King, C. A. Butler, C. R. Rothrock, R. S. Eapen, K. Hirayama-Yamada, K. Joo, R. Matsuoka, J. C. Cohen, and D. Srivastava. 2003. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424:443-447. [DOI] [PubMed] [Google Scholar]
- 14.Gessler, M., K. P. Knobeloch, A. Helisch, K. Amann, N. Schumacher, E. Rohde, A. Fischer, and C. Leimeister. 2002. Mouse gridlock: no aortic coarctation or deficiency, but fatal cardiac defects in Hey2-/- mice. Curr. Biol. 12:1601-1604. [DOI] [PubMed] [Google Scholar]
- 15.Gu, H., Y. R. Zou, and K. Rajewsky. 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73:1155-1164. [DOI] [PubMed] [Google Scholar]
- 16.Habets, P. E., A. F. Moorman, D. E. Clout, M. A. van Roon, M. Lingbeek, M. van Lohuizen, M. Campione, and V. M. Christoffels. 2002. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 16:1234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heicklen-Klein, A., and T. Evans. 2004. T-box binding sites are required for activity of a cardiac GATA-4 enhancer. Dev. Biol. 267:490-504. [DOI] [PubMed] [Google Scholar]
- 18.Henderson, A. M., S. J. Wang, A. C. Taylor, M. Aitkenhead, and C. C. Hughes. 2001. The basic helix-loop-helix transcription factor HESR1 regulates endothelial cell tube formation. J. Biol. Chem. 276:6169-6176. [DOI] [PubMed] [Google Scholar]
- 19.Hirai, M., K. Ono, T. Morimoto, T. Kawamura, H. Wada, T. Kita, and K. Hasegawa. 2004. FOG-2 competes with GATA-4 for transcriptional coactivator p300 and represses hypertrophic responses in cardiac myocytes. J. Biol. Chem. 279:37640-37650. [DOI] [PubMed] [Google Scholar]
- 20.Huang, Q., A. Raya, P. DeJesus, S. H. Chao, K. C. Quon, J. S. Caldwell, S. K. Chanda, J. C. Izpisua-Belmonte, and P. G. Schultz. 2004. Identification of p53 regulators by genome-wide functional analysis. Proc. Natl. Acad. Sci. USA 101:3456-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell Physiol. 194:237-255. [DOI] [PubMed] [Google Scholar]
- 22.Iso, T., V. Sartorelli, C. Poizat, S. Iezzi, H. Y. Wu, G. Chung, L. Kedes, and Y. Hamamori. 2001. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol. Cell. Biol. 21:6080-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh, F., S. Itoh, M. J. Goumans, G. Valdimarsdottir, T. Iso, G. P. Dotto, Y. Hamamori, L. Kedes, M. Kato, and P. Dijke Pt. 2004. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 23:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kathiriya, I. S., I. N. King, M. Murakami, M. Nakagawa, J. M. Astle, K. A. Gardner, R. D. Gerard, E. N. Olson, D. Srivastava, and O. Nakagawa. 2004. Hairy-related transcription factors inhibit GATA-dependent cardiac gene expression through a signal-responsive mechanism. J. Biol. Chem. 279:54937-54943. [DOI] [PubMed] [Google Scholar]
- 25.Kim, T. G., J. Chen, J. Sadoshima, and Y. Lee. 2004. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol. Cell. Biol. 24:10151-10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kokubo, H., S. Miyagawa-Tomita, H. Tomimatsu, Y. Nakashima, M. Nakazawa, Y. Saga, and R. L. Johnson. 2004. Targeted disruption of hesr2 results in atrioventricular valve anomalies that lead to heart dysfunction. Circ. Res. 95:540-547. [DOI] [PubMed] [Google Scholar]
- 27.Krebs, L. T., Y. Xue, C. R. Norton, J. R. Shutter, M. Maguire, J. P. Sundberg, D. Gallahan, V. Closson, J. Kitajewski, R. Callahan, G. H. Smith, K. L. Stark, and T. Gridley. 2000. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14:1343-1352. [PMC free article] [PubMed] [Google Scholar]
- 28.Leimeister, C., A. Bach, and M. Gessler. 1998. Developmental expression patterns of mouse sFRP genes encoding members of the secreted frizzled related protein family. Mech. Dev. 75:29-42. [DOI] [PubMed] [Google Scholar]
- 29.Leimeister, C., A. Externbrink, B. Klamt, and M. Gessler. 1999. Hey genes: a novel subfamily of hairy- and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mech. Dev. 85:173-177. [DOI] [PubMed] [Google Scholar]
- 30.Lepore, J. J., T. P. Cappola, P. A. Mericko, E. E. Morrisey, and M. S. Parmacek. 2005. GATA-6 regulates genes promoting synthetic functions in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 25:309-314. [DOI] [PubMed] [Google Scholar]
- 31.Liang, Q., L. J. De Windt, S. A. Witt, T. R. Kimball, B. E. Markham, and J. D. Molkentin. 2001. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 276:30245-30253. [DOI] [PubMed] [Google Scholar]
- 32.Lu, J. R., T. A. McKinsey, H. Xu, D. Z. Wang, J. A. Richardson, and E. N. Olson. 1999. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol. Cell. Biol. 19:4495-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maier, M. M., and M. Gessler. 2000. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem. Biophys. Res. Commun. 275:652-660. [DOI] [PubMed] [Google Scholar]
- 34.McBride, K., and M. Nemer. 2001. Regulation of the ANF and BNP promoters by GATA factors: lessons learned for cardiac transcription. Can. J. Physiol. Pharmacol. 79:673-681. [PubMed] [Google Scholar]
- 35.Morrisey, E. E., H. S. Ip, M. M. Lu, and M. S. Parmacek. 1996. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev. Biol. 177:309-322. [DOI] [PubMed] [Google Scholar]
- 36.Nagy, A., M. Gertsenstein, K. Vintersten, and R. Behringer. 2003. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Nakagawa, O., D. G. McFadden, M. Nakagawa, H. Yanagisawa, T. Hu, D. Srivastava, and E. N. Olson. 2000. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc. Natl. Acad. Sci. USA 97:13655-13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peitz, M., K. Pfannkuche, K. Rajewsky, and F. Edenhofer. 2002. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. USA 99:4489-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterkin, T., A. Gibson, M. Loose, and R. Patient. 2005. The roles of GATA-4, -5 and -6 in vertebrate heart development. Semin. Cell. Dev. Biol. 16:83-94. [DOI] [PubMed] [Google Scholar]
- 40.Pichon, B., V. Taelman, E. J. Bellefroid, and D. Christophe. 2004. Transcriptional repression by the bHLH-Orange factor XHRT1 does not involve the C-terminal YRPW motif. Biochim. Biophys. Acta 1680:46-52. [DOI] [PubMed] [Google Scholar]
- 41.Pikkarainen, S., H. Tokola, R. Kerkela, and H. Ruskoaho. 2004. GATA transcription factors in the developing and adult heart. Cardiovasc. Res. 63:196-207. [DOI] [PubMed] [Google Scholar]
- 42.Rockman, H. A., R. S. Ross, A. N. Harris, K. U. Knowlton, M. E. Steinhelper, L. J. Field, J. Ross, Jr., and K. R. Chien. 1991. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 88:8277-8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakamoto, M., H. Hirata, T. Ohtsuka, Y. Bessho, and R. Kageyama. 2003. The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J. Biol. Chem. 278:44808-44815. [DOI] [PubMed] [Google Scholar]
- 44.Sakata, Y., C. N. Kamei, H. Nakagami, R. Bronson, J. K. Liao, and M. T. Chin. 2002. Ventricular septal defect and cardiomyopathy in mice lacking the transcription factor CHF1/Hey2. Proc. Natl. Acad. Sci. USA 99:16197-16202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakata, Y., F. Xiang, Z. Chen, Y. Kiriyama, C. N. Kamei, D. I. Simon, and M. T. Chin. 2004. Transcription factor CHF1/Hey2 regulates neointimal formation in vivo and vascular smooth muscle proliferation and migration in vitro. Arterioscler. Thromb. Vasc. Biol. 24:2069-2074. [DOI] [PubMed] [Google Scholar]
- 46.Satow, T., S. K. Bae, T. Inoue, C. Inoue, G. Miyoshi, K. Tomita, Y. Bessho, N. Hashimoto, and R. Kageyama. 2001. The basic helix-loop-helix gene hesr2 promotes gliogenesis in mouse retina. J. Neurosci. 21:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin, C. H., Z. P. Liu, R. Passier, C. L. Zhang, D. Z. Wang, T. M. Harris, H. Yamagishi, J. A. Richardson, G. Childs, and E. N. Olson. 2002. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 110:725-735. [DOI] [PubMed] [Google Scholar]
- 48.Steidl, C., C. Leimeister, B. Klamt, M. Maier, I. Nanda, M. Dixon, R. Clarke, M. Schmid, and M. Gessler. 2000. Characterization of the human and mouse HEY1, HEY2, and HEYL genes: cloning, mapping, and mutation screening of a new bHLH gene family. Genomics 66:195-203. [DOI] [PubMed] [Google Scholar]
- 49.Sun, J., C. N. Kamei, M. D. Layne, M. K. Jain, J. K. Liao, M. E. Lee, and M. T. Chin. 2001. Regulation of myogenic terminal differentiation by the hairy-related transcription factor CHF2. J. Biol. Chem. 276:18591-18596. [DOI] [PubMed] [Google Scholar]
- 50.Takata, T., and F. Ishikawa. 2003. Hum. Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 301:250-257. [DOI] [PubMed] [Google Scholar]
- 51.Tevosian, S. G., A. E. Deconinck, A. B. Cantor, H. I. Rieff, Y. Fujiwara, G. Corfas, and S. H. Orkin. 1999. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc. Natl. Acad. Sci. USA 96:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tevosian, S. G., A. E. Deconinck, M. Tanaka, M. Schinke, S. H. Litovsky, S. Izumo, Y. Fujiwara, and S. H. Orkin. 2000. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell 101:729-739. [DOI] [PubMed] [Google Scholar]
- 53.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujiwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 54.Van Wayenbergh, R., V. Taelman, B. Pichon, A. Fischer, S. Kricha, M. Gessler, D. Christophe, and E. J. Bellefroid. 2003. Identification of BOIP, a novel cDNA highly expressed during spermatogenesis that encodes a protein interacting with the orange domain of the hairy-related transcription factor HRT1/Hey1 in Xenopus and mouse. Dev. Dyn. 228:716-725. [DOI] [PubMed] [Google Scholar]
- 55.Vittet, D., M. H. Prandini, R. Berthier, A. Schweitzer, H. Martin-Sisteron, G. Uzan, and E. Dejana. 1996. Embryonic stem cells differentiate in vitro to endothelial cells through successive maturation steps. Blood 88:3424-3431. [PubMed] [Google Scholar]
- 56.Zamurovic, N., D. Cappellen, D. Rohner, and M. Susa. 2004. Coordinated activation of notch, Wnt, and transforming growth factor-beta signaling pathways in bone morphogenic protein 2-induced osteogenesis. Notch target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J. Biol. Chem. 279:37704-37715. [DOI] [PubMed] [Google Scholar]
- 57.Zavadil, J., L. Cermak, N. Soto-Nieves, and E. P. Bottinger. 2004. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 23:1155-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeller, R., K. D. Bloch, B. S. Williams, R. J. Arceci, and C. E. Seidman. 1987. Localized expression of the atrial natriuretic factor gene during cardiac embryogenesis. Genes Dev. 1:693-698. [DOI] [PubMed] [Google Scholar]
- 59.Zhong, T. P., S. Childs, J. P. Leu, and M. C. Fishman. 2001. Gridlock signalling pathway fashions the first embryonic artery. Nature 414:216-220. [DOI] [PubMed] [Google Scholar]