Abstract
The tumor-selective, proapoptotic, death receptor ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a mediator of antitumor drug activity and in itself is a promising agent for the treatment of human malignancies. Like many tumors, however, glioblastoma multiforme (GBM), the most fatal form of glioma, exhibits a range of TRAIL sensitivity, and only a small percentage of GBM tumors undergo TRAIL-induced apoptosis. We here show that TRAIL resistance in GBM is a consequence of overexpression of the short isoform of the caspase-8 inhibitor, c-FLICE inhibitory protein (FLIPS), and that FLIPS expression is in turn translationally enhanced by activation of the Akt-mammalian target of rapamycin (mTOR)-p70 S6 kinase 1 (S6K1) pathway. Conversely, pharmacologic or genetic inhibition of mTOR, or the mTOR target S6K1, suppresses polyribosomal accumulation of FLIPS mRNA, FLIPS protein expression, and TRAIL resistance. In archived material from 12 human GBM tumors, PTEN status was a predictor of activation of the Akt-mTOR-S6K1 pathway and of FLIPS levels, while in xenografted human GBM, activation status of the PTEN-Akt-mTOR pathway distinguished the tumors inherently sensitive to TRAIL from those which could be sensitized by the mTOR inhibitor rapamycin. These results define the mTOR pathway as a key limiter of tumor elimination by TRAIL-mediated mechanisms, provide a means by which the TRAIL-sensitive subset of GBM can be identified, and provide rationale for the combined use of TRAIL with mTOR inhibitors in the treatment of human cancers.
Mammalian target of rapamycin (mTOR) is a phosphatidylinositol 3 kinase (PI3K)-related serine/threonine kinase that regulates a range of cellular functions. mTOR-dependent signaling is regulated by a variety of means including the PI3K/Akt pathway (14, 24), amino acids (26), and ATP (12). In the simplest sense, PI3K-mediated activation of Akt leads to phosphorylation of TSC2, which in turn inhibits the function of the TSC1/2 complex (31, 54). In the absence of TSC1/2, the small GTPase Rheb enhances mTOR activity, stimulating activation of downstream targets of mTOR (7).
The best-characterized downstream effectors of mTOR are the 70-kDa ribosomal S6 kinase 1 (S6K1) and the eukaryotic translation initiation factor 4E (eIF4E) binding protein 1 (4E-BP1) (18-20, 23, 25). S6K1 directly phosphorylates the 40S ribosomal protein S6, which in turn has been proposed to increase the translation of mRNAs that possess a 5′ terminal oligopyrimidine sequence (5′ TOP), most notably ribosomal proteins and translation elongation factors (18-20, 23). 4E-BP1, in contrast, binds to and sequesters the rate-limiting translation initiation factor eIF4E, which is involved in binding of the 5′ mRNA cap structure and the initiation of cap-dependent translation (18-20, 23, 25). mTOR-mediated 4E-BP1 phosphorylation alters the properties of eIF4E, allowing interaction of eIF4E with eIF4A, eIF4G, and eIF4B (23, 25), binding of the mRNA 5′ cap structure, unwinding of the cap-proximal mRNA secondary structure, 40S ribosomal subunit recruitment to mRNA, and initiation of translation (23). The net result of mTOR-mediated stimulation of the S6K1 and 4E-BP1/eIF4E pathways, therefore, is considered to be enhanced translation of mRNAs encoding proteins involved with cell growth/size and cell cycle progression (17-20, 22, 23, 33). Other studies, however, suggest that translational control of TOP mRNAs is independent of S6K1 (66). mTOR has also been shown to contribute in a non-translation-dependent manner to the regulation of apoptosis and autophagy (5, 15), suggesting that the full range of mTOR function remains to be defined.
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a 281-amino-acid proapoptotic ligand of the tumor necrosis factor superfamily. TRAIL binds to the death receptors DR4/DR5, which, in their bound form, interact with the adaptor protein FADD and procaspase-8, forming the death inducing signaling complex in target cells (1, 40). Procaspase-8 activation in the death inducing signaling complex leads to cleavage/activation of procaspase-3 and engagement of the cellular machinery associated with the type I extrinsic apoptotic pathway (6, 44). Activation of the intrinsic, mitochondrial-associated type II apoptotic pathway also appears to play some role in TRAIL-induced cell death because TRAIL, through caspase-8, activates Bid and synergizes with agents that induce apoptosis exclusively through a type II mechanism (11, 52). The normal function of TRAIL remains unclear, although studies suggest that TRAIL plays a key role in tumor surveillance by the immune system (60). Up-regulation of TRAIL expression has also been noted following cellular exposure to histone deacetylase inhibitors (2, 47), suggesting that in addition to endogenous functions, TRAIL plays a role in chemotherapy-based tumor elimination. Consistent with this idea, TRAIL has been shown to induce apoptosis in a wide variety of tumor cells but not in normal cells (21, 67). Although the tumor selectivity of TRAIL suggested its use in the treatment of various malignancies, including glioma (45), early reports showed that some forms of recombinant TRAIL were hepatotoxic (36, 42, 48). Separate studies, however, have shown that intracranial delivery of native human TRAIL suppresses the growth of human glioma xenografts in mice without host toxicity (57, 58). These newer studies suggest that TRAIL, as one of a very few truly cancer cell-specific inducers of cell death, has significant potential for glioma therapy.
A limiting factor in the success of TRAIL and TRAIL-dependent regimens, however, is the suggestion that a significant percentage of human tumors may be insensitive to TRAIL-induced apoptosis. As an example, compiled data suggest that less than 50% of glioma cell lines undergo TRAIL-induced apoptosis (59, 65, 69). Studies in nonglioma cells have suggested that mutation of TRAIL receptors, silencing of TRAIL receptor expression, and/or upregulation of TRAIL decoy receptors are underlying mechanisms of TRAIL resistance (30, 49, 59). No clear correlation, however, exists between TRAIL sensitivity and expression of TRAIL receptors or TRAIL decoy receptors in the glioma cell lines examined (56). These studies suggest that TRAIL resistance in glioma cells is not receptor based but, rather, is the result of alterations in the pathway that links TRAIL receptor activation to the apoptotic machinery.
There are a number of means by which the connection between TRAIL receptor activation and the apoptotic machinery could be altered in TRAIL-resistant gliomas. The Akt pathway is of particular interest as it is known to be activated in a majority of human gliomas as a consequence of loss of PTEN function (63). Additionally, because Akt is known to play a role in the control of apoptosis (3), we considered the possibility that Akt and/or downstream targets of Akt might contribute to the linkage of TRAIL receptors to the apoptotic machinery and to the regulation of TRAIL sensitivity in glioma. The results of these studies show that the Akt target, mTOR, alters ribosomal distribution and translation of the mRNA encoding FLIPS, a FLIP splice variant that blocks caspase-8 activation (41), and in doing so confers TRAIL resistance to glioblastoma multiforme (GBM) cells. These results define a novel means by which mTOR and its downstream targets regulate apoptosis, define the PTEN-Akt-mTOR pathway as a key limiter of tumor elimination by TRAIL-mediated mechanisms, provide a means by which the TRAIL-sensitive subset of GBM can be identified, and provide a rationale for the combined use of TRAIL with rapamycin in the treatment of human cancers.
MATERIALS AND METHODS
Cell culture, drug treatment, and tumor material.
Human GBM cell lines were cultured in Dulbecco's modified Eagle's medium (H-21) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere. Human recombinant TRAIL was purchased from Sigma and was dissolved in dimethyl sulfoxide (DMSO). Rapamycin was purchased from Cell Signaling Technology and dissolved in DMSO. Wortmannin and 4-hydroxytamoxifen (4HT) were purchased from Sigma and were dissolved in DMSO. Cells were exposed to TRAIL (0 to 1,000 ng/ml) for 24 h prior to harvest, except for caspase activation studies in which cells were harvested 0 to 7 h after TRAIL exposure. In some cases cells were preincubated with 100 nM rapamycin, 4HT (10 nM), or 4HT plus rapamycin (100 nM) for 30 min, after which TRAIL was added. Protein extracts from resected primary GBM tissue were obtained from the University of California San Francisco Brain Tumor Research Center Tissue Bank. Freshly resected human GBM xenografts were obtained from the Mayo Clinic (Rochester, MN), dissected into small (<1-mm diameter) pieces, passed through a 100-μm-pore-size tissue culture sieve (Fisher Scientific, Santa Clara, CA), and grown (48 h) on reduced matrigel-coated dishes, after which cells were exposed to TRAIL (800 ng/ml; 24 h), rapamycin (100 nM; 24 h), or rapamycin plus TRAIL. The PTEN status of GBM tissues was determined as previously described (16).
Immunoblot analysis and analysis of apoptosis by flow cytometry.
Cells were washed with ice-cold phosphate-buffered saline, scraped from the culture dish, and incubated in tissue lysis buffer containing 10 mM KCl, 1 mM sucrose, 2 mM MgCl2, 0.5% Igepal CA-630, 1 mM EDTA, 1 mM dithiothreitol, 10 mM β-glycerophosphate, 1 mM Na3VO4, 10 mM NaF, 100 μg/ml phenylmethylsulfonyl fluoride, and 10 μg/ml aprotinin (all reagents were purchased from Sigma) for 30 min on ice. The cell lysate was centrifuged, and the supernatant was stored at −80°C until use. The protein concentration of extracts was measured using a Protein Assay reagent (Bio-Rad Laboratories). Protein (30 μg) was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted onto Immobilon-P membrane (Millipore). The membrane was blocked in 5% nonfat skim milk-20 mM Tris-HCl (pH 7.4)-150 mM NaCl-0.1% Tween 20 at 4°C overnight and incubated for 1 h at room temperature with the following: rabbit polyclonal antibody against the N terminus of DR4, DR5, DcR1, or DcR2 (Chemicon); mouse polyclonal antibody against alpha tubulin, p70 S6K1, phospho-S6K1 (Thr-389), or 4E-BP1 (Santa Cruz Biotechnology); rabbit antibody against AU1 tags (Abcam); rabbit polyclonal antibody against eIF4E, phospho-S6, phospho-Akt (Ser-473), or glycogen sythase kinase 3α/β (GSK-3α/β; Cell Signaling Technology); goat polyclonal antibody against the N terminus of procaspase-8 or -3 (Chemicon); rabbit polyclonal antibody against the C terminus of procaspase-8 or -3 (Cell Signaling Technology); or goat monoclonal antibodies against FLIPL and FLIPS (Santa Cruz Biotechnology). Bound antibody was detected with mouse anti-goat immunoglobulin G (IgG), goat anti-rabbit IgG, or goat anti-mouse IgG (Santa Cruz Biotechnology) using enhanced chemiluminescence Western blotting detection regents (Amersham Pharmacia Biotech, Inc). Densitometric measurements of immunoreactive bands were acquired using an AlphaImager 2200 (Alpha Innotech Corporation, San Leandro, CA). The expression of α-tubulin was used to verify equal loading in all studies. The extent of apoptosis in cultures (attached and floating cells) was determined by fluorescence-activated cell sorting (FACS) analysis (sub-G1 DNA content), with measurements verified by annexin V/propidium iodide staining as previously described (9).
Retroviral infection, transfection of plasmids, and siRNA.
The pFB retroviral constructs encoding FLIPS or FLIPL protein were kindly provided by L. Bin (University of Colorado) (4). The 4HT-inducible Akt construct was a generous gift from M. McMahon (University of California San Francisco) (28). The pcDNA3 expression vectors encoding rapamycin resistant (RR) mTOR (Ser2035Ile) or kinase dead (KD) mTOR (Asp2338Ala) were kindly provided by Robert Abraham (Burnham Institute, San Diego, CA), as were the pACTAG2/HA-WT-4E-BP1 expression vector, encoding hemagglutinin (HA)-tagged wild-type (WT) 4E-BP1, and the related pRK7/HA-S6K1 and pCAN/HA-eIF4E expression vectors, encoding HA-tagged WT p70 S6K1 and HA-tagged WT eIF4E, respectively. The retroviral pBABE/K100R-S6K1 constructs encoding inactive (K100R) S6K1 or rapamycin-resistant S6K1 (pBABE/F5A-E389) were kindly provided by J. Blenis (Harvard University, Boston, MA) (10). The pMV7/W73A-eIF4E retroviral construct encoding an inactive (W73A) eIF4E was kindly provided by Kathy Borden (University of Montreal) (61). Retroviral vectors were used to infect cells as previously described, while expression constructs were transfected into target cells (62). Pools of productively infected cells (obtained by selection with neomycin [1 mg/ml; 7 days] or puromycin [9 μg/ml; 7 days]) were used for further analysis. In cells expressing multiple constructs, all retroviral infections and selections were done serially. The small interfering RNA (siRNA) sequences targeting FLIPL or FLIPS were previously described (51). For siRNA studies, 200 nM FLIP-targeted siRNA (Ambion) or 1 μM p70 S6 kinase SMARTpool siRNAs or scramble siRNA (Dharmacon, Lafayette, CO) was transfected into cells, and protein levels were analyzed 1 to 4 days later. Four days after siRNA introduction, cells were incubated with TRAIL (800 ng/ml; 24 h) and assessed for apoptosis by FACS analysis.
In vitro kinase assay.
For Akt-kinase assays, cells were allowed to reach 70% confluence in Dulbecco's modified Eagle's medium. Cells were washed once with ice-cold phosphate-buffered saline and harvested using 1× ice-cold cell lysis buffer supplemented with 1 mM phenylmethylsulfonyl fluoride and incubated on ice for 10 min. Akt was selectively immunoprecipitated from 200 μg of protein (whole-cell lysates) by combining the cell lysate with 20 μl of Akt monoclonal antibody (Cell Signaling Technology) conjugated to agarose A/G beads (Santa Cruz Biotechnology), followed by gentle rotation for 4 h at 4°C. Samples were then centrifuged briefly (30 sec at 2,000 × g), and pellets were washed twice with 1× lysis buffer and once with 1× kinase buffer. Immunocomplexes (pellets) were resuspended in 40 μl of 1× kinase buffer (composed of 25 mM Tris [pH 7.5], 5 mM β-glycerolphosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, and 10 mM MgC12 supplemented with 200 μM ATP and 1 μg of the GSK-3 glutathione transferase fusion protein [GSK-3α/β, a well-characterized Akt/protein kinase B substrate; Cell Signaling Technology]) and incubated for 30 min at 30°C, allowing immunoprecipitated Akt (if activated) to phosphorylate GSK-3. The kinase reaction was terminated by adding 20 μl of 3× sodium dodecyl sulfate sample buffer. Phosphorylated GSK-3 was then detected by Western blot analysis using phospho-GSK-3α/β (for Ser-21 of GSK-3α and Ser-9 of GSK-3β) antibody (Cell Signaling Technology). The total amount of the GSK-3α/β glutathione transferase fusion protein in each reaction was used to verify equal loading.
Sucrose density gradient fractionation and RNA isolation/analysis.
Fractionation of cells by sucrose density gradient centrifugation was performed as previously described (37). The gradient was divided into 48 fractions (250 μl each), each of which was analyzed for absorbance at 260 nm and then pooled into a total of 12 fractions (4 fractions per group). RNA from each fraction, or from total cell lysates, was spiked with 0.5 μg of exogenous Drosophila ribosomal protein L3 (RPL3) mRNA (Ambion) (to control for losses of mRNA during purification) before purification using Trizol reagent (Invitrogen). A total of 100 ng of RNA sample was then reverse transcribed in triplicate, and quantitative PCR was performed (TaqMan EZ-RT PCR kit; Applied Biosystems, Foster City, CA). Primers and probes were designed for c-FLIPL and c-FLIPS by Integrated DNA Technology, Inc. (Coralville, IA). c-FLIPL and c-FLIPS probes were labeled with the 5′ fluorescent reporter dye 6-carboxy-fluorescin (FAM), while glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes were labeled with JOE (2,7-dimethoxy-4,5-dichloro-6-carboxy-fluorescein). All reporter dyes were quenched with the 3′ quencher 6-carboxy-tetramethylrhodamine (TAMRA). The following primer and probe sequences were used. For c-FLIPL, the primers were 5′-TTGGCCAATTTGCCTGTATG-3′ and 5′-TCGGCTCACCAGGACACA-3′, and the probe was 6FAM-CGAGCACCGAGACTACGACAGCTTTGT-TAMRA. For c-FLIPS, the primers were 5′-CAGTCTGTTCAAGGAGCAGGG-3′ and 5′-TTTCAGATCAGGACAATGGGC-3′, and the probe was 6FAM-CTCCAAGCAGCAATCCAAAAGAGTCTCAAG-TAMRA (51). GAPDH control primers and probes were obtained from Applied Biosystems. Thermal cycling conditions consisted of an initial uracil N-glycosylase incubation at 50°C for 2 min, AmpliTaq Gold activation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s, and annealing and extension at 60°C for 1 min. The amounts of FLIPL and FLIPS transcripts were divided by the endogenous reference (GAPDH) amount, after which the ratio was normalized to the ratio in standard cells (U373 or appropriate control cells) to obtain a normalized target value.
For Northern blot analysis, total or fractionated RNAs were purified (RNeasy; QIAGEN) following the addition of 0.5 μg of Drosophila RPL3 mRNA (Ambion) (to control for losses of mRNA during purification). Northern blots were carried out as previously described (28) using probes generated by reverse transcription-PCR (RT-PCR).
RESULTS
Characterization of TRAIL-sensitive and TRAIL-resistant GBM cell lines.
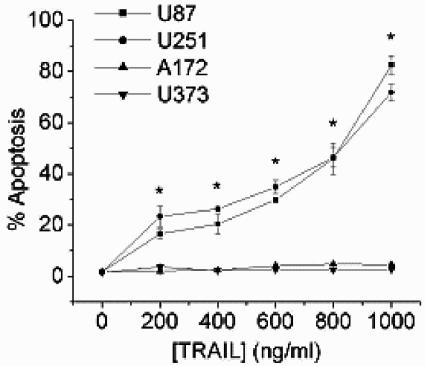
To define GBM cell lines in which to examine the basis for TRAIL sensitivity/resistance, four GBM cell lines were initially examined for TRAIL sensitivity. The U87 and U251 cells were previously reported to be susceptible to TRAIL-induced apoptosis, while the A172 and U373 cells were previously reported to be TRAIL insensitive (21). A 24-h exposure of U87 or U251 cells to as little as 200 ng/ml TRAIL resulted in the appearance of cells with surface expression of annexin V (not shown) and <2N sub-G1 DNA content as assessed by FACS analysis. The percentage of apoptotic cells increased in a dose-dependent manner (Fig. 1). In contrast, neither the A172 or U373 cells displayed the characteristics of apoptosis after 24-h exposures of up to 1,000 ng/ml TRAIL. No significant difference, however, existed in expression of either of the TRAIL receptors or TRAIL decoy receptors among the glioma cell lines examined, and no correlation existed between TRAIL receptor expression and TRAIL sensitivity (not shown).
FIG. 1.
Identification of TRAIL-sensitive and TRAIL-resistant GBM cell lines. Human GBM cells were incubated with TRAIL (0 to 1,000 ng/ml; 24 h), stained with propidium iodide, and analyzed by flow cytometry for the percentage of cells having a <2N DNA content (apoptotic cells). Data shown are the means ± standard errors derived from three independent experiments. *, P < 0.05.
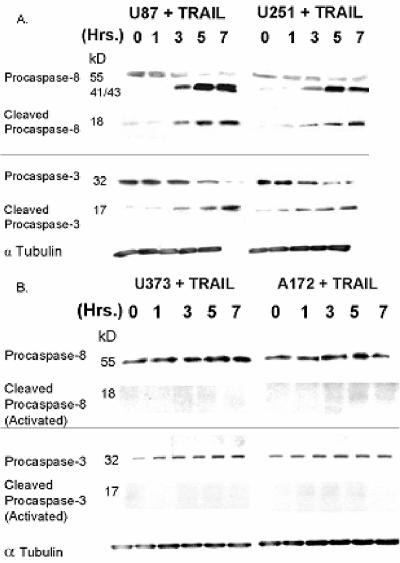
To more clearly define the point at which the apoptotic cascade was blocked in the TRAIL-resistant A172/U373 cells, the ability of GBM cells to initiate TRAIL-induced apoptosis by cleavage of procaspase-3 and its upstream activator procaspase-8 was measured. Exposure of cells to TRAIL (800 ng/ml for 24 h) resulted in a time-dependent disappearance of the full-length 55-kDa procaspase-8 and 32-kDa procaspase-3 (top bands) and a time-dependent appearance of intermediate (41 and 43 kDa) forms of procaspase-8 and active (cleaved) p18/17 subunits of caspase-8 and -3 in TRAIL-sensitive U87 and U251 cells (Fig. 2A) but not in similarly treated U373 and A172 cells (Fig. 2B). These results suggest that the TRAIL resistance noted in the GBM cell lines studied is a result of alterations in the pathway that connects activated TRAIL receptors to the apoptotic machinery.
FIG. 2.
TRAIL-resistant GBM cells fail to activate caspase-8 and caspase-3 in response to TRAIL. TRAIL-sensitive (A) or TRAIL-resistant (B) GBM cells were incubated with TRAIL (800 ng/ml; 0 to 7 h) and assayed for procaspase-8 and procaspase-3 cleavage by Western blotting. Data presented are representative of three independent experiments.
TRAIL resistance is associated with overexpression of FLIP.
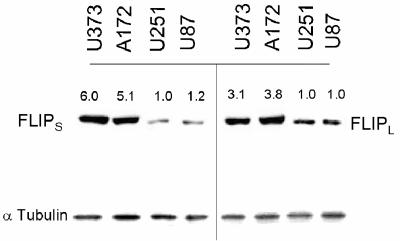
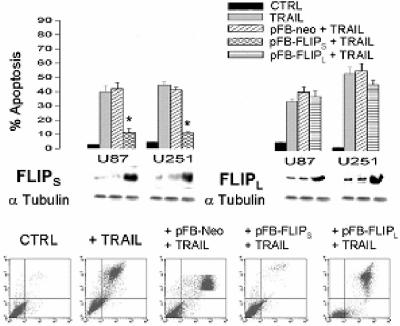
A potential modulator of the connection between TRAIL receptors and downstream caspase activation is FLIP. The FLIP protein exists in two forms in GBM, a short (28 kDa) FLIPS protein and a long (55 kDa) FLIPL protein that contains C-terminal sequences lacking in FLIPS. Both FLIP proteins are derived from alternate splicing of the same primary c-FLIP transcript (32). Although both FLIP isoforms are capable of blocking caspase-8 activation, FLIPS has antiapoptotic functions while FLIPL has been reported to have both pro- and antiapoptotic actions (8). In order to determine if FLIP expression was associated with the differential response of GBM cells to TRAIL, we assessed the expression of both isoforms of FLIP in TRAIL-sensitive and -resistant cell lines. As shown in Fig. 3, TRAIL-resistant cell lines exhibited significantly increased levels of both FLIPL and FLIPS relative to TRAIL-sensitive U87 and U251 cells. Because of the potential importance of both forms of FLIP in TRAIL resistance, TRAIL-sensitive U87 and U251 cells were infected with a blank retrovirus or with blank retroviral constructs encoding FLIPS or FLIPL. Following selection for stable expression, cells were assessed for effects of FLIPS or FLIPL expression on TRAIL sensitivity. Infection of TRAIL-sensitive U87 and U251 cells with a FLIPS-encoding construct significantly increased levels of FLIPS but did not affect levels of FLIPL, while infection with a FLIPL-encoding construct significantly increased levels of FLIPL but did not affect levels of FLIPS (Fig. 4, middle panels, and not shown). Only introduction of the FLIPS-encoding construct, however, significantly reduced the sensitivity of these cells to TRAIL-induced apoptosis and resulted in a decreased appearance of cells with surface expression of annexin V (relative to cells receiving an empty construct) (Fig. 4, top and bottom panels). These results show that overexpression of FLIPS as is noted in TRAIL-resistant GBM cells can confer TRAIL resistance to otherwise TRAIL-sensitive GBM cells and that FLIPS has the potential to modulate TRAIL sensitivity in GBM.
FIG. 3.
TRAIL sensitivity correlates with FLIP protein expression in GBM cells. TRAIL-resistant (U373/A172) or TRAIL-sensitive (U87/U251) cells were analyzed for FLIPL and FLIPS expression by Western blotting. Values listed are the means derived from three independent experiments. *, P < 0.05.
FIG. 4.
Overexpression of FLIPS, but not FLIPL, suppresses TRAIL-induced apoptosis in GBM cells. TRAIL-sensitive (U87/U251) GBM cells were sham infected (CTRL) or stably infected with blank (pFB-Neo), FLIPS- or FLIPL-encoding constructs. Following selection, cells were either analyzed for FLIPS and FLIPL expression by Western blot analysis (middle panels) or were exposed to TRAIL (800 ng/ml; 24 h), incubated with propidium iodide and a fluorescein isothiocyanate-conjugated annexin V antibody, and analyzed by flow cytometry for the percentage of propidium iodide (+) (y axis)/annexin V(+) (x axis) apoptotic cells (bottom panels). Data shown are representative of three independent experiments. Top panels are graphic representations of the data in the bottom panels and from similar studies in U251 cells (not shown). Values listed are the means ± standard errors. *, P < 0.05.
Translational regulation of FLIPS.
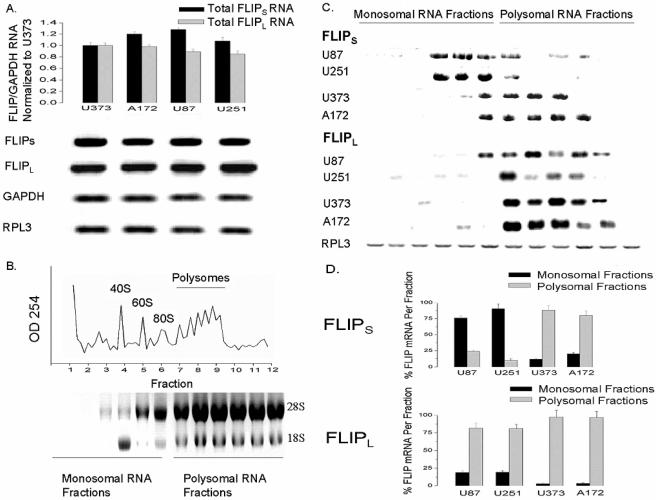
To address the basis for FLIPS overexpression in TRAIL-resistant GBM, we first examined FLIPS and FLIPL mRNA levels in GBM cell lines. Despite differing in FLIPS and FLIPL protein expression, all GBM cell lines had comparable levels of FLIPL and FLIPS mRNA (as determined by Northern blot analysis and quantitative RT-PCR) (Fig. 5A). We therefore employed sucrose gradient density centrifugation in combination with Northern blot analysis to assess the contribution of ribosomal distribution of the FLIPS and FLIPL mRNA to TRAIL resistance. As shown in Fig. 5B, unassembled ribosome subunits (monosomes) existed in the slowest sedimenting fractions of the gradient derived from centrifugation of U87 cells (fractions 1 to 6), while assembled translating polysomes existed in the fastest sedimenting fractions of the gradient (fractions 7 to 12). Northern blot analysis of FLIPS and FLIPL RNA content showed that FLIPS mRNA was preferentially associated with the nontranslating monosomes in cells with low-level expression of FLIPS (TRAIL-sensitive U87 and U251) and with the translating polysomes in cells with high-level expression of FLIPS (TRAIL-resistant U373 and A172) (Fig. 5C and D). In contrast, FLIPL mRNA was associated with the polysomal fractions in all GBM cell lines, regardless of FLIPL protein expression (Fig. 5C and D). These results show that the expression of FLIPS, a key regulator of TRAIL sensitivity, is associated with changes in ribosomal distribution and translation efficiency of the FLIPS mRNA in GBM cells.
FIG. 5.
Translational regulation of FLIPS expression. TRAIL-sensitive (U87/U251) or TRAIL-resistant (U373/A172) cells were lysed, spiked with Drosophila RPL3 RNA (to normalize for equal isolation/loading), and analyzed by RT-PCR (A, top) or Northern blotting (A, bottom) for levels of FLIPS, FLIPL, GAPDH, and RPL3 mRNA. Lysed cells were also subjected to sucrose density gradient (5 to 70%) centrifugation, with subsequent RNA from fractions containing unassembled ribosomal subunits (fractions 1 to 6) or assembled polyribosomes (fractions 7 to 12) (B), spiked with RPL3 RNA and analyzed for FLIPS, FLIPL, and RPL3 content by Northern blotting (C). (D) Graphic representation of the percentage of total FLIPS or FLIPL mRNA found in the combined monosomal or polysomal fractions. Data shown are representative of three independent experiments. Values listed are the means ± standard errors. *, P < 0.05.
FLIPS levels are translationally regulated by the Akt-mTOR pathway.
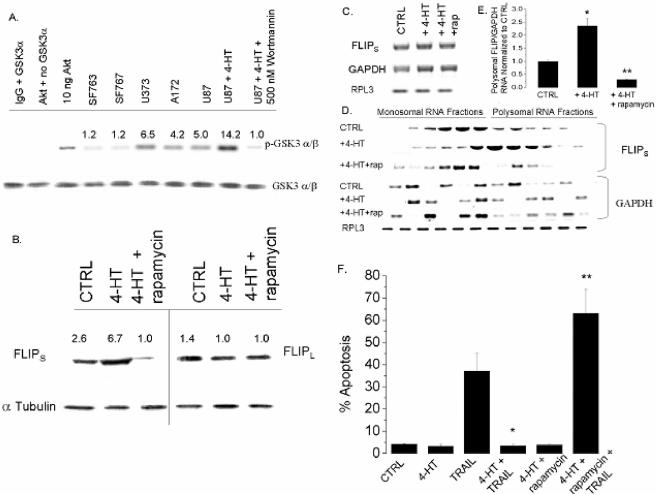
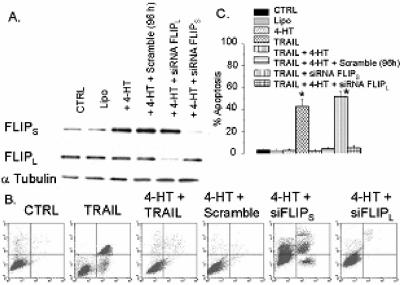
Because Akt activity is elevated in gliomas and can control translation via its downstream target mTOR, we assessed the possible role of the Akt-mTOR pathway in FLIPS expression and TRAIL sensitivity in GBM. For initial studies, TRAIL-sensitive U87 cells stably infected with a 4HT-inducible, Akt-encoding construct were exposed to vehicle or to 4HT concentrations previously shown to induce Akt expression in these cells (29), after which the effects of Akt activation on FLIPS mRNA levels and distribution, FLIPS protein levels, and TRAIL sensitivity were measured. As shown in Fig. 6A, incubation of U87 cells containing an inducible Akt construct with 4HT resulted in a 2.5-fold increase in Akt kinase activity (as measured by the ability of cellular extracts to phosphorylate the Akt substrate GSK-3α/β) relative to uninduced U87 cells and to PTEN mutant U373/A172 cells. While 4HT-mediated induction of Akt activity in U87 cells did not change FLIPL protein levels (Fig. 6B), expression of FLIPS protein was significantly increased relative to uninduced cells. Akt-mediated increases in FLIPS protein expression were not associated with changes in total FLIPS mRNA levels (Fig. 6C, Northern blot) but, rather, were accompanied by a pronounced shift in FLIPS mRNA distribution from the unassembled ribosomal subunits to the assembled ribosomes found in the polysomal fractions (Fig. 6D and E). Distribution of GAPDH and FLIPL mRNAs, however, was not altered by Akt activation (Fig. 6E and data not shown), suggesting that Akt-mediated effects on FLIPS mRNA distribution were not a consequence of global effects of Akt activation. Consistent with its ability to increase FLIPS expression, 4HT-mediated activation of Akt also significantly increased TRAIL resistance (Fig. 6F). Furthermore, as shown in Fig. 7A, siRNA targeting FLIPS blocked Akt-induced up-regulation of FLIPS protein expression, while siRNA targeting FLIPL or a scramble siRNA had no effect. The siRNA-mediated suppression of FLIPS levels conferred TRAIL sensitivity to otherwise TRAIL-resistant Akt overexpressing cells, increasing the percentage of cells expressing annexin V on their surface following TRAIL exposure to levels comparable to TRAIL-exposed parental U87 cells not induced to express Akt (Fig. 7B and C). In contrast, siRNA targeting FLIPL selectively reduced expression of the FLIPL protein in Akt-overexpressing U87 cells but had no effect on TRAIL sensitivity (Fig. 7B and C, compare the siRNA FLIPL data to that of the TRAIL plus 4HT, scramble, and siRNA FLIPS groups). These combined results show that Akt activation suppresses TRAIL sensitivity in GBM cells by redistributing FLIPS mRNA to polyribosomes and increasing translation of the FLIPS mRNA, thereby increasing expression of the antiapoptotic FLIPS protein.
FIG. 6.
The Akt-mTOR pathway plays a role in the translational regulation of FLIPS expression and TRAIL sensitivity. (A) U87 cells retrovirally infected with a 4HT-inducible Akt-encoding construct were incubated with 4HT (0 or 10 nM; 24 h), lysed, and subjected to an Akt in vitro kinase assay using GSK-3α/β as the substrate and endogenous GSK-3α/β protein to normalize for equal protein loading. (B to E) U87 cells infected with a 4HT-inducible Akt-encoding construct were incubated with 4HT (0 or 10 nM) or 4HT plus rapamycin (100 nM) for 30 min, after which TRAIL was added (800 ng/ml; 24 h). Cells were then lysed, spiked with Drosophila RPL3 RNA, and either analyzed by Western blotting for FLIPL and FLIPS expression (B); analyzed by Northern blotting for levels of FLIPS, GAPDH, and RPL3 mRNA (C); or separated into monosomal and polysomal fractions, after which levels of FLIPS, FLIPL, GAPDH, and RPL3 mRNA in each fraction were assessed by Northern blot analysis (D). (E) A graphic representation of data derived in panel D, with FLIPS values normalized to GAPDH RNA levels and compared to the FLIPS/GAPDH RNA ratio in control cells. (F) The percentage of cells having a <2N DNA content (apoptotic cells) following TRAIL exposure (800 ng/ml; 24 h). Data shown are representative of three independent experiments. Values listed are the means ± standard errors (where shown). *, P < 0.05.
FIG. 7.
Overexpression of FLIPS, but not FLIPL, sensitizes Akt-overexpressing U87 cells to TRAIL-induced apoptosis. U87 cells infected with a 4HT-inducible Akt-encoding construct were incubated with 4HT (0 or 10 nM) for 30 min, after which either a scramble siRNA or an siRNA targeting FLIPS or FLIPL was added for 96 h. Cells were then either analyzed by Western blotting for FLIPL and FLIPS expression (A) or exposed to TRAIL (800 ng/ml; 24 h), incubated with propidium iodide, and a fluorescein isothiocyanage-conjugated annexin V antibody and analyzed by flow cytometry for the percentage of propidium iodide (+)/annexin V (+) apoptotic cells (B). (C) A graphic representation of the data derived in panel B. Data shown are representative of three independent experiments. Values listed are the means ± standard errors (where shown). *, P < 0.05.
Because Akt overexpression increased TRAIL resistance in U87 GBM cells in association with translational up-regulation of FLIPS, we considered the possible involvement of the translational regulator and target of Akt, mTOR. U87 cells induced to overexpress Akt were simultaneously exposed to the mTOR inhibitor rapamycin, after which effects of mTOR inhibition on FLIPS expression and TRAIL sensitivity were monitored. As shown in Fig. 6B, exposure of cells to a concentration of rapamycin that suppressed phosphorylation of the downstream targets S6K1 and 4E-BP1 (not shown) blocked Akt-mediated upregulation of FLIPS, but not FLIPL, expression in Akt-overexpressing cells. Furthermore, rapamycin exposure, while not altering total FLIPS or FLIPL mRNA levels (Fig. 6C) and while not altering the monosomal/polysomal distribution of an unrelated mRNA (GAPDH), blocked the 4HT-induced shift of FLIPS mRNA to polysomes (Fig. 6D and E). Although rapamycin exposure itself had no effect on TRAIL sensitivity in U87 cells induced to overexpress Akt (Fig. 6F), it conferred TRAIL sensitivity to otherwise TRAIL-resistant Akt-overexpressing U87 cells, making them as sensitive as the parental U87 cells to TRAIL-induced apoptosis.
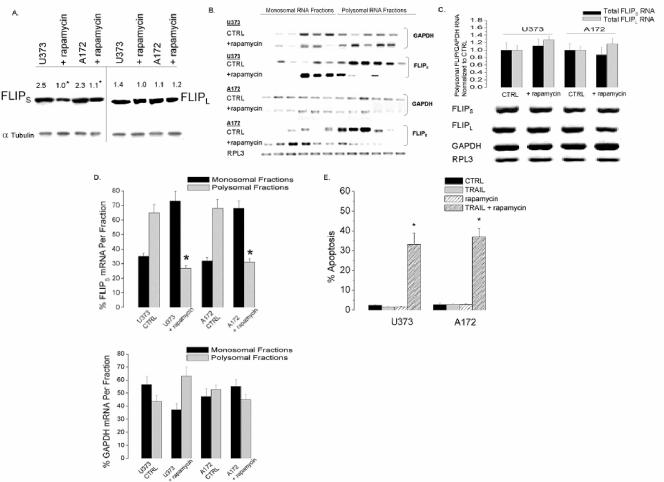
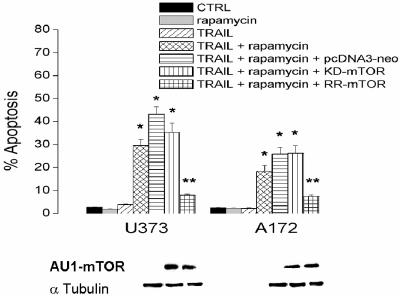
To expand these observation, cells that constitutively express high levels of pAkt and FLIPS (A172 and U373) were also exposed to rapamycin, after which FLIPS protein levels, FLIPS mRNA distribution, and TRAIL sensitivity were assessed. As in U87 cells induced to overexpress Akt, rapamycin exposure significantly reduced protein levels of FLIPS, but not FLIPL, in U373 and A172 cells without altering total FLIPS or FLIPL mRNA levels (Fig. 8A and B). Rapamycin exposure also significantly decreased the amount of FLIPS (but not GAPDH or FLIPL) mRNA associated with the translating polyribosomal fraction (Fig. 8C and D) and sensitized both cell lines to TRAIL-induced apoptosis (Fig. 8E). As shown in Fig. 9, rapamycin-induced sensitization could not be reversed by expression of AU1-tagged KD-mTOR in U373 or A172 cells but could be completely reversed by expression of AU1-tagged RR-mTOR. These results suggest that the effects of rapamycin on TRAIL sensitivity are the result of mTOR inhibition, and not inhibition of other targets, and that the Akt-mTOR pathway controls TRAIL sensitivity in GBM via translational control of FLIPS expression.
FIG. 8.
Rapamycin reduces FLIPS protein levels and sensitizes GBM cells to TRAIL-induced cell death. TRAIL-resistant (U373 and A172) GBM cells were incubated with rapamycin (0 or 100 nM; 30 min), after which the cells were exposed to TRAIL (0 or 800 ng/ml; 24 h). Cells were then either analyzed by Western blotting for FLIPL and FLIPS expression in the absence of TRAIL (A) or lysed, spiked with Drosophila RPL3 RNA, and analyzed for the effects of rapamycin (in the absence of TRAIL) on total levels of FLIPS, FLIPL, GAPDH, and RPL3 mRNA by Northern blot analysis (B, bottom panel) and quantitative RT-PCR (B, top panel). Cellular lysates were also separated into monosomal and polysomal fractions and spiked with Drosophila RPL3 RNA, after which RNA was isolated and levels of FLIPS, FLIPL, GAPDH, and RPL3 mRNA in each ribosomal fraction of control or rapamycin-treated U373 and A172 cells was assessed by Northern blot analysis (C). (D) A graphic representation of the data derived in panel C, with values expressed as the percentage of GAPDH, FLIPS, or FLIPL mRNA detected in the pooled monosomal or polysomal fractions. (E) Cells were stained with propidium iodide and analyzed by flow cytometry for the percentage of cells having <2N DNA content (apoptotic cells) following TRAIL exposure (800 ng/ml; 24 h). Data shown are representative of three independent experiments. All values listed are the means ± standard errors (where shown). *, P < 0.05.
FIG. 9.
Rapamycin sensitizes GBM cells to TRAIL-induced apoptosis through an mTOR-dependent mechanism. TRAIL-resistant (U373 and A172) GBM cells were infected with a blank retroviral construct or a construct encoding either an AU1-tagged KD or RR mTOR. Following selection, cells were lysed and expression of the AU1-tagged proteins of interest were verified by Western blotting (bottom panels). Cells were then incubated with 100 nM rapamycin and 800 ng/ml TRAIL for 24 h, after which the cells were stained with propidium iodide and analyzed by flow cytometry for the percentage of cells having <2N DNA content (apoptotic cells). The data shown are the means ± standard errors derived from three independent experiments. *, P < 0.05 in comparison to control cells; **, P < 0.05 in comparison to cells expressing either KD or RR mTOR constructs.
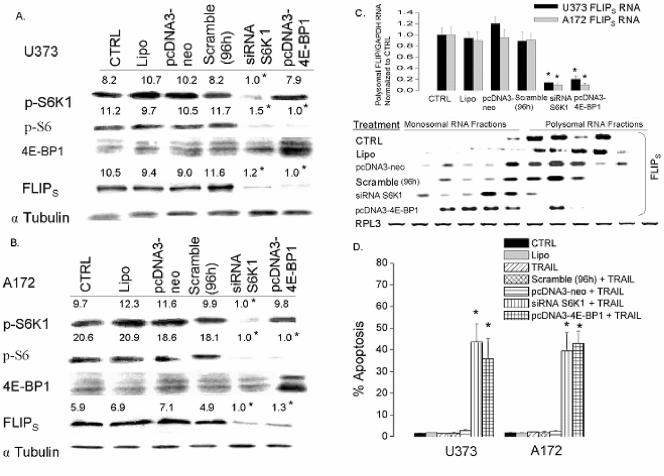
Downstream targets of mTOR contribute to effects on FLIPS translation and TRAIL sensitivity.
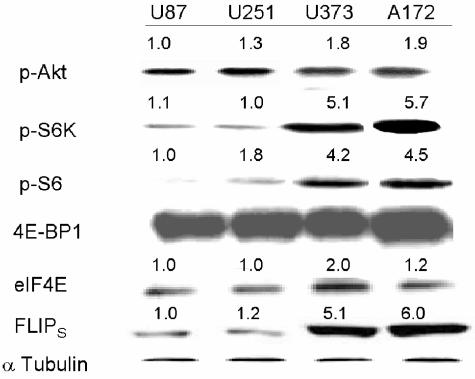
Because inhibition of mTOR blocks Akt-induced increases in FLIPS translation and TRAIL resistance and because mTOR-mediated effects on translation are thought to be mediated through effects on S6K1 and 4E-BP1/eIF4E, we compared the expression of pAkt and the downstream targets of mTOR in TRAIL-sensitive cells to that in TRAIL-resistant cells. As shown in the Western blot shown in Fig. 10, there was no significant difference in levels of pAkt, 4E-BP1, or eIF4E among the TRAIL-sensitive and -resistant cell lines. TRAIL-sensitive U87/U251 cells, however, had decreased activation of both S6K1 and its downstream effector S6 ribosomal protein relative to the TRAIL-resistant cells, suggesting a potential role for this pathway in FLIPS regulation. In order to determine how activation of the downstream targets of mTOR related to expression of the FLIPS protein, we generated constructs designed to enhance or suppress mTOR target activation, expressed these constructs in cells with low or high levels of FLIPS, respectively, and monitored their effects on FLIPS translation and TRAIL sensitivity. To inhibit pathways downstream of mTOR in cells with high levels of FLIPS, A172 and U373 cells were transfected with siRNA targeting S6K1 or were stably transfected with an expression construct encoding the eIF4E inhibitor 4E-BP1. Introduction of siRNA targeting S6K1 reduced levels of pS6K1 and the S6K1 target pS6 to levels comparable to those in TRAIL-sensitive U87/U251 cells, while not altering the expression or phosphorylation status of 4E-BP1 (Fig. 11A and B and data not shown). Exogenous expression of 4E-BP1 increased 4E-BP1 levels in U373 and A172 cells, while not altering levels of pS6K1. 4E-BP1 overexpression did, however, also reduce levels of pS6, suggesting cross talk between the S6K1 and 4E-BP1 pathways at the levels of S6 phosphorylation. Expression of either S6K1 siRNA or 4E-BP1 reduced expression of FLIPS in both A172 and U373 cells to levels comparable to those in TRAIL-sensitive U87 and U251 cells (Fig. 11A and B), shifted FLIPS mRNA from the translating polysomal fraction to the nontranslating monosomal fraction (Fig. 11C), and significantly sensitized U373 and A172 cells to TRAIL-induced apoptosis (Fig. 11D), with the extent of sensitization mediated by S6K1 siRNA no different from that mediated by overexpression of 4E-BP1, or by rapamycin (Fig. 9).
FIG. 10.
Differences in the activation of the S6K1 arm of the mTOR signaling pathway in TRAIL-sensitive versus TRAIL-resistant GBM cells. TRAIL-sensitive (U87 and U251) or TRAIL-resistant (U373 and A172) cells were analyzed for expression of phosphorylated (activated) Akt, phospho-S6K1, phospho-S6, 4E-BP1, eIF4E, and FLIPS by Western blotting. Values listed are the means derived from three independent experiments. *, P < 0.05.
FIG. 11.
Genetic suppression of the S6K1 pathway reduces expression of FLIPS protein and sensitizes GBM cells to TRAIL-induced apoptosis. TRAIL-resistant (U373 and A172) GBM cells were sham transfected, transiently transfected with either siRNA targeting S6K1 or a nonspecific scrambled siRNA (96 h), or stably transfected with an empty vector control (pcDNA3-Neo) or a construct encoding the eIF4E inhibitor 4E-BP1. Cells were then analyzed by Western blotting for levels of phospho-S6K1, phospho-S6 protein, 4E-BP1, and FLIPS (A and B) or lysed, separated into monosomal and polysomal fractions, and spiked with Drosophila RPL3 RNA, after which RNA was isolated and analyzed for FLIPS mRNA levels by quantitative RT-PCR (C, top) and Northern blot analysis (C, bottom). (D) Cells were stained with propidium iodide and analyzed by flow cytometry for the percentage of cells having <2N DNA content (apoptotic cells) after exposure to TRAIL (800 ng/ml; 24 h). Data shown are representative of three independent experiments. All values listed are the means ± standard errors (where shown). In panel D, polysomal FLIPS mRNA levels were normalized to GAPDH mRNA levels and then to FLIP/GAPDH mRNA ratios in control (CTRL) cells for each cell line to generate the mean ± standard error values presented. *, P < 0.05.
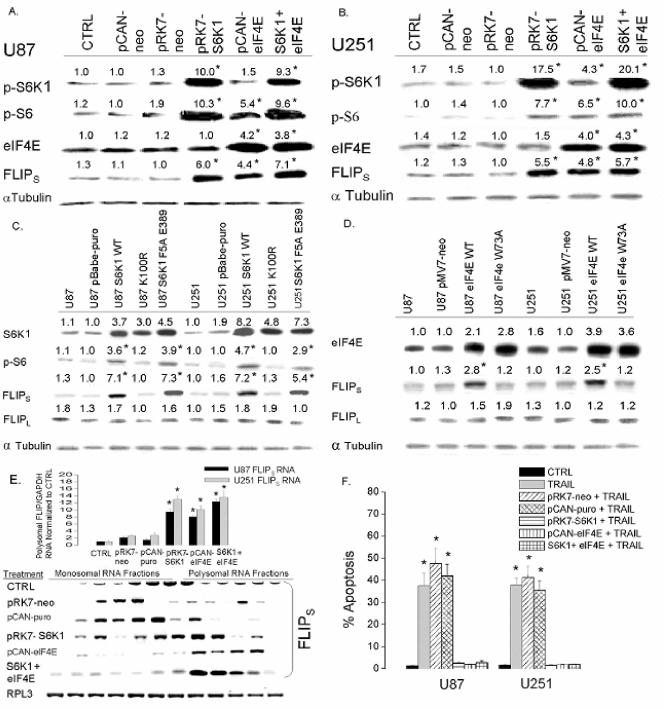
In converse experiments, cells with low levels of FLIPS protein (U87 and U251) were stably transfected with constructs encoding either S6K1, eIF4E, or both in a serial fashion, and the effects on FLIPS expression, TRAIL sensitivity, and FLIPS mRNA distribution were then monitored. While transfection of cells with a construct encoding S6K1 (pRK7-S6K1) had no effect on eIF4E levels, it increased levels of pS6K1 and pS6 protein approximately 10-fold relative to cells receiving an empty construct and to uninfected controls and approximately twofold relative to TRAIL-resistant U373 and A172 cells (Fig. 12A and B and data not shown). Transfection of cells with a construct encoding eIF4E (pCAN-eIF4E) increased levels of eIF4E and of pS6 (a presumed consequence of pathway cross talk) to levels that were approximately three times that in uninfected cells, in cells receiving empty construct controls, or in TRAIL-resistant U373 and A172 cells (Fig. 12A and B and data not shown). Overexpression of S6K1, eIF4E, or the combination also increased FLIPS expression to levels comparable to those in TRAIL-resistant U373 and A172 cells (12A and B and data not shown). Levels of pS6 and FLIPS (but not FLIPL) could be similarly increased by introduction of retroviral constructs encoding WT or RR (F5A E389) S6K1, but not a KD S6K1 (K100R) (Fig. 12C). Similarly, FLIPS but not FLIPL levels could be increased by introduction of a retroviral construct encoding WT eIF4E but not a KD eIF4E (W73A) (Fig. 12D). As expected, overexpression of S6K1 or eIF4E significantly shifted FLIPS mRNA from the monosomal to the polysomal fractions (Fig. 12E) and protected both U87 and U251 cells from TRAIL-induced apoptosis, with the extent of protection mediated by S6K1 no different from that mediated by expression of eIF4E or by the combined expression of S6K1 and eIF4e (Fig. 12F). Taken as a whole, these results show that activation of the S6K1 arm of the Akt-mTOR pathway, either directly or by eIF4E-mediated phosphorylation of S6, causes redistribution of FLIPS mRNA to the polyribosomal fractions, increasing FLIPS protein levels and TRAIL resistance.
FIG. 12.
Overexpression of S6K1 enhances FLIPS expression and confers resistance to TRAIL-induced apoptosis. TRAIL-sensitive (U87 and U251) cells were either sham transfected or stably transfected with an empty vector (pCAN-neo or pRK7-neo) or constructs encoding either wild-type S6K1, wild-type eIF4E, or both. Following selection, cells were analyzed by Western blotting for levels of phospho-S6K1, phospho-S6 protein, eIF4E, and FLIPS (A and B), stained with propidium iodide, and analyzed by flow cytometry for the percentage of cells having <2N DNA content (apoptotic cells) 24 h after exposure to 800 ng/ml TRAIL (F); or lysed, separated into monosomal and polysomal fractions, and spiked with Drosophila RPL3 RNA, after which RNA was isolated and analyzed for FLIPS mRNA levels by quantitative RT-PCR (E, top) and Northern blot analysis (E, bottom). TRAIL-sensitive U87 and U251 cells were also sham infected with empty control vectors (pBabe-puro or pMV7-neo) or a construct encoding wild-type S6K1, an inactive S6K1 (K100R), a rapamycin-resistant S6K1, a wild-type eIF4E, or an inactive eIF4E (W73A). Following selection, cells were analyzed for levels of S6K1 protein, phospho-S6 protein, eIF4E, and FLIPS/FLIPL expression by Western blot analysis (C and D). All values are the means ± standard errors (where shown) derived from three independent experiments. *, P < 0.05.
Conservation of the linkage between PTEN, Akt, pS6K1, and FLIPS in primary human GBM cells.
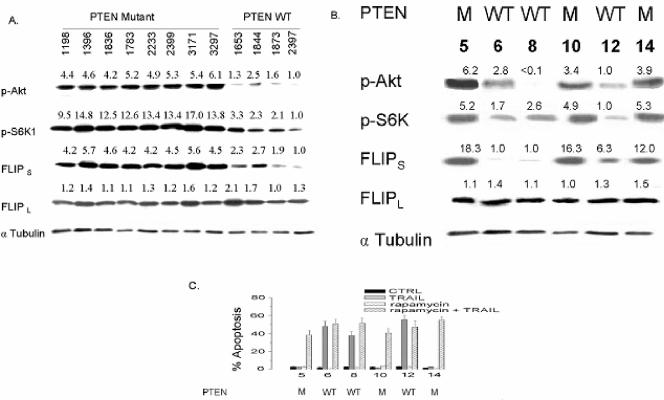
Because the data presented suggest that Akt, mTOR, and FLIPS may be linked to each other, and perhaps to PTEN (an upstream regulator of Akt activity) in a manner that controls TRAIL resistance in GBM, we addressed this potential linkage, first in archived primary GBM samples and then in human GBM xenografts. In archived primary GBM tissue, FLIPL levels exhibited little variability across all tumors and showed no association with other variables (Fig. 13A). Levels of pAkt, pS6K1, and FLIPS, however, were consistently elevated in tumors with compromised PTEN relative to tumors with WT PTEN. In these tumors, PTEN status was therefore closely linked to levels of pAkt, pS6K1, and FLIPS. Although GBM with WT PTEN function (and low levels of pAkt) would, therefore, be predicted to represent a TRAIL-sensitive subpopulation, TRAIL sensitivity could not be directly examined in this archived material. We therefore examined the integrity of the PTEN-Akt-mTOR-FLIPS pathway, and its linkage to TRAIL sensitivity, in human GBM xenografts. As in archived GBM material, PTEN status in these human tumors grown exclusively in vivo was closely linked to levels of pAkt, pS6K1, and FLIPS (Fig. 13B). Additionally, as predicted from cell line data, only PTEN-WT GBM xenografts with corresponding low levels of FLIPS underwent apoptosis in response to TRAIL exposure in short-term culture (Fig. 13C). Consistent with the involvement of mTOR in the pathway controlling FLIPS expression in GBM, all four PTEN mutant GBM xenografts that displayed TRAIL resistance could also be made TRAIL sensitive by preincubation with the mTOR inhibitor rapamycin (Fig. 13C). These results show that in GBM, a pathway exists linking PTEN status to Akt, mTOR, S6K1, FLIPS, and TRAIL sensitivity and that this pathway provides a means to both identify TRAIL-sensitive GBM and to sensitize otherwise TRAIL-resistant tumors to TRAIL-induced apoptosis.
FIG. 13.
Analysis of the linkage between PTEN, Akt, pS6K, FLIPS, and TRAIL sensitivity in primary human GBM and GBM cell lines. Cellular lysates from surgically resected primary GBM (A) or xenografted human GBM (B) of known PTEN status were assessed for levels of pAkt, pS6K1, FLIPS, and FLIPL by Western blot analysis. (C) Cells from GBM xenografts were incubated with 0 or 100 nM rapamycin and 0 or 800 ng/ml TRAIL for 24 h, after which the cells were stained with propidium iodide and analyzed by flow cytometry for the percentage of apoptotic cells. The data shown are the means ± standard errors derived from three independent experiments. Values listed are the means derived from three independent experiments.
DISCUSSION
The mammalian target of rapamycin is a key controller of translation and gene expression. While mTOR's ability to alter gene expression in response to nutrient and growth factor status is thought to play a key role in its integration of these signals with cell growth and cell cycle progression, accumulating evidence suggests that mTOR may also contribute to a variety of other cellular processes. The present study provides evidence that mTOR, via S6K1-mediated effects on FLIPS translation, controls the sensitivity of human glioma cells to apoptosis induced by the death ligand TRAIL. This observation has implications for the identification of tumors potentially sensitive to TRAIL and TRAIL-dependent therapies, for strategies to sensitize tumors to TRAIL-mediated cell death, and for the larger role of mTOR in controlling cellular response to death ligands.
The basis for TRAIL sensitivity or resistance in human gliomas has been examined previously, and in most cases the defect conferring TRAIL resistance has been localized to an inability to activate the apoptotic cascade (21, 70, 71). The data presented here are consistent with the idea that TRAIL resistance in GBM is not a result of altered TRAIL receptor expression (DR4/5 or TRAIL decoy receptors) but, rather, is the result of lack of caspase activation. While we cannot formally rule out mutations of TRAIL receptors as a basis for TRAIL resistance, the localization of the defect in TRAIL-resistant cells in the present study to the level of caspase-8 activation is consistent with a block in the activation process. Both FLIPL and FLIPS were overexpressed in TRAIL-resistant cells, and both proteins can interact with caspase-8 and block its activation. Although both FLIPL and FLIPS have been reported to regulate TRAIL sensitivity in various tumor cell lines (32, 70, 71), data presented show that only FLIPS overexpression blocked TRAIL-induced apoptosis, only FLIPS suppression enhanced TRAIL-induced apoptosis, and only FLIPS levels were affected by various genetic manipulations of the Akt-mTOR pathway that also altered TRAIL sensitivity. These observations suggest that FLIPS, by blocking activation of the apoptotic cascade, plays a unique and key role in controlling TRAIL-induced apoptosis in glioma. Previous studies have suggested that FLIPS levels can be regulated at the level of transcription initiation by c-myc or Akt (46, 55) and at the level of protein phosphorylation by calcium/calmodulin-dependent protein kinase II (71). The present study, however, clearly defines translational control as a new mechanism by which FLIPS expression is regulated and defines a new role for mTOR, namely, control of death ligand-induced apoptosis.
Because mTOR translationally regulates FLIPS levels and TRAIL sensitivity, we further addressed the contribution of known downstream targets of mTOR to FLIPS regulation and TRAIL sensitivity. Although mTOR has many downstream targets, two, S6K1 and 4E-BP1, are recognized to play key roles in translational regulation. S6K1 and 4E-BP1 (via its partner eIF4E) are both thought to be critical in mTOR-mediated enhanced translation of mRNAs with 5′ TOP or capped mRNAs with complex 5′ untranslated regions, respectively. Furthermore, S6K1, by blocking BAD phosphorylation, blocks apoptosis induced by growth factor withdrawal (27), while eIF4E, by transcriptionally and translationally upregulating Bcl-XL, blocks myc-induced apoptosis (43, 53). Additionally, both arms of the mTOR pathway have been shown to contribute to the effects of mTOR on cell cycle progression (18). Many questions remain, however, as to how S6K1 and eIF4E/4E-BP1 regulate translation, and recent studies showing that translational activation of TOP mRNAs is independent of S6K1 have cast doubt on previously accepted explanations. The relative contributions of the two arms of the mTOR pathway to translational control of apoptosis have also not been examined. The present data show that TRAIL-induced apoptosis can be suppressed by activation of either arm of the mTOR pathway and that inhibition of either arm sensitizes cells to TRAIL-induced apoptosis as effectively as mTOR inhibition by rapamycin. The ability of eIF4E expression to translationally upregulate FLIPS and protect cells from TRAIL-induced apoptosis is similar to its ability to translationally upregulate another inhibitor of apoptosis (Bcl-XL) following different apoptotic stimuli (43). The effects of eIF4E on Bcl-XL, however, are only partially mediated at the translational level and only partially able to block apoptosis, while the effects of eIF4E on FLIPS expression do not involve alterations in FLIPS mRNA levels and are sufficient in themselves to completely block TRAIL-induced apoptosis. The apparent involvement of 4E-BP1/eIF4E in FLIPS regulation is also somewhat misleading as the effects of eIF4E overexpression on FLIPS expression appear at least in part to involve cross talk between the eIF4E/4E-BP1 and S6K1 pathways. Overexpression of eIF4E increased pS6 levels (but not pS6K1 levels) while overexpression of the eIF4E inhibitor 4E-BP1 suppressed pS6 levels (but not pS6K1) as effectively as siRNA targeting pS6K1. Because overexpression of neither eIF4E nor 4E-BP1 altered pS6K1 levels, cross talk between the pathways appears to occur at the level of S6 phosphorylation. The suspected cross talk between the pathways is further supported by the observation that activation of the S6K1 pathway is no more effective at increasing FLIPS levels than activation of eIF4E or activation of both pathways combined, while suppression of either the S6K1 or 4E-BP1/eIF4E pathways in TRAIL-resistant cells suppresses FLIPS levels and increases TRAIL sensitivity to an extent comparable to that mediated by rapamycin (compare Fig. 9 and 12F). Cross talk between the eIF4E/4E-BP1 and S6K1 pathways has been reported (39), although in these studies in NIH 3T3 cells, overexpression of eIF4E led to suppression of pS6K1 levels (and presumably also pS6 levels) rather than enhancement, as noted in the present study. While the basis for this difference is not clear and may represent cell type-specific differences in pathway connections, it is clear that the eIF4E/4E-BP1 pathway contributes indirectly via the S6K1/S6 pathway to control FLIPS expression.
In contrast to the apparent indirect actions of eIF4E on FLIPS translation and TRAIL sensitivity, genetic manipulations of S6K1 levels altered FLIPS translation and TRAIL sensitivity without apparent effects on eIF4E or 4E-BP1 expression/phosphorylation. The direct effects of S6K1 on FLIPS translation and TRAIL-induced apoptosis are novel and differ from previous studies which focused on the ability of S6K1 to suppress growth factor withdrawal-induced apoptosis via effects on Bad phosphorylation (27). Although the S6K1 pathway has been suggested to play a key role in the translation regulation of TOP mRNAs (34, 38), the basis for its effects on FLIPS, but not FLIPL, mRNA translation, are less apparent. While both the FLIPS and FLIPL mRNAs are derived from the same primary transcript (32), the FLIP mRNAs differ in their 5′ untranslated regions, and these differences might account for the differential regulation of the FLIPS mRNA. Neither the FLIPL nor FLIPS mRNA, however, has 5′ TOP sequences, and coupled with the recent observation that TOP mRNAs are appropriately translationally regulated in S6K1 knockout cells (64), it seems likely that undefined, non-TOP-dependent mechanisms control FLIPS translation. Such regulation might involve the previously identified regulation of elongation factor phosphorylation by S6K1 (68), although this remains to be examined.
In addition to clearly defining the role of mTOR in regulating the response of glioma cells to TRAIL-induced apoptosis, the present studies also have larger clinical implications. Only a percentage of human gliomas are likely to respond to TRAIL or to chemotherapeutic regimens dependent on endogenous TRAIL activation (35), and to date there has been no way of identifying these vulnerable tumors. Our data suggest that FLIPS levels are a reliable indicator of TRAIL sensitivity in GBM, and that because FLIPS levels are controlled by mTOR, Akt, and the Akt regulator PTEN, a number of variables might be suitable for stratification of GBM into inherently sensitive tumors suitable for TRAIL-based therapy and tumors which, by virtue of activation of the Akt-mTOR-FLIPS pathway, might be more effectively treated by TRAIL-based combination regimens that also suppress TRAIL resistance pathways. A logical combination therapy based on these ideas could employ the mTOR inhibitor CCI-779, which is already widely used clinically and has activity against PTEN-deficient tumors (including GBM) (63), in combination with TRAIL or other TRAIL-inducing agents. The knowledge that mTOR and the downstream targets of mTOR play a role in TRAIL sensitivity in gliomas should allow the design of clinical studies to test these ideas. Finally, it is worth noting that Akt/mTOR activation contributes to the ability of tumors to evade immune surveillance by enhancing tumor-specific expression of B7-H1, a member of the B7 family of costimulatory molecules that promotes interleukin-10/Fas ligand-mediated apoptotic death of activated tumor antigen-specific human T cells (13, 50). Because various members of the tumor necrosis family, including perhaps TRAIL, are contributors to the immune response (60), the actions of mTOR in suppressing TRAIL-induced apoptosis may be part of a broader function of mTOR in suppressing cellular elimination. If this is true, the use of mTOR inhibitors could sensitize tumors to a broad range of apoptosis-inducing death ligands secreted by responding cells of the immune system. The identification of mTOR as a regulator of TRAIL sensitivity, therefore, may also be an indicator of a larger and potentially exploitable function of this increasingly important molecule.
Acknowledgments
This work was supported by National Institutes of Health Awards RO1 CA94989 and P50 CA97257 to R.O.P. and M.S.B.
REFERENCES
- 1.Almasan, A., and A. Ashkenazi. 2003. Apo2L/TRAIL: apoptosis signaling, biology, and potential for cancer therapy. Cytokine Growth Factor Rev. 14:337-348. [DOI] [PubMed] [Google Scholar]
- 2.Altucci, L., A. Rossin, W. Raffelsberger, A. Reitmair, C. Chomienee, and H. Gronemeyer. 2001. Retinoic acid-induced apoptosis in leukemia cells is mediated by paracrine action of tumor-selective death ligand TRAIL. Nat. Med. 7:680-686. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, M., O. Batista, A. Bellacosa, P. Tsichlis, and P. K. Vogt. 1998. The Akt kinases: molecular determinants of oncogenicity. Proc. Natl. Acad. Sci. USA 95:14950-14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bin, L., X. Li, L.-G. Xu, and H.-B. Shu. 2002. The short splice form of Casper/c-FLIP is a major cellular inhibitor of TRAIL-induced apoptosis. FEBS Lett. 510:37-40. [DOI] [PubMed] [Google Scholar]
- 5.Blommaart, E. F., J. J. Luiken, P. J. Blommaart, G. M. van Woerkom, and A. J. Meijer. 1995. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270:2320-2326. [DOI] [PubMed] [Google Scholar]
- 6.Boldin, M. P., T. M. Goncharov, Y. V. Goltsev, and D. Wallach. 1996. Involvement of MACH, a novel MORT1/FADD-interacting proteinase, in Fas/APO-1- and TNF receptor-induced cell death. Cell 85:803-815. [DOI] [PubMed] [Google Scholar]
- 7.Castro, A. F., J. F. Rebhun, G. J. Clark, and L. A. Quilliam. 2003. Rheb promotes tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J. Biol. Chem. 278:32493-32496. [DOI] [PubMed] [Google Scholar]
- 8.Chang, D. W., Z. Xing, V. L. Capacio, M. E. Peter, and X. Yang. 2003. Interdimer processing mechanism of procaspase-8 activation. EMBO J. 22:4132-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, G. H.-F., N. M. Barbaro, and R. O. Pieper. 2000. Phosphatidylserine-dependent phagocytosis of apoptotic glioma cells by normal human microglia, astrocytes, and glioma cells. Neuro-Oncol. 2:174-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheatham, L., M. Monfar, M. M. Chou, and J. Blenis. 1995. Structural and functional analysis of pp70S6K. Proc. Natl. Acad. Sci. USA 92:11696-11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, Y., Y. Lin, and X. Wu. 2002. TRAIL-induced apoptosis requires Bax-dependent mitochondrial release of Smac/DIABLO. Genes Dev. 16:33-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis, P. B., A. Jaeschke, M. Saitoh, B. Fowler, S. C. Kozma, and G. Thomas. 2001. Mammalian TOR: a homeostatic ATP sensor. Science 294:1102-1105. [DOI] [PubMed] [Google Scholar]
- 13.Dong, H., and L. Chen. 2003. B7-H1 and its role in the evasion of tumor immunity. J. Mol. Med. 81:281-287. [DOI] [PubMed] [Google Scholar]
- 14.Duffner, A., and G. Thomas. 1999. Ribosomal S6 kinase signaling and the control of translation. Exp. Cell. Res. 253:100-109. [DOI] [PubMed] [Google Scholar]
- 15.Edinger, A. L., and C. B. Thompson. 2002. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Cell. Biol. 13:2276-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ermoian, R. P., C. S. Furniss, K. R. Lamborn, D. Basila, M. S. Berger, A. R. Gottschalk, M. K. Nicholas, D. Stokoe, and D. A. Haas-Kogan. 2002. Dysregulation of PTEN and protein kinase B is associated with glioma histology and patient survival. Clin. Cancer Res. 8:1100-1106. [PubMed] [Google Scholar]
- 17.Fang, Y., I.-H. Park, A.-L. Wu, G. Du, P. Huang, M. A. Frohman, S. J. Walker, H. A. Brown, and J. Chen. 2003. PLD1 regulates mTOR signaling and mediates Cdc42 activation of S6K1. Curr. Biol. 13:2037-2044. [DOI] [PubMed] [Google Scholar]
- 18.Fingar, D. C., C. J. Richardson, A. R. Tee, L. Cheatham, C. Tsou, and J. Blenis. 2004. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 24:200-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fingar, D. C., and J. Blenis. 2004. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23:3151-3171. [DOI] [PubMed] [Google Scholar]
- 20.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4E-BP11/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fulda, S., W. Wick, M. Weller, and K.-M. Debatin. 2002. Smac agonists sensitize for Apo2L/TRAIL- or anticancer drug-induced apoptosis and induce regression of malignant glioma in vivo. Nat. Med. 8:807-815. [DOI] [PubMed] [Google Scholar]
- 22.Fumagelli, S., and G. Thomas. 2000. S6 phosphorylation and signal transduction, p. 695-718. In N. Sonenberg, J. W. B. Hershey, and M. B. Matthews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Gingras, A. C., B. Raught., and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807-826. [DOI] [PubMed] [Google Scholar]
- 24.Gingras, A. C., S. G. Kennedy, M. A. O'Leary, N. Sonenberg, and N. Hay. 1998. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt (PKB) signaling pathway. Genes Dev. 12:502-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gingras, A. C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R.Abersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hara, K., K. Yonezawa, Q. P. Weng, M. T. Lozlowski, C. Belham, and J. Avruch. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273:14484-14494. [DOI] [PubMed] [Google Scholar]
- 27.Harada, H., J. S. Andersen, M. Mann, N. Terada, and S. J. Korsmeyer. 2001. p70S6 kinase signals cell survival as well as growth, inactivating the pro-apoptotic molecule BAD. Proc. Natl. Acad. Sci. USA 98:9666-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirose, Y., M. Katayama, D. Stokoe, D. A. Haas-Kogan, M. S. Berger, and R. O. Pieper. 2003. The p38 mitogen-activated protein kinase pathway links the DNA mismatch repair system to the G2 checkpoint and to resistance to chemotherapeutic DNA-methylating agents. Mol. Cell. Biol. 23:8306-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirose, Y., M. Katayama, O. K. Mirzoeva, M. S. Berger, and R. O. Pieper. 2005. Akt activation suppresses Chk2-mediated methylating agent-induced G2 arrest and protects from temozolomide-induced mitotic catastrophe and cellular senescence. Cancer Res. 65:4861-4869. [DOI] [PubMed] [Google Scholar]
- 30.Hopkins-Donaldson, S., A. Ziegler, S. Kurtz, C. Bigosch, D. Kandioler, C. Ludwig, U. Zangemeister-Wittke, and R. Stahel. 2003. Silencing of death receptors and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ. 10:356-364. [DOI] [PubMed] [Google Scholar]
- 31.Inoke, K., Y. Li, T. Zhu, J. Wu, and K. L. Guan. 2002. TSC2 is phosphorylated and inhibited by AKT and suppresses mTOR signaling. Nat. Cell Biol. 4:648-657. [DOI] [PubMed] [Google Scholar]
- 32.Irmler, M., M. Thome, M. Hahne, P. Schneider, K. Hofmann, V. Steiner, J.-L. Bodmer, M. Schroter, K. Burns, C. Mattman, D. Rimoldi, L. E. French, and J. Tschopp. 1997. Inhibition of death receptor signals by FLIP. Nature 388:190-195. [DOI] [PubMed] [Google Scholar]
- 33.Jacinto, E., and M. N. Hall. 2003. TOR signaling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 34.Jefferies, H. B., S. Fumagalli, P. B. Dennis, C. Reinhard, R. B. Pearson, and G. Thomas. 1997. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 16:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeremias, I., H. H. Steiner, A. Benner, K. M. Debatin, and C. Herold-Mende. 2004. Cell death induction by betulinic acid, ceramide and TRAIL in primary glioblastoma multiforme cells. Acta Neurochir. (Vienna) 146:721-729. [DOI] [PubMed] [Google Scholar]
- 36.Jo, M., T. H. Kim, D. W. Seol, J. E. Esplen, K. Dorko, T. R. Billiar, and S. C. Strom. 2000. Apoptosis induced in normal human hepatocytes by tumor necrosis factor-related apoptosis-related apoptosis-inducing ligand. Nat. Med. 6:564-567. [DOI] [PubMed] [Google Scholar]
- 37.Johannes, G., and P. Sarnow. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4:1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawasome, H., P. Papst, S. Webb, G. M. Keller, G. L. Johnson, E. W. Gelfand, and N. Terada. 1998. Targeted disruption of p70S6K defines its role in protein synthesis and rapamycin sensitivity. Proc. Natl. Acad. Sci. USA 95:5033-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khaleghpour, K., S. Pyronnet, A. C. Gingras, and N. Sonenberg. 1999. Translational homeostasis: eukaryotic translation initiation factor 4E control of 4E-binding protein 1 and p70 S6 kinase activities. Mol. Cell. Biol. 19:4302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kischel, F. C., S. Helbardt, I. Behrmann, M. Germer, M. Pawlita, P. H. Krammer, and M. E. Peter. 1995. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14:5579-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krueger, A., I. Schmitz, P. H. Krammer, and S. Kirchhoff. 2001. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J. Biol. Chem. 276:20633-20640. [DOI] [PubMed] [Google Scholar]
- 42.Lawrence, D., Z. Shahrokh, S. Marsters, K. Achilles, D. Shih, B. Mounho, K. Hillan, K. Totpal, L. DeForge, P. Schow, J. Hooley, S. Sherwood, R. Pai, S. Leung, L. Khan, B. Gliniak, J. Bussiere, C. A. Smith, S. S. Strom, S. Kelley, J. A. Fox, D. Thomas, and A. Ashkenazi. 2001. Differential hepatocyte toxicity of recombinant Apo2L/TRAIL versions. Nat. Med. 7:383-385. [DOI] [PubMed] [Google Scholar]
- 43.Li, S., T. Takasu, D. M. Perlman, M. S. Peterson, D. Burrichter, S. Avdulov, P. B. Bitterman, and V. A. Polunovsky. 2003. Translation factor eIF4E rescues cells from myc-dependent apoptosis by inhibiting cytochrome c release. J. Biol. Chem. 278:3015-3022. [DOI] [PubMed] [Google Scholar]
- 44.Muzio, M., A. M. Chinnaiyan, F. C. Kischel, K. O'Rourke, A. Shevchenko, J. Ni, C. Scaffidi, J. D. Bretz, M. Zhang, R. Gentz, M. Mann, P. H. Krammer, M. E. Peter, and V. M. Dixit. 1996. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85:817-827. [DOI] [PubMed] [Google Scholar]
- 45.Nagane, M., H.-J. S. Huang, and W. K. Cavenee. 2001. The potential of TRAIL for cancer chemotherapy. Apoptosis 6:191-197. [DOI] [PubMed] [Google Scholar]
- 46.Nam, S. Y., G. A. Jung, G. C. Hur, H. Y. Chung, W. H. Kim, D. W. Seol, and B. L. Lee. 2003. Upregulation of FLIPS by Akt, a possible inhibition mechanism of TRAIL-induced apoptosis in human gastric cancers. Cancer Sci. 94:1066-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nebbioso, A., N. Clarke, E. Voltz, E. Germain, C. Ambriosino, P. Bontempo, R. Alvarez, E. M. Schiavone, F., Ferrara, F. Bresciana, A. Weisz, A. R. de Lera, H. Gronemeyer, and L. Altucci. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. 2005. Nat. Med. 11:77-84. [DOI] [PubMed] [Google Scholar]
- 48.Nitsch, R., I. Bechmann, R. A. Deisz, D. Haas, T. N. Kehmann, U. Wendling, and F. Zipp. 2000. Human brain cell death induced by tumor-necrosis factor-related apoptosis inducing ligand (TRAIL). Lancet 356:827-828. [DOI] [PubMed] [Google Scholar]
- 49.Pai, S. I., G. S. Wu, N. Ozoren, L. Wu, J. Jen, D. Sidransky, and W. S. El-Deiry. 1998. Rare loss-of-function mutation of a death receptor gene in head and neck cancer. Cancer Res. 58:3513-3518. [PubMed] [Google Scholar]
- 50.Parsa, A. T., and E. C. Holland. 2004. Cooperative translational control of gene expression by Ras and Akt in cancer. Trends Mol. Med. 10:607-613. [DOI] [PubMed] [Google Scholar]
- 51.Perez, D., and E. White. 2003. E1A sensitizes cells to tumor necrosis factor alpha by downregulating c-FLIP S. J. Virol. 77:2651-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petak, I., and J. A. Houghton. 2001. Shared pathways: death receptors and cytotoxic drugs in cancer therapy. Path. Oncol. Res. 7:95-106. [DOI] [PubMed] [Google Scholar]
- 53.Polunovsky, V. A., I. B. Rosenwald, A. T. Tan, J. White, L. Chiang, N. Sonenberg, and P. B. Bitterman. 1996. Translational control of cell death: eukaryotic translation initiation factor 4E blocks apoptosis in growth-factor restricted fibroblasts with physiologically expressed or deregulated Myc. Mol. Cell. Biol. 16:6573-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potter, C. J., L. G. Pedraza, and T. Xu. 2002. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4:658-665. [DOI] [PubMed] [Google Scholar]
- 55.Ricci, M. S., Z. Jin, M. Dews, D. Yu, A. Thomas-Tikhonenko, D. T. Dicker, and W. S. El-Diery. 2004. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol. Cell. Biol. 24:8541-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rieger, J., U. Naumann, T. Glaser, A. Ashkenazi, and M. Weller. 1998. APO2 ligand: a novel lethal weapon against malignant glioma? FEBS Lett. 427:124-128. [DOI] [PubMed] [Google Scholar]
- 57.Roth, W., S. Isenmann, U. Naumann, S. Kugler, M. Bahr, J. Dichgans, A. Ashkenazi, and M. Weller. 1999. Locoregional Apo2L/TRAIL eradicates intracranial human malignant glioma xenografts in athymic mice in the absence of neurotoxicity. Biochem. Biophys. Res. Commun. 265:479-483. [DOI] [PubMed] [Google Scholar]
- 58.Saito, R., J. R. Bringas, A. Panner, M. Tamas, R. O. Pieper, M. S. Berger, and K. Bankiewicz. 2004. Convection-enhanced delivery of tumor necrosis factor-related apoptosis-inducing ligand with systemic administration of temozolomide prolongs survival in an intracranial glioblastoma xenograft model. Cancer Res. 64:6095-6101. [DOI] [PubMed] [Google Scholar]
- 59.Sheridan, J. P., S. A. Marsters, R. M. Pitti, A. Gurney, M. Skubatch, D. Baldwin, L. Ramakrishnan, C. L. Gray, K. Baker, W. I. Wood, A. D. Goddard, P. Godowski, and A. Ashkenazi. 1997. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science 277:818-821. [DOI] [PubMed] [Google Scholar]
- 60.Smyth, M. J., K. Takeda, Y. Hayakawa, J. J. Peschon, M. R. M. van der Brink, and H. Yagita. 2003. Nature's TRAIL: on a path to cancer immunity. Immunity 18:1-6. [DOI] [PubMed] [Google Scholar]
- 61.Sonenberg, N., and A. C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10:268-275. [DOI] [PubMed] [Google Scholar]
- 62.Sonoda, Y., T. Ozawa, Y. Hirose, K. D. Aldape, M. McMahon, M. S. Berger, and R. O. Pieper. 2001. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 61:4956-4960. [PubMed] [Google Scholar]
- 63.Steck, P. A., M. A. Pershouse, S. A. Jasser, W. K. Yung, H. Lin, A. H. Ligon, L. A. Langford, M. L. Baumgard, T. Hattier, T. Davis, C. Frye, R. Hu, B. Swedlund, D. H. Teng, and S. V. Tavtigian. 1997. Identification of a candidate tumor suppressor gene, MMAC1, at chromosome 10q233 that is mutated in multiple advanced cancers. Nat. Genet. 15:356-362. [DOI] [PubMed] [Google Scholar]
- 64.Stolovich M., H. Tang, E. Hornstein, G. Levy, R. Cohen, S. S. Bae, M. J. Birnbaum, and O. Meyuhas. 2002. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol. Cell. Biol. 22:8101-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas, W. D., and P. Hersey. 1998. TNF-related apoptosis-inducing ligand (TRAIL) induces apoptosis in Fas ligand-resistant melanoma cells and mediates CD4 T cell killing of target cells. J. Immunol. 161:2195-2200. [PubMed] [Google Scholar]
- 66.von Manteuffel, S. R., P. B. Dennis, N. Pullen, A. C. Gingras, N. Sonenberg, and G. Thomas. 1997. The insulin-induced signaling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol. Cell. Biol. 9:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walczak, H., R. E. Miller, K. Ariail, B. Gliniak, T. S. Griffith, M. Kubin, W. Chin, J. Jones, A. Woodward, T. Le, C. Smith, P. Smolak, R. G. Goodwin, C. T. Rauch, J. C. Schuh, and D. H. Lynch. 1999. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 5:157-1163. [DOI] [PubMed] [Google Scholar]
- 68.Wang, X., W. Li, M. Williams, N. Terada, D. Alessi, and C. Proud. 2001. Regulation of elongation factor 2 kinase by p90 (RSK1) and p70 S6 kinase. EMBO J. 20:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiley, S. R., K. Schooley, P. J. Smolak, W. S. Din, C. P. Huang, J. K. Nicholl, G. R. Sutherland, T. D. Smith, C. Rauch, and C. A. Smith. 1995. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3:673-682. [DOI] [PubMed] [Google Scholar]
- 70.Xiao, C., B. F. Yang, N. Asadi, F. Beguinot, and C. Hao. 2002. Tumor necrosis factor-related apoptosis-inducing ligand induces death-inducing signaling complex and its modulation by c-FLIP and PED/PEA-15 in glioma cells. J. Biol. Chem. 277:25020-25025. [DOI] [PubMed] [Google Scholar]
- 71.Yang, B. F., C. Xiao, W. H. Roa, P. H. Krammer, and C. Hao. 2003. Calcium/calmodulin-dependent protein kinase II regulation of c-FLIP expression and phosphorylation in modulation of Fas-mediated signaling in malignant cells. J. Biol. Chem. 278:7043-7050. [DOI] [PubMed] [Google Scholar]