Abstract
During cell division, chromatin undergoes structural changes essential to ensure faithful segregation of the genome. Condensins, abundant components of mitotic chromosomes, are known to form two different complexes, condensins I and II. To further examine the role of condensin I in chromosome structure and in particular in centromere organization, we depleted from S2 cells the Drosophila CAP-H homologue Barren, a subunit exclusively associated with condensin I. In the absence of Barren/CAP-H the condensin core subunits DmSMC4/2 still associate with chromatin, while the other condensin I non-structural maintenance of chromosomes family proteins do not. Immunofluorescence and in vivo analysis of Barren/CAP-H-depleted cells showed that mitotic chromosomes are able to condense but fail to resolve sister chromatids. Additionally, Barren/CAP-H-depleted cells show chromosome congression defects that do not appear to be due to abnormal kinetochore-microtubule interaction. Instead, the centromeric and pericentromeric heterochromatin of Barren/CAP-H-depleted chromosomes shows structural problems. After bipolar attachment, the centromeric heterochromatin organized in the absence of Barren/CAP-H cannot withstand the forces exerted by the mitotic spindle and undergoes irreversible distortion. Taken together, our data suggest that the condensin I complex is required not only to promote sister chromatid resolution but also to maintain the structural integrity of centromeric heterochromatin during mitosis.
The genome of eukaryotic proliferating cells undergoes programmed structural changes in order to ensure the integrity of genetic material and cell viability during cell division. First, during S phase, when DNA is duplicated, sister chromatid cohesion is established along the entire length of DNA molecules and is maintained until entry into mitosis. Subsequently, during the early stages of mitosis, chromosomes condense into higher-order levels of chromatin organization, leading to the resolution of chromosome arms, a prerequisite for genome stability. Although mitotic chromosomes were one of the first subcellular structures observed (10), the mechanisms underlying their establishment have only recently begun to be unveiled.
A major contribution was the identification of the multiprotein condensin complex, initially purified and characterized from Xenopus extracts (17) and later shown to be highly conserved (reviewed in reference 45). Condensin is composed of two subcomplexes: a core heterodimer formed by the chromosomal ATPase SMC family (structural maintenance of chromosomes) proteins SMC2 and SMC4 (reviewed in reference 21) and a regulatory subcomplex formed by three non-SMC proteins, CAP-D2, CAP-G, and CAP-H (reviewed in reference 16). All the members of the condensin complex have been shown to be essential for cell and organism viability (reviewed in reference 45).
Mutation analysis of condensin subunits in both Saccharomyces cerevisiae and Schizosaccharomyces pombe show defects in chromosome condensation and segregation (11, 25, 33, 35, 42). However, genetic analyses in multicellular organisms such as Drosophila revealed that loss of condensin subunits leads to strong defects in segregation but had only partial effects on chromosome condensation. Mutation of Drosophila SMC4/gluon was shown to severely compromise sister chromatid resolution but not longitudinal axis shortening (40). Mutation of barren, the Drosophila CAP-H orthologue, does not affect chromosome condensation but impairs sister chromatid segregation (4). More recently, genetic analysis of Drosophila CAP-G shows that chromosome condensation is perturbed in prometaphase but normal condensation levels can be achieved at metaphase (9). Consistently, depletion of scII/SMC2 in DT40 chicken cells showed that chromosome condensation is delayed, however, normal levels are eventually reached (19). Similar results were obtained after depletion of SMC4 and MIX-1 in Caenorhabditis elegans (13). These data suggest that the condensin complex might not be the major factor required for the organization of the mitotic chromosome.
Indeed, recent studies have identified a new condensin complex in HeLa cell extracts named condensin II (32). Condensin II shares the core SMC proteins with condensin I but has different regulatory subunits. It has been suggested both condensin complexes contribute distinctly to the metaphase chromosome architecture in vertebrate cells. However, not all organisms appear to have the two types of complexes and different condensin complexes might be required for different tissues or at different developmental stages (32). Condensins I and II were shown to display different spatial and temporal chromatin localizations (18, 31). Condensin II was shown to be predominantly nuclear during interphase, and it was suggested to contribute to early stages of chromosome assembly in prophase, whereas condensin I was described to access chromatin only after nuclear envelope breakdown. Moreover, in HeLa cell chromosomes at metaphase, condensin II is enriched at the primary constriction. Previously, studies in Drosophila melanogaster revealed a strong localization of condensin I at the centromere (40). These findings raise the hypothesis that condensin complexes play a specific role in the organization of centromeric chromatin.
The centromere plays an essential role in chromosome segregation. First, it underlies the organization of the kinetochore and thereby the attachment and movement of chromosomes along spindle microtubules. Second, it ensures sister chromatid cohesion until metaphase-anaphase transition. In that way centromeres contribute to bipolar attachment of chromosomes, essential for the proper partitioning of the genome in cell division. In most higher eukaryotes, centromeres are formed by large arrays of tandem repeated sequences (reviewed in reference 43). Moreover, centromere inheritance appears to be dependent on the presence of specialized centromeric nucleosomes containing CENP-A (centromere protein A), a specific histone isoform that belongs to the histone H3 protein family (reviewed in reference 38). In the holocentric chromosome of C. elegans, several studies indicate that CENP-A colocalizes with the condensin subunits along the entire chromosome length and can play a role in centromere organization. Indeed, it has been shown that SMC-4 and MIX-1 are required for proper centromere biorientation and segregation (13).
These results could be attributed to the particular features of C. elegans holocentric chromosomes. However, there is increasing evidence that condensin might have a role at the centromeres of monocentric chromosomes. In agreement, a genetic and physical interaction between Drosophila CAP-G and the centromere-specific CID/CENP-A has recently been reported (20). Also, in S. pombe, chromatin immunoprecipitation assays showed that condensin localizes to CEN DNA (3). However, little is known about the role of condensins in the centromere structure.
In this study we have evaluated the role of condensin I upon the organization and segregation of mitotic chromosomes by depleting Barren/CAP-H from Drosophila S2 cells. We show that depletion of Barren/CAP-H compromises the binding to chromatin of the other condensin I regulatory subunits, DmCAP-D2 and DmCAP-G. However, the absence of Barren does not interfere with the binding to chromatin of the DmSMC4/2 core heterodimer, demonstrating the ability of the heterodimer to associate with chromatin independently of the regulatory subcomplex. We also show that S2 cells depleted of Barren/CAP-H display abnormal sister chromatid resolution and segregation.
In vivo analysis of Barren/CAP-H-depleted cells expressing green fluorescent protein (GFP)-histone H2B shows that chromosomes are unable to align at the metaphase plate and exhibit chromatin bridges as soon as anaphase starts. Immunofluorescence analysis also indicates that although chromosomes show bipolar attachment, intercentromere distances are unusually large. Moreover, centromeric markers appear distorted and the cohesin protein DRAD21 shows an abnormally broad distribution. Furthermore, we find that the heterochromatin- specific K9 dimethylated histone H3 is also abnormally distributed. Taken together, our results suggest that condensin I plays a major role in the organization of centromeric heterochromatin in order to maintain its elastic properties, which are essential to withstand the forces exerted by the mitotic spindle.
MATERIALS AND METHODS
Double-stranded RNA interference in Drosophila S2 cells.
To deplete Barren/CAP-H from Drosophila S2 tissue culture cells, a 1,445-bp EcoRI-AccI fragment spanning the 5′ untranslated region and including the ATG initiation codon of Barren/CAP-H cDNA (RE48802) was cloned into pSPT18 and pSPT19 expression vectors (Roche). RNA synthesis was performed using the T7 Megascript kit (Ambion) using the recombinant vectors as templates. In all RNA interference (RNAi) experiments, 15 μg of double-stranded RNA was added to 106 cells in 1 ml Schneider's medium (Gibco BRL) and incubated for 1 h at 25°C. Cells were then supplemented with 2 ml medium with 10% fetal bovine serum (Gibco BRL). At each time point, cells were collected and processed for both immunofluorescence and immunoblotting. For immunoblotting, cells were collected by centrifugation at 10,000 rpm for 10 min, washed with phosphate-buffered saline supplemented with protease inhibitors (Roche) and resuspended in 20 μl of sodium dodecyl sulfate sample buffer. Samples were boiled for 5 min before loading on a 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. When required, cells were incubated with 20 μM MG132 (Calbiochem) or/and with 30 μM colchicine (Sigma). Hypotonic shock was performed by resuspending cells in a 0.1% sodium citrate solution for 10 seconds.
Immunofluorescence in S2 cells.
Cells were centrifuged onto slides, fixed in 3.7% methanol free formaldehyde, 0.5% Triton X-100 an 1 × phosphate-buffered saline for 10 min followed by three washes in PBS-T (1x phosphate-buffered saline, 0.05% Tween 20) for 5 min. Blocking was performed in PBS-TF (PBS-T, 10% fetal bovine serum) for 30 min at room temperature. Primary antibody incubations were performed in PBS-TF for 1 h at room temperature followed by PBS-T wash (three times for 5 min). Incubation with fluorescent labeled secondary antibodies was as previously described according to manufacturer's instructions (Molecular Probes, The Netherlands). Slides were washed again three times with PBS-T for 5 min and mounted in Vectashield with 1 μg/ml of 4′,6′-diamidino-2-phenylindole (DAPI) (Vector, United Kingdom). Calcium treatment was performed as previously described (22). Briefly, cells were permeabilized for 90 s in a buffer containing 100 mM piperazine-bis(ethanesulfonic acid) (pH 6.8), 1 mM MgCl2, 0.1 mM CaCl2, and 0.1% Triton X-100 and then fixed for 10 min in the same buffer supplemented with 4% formaldehyde. Immunofluorescence was performed as described above using Tris-buffered saline instead of phosphate-buffered saline.
Images were collected either in the Zeiss Axiovert 200 M microscope (Carl Zeiss, Germany) using an Axiocam (Carl Zeiss, Germany) or the Leica Confocal SP2 (Leica Microsystems, Germany). Data stacks were deconvolved, using the Huygens Essential version 3.0.2p1 (Scientific Volume Imaging B.V., The Netherlands).
Intercentromere distances measurements were performed analyzing each image stack by stack. CID-labeled centromeres found in the same stack flanking a brighter DAPI-stained region (heterochromatin) of a chromosome were considered as sister centromeres and the distance was measured using AxioVision4.3 software (Carl Zeiss, Germany).
Antibodies.
The primary antibodies were anti-α-tubulin mouse B512 (Sigma-Aldrich) used at 1:4,000 for immunofluorescence and 1:10,000 for immunoblotting; anti-phospho-histone H3 rabbit polyclonal (Upstate Biotechnology) used at 1:1,000; anti-POLO mouse monoclonal MA294 (26) used at 1:30; anti-Barren/CAP-H rabbit polyclonal (4) used at 1:1,500 (immunofluorescence) and 1:3,000 (immunoblotting); anti-DmSMC4 (40) rabbit polyclonal used at 1:500 (immunoblotting) and sheep polyclonal used at 1:500 (immunofluorescence); anti-SMC2 rabbit polyclonal used at 1:1,000; anti-CAP-D2 rabbit polyclonal used at 1:10,000 (immunoblotting) and 1:2,000 (immunofluorescence); immunopurified anti-CAP-G rabbit polyclonal used at 1:5; anti-CID chicken polyclonal (6) used at 1:100; anti-CID rabbit polyclonal (15) used at 1:1,500; anti-dimethylated K9 histone H3 rabbit polyclonal (Upstate Biotechnology) used according to the manufacturer's instructions; anti-DRAD21 rabbit polyclonal used at 1:1,000 (47); anti-INCENP rabbit polyclonal used at 1:1,500 (1); and anti-ZW10 rabbit polyclonal used at 1:500 (49).
Time-lapse fluorescence imaging.
Live analysis of mitosis was performed on S2 cells stably expressing green fluorescent protein (GFP)-histone H2B. Control or Barren/CAP-H RNAi-treated cells were incubated for 72 h and plated on glass coverslips treated with 100 μg/ml concanavalin A (Sigma). Time-lapse images were collected at 1.5-min intervals, starting from the time mitotic chromosomes could be visualized, using a Cell Observer System (Carl Zeiss, Germany). Image processing and movie assembly was processed using AxioVision4.3 software (Carl Zeiss, Germany).
FACS analysis.
For fluorescence-activated cell sorting (FACS) analysis 106 cells were spun at 3,000 rpm for 5 min and resuspended in 200 μl phosphate-buffered saline. Cells were fixed with 2 ml 70% ice-cold ethanol in phosphate-buffered saline added drop by drop with continuous vortexing. Samples were kept on ice for 30 min before being spun at 3,000 rpm for 5 min and resuspended in 200 μl phosphate-buffered saline with 100 μg/ml RNase and 100 μg/ml propidium iodide. Samples were incubated at 37°C for 30 min. To analyze DNA content we used a FACSCalibur (Becton Dickinson) flow cytometer and data from 25,000 cells were obtained. Results were analyzed using CellQuest data acquisition software.
RESULTS
Localization and stability of condensin after depletion of Barren/CAP-H.
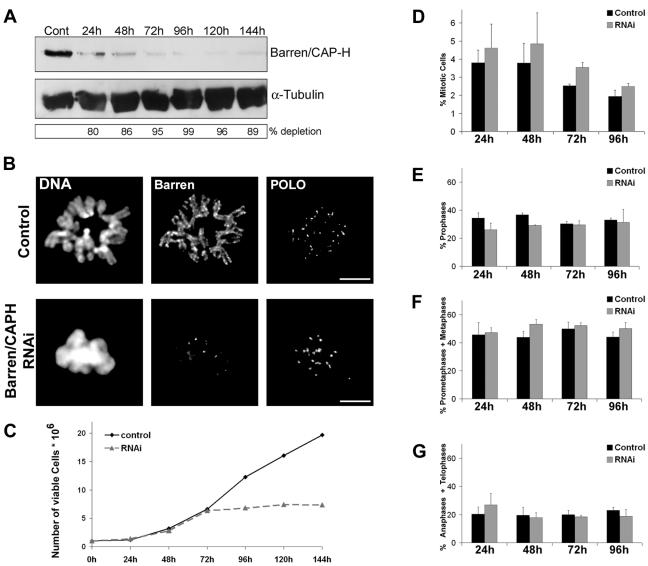
We performed double-stranded RNA interference in S2 Drosophila cells to deplete the regulatory subunit Barren/CAP-H of the condensin I complex. Western blot (Fig. 1A) and immunofluorescence (Fig. 1B) show that 96 h after double-stranded RNA addition, Barren/CAP-H is severely depleted (99% depletion), and the majority of the analysis was performed at this time point. By 96 h the culture doubling time is severely affected, suggesting that Barren/CAP-H is essential for cell viability (Fig. 1C). Nevertheless, Barren/CAP-H depletion did not significantly affect the mitotic index of the culture or mitotic progression among mitotic cells (Fig. 1D to G).
FIG. 1.
Depletion of Barren/CAP-H from Schneider 2 cells by double-stranded RNA interference. (A) Total protein extracts from 5 × 105 cells were run on a 7.5% SDS-PAGE and Barren/CAP-H protein levels were monitored at different times of the experiment by Western blot. α-Tubulin was used as loading control. (B) Depletion of Barren/CAP-H was confirmed by immunofluorescence. Metaphase chromosomes from double-stranded RNA-treated cells show no accumulation of Barren/CAP-H in contrast to control cells, where Barren/CAP-H is localized at the axis of each chromatid. Scale bars are 5 μm. (C) Proliferation profiles of control and Barren/CAP-H double-stranded RNA interference-treated S2 cells throughout the experiment. (D) Mitotic index and (F to H) mitotic progression throughout the experiment were quantified using either POLO/phospho-histone 3 or tubulin/phospho-histone 3 double staining. Approximately 6,500 cells were counted for each time point. Note that there is no significant change in both the mitotic index and mitotic progression between control and Barren/CAP-H-depleted cells meaning that Barren/CAP-H-depleted cells are able to undergo mitosis with normal timing.
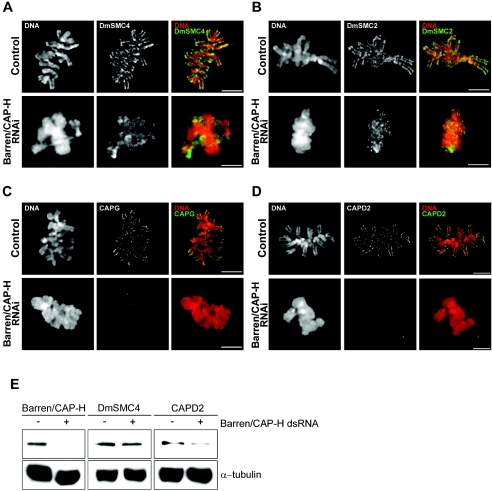
In order to determine whether Barren/CAP-H is necessary for the localization of other condensin I components, we carried out immunolocalization of both core and regulatory subunits of this complex (Fig. 2). In control cells, condensin I subunits DmSMC2, DmSMC4, DmCAP-D2, and DmCAP-G localize at the axis of metaphase chromosomes (Fig. 2A to D). In Barren/CAP-H-depleted cells, the two core proteins, DmSMC2 and DmSMC4, are able to localize on chromatin but the staining is no longer confined to an axis (Fig. 2A and B). In contrast, the other non-SMC proteins, DmCAP-D2 and DmCAP-G, are completely absent from Barren/CAP-H-depleted chromosomes (Fig. 2C and D). Furthermore, Western blot analysis of total protein extracts shows that the levels of DmSMC4 do not change significantly after Barren/CAP-H depletion while those of DmCAP-D2 are reduced by half relative to control levels (Fig. 2E). This suggests that the stability and the chromosomal localization of the non-SMC regulatory subunits of condensin I require all non-SMC subunits to be present. Furthermore, these results show the ability of the core DmSMC2 and DmSMC4 proteins to bind mitotic chromatin in the absence of regulatory proteins.
FIG. 2.
Stability of condensin I after depletion of Barren/CAP-H. (A to D) Immunolocalization of condensin subunits in control and Barren/CAP-H-depleted (96 h) S2 cells. In control metaphase cells, all other condensin I subunits, including (A) DmSMC4, (B) DmSMC2, (C) DmCAP-D2, and (D) DmCAP-G, localize at the axis of sister chromatids. After depletion of Barren/CAP-H, DmSMC4 and DmSMC2 still localize to chromatin but now appear diffused and are no longer confined to the axis of chromosomes. However, the non-SMC subunits DmCAP-D2 and DmCAP-G are not able to localize to Barren/CAP-H-depleted chromosomes. Scale bars are 5 μm. (E) Total protein extracts from 106 cells were assayed by Western blot to determine the levels of DmSMC4 and CAP-D2 in control (−) or Barren/CAP-H RNAi-treated (+) cells. Note that the levels of DmSMC4 do not change significantly after Barren/CAP-H depletion compared to the controls. In contrast, DmCAP-D2 levels are significantly reduced (45%) compared to the controls. Tubulin was used as the loading control and quantifications were performed using Image J Software.
Depletion of Barren/CAP-H affects chromosome resolution and sister chromatid segregation.
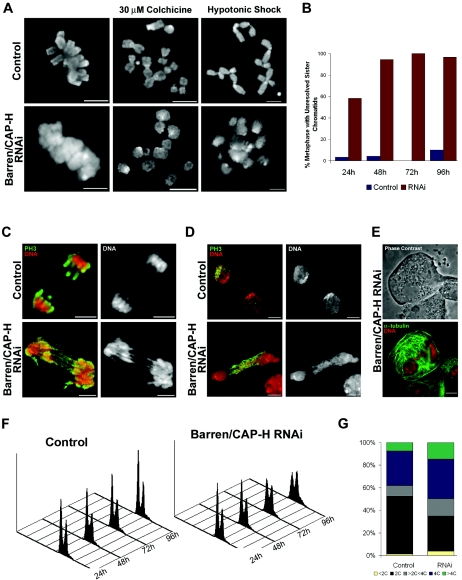
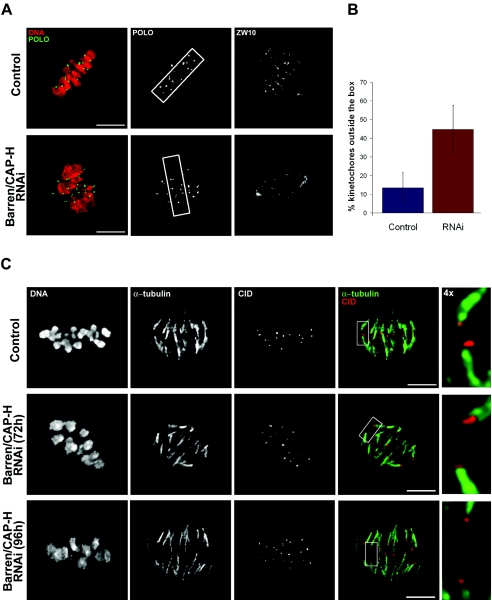
In order to define the specific contribution of condensin I to chromosome structure, we analyzed the mitotic phenotype of Barren/CAP-H-depleted cells (Fig. 3). Mitotic chromosomes from depleted cells were unable to resolve their sister chromatids but were able to condense along their longitudinal axis (Fig. 3A and B). The frequency of cells at metaphase with unresolved sister chromatids increased substantially during the double-stranded RNA interference experiment (Fig. 3B). Colchicine treatment, used to depolymerize microtubules and extend the period in prometaphase, did not allow better resolution of sister chromatids in Barren/CAP-H-depleted chromosomes (Fig. 3A). Moreover, Barren/CAP-H-depleted chromosomes were unable to sustain stress induced by hypotonic shock (Fig. 3A).
FIG. 3.
Characterization of S2 cells after depletion of Barren/CAP-H. (A) Control or Barren/CAP-H-depleted cells were fixed directly after 96 h or treated with colchicine (2 h) or hypotonic shock before fixation and then stained with DAPI to reveal chromosome structure. Unlike the chromosome morphology in control cells, Barren/CAP-H-depleted chromosomes show unresolved sister chromatids that become fuzzy after hypotonic shock. (B) Quantification of the percentage of cells in metaphase with unresolved sister chromatids shows that at 96 h virtually all cells in metaphase show unresolved sister chromatids. (C and D) Depletion of Barren/CAP-H leads to the formation of DNA bridges observed in anaphase and telophase. Cells were immunostained with an anti-phospho-histone H3 antibody (PH3) and DNA. (E) Representative image of a giant binucleated cell found after Barren/CAP-H depletion. Cells were immunostained for α-tubulin and DNA. Scale bars are 5 μm. (F) FACS profiles of both control and Barren/CAP-H-depleted cells, showing DNA content and cell number at different time points. (G) Graphic representation of the frequency of cells with different DNA content obtained from FACS analysis at 96 h after double-stranded RNA addition.
Despite the misresolution in Barren/CAP-H-depleted chromosomes, the cells are able to enter anaphase, displaying extensive DNA bridges even during very late telophase (Fig. 3C and D). Immunofluorescence analysis shows the presence of binucleated cells after Barren/CAP-H depletion (Fig. 3E). The frequency of these binucleated cells is higher in Barren/CAP-H-depleted cells compared to controls, while the frequency of cells undergoing cytokinesis is reduced (see Table S1 in the supplemental material). FACS analysis of both control and Barren/CAP-H-depleted cells suggests that depletion of Barren/CAP-H causes increased aneuploidy and the formation of highly polyploid cells (Fig. 3F and G). These results suggest that Barren/CAP-H depletion results in cytokinesis failure. However, this does not appear to be due to mislocalization of essential factors, as INCENP localizes normally to the spindle midzone during telophase in Barren/CAP-H-depleted cells (data not shown).
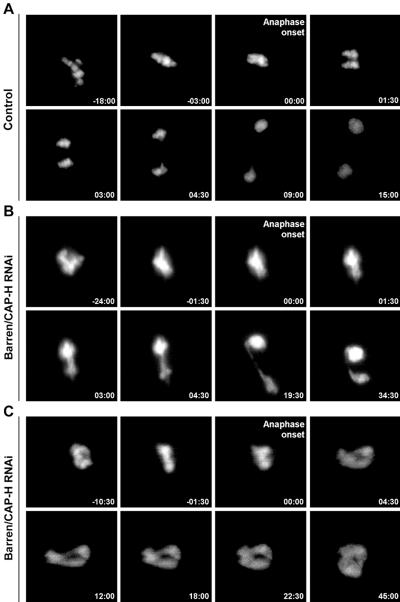
To highlight Barren/CAP-H-depleted chromosome dynamics during mitosis we performed time-lapse fluorescence imaging in S2 cells stably expressing GFP-histone H2B (Fig. 4). In control cells we can clearly observe chromosome congression to the metaphase plate, sister chromatid separation, and segregation to opposite poles (Fig. 4A; see also Movie S1 in the supplemental material). However, in Barren/CAP-H-depleted cells we consistently observed persistent oscillation of chromosomes during an extended prometaphase. Indeed, a well-defined metaphase plate was never observed before anaphase onset, suggesting that chromosomes fail to align properly (Fig. 4B; see also Movie S2 in the supplemental material). Furthermore, in Barren/CAP-H-depleted cells, DNA bridges are detected after anaphase onset. DNA bridges were found in 92.3% (n = 13) of Barren/CAP-H-depleted cells whereas in control cells only one cell showed DNA bridging in anaphase (n = 11). In 15.4% (n = 13) of Barren/CAP-H-depleted cells we observed the formation of massive DNA bridges that after the initial separation at anaphase onset fused back into a single large nucleus (Fig. 4C; see also Movie S3 in the supplemental material). These results further support a cytokinesis failure as inferred by both FACS analysis and immunofluorescence.
FIG. 4.
In vivo analysis of mitotic progression after depletion of Barren/CAP-H. Selected images from time-lapse movies of control and Barren/CAP-H-depleted (72 h) S2 cells stably expressing GFP-histone H2B acquired every 2 min from the time mitotic chromosomes could be identified. To align the movies anaphase onset was defined as time zero. (A) In control cells, prometaphase is followed by a tight organization of the chromosome at the metaphase plate, which after a few minutes initiates sister chromatid separation. (B) Analysis of Barren/CAP-H-depleted cells shows that these cells never appear to define a normal tight metaphase plate. Furthermore, chromatin bridges are observed as soon as anaphase is initiated. (C) In some cases we also observed cells depleted of Barren/CAP-H undergo anaphase onset but extensive chromatin bridges form, and after an initial attempt to segregate, the chromatin collapses back into a single large nucleus.
Chromosomes depleted of Barren/CAP-H have functional kinetochores but fail to congress normally.
Time-lapse fluorescence imaging of Barren/CAP-H-depleted cells in mitosis shows that chromosome alignment at the metaphase plate is not achieved, which suggests a failure in chromosome congression. To address this further, we arrested cells in metaphase with the proteosome inhibitor MG123 (12). Control and Barren/CAP-H-depleted cells were incubated in MG132 (up to 2 h) and immunostained for POLO (kinetochore marker) and ZW10, which migrates to spindle microtubules when the chromosomes reach bipolar attachment (49) (Fig. 5).
FIG. 5.
Kinetochore microtubule attachment and congression after depletion of Barren/CAP-H. (A) Both control and Barren/CAP-H-depleted cells (96 h) were arrested in metaphase by incubation with the proteasome inhibitor MG132 for 2 h. In order to evaluate kinetochore congression, a box comprising 85% of aligned kinetochores in a control metaphase was defined. The same box was placed over Barren/CAP-H-depleted metaphases perpendicular to the spindle (indicated by ZW10 spindle staining). (B) Quantification of the percentage of kinetochores placed outside the box in both control and Barren/CAP-H-depleted metaphase cells (for the control 14 cells were analyzed, n = 292 kinetochores, and for Barren/CAP-H depletion 14 cells were analyzed, n = 268 kinetochores). Note that depletion of Barren/CAP-H causes a severe increase in the frequency of misaligned kinetochores. (C) Control or Barren/CAP-H-depleted cells arrested with MG132 for 2 h were incubated with calcium to remove all microtubules except the kinetochore fiber. Cells were fixed and immunostained for α-tubulin and CID. Higher magnification images (4×) show that in both control and Barren/CAP-H-depleted cells, metaphase chromosomes are under bipolar attachment. Scale bars are 5 μm.
First, we carried out a quantification to determine the number of misaligned kinetochores after Barren/CAP-H depletion. A box perpendicular to the spindle, comprising approximately 85% of the kinetochores from control cells (10 by 3 μm in area) was used to quantify congression (Fig. 5A and B). In Barren/CAP-H-depleted cells more than 45% of kinetochores localize outside the box, suggesting that Barren/CAP-H-depleted chromosomes are unable to congress properly. As chromosome misalignment is usually associated with defective microtubule-kinetochore interaction (1, 24, 50), we investigated whether Barren/CAP-H-depleted chromosomes are able to establish stable microtubule attachment. In order to detect only kinetochore microtubules, control and Barren/CAP-H-depleted cells were treated with calcium (Fig. 5C), which specifically destabilizes nonkinetochore microtubules (22, 28). Similar to the controls, Barren/CAP-H-depleted chromosomes were found to be mostly under bipolar attachment, with kinetochores located at the end of well-defined microtubule bundles. Also, ZW10 was observed along kinetochore fibers, suggesting a normal kinetochore microtubule interaction (Fig. 5A). Finally, immunofluorescence analysis failed to detect Mad2 (data not shown), a checkpoint protein known to leave kinetochores only after spindle attachment (27). These results strongly suggest that Barren/CAP-H-depleted chromosomes, although unable to resolve their sister chromatids, organize well-defined kinetochores that can bind spindle microtubules. Therefore, the inability of Barren/CAP-H-depleted chromosomes to congress to the metaphase plate is not due to abnormal kinetochore-microtubule attachment.
Barren/CAP-H-depleted chromosomes show unusually large distances between sister centromeres after bipolar attachment.
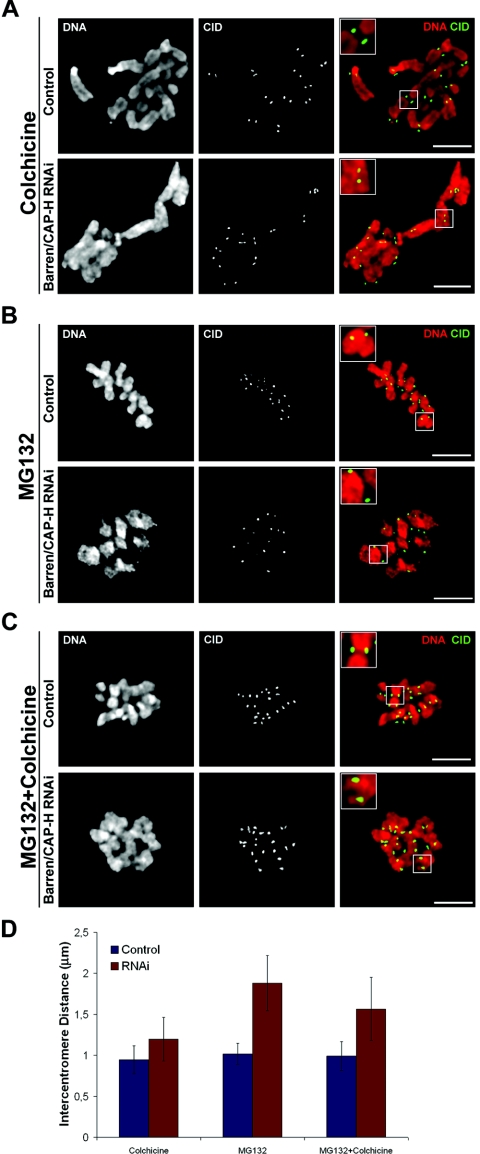
As the congression defect observed in Barren/CAP-H-depleted cells cannot be explained by an incorrect microtubule-kinetochore interaction, we considered the possibility that this could result from structural alterations in the centromeric region of these chromosomes. To address this hypothesis we analyzed the centromeric region by determining the distance between CID-labeled centromeres of sister chromatids (Fig. 6). First, we measured the intercentromere distance after colchicine incubation, when no microtubules are present, in order to avoid the spindle-pulling forces exerted upon kinetochores. The intercentromere distance was 0.95 ± 0.17 μm (n = 62) for control and 1.20 ± 0.24 μm (n = 62) for Barren/CAP-H-depleted cells (Fig. 6A and D). This indicates that already in the absence of pulling forces, the centromeres appear slightly further apart than in control cells. To evaluate the effect of the opposite pulling forces exerted by the spindle on centromere structure, we determined the intercentromere distance in cells arrested in metaphase by incubation with MG132 only (Fig. 6 B and D). Under these conditions, the centromeres from control chromosomes show a distance of 1.02 ± 0.13 μm (n = 33) whereas in Barren/CAP-H-depleted chromosomes sister centromeres are 1.88 ± 0.34 μm (n = 44) apart, significantly different from controls (P < 0.0001) (Fig. 6B and D). These results suggest that condensin I is required for centromeres to maintain a rather rigid structure capable of withstanding the extreme pulling forces exerted by the spindle without being deformed.
FIG. 6.
Analysis of intercentromere distances after depletion of Barren/CAP-H. (A to C) Both control and Barren/CAP-H-depleted (72 h) cells were immunostained for CID and DNA. Cultures were (A) incubated with 30 μM colchicine for 2 h to depolymerize all microtubules before entering mitosis, (B) incubated for 2 h with 20 μM MG132 to arrest cells in metaphase, (C) incubated with 20 μM MG132 to arrest cells in metaphase followed by a 30-min incubation with 30 μM colchicine to depolymerize all microtubules that were previously attached to the kinetochores. Scale bars are 5 μm. Higher magnifications (2×). (D) Quantification of intercentromere distances of control and Barren/CAP-H-depleted cells after the indicated experimental conditions.
If the deformation of the centromeric region was within its elastic limit, we would expect the centromeres to return to their original position once the force applied by the spindle is released. To address this, cells were first incubated with MG132 to arrest them in metaphase, under bipolar attachment, and then with colchicine to induce subsequent microtubule depolymerization (Fig. 6C). Under these conditions, in Barren/CAP-H-depleted chromosomes the distance across centromeres remains significantly higher (P < 0.0001) than that of controls (1.57 ± 0.27 μm, n = 40 versus 0.99 ± 0.13 μm n = 51). Indeed, this distance is even higher than the one observed in the absence of microtubule attachment (Fig. 6A and D). Similar results were obtained after inhibition of microtubule dynamics by treatment with low doses of taxol (data not shown). These results show that the removal of the pulling forces exerted by the spindle did not allow sister centromeres to recover their original organization, indicating that the elastic properties of the centromere proximal chromatin are severely compromised in the absence of Barren/CAP-H.
Barren/CAP-H-depleted chromosomes under bipolar attachment show abnormal localization of the SCC1/DRAD21 cohesin subunit.
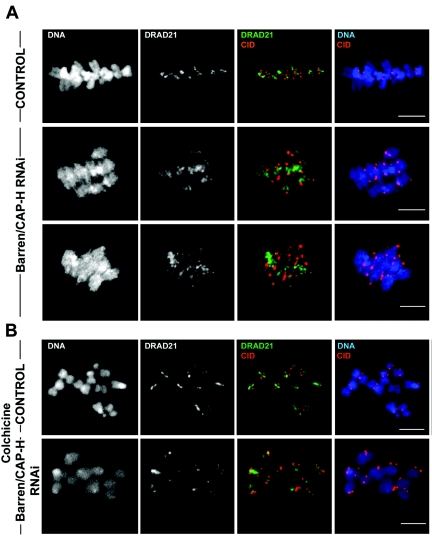
We have shown that centromeric region from Barren/CAP-H-depleted chromosomes is unable to maintain its integrity after bipolar attachment. Previous in vivo studies in yeast reveal that SCC1/RAD21 cohesin subunit displays a dynamic localization at the centromere after bipolar attachment (46). Therefore, we wanted to evaluate whether the elongation of the centromeric region observed in Barren/CAP-H-depleted chromosomes could affect the maintenance of cohesion of separated sister centromeres. To address this, we performed immunolocalization of SCC1/DRAD21 in metaphase-arrested Barren/CAP-H-depleted chromosomes (Fig. 7A). The results showed that SCC1/DRAD21 localizes between the stretched sister centromeres as cells reach a metaphase-like configuration. However, in contrast to control cells, in which SCC1/DRAD21 localizes as a thin line between sister centromeres, in Barren/CAP-H-depleted chromosomes, SCC1/DRAD21 distribution is very broad, occupying a large area between the two separated centromeres (Fig. 7A). These observations demonstrate that cohesin is still present in Barren/CAP-H-depleted chromosomes despite the distortion of centromeric region.
FIG. 7.
Analysis of the localization of SCC1/DRAD21 on chromosomes after depletion of Barren/CAP-H. Both control and Barren/CAP-H-depleted (96 h) cells were immunostained for POLO and DRAD21. (A) Cells arrested at metaphase by 2 h incubation with 20 μM MG132. In control cells SCC1/DRAD21 localizes between sister chromatids as a tight line between sister centromeres. However, after depletion of Barren/CAP-H, SCC1/DRAD21 is distributed over a broad area between sister centromeres. (B) Cells were incubated with 30 μM colchicine for 2 h to arrest them at prometaphase before microtubules could bind kinetochores. In these cells, SCC1/DRAD21 localizes to a thin line between sister centromeres in both control and Barren/CAP-H-depleted chromosomes. Scale bars are 5 μm.
To clarify if the broad SCC1/DRAD21 staining results from chromatin stretch induced by bipolar attachment, we immunostained both control and Barren/CAP-H-depleted cells that were treated with colchicine for a long period so that kinetochore-microtubule interactions were never established. Under these conditions the localization of SCC1/DRAD21 in Barren/CAP-H-depleted chromosomes now appears confined to the centromeric and pericentromeric regions, resembling the staining obtained in control cells (Fig. 7B). These results indicate that the abnormally broad distribution of SCC1/DRAD21 in Barren/CAP-H-depleted chromosome is only observed after spindle bipolar attachment.
Pericentromeric heterochromatin of Barren/CAP-H-depleted chromosomes is disorganized after bipolar attachment.
It is known that SCC1/DRAD21 enrichment at centromere region is dependent on HP1/Su(Var)2-5 and on the dimethylation of lysine 9 (K9) performed by Su(Var)3-9 on histone H3 (30). Indeed, histone H3 di- or trimethylated on K9 is known to localize specifically to heterochromatin (36). Therefore, we wanted to determine whether the broad distribution of SCC1/DRAD21 could be due to an altered structure of the template centromeric and pericentromeric heterochromatin.
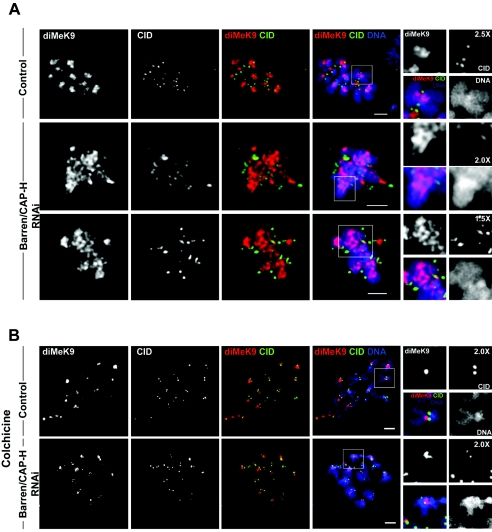
In order to address this question we immunostained both control and Barren/CAP-H-depleted cells arrested with MG132 at metaphase with an antibody that specifically detects dimethylated K9 histone H3 (diMeK9) and the centromere protein CID. In control cells, diMeK9 staining was mainly observed at the centromeric and pericentromeric region, as shown by the localization of the centromere marker CID (Fig. 8A). However, after depletion of Barren/CAP-H, diMeK9 staining appears to occupy a much broader area extending significantly beyond the stretched centromere, as defined by CID (Fig. 8A).
FIG. 8.
Immunolocalization of dimethylated K9 histone H3 in the heterochromatin of Barren/CAP-H RNAi-depleted chromosomes. Control and Barren/CAP-H cells were immunostained for CID and K9 dimethylated histone H3 (diMeK9). (A) In control cells at metaphase, diMeK9 is confined to heterochromatin. However, after depletion of Barren/CAP-H the pattern of diMeK9 appears to be significantly altered under bipolar attachment. diMeK9 is now detected over a broad area of chromatin localized between the two CID-labeled centromeres. (B) Both control and Barren-CAP-H-depleted cells were incubated with 30 μM colchicine for 2 h to depolymerize microtubules and the pattern of diMeK9 was determined. Note that under these conditions diMeK9 is confined to a tight region between sister centromeres in both control and Barren/CAP-H-depleted cells. Scale bars are 5 μm.
One possible interpretation for the broader diMeK9 staining is that methylation of histone H3 is altered in the absence of Barren/CAP-H. To rule out this hypothesis, we analyzed the localization of diMeK9 in chromosomes of cells treated with colchicine for 3 h and submitted to hypotonic shock, to avoid bipolar attachment and to force structural alterations. We observed that under these conditions the staining of diMeK9 and CID are almost identical in control and Barren/CAP-H-depleted chromosomes (Fig. 8B), which suggests that dimethylation of histone H3 can occur normally in the absence of Barren/CAP-H.
Therefore, the distinct diMeK9 and CID staining patterns reported for control and Barren/CAP-H-depleted cells after MG132 arrest can only reflect a difference in the organization of the centromeric and pericentromeric heterochromatin, which, after bipolar attachment, undergoes irreversible distortion.
We also performed a comparative analysis between control and Barren/CAP-H-depleted dimethylated K4 (diMeK4) histone H3-stained euchromatic chromosome arms (7) (see Fig. S1 in the supplemental material). Despite a broader staining observed in Barren/CAP-H-depleted chromosomes, as a result of an overall altered chromosome structure, the diMeK4 pattern remains confined to the chromosome arms and chromatin distortion is not as severe as found at the centromeric and pericentromeric regions. These data further indiate that severe structural alterations occur specifically at the centromeric and pericentromeric heterochromatin as a result of the opposite pulling forces exerted by the spindle.
DISCUSSION
Here we provide evidence that condensin I is absolutely required for mitotic chromosome resolution and cell viability. In the absence of condensin I DNA bridges are observed during anaphase and telophase. Importantly, we show for the first time that condensin I depletion results in congression defects associated with alterations in the structural integrity of the centromere-proximal chromatin.
We have shown that depletion of Barren/CAP-H, a condensin I-specific subunit in Drosophila S2 cells, leads to the formation of chromosomes that cannot resolve their sister chromatids. Nevertheless, DmSMC2 and DmSMC4, the two core proteins shared by both condensins I and II, are able to localize to Barren/CAP-H-depleted chromosomes. However, DmSMC2/4 subunits were found diffuse over the chromatin and are not confined to a well-defined central axis. This strongly suggests that condensin I depletion results in mitotic chromosomes with a poorly defined axial organization.
The identification of a second condensin complex in HeLa cells, named condensin II, suggested the two condensin complexes could play distinct roles in mitotic chromosome organization (32). The authors reported that complete disruption of chromatid axial organization was only achieved when both condensin I and condensin II were absent. While the contribution of condensin II to mitotic chromosome structure in Drosophila cells remains undetermined, previous studies in S2 cells have shown that if both condensin complexes are removed by depleting one core subunit (DmSMC4), sister chromatid resolution is specifically affected (8). Accordingly, depletion of Barren/CAP-H results in a chromosome structure phenotype similar to that described previously for depletion of DmSMC4. These observations suggest that in S2 tissue culture cells, if a condensin II complex does exist, it does not play a significant role in mitotic chromosome organization.
Additionally, we have also observed that in the absence of a regulatory subunit of condensin I, the DmSMC2 and DmSMC4 core proteins are able to localize to chromatin. This suggests that the DmSMC2/4 heterodimer binds DNA independently of the regulatory subunits. In agreement, in vitro studies have shown that the core SMC heterodimer alone has DNA binding properties (23, 41). Furthermore, short interfering RNAi depletion of human CAP-D2 from HeLa cells does not alter the levels of human CAP-E/SMC2 on mitotic chromosomes (48). In contrast, studies in budding yeast revealed that only the entire condensin complex is able to associate with DNA (25). This diversity probably results from species differences in the mechanism responsible for loading condensin to mitotic chromosomes.
We have also shown that the condensin I regulatory subunits DmCAP-D2 and DmCAP-G do not localize to Barren/CAP-H-depleted mitotic chromosomes. These data indicate that loading of the regulatory subcomplex to mitotic chromosomes requires all non-SMC subunits to be present. Interestingly, a homologue for CAP-G2 was not found in Drosophila melanogaster (32), and it has been suggested that the DmCAP-G subunit could be shared by both condensins I and II in this organism. The absence of DmCAP-G in Barren/CAP-H-depleted chromosomes clearly reveals that either the Drosophila DmCAP-G equivalent in condensin II has not been identified yet or the condensin II complex is totally absent from mitotic chromosomes in S2 cells.
One of the consequences of depleting condensins from mitotic chromosomes is the consistent presence of DNA bridges formed during anaphase that remain unresolved until telophase or even further. Here we show for the first time by in vivo time-lapse microscopy that the chromatin bridges observed in Barren/CAP-H-depleted cells must already be present at the metaphase-anaphase transition and are likely to be due to the inability to resolve catenated sister chromatids. Moreover, we have shown Barren/CAP-H depletion can disrupt cytokinesis. Cytokinesis failure has already been correlated with condensin depletion in other studies (4, 19). This correlation is more likely related to a physical incapacity in completing cell division due to DNA bridges at the cleavage furrow than to a direct role of condensin in cytokinesis.
In vivo analysis of condensin I-depleted cells in mitosis also revealed that chromosome congression is abnormal. Indeed, we have found that Barren/CAP-H-depleted chromosomes are unable to align at the metaphase plate even when extra time is provided by preventing anaphase onset with the proteosome inhibitor MG132. Studies in HeLa cells have also pointed out abnormal chromosome alignment after depletion of condensin I (31, 48) and it has been suggested that condensin is required for normal centromere/kinetochore function. However, our results show that in the absence of condensin I, the centromere supports the formation of a functional kinetochore as revealed by the normal localization of POLO and the correct kinetochore-microtubule bipolar attachment. Previous experiments in which DmSMC4 was depleted in S2 cells also reported a normal kinetochore organization and function (8). These findings show that in Drosophila melanogaster, the organization of the kinetochore does not require the underlying chromatin to contain condensins.
Indeed, our results demonstrate that the abnormal chromosome congression observed in Barren/CAP-H-depleted cells is likely to be related to the loss of centromere elasticity rather than kinetochore malfunction. Here we showed that in the absence of Barren/CAP-H, after bipolar attachment is established, the centromeric region elongates nearly twice the distance observed in control chromosomes. In agreement, abnormal centromere separation has also been recently reported when CAP-G is mutated in Drosophila melanogaster (9). Also, several studies in C. elegans have suggested a role for condensin II (the sole condensin complex in this organism) in centromere resolution and integrity (13, 29, 39). Our observations also show that the normal organization of pericentric heterochromatin is not restored after removal of microtubules. This suggests that Barren/CAP-H is essential to prevent irreversible loss of centromere integrity after bipolar attachment.
A possible explanation for the abnormal separation of sister centromeres could be due to an altered cohesion between sister chromatids in the absence of Barren/CAP-H. However, we have clearly shown here that despite its broad distribution pattern, SCC1/DRAD21 is still present between the abnormally apart sister centromeres in metaphase arrested cells. Additionally, it was previously described that cohesin follows a normal dynamics during mitosis in DmSMC4-depleted cells (8). Thus, the structural alterations we observed after depletion of Barran/CAP-H are unlikely to result from abnormal cohesin distribution. Moreover, we found not only that the pairing domain of sister chromatid is altered, but also that the pericentric heterochromatin-associated dimethylated K9 histone H3 is irregularly distributed and the centromere marker CID appears distorted.
It has been demonstrated that centric and pericentric heterochromatin shows stronger attachment to a central proteinaceous scaffold or matrix (5, 44). Reciprocally, chromatin immunoprecipitation experiments in S. pombe revealed a preferential association of condensin subunits with central centromeric sequences (3). Recently, a genetic and direct interaction between Drosophila CAP-G and the centromere-specific histone H3 variant CID was reported (20). These observations, taken together with our data, strongly support the idea that the association between the centromere/pericentromere chromatin and the chromosome axis is required for the establishment of an elastic but rigid structure able to resist the forces exerted by the spindle upon sister centromeres during congression.
Several studies regarding the longitudinal elastic properties of mitotic chromosomes have shown that the elastic behavior depends strongly on the continuity of the DNA chain (2, 34). However, the contribution of the protein scaffold to the elastic response of chromatin is controversial. It has been shown that the elastic and bending properties of mitotic chromosomes are inconsistent with the existence of a well-defined central chromosome scaffold, and alternatively, it has been suggested that the mitotic chromosome is essentially a chromatin network (34). Other studies revealed that the elastic properties depend on a mitotic chromosome protein scaffold, in particular on SMC proteins, as domains containing SMC proteins were shown to exhibit a higher elastic response (2).
While most studies have concentrated on the elastic properties of the arms, much less is known about the elasticity of the centromere region. Several studies pointed out the elastic properties of the centromere-proximal chromatin (14, 37). Indeed, we showed that the absence of condensin I compromises the elastic properties of centromeric chromatin. Therefore, our data favor the hypothesis that the elastic properties of the chromosome are indeed dependent on a proteinaceous structure, at least at the centromeric region.
In summary, we have shown that Barren-CAP-H is essential to allow the organization of a defined chromosome axis and to resolve sister chromatids. Furthermore, condensin I is not required for the organization of functional kinetochores but is essential to maintain the structural integrity of the centromeric region during mitosis.
Supplementary Material
Acknowledgments
We thank Hugo Bellen, Gary Karpen, Steve Henikoff, Mar Carmena, Michael Goldberg, and Margarete Heck for antibodies. Special thanks go to Patrick O'Farrell for the AFP-histone H2B-expressing cells, Andre Maia for help with the MG132 incubations, Miguel Monteiro for FACS analysis, Augusta Monteiro for technical support, Soren Steffensen for DmCAP-G antibody, and the members of the Sunkel lab for helpful comments in the preparation of the manuscript.
R.A.O. is a Ph.D. student at the Programa Doutoral em Biologia Experimental e Biomedicina of the Universidade de Coimbra supported by a Ph.D. fellowship from the Fundação para a Ciência e a Tecnologia of Portugal. The laboratory of C.E.S. is supported by grants from the Fundação para a Ciência e Tecnologia of Portugal and the European Union.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Adams, R. R., H. Maiato, W. C. Earnshaw, and M. Carmena. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almagro, S., D. Riveline, T. Hirano, B. Houchmandzadeh, and S. Dimitrov. 2004. The mitotic chromosome is an assembly of rigid elastic axes organized by structural maintenance of chromosomes (SMC) proteins and surrounded by a soft chromatin envelope. J. Biol. Chem. 279:5118-5126. [DOI] [PubMed] [Google Scholar]
- 3.Aono, N., T. Sutani, T. Tomonaga, S. Mochida, and M. Yanagida. 2002. Cnd2 has dual roles in mitotic condensation and interphase. Nature 417:197-202. [DOI] [PubMed] [Google Scholar]
- 4.Bhat, M. A., A. V. Philp, D. M. Glover, and H. J. Bellen. 1996. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell 87:1103-1114. [DOI] [PubMed] [Google Scholar]
- 5.Bickmore, W. A., and K. Oghene. 1996. Visualizing the spatial relationships between defined DNA sequences and the axial region of extracted metaphase chromosomes. Cell 84:95-104. [DOI] [PubMed] [Google Scholar]
- 6.Blower, M. D., and G. H. Karpen. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3:730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrd, K. N., and A. Shearn. 2003. ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc. Natl. Acad. Sci. USA 100:11535-11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coelho, P. A., J. Queiroz-Machado, and C. E. Sunkel. 2003. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J. Cell Sci. 116:4763-4776. [DOI] [PubMed] [Google Scholar]
- 9.Dej, K. J., C. Ahn, and T. L. Orr-Weaver. 2004. Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics 168:895-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flemming, W. Z. 1965. Historical paper. Contributions to the knowledge of the cell and its vital processes. J. Cell Biol. 25:1-69. [PubMed] [Google Scholar]
- 11.Freeman, L., L. Aragon-Alcaide, and A. Strunnikov. 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149:811-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genschik, P., M. C. Criqui, Y. Parmentier, A. Derevier, and J. Fleck. 1998. Cell cycle-dependent proteolysis in plants. Identification Of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor mg132. Plant Cell. 10:2063-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagstrom, K. A., V. F. Holmes, N. R. Cozzarelli, and B. J. Meyer. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16:729-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, X., S. Asthana, and P. K. Sorger. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101:763-775. [DOI] [PubMed] [Google Scholar]
- 15.Henikoff, S., K. Ahmad, J. S. Platero, and B. van Steensel. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirano, T. 2005. Condensins: organizing and segregating the genome. Curr. Biol. 15:R265-275. [DOI] [PubMed] [Google Scholar]
- 17.Hirano, T., R. Kobayashi, and M. Hirano. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89:511-521. [DOI] [PubMed] [Google Scholar]
- 18.Hirota, T., D. Gerlich, B. Koch, J. Ellenberg, and J.-M. Peters. 2004. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117:6435-6445. [DOI] [PubMed] [Google Scholar]
- 19.Hudson, D. F., P. Vagnarelli, R. Gassmann, and W. C. Earnshaw. 2003. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell 5:323-336. [DOI] [PubMed] [Google Scholar]
- 20.Jager, H., M. Rauch, and S. Heidmann. 2005. The Drosophila melanogaster condensin subunit Cap-G interacts with the centromere-specific histone H3 variant CID. Chromosoma 113:350-361. [DOI] [PubMed] [Google Scholar]
- 21.Jessberger, R. 2002. The many functions of SMC proteins in chromosome dynamics. Nat Rev. Mol. Cell Biol. 3:767-778. [DOI] [PubMed] [Google Scholar]
- 22.Kapoor, T. M., T. U. Mayer, M. L. Coughlin, and T. J. Mitchison. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura, K., and T. Hirano. 2000. Dual roles of the 11S regulatory subcomplex in condensin functions. Proc. Natl. Acad. Sci. USA 97:11972-11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kline-Smith, S. L., A. Khodjakov, P. Hergert, and C. E. Walczak. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol Cell 15:1146-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavoie, B. D., E. Hogan, and D. Koshland. 2002. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell Biol. 156:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llamazares, S., A. Moreira, A. Tavares, C. Girdham, B. A. Spruce, C. Gonzalez, R. E. Karess, D. M. Glover, and C. E. Sunkel. 1991. POLO encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 5:2153-2165. [DOI] [PubMed] [Google Scholar]
- 27.Logarinho, E., H. Bousbaa, J. M. Dias, C. Lopes, I. Amorim, A. Antunes-Martins, and C. E. Sunkel. 2004. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J. Cell Sci. 117:1757-1771. [DOI] [PubMed] [Google Scholar]
- 28.Mitchison, T., L. Evans, E. Schulze, and M. Kirschner. 1986. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 45:515-527. [DOI] [PubMed] [Google Scholar]
- 29.Moore, L. L., G. Stanvitch, M. B. Roth, and D. Rosen. 2005. HCP-4/CENP-C promotes the prophase timing of centromere resolution by enabling the centromere association of HCP-6 in Caenorhabditis elegans. Mol. Cell. Biol. 25:2583-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nonaka, N., T. Kitajima, S. Yokobayashi, G. Xiao, M. Yamamoto, S. I. Grewal, and Y. Watanabe. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4:89-93. [DOI] [PubMed] [Google Scholar]
- 31.Ono, T., Y. Fang, D. L. Spector, and T. Hirano. 2004. Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell 15:3296-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono, T., A. Losada, M. Hirano, M. P. Myers, A. F. Neuwald, and T. Hirano. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115:109-121. [DOI] [PubMed] [Google Scholar]
- 33.Ouspenski, I. I., O. A. Cabello, and B. R. Brinkley. 2000. Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol. Biol. Cell 11:1305-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirier, M. G., and J. F. Marko. 2002. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc. Natl. Acad. Sci. USA 99:15393-15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saka, Y., T. Sutani, Y. Yamashita, S. Saitoh, M. Takeuchi, Y. Nakaseko, and M. Yanagida. 1994. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 13:4938-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann, S. Rea, T. Jenuwein, R. Dorn, and G. Reuter. 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shelby, R. D., K. M. Hahn, and K. F. Sullivan. 1996. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 135:545-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, M. M. 2002. Centromeres and variant histones: what, where, when and why? Curr. Opin. Cell Biol. 14:279-285. [DOI] [PubMed] [Google Scholar]
- 39.Stear, J. H., and M. B. Roth. 2002. Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes Dev. 16:1498-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steffensen, S., P. A. Coelho, N. Cobbe, S. Vass, M. Costa, B. Hassan, S. N. Prokopenko, H. Bellen, M. M. Heck, and C. E. Sunkel. 2001. A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr. Biol. 11:295-307. [DOI] [PubMed] [Google Scholar]
- 41.Stray, J. E., and J. E. Lindsley. 2003. Biochemical analysis of the yeast condensin Smc2/4 complex: an ATPase that promotes knotting of circular DNA. J. Biol. Chem. 278:26238-26248. [DOI] [PubMed] [Google Scholar]
- 42.Strunnikov, A. V., E. Hogan, and D. Koshland. 1995. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 9:587-599. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan, B. A., M. D. Blower, and G. H. Karpen. 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2:584-596. [DOI] [PubMed] [Google Scholar]
- 44.Sumer, H., J. M. Craig, M. Sibson, and K. H. Choo. 2003. A rapid method of genomic array analysis of scaffold/matrix attachment regions (S/MARs) identifies a 2.5-Mb region of enhanced scaffold/matrix attachment at a human neocentromere. Genome Res. 13:1737-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swedlow, J. R., and T. Hirano. 2003. The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell 11:557-569. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka, T., J. Fuchs, J. Loidl, and K. Nasmyth. 2000. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2:492-499. [DOI] [PubMed] [Google Scholar]
- 47.Warren, W. D., S. Steffensen, E. Lin, P. Coelho, M. Loupart, N. Cobbe, J. Y. Lee, M. J. McKay, T. Orr-Weaver, M. M. Heck, and C. E. Sunkel. 2000. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr. Biol. 10:1463-1466. [DOI] [PubMed] [Google Scholar]
- 48.Watrin, E., and V. Legagneux. 2005. Contribution of hCAP-D2, a non-SMC subunit of condensin I, to chromosome and chromosomal protein dynamics during mitosis. Mol. Cell. Biol. 25:740-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams, B. C., T. L. Karr, J. M. Montgomery, and M. L. Goldberg. 1992. The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J. Cell Biol. 118:759-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood, K. W., R. Sakowicz, L. S. Goldstein, and D. W. Cleveland. 1997. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell 91:357-366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.