Abstract
The M26 hot spot of meiotic recombination in Schizosaccharomyces pombe is the eukaryotic hot spot most thoroughly investigated at the nucleotide level. The minimum sequence required for M26 activity was previously determined to be 5′-ATGACGT-3′. Originally identified by a mutant allele, ade6-M26, the M26 heptamer sequence occurs in the wild-type S. pombe genome approximately 300 times, but it has been unclear whether any of these are active hot spots. Recently, we showed that the M26 heptamer forms part of a larger consensus sequence, which is significantly more active than the heptamer alone. We used this expanded sequence as a guide to identify a smaller number of sites most likely to be active hot spots. Ten of the 15 sites tested showed meiotic DNA breaks, a hallmark of recombination hot spots, within 1 kb of the M26 sequence. Among those 10 sites, one occurred within a gene, cds1+, and hot spot activity of this site was confirmed genetically. These results are, to our knowledge, the first demonstration in any organism of a simple, defined nucleotide sequence accurately predicting the locations of natural meiotic recombination hot spots. M26 may be the first example among a diverse group of simple sequences that determine the distribution, and hence predictability, of meiotic recombination hot spots in eukaryotic genomes.
Homologous recombination occurs at high frequency during meiosis in most sexually reproducing organisms (4). Crossovers resulting from recombination create connections between homologous chromosomes (chiasmata), which are critical for the proper segregation of chromosomes at the first meiotic division. In the absence of recombination, homologs segregate nearly randomly in most organisms, often resulting in aneuploid meiotic products (gametes or spores). Aneuploid progeny in multicellular eukaryotes, such as humans, are usually inviable or suffer from a number of physical and mental abnormalities, as seen, for example, in human trisomy 21 (Down syndrome). In addition to its role in the proper segregation of chromosomes, meiotic recombination also results in the formation of new genetic combinations and is therefore an important mechanism of increasing genetic diversity within a species.
Recombination does not occur at a uniform frequency throughout the genomes of the organisms examined. Rather, there are sites that recombine at a significantly higher or lower frequency than the genomic average, termed hot spots and cold spots, respectively. Hot spots have been described in organisms as diverse as bacteria and their phages, mice, and humans (9, 26, 30). In the two distantly related yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, meiotic hot spots of recombination are sites of programmed double-strand DNA breaks (DSBs) made by the Spo11 protein (Rec12 in S. pombe) (11, 26, 31, 33). Spo11 is a meiotically induced protein with amino acid sequence similarity to an archaeal type II topoisomerase, TopoVI. Spo11 homologs are widely conserved in eukaryotes and are essential for meiotic recombination in several diverse organisms (20, 21). Thus, it is likely that programmed DSBs are a universal mechanism for the initiation of meiotic recombination.
Though much progress has been made in elucidating the mechanism of recombination, it is less clear how this process is regulated in eukaryotes. In particular, little is known about the factors controlling the locations of prominent DNA break sites, i.e., hot spots of recombination. In a global analysis of meiotic DSB sites in S. cerevisiae, Gerton et al. (15) found a significant correlation between DSB sites and the local G+C content. When viewed in 5-kb windows, >90% of all DSBs were found within 2.5 kb of regions where the G+C content exceeded the genomic average. The molecular basis of this correlation is not clear. Blumental-Perry et al. (6) reported that DSBs in S. cerevisiae were correlated with a 50-bp degenerate motif termed the common homology region or CoHR. However, a recent test of this hypothesis found no significant association of the CoHR with DSB sites, and deletion of the CoHR at HIS2, one of the hot spots used to identify the CoHR sequence, affected neither the position nor the intensity of meiotic DNA breakage at that site (19).
In prokaryotes, hot spots of recombination are often determined by short DNA sequence motifs. For example, in Escherichia coli a unique eight-base sequence, Chi, is recognized and activated by the RecBCD enzyme at an early step in recombination (30). Chi hot spots are active in other enteric bacteria (24), and short (5- to 7-bp) sequences also appear to be hot spots in the distantly related gram-positive bacteria Bacillus subtilis and Lactococcus lactis. Both of these species contain functional homologs of the RecBCD enzyme, whose activity is regulated by their respective Chi sequences in a manner comparable to that of the E. coli RecBCD enzyme (7, 10). Thus, discrete sequence motifs defining hot spots of recombination may be a common feature of prokaryotes.
The M26 hot spot of S. pombe is the only reported eukaryotic hot spot comparable to the Chi hot spot in that it is defined by a simple sequence motif. This hot spot was first identified as a mutation in the ade6 gene, ade6-M26, that elevated recombination up to 20-fold compared to other ade6 alleles (17). The hot spot results from a single G→T nonsense mutation near the 5′ end of ade6 (34). This mutation creates a 7-bp sequence, 5′-ATGACGT-3′ (M26 mutation underlined), essential for hot spot activity (28). This sequence is at least part of the binding site for a heterodimeric transcription factor, Atf1-Pcr1, which is also essential for M26 hot spot activity (22, 36). When created by mutation of 1 to 4 bp, the M26 heptamer generates hot spots at multiple positions within the ade6 and ura4 genes (12), and all of those sites tested stimulate the formation of Rec12-dependent meiosis-specific DSBs near the site of the M26 heptamer (31). In addition to M26, Fox et al. (13) showed that a closely related sequence, 5′-TGACGTC/A-3′, termed CRE (cyclic AMP response element), is also an Atf1-Pcr1-dependent hot spot at one site within the ade6 gene. M26 and CRE overlap at six of their seven base pairs, differing only at the ends (Fig. 1).
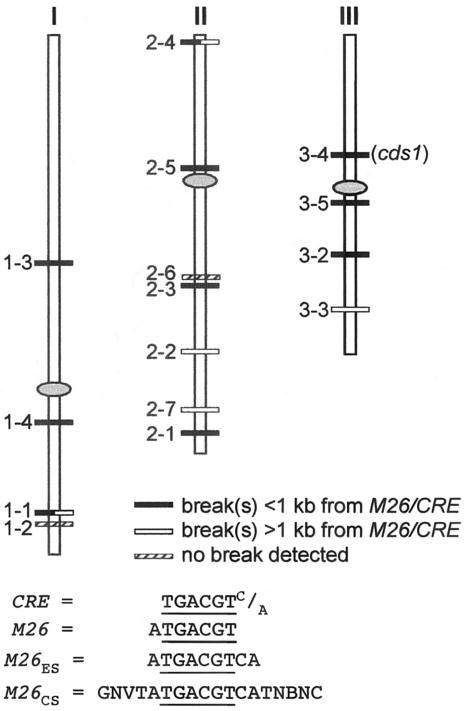
FIG. 1.
Positions of M26 or CRE sites (M26/CRE) tested for meiotic DNA breakage. Site numbers correspond to those described in the text. Chromosomes are drawn to scale, and centromeres are indicated by ovals. Below the chromosomes are definitions of the sites studied here.
Collectively, the M26 and CRE heptamers occur 879 times in the sequenced S. pombe genome (39), but hot spot activity of these sites has not been reported. One M26 heptamer that occurs in the ura1+ gene was tested for its potential as a hot spot, but mutation of this site had no significant effect on recombination within ura1 (M. Fox and G. Smith, unpublished observations). Thus, not all M26 or CRE heptamers, henceforth referred to as M26/CRE, are hot spots. Recently, however, we showed that the M26/CRE heptamer alone is not sufficient for full hot spot activity (32). In at least one location within ade6, this sequence showed no significant activity and bound Atf1-Pcr1 weakly compared to an overlapping active heptamer in the opposite orientation. This observation led to the discovery that M26/CRE forms part of a larger consensus binding sequence, 5′-GNVTATGACGTCATNBNC-3′ (V is A, C, or G; B is C, G, or T; N is any nucleotide), termed M26CS, required for optimal binding of Atf1-Pcr1. When M26CS was created at the site of the inactive ade6 heptamer, an exceptionally strong hot spot resulted. In addition, creation of M26CS at the site of an already active M26 heptamer, ade6-M26, increased recombination about threefold compared to the heptamer alone. This observation suggested that M26/CRE is most likely to be active when it forms part of an expanded sequence containing at least some of the additional nucleotides found in the consensus binding sequence. We used this information to narrow our survey of genomic M26/CRE sites and found that this simple sequence motif accurately predicted the locations of numerous natural recombination hot spots.
MATERIALS AND METHODS
S. pombe strains, growth media, and meiotic crosses.
Solid and liquid growth media were made as previously described (18, 32). For media, 5S refers to the supplements adenine, leucine, lysine (100 μg/ml each), uracil, and histidine (50 μg/ml each). 4S refers to the same supplements minus adenine. Guanine (G; 100 μg/ml) was added to media as necessary to select for Ade+ recombinants (16).
The strains used in this study are listed in Table 1. The ade6 alleles M26, M375, and 469 have been described previously (17, 34). The other ade6 alleles, pcr1::his7+, and atf1::ura4+ were described by Steiner and Smith (32); pat1-114 and rad50S were described by Young et al. (40); h+ mat1PΔ17::LEU2 was described by Arcangioli and Klar (2). The cds1 alleles contain the following nucleotide substitutions (numbered from the start of the coding sequence): cds1-T11A, A31G (35); cds1-1, G543T (nonsense mutation); cds1-2, C533T; cds1-3, C533T G534T; cds1-4, T192C. All cds1 mutations were generated with a site-directed mutagenesis kit (QuikChange; Stratagene) and, as the template, a plasmid containing a 3.3-kb PstI-SpeI fragment of cds1 (pBS-cds1) (35). Mutations were confirmed by nucleotide sequencing. The mutant plasmids were digested with PstI and SpeI and used for LiCl-mediated transformation (3) of GP4279, containing Δcds1::ura4+ (25). After overnight recovery in YEL-5S, linear transformants were selected on NBA plates (0.67% Difco yeast nitrogen base without amino acids) supplemented with uracil, leucine, and 1 mg/ml 5-fluoroorotic acid (32). Homologous integration was confirmed by Southern blot hybridization, and mutations were confirmed by sequencing or by the expected restriction site alteration.
TABLE 1.
Strains used in this study
| Strain | Genotypea |
|---|---|
| GP3111 | h+ade6-M26 his7-366 ura4+pcr1::his7+pat1-114 rad50S |
| GP3307 | h−ade6-3044 leu1-32 |
| GP3316 | h+ade6-M375 leu1-32 |
| GP3667 | h+ade6-3044 leu1-32 his7-366 |
| GP3698 | h+ade6-M375 leu1-32 his7-366 pcr1::his7+ |
| GP3700 | h+ade6-3044 leu1-32 his7-366 pcr1::his7+ |
| GP4279 | h−leu1-32 cds1::ura4+ |
| GP4376 | h−cds1-1 leu1-32 |
| GP4378 | h+cds1-T11A leu1-32 |
| GP4451 | h+mat1PΔ17::LEU2 cds1-T11A leu1-32 atf1::ura4+ |
| GP4452 | h−cds1-1 leu1-32 atf1::ura4+ |
| GP4453 | h−cds1-1 his7-366 pcr1::his7+ |
| GP4454 | h−cds1-T11A leu1-32 his7-366 pcr1::his7+ |
| GP4457 | h−ade6-3070 leu1-32 |
| GP4505 | h−ade6-3083 leu1-32 |
| GP4630 | h−leu1-32 cds1-1,2 |
| GP4669 | h+mat1P Δ17::LEU2 leu1-32 cds1-T11A,2 |
| GP4872 | h+mat1P Δ17::LEU2 leu1-32 cds1-T11A,3 |
| GP4873 | h+mat1P Δ17::LEU2 leu1-32 cds1-T11A,2,4 |
| GP4874 | h−leu1-32 cds1-1,3 |
| GP4875 | h−leu1-32 cds1-1,2,4 |
| GP4879 | h+ade6-3074 ura4+pat1-114 rad50S end1-458 |
| GP4880 | h+ade6-3083 ura4+pat1-114 rad50S end1-458 |
| GP4937 | h−ade6-3070 leu1-32 pcr1::his7+ |
| GP4938 | h−ade6-3083 leu1-32 pcr1::his7+ |
| WS101 | h+ade6-M375 + his7-366 |
| h−ade6-3070 leu1-32 + | |
| WS102 | h+ade6-469 + his7-366 |
| h−ade6-3083 leu1-32 + | |
| WS103 | h+ade6-3070 leu1-32 + atf1::ura4+ |
| h−ade6-M375 + his7-366 atf1::ura4+ | |
| WS111 | h+ade6-469 + his7-366 |
| h−ade6-M375 leu1-32 + | |
| WS118 | h+ade6-M375 his7-366 + |
| h−ade6-3044 + leu1-32 | |
| WS119 | h+ade6-469 + his7-366 atf1::ura4+ |
| h−ade6-3083 leu1-32 + atf1::ura4+ |
All strains are also ura4-D18 unless otherwise indicated. See Materials and Methods for references to specific alleles.
Diploid strains were constructed by mating appropriate haploid strains for 6 to 12 h on SPA-5S (18) at 25°C and streaking them onto NBA containing appropriate supplements but lacking leucine and histidine to select for diploids. Alternatively, diploid strain WS119 was constructed by protoplast fusion (1).
Ade+ recombinant frequencies from zygotic and azygotic meioses (see Tables 3 and 4) were determined by random spore analysis as previously described (32). For diploid strains, freshly grown single colonies were used to inoculate 5-ml YEL-5S cultures, which were grown overnight at 32°C. Appropriate dilutions of these cultures were plated onto YEA-5S or YEA-4SG in order to determine the frequency of Ade+ mitotic recombinants prior to sporulation. The frequency of Ade+ cells in these cultures was <50/106 in all cultures tested. One milliliter of each diploid culture was centrifuged; the cells were washed twice with 0.85% NaCl and plated onto SPA supplemented with adenine and/or uracil. After 2 days of incubation at 25°C, spores were treated with Glusulase and ethanol to kill the remaining vegetative cells as previously described (32).
TABLE 3.
The ade6-M26CS hot spot is only partially Pcr1 dependent
| Cross | ade6 allelesa | pcr1 | No. Ade+/106b |
|---|---|---|---|
| GP4505 × GP3667 | 3083 × 3044 | + | 16,900 ± 1,600 |
| GP4938 × GP3700 | 3083 × 3044 | Δc | 6,140 ± 560 |
| GP3316 × GP3307 | M375 × 3044 | + | 530 ± 50 |
| GP3316 × GP4457 | 3070 × M375 | + | 23,500 ± 1,100 |
| GP3698 × GP4937 | 3070 × M375 | Δ | 10,600 ± 1,300 |
See Fig. 3 for allele descriptions. M26CS hot spot alleles are in bold.
Mean frequency ± the standard error of the mean of Ade+ recombinants per million viable spores from three crosses.
Δ, pcr1::his7+.
TABLE 4.
The ade6-M26CS hot spot is strongly Atf1 dependent
| Diploida | ade6 allelesb | atf1 | No. Ade+/106c |
|---|---|---|---|
| WS102 | 3083 × 469 | + | 12,300 ± 1,800 |
| WS111 | M375 × 469 | + | 300 ± 40 |
| WS119 | 3083 × 469 | Δd | 1,000 ± 90 |
| WS101 | 3070 × M375 | + | 12,100 ± 870 |
| WS118 | 3044 × M375 | + | 320 ± 30 |
| WS103 | 3070 × M375 | Δ | 310 ± 10 |
atf1 mutants show severe defects in mating and meiosis (23, 29, 38). Thus, in order to obtain adequate numbers of viable spores, unstable diploids were used for these experiments.
See Fig. 3 for allele descriptions. M26CS hot spot alleles are in bold.
Mean frequency ± the standard error of the mean of Ade+ recombinants per million viable spores from three crosses.
Δ, atf1::ura4+.
cds1 mutants were also mated and sporulated on SPA and treated as described above. Spores were plated on YEA-5S and YEA-5S plus 5 mM hydroxyurea (HU) (35) to determine total and cds1+ spore yields, respectively.
Meiotic DNA break analysis.
Meiotic DNA breaks were assayed in pat1-114 rad50S strains GP4879, GP4880 (pcr1+), and GP3111 (pcr1Δ). Meiotic inductions, preparation of genomic DNA, restriction enzyme digestion, pulsed-field gel electrophoresis, and Southern blot hybridizations were performed as previously described (31, 40). Under the conditions used, premeiotic DNA replication occurs 2 to 3 h after induction; meiosis-specific DSBs are usually visible by 3.5 to 4 h, and these breaks are not repaired due to the presence of the rad50S allele. Samples were taken at multiple time points after meiotic induction, and DNA was purified in agarose plugs to minimize mechanical breakage. Restriction enzymes were chosen that produced fragments of 8 to 29 kb containing the M26 or CRE sites, and DSBs were assayed by Southern blot hybridization with probes specific to one end of each fragment.
DNA breakage was quantitated with a Molecular Dynamics PhosphorImager. Volume reports were determined for broken and unbroken fragments with the local average background subtracted. After quantitation, blots were stripped by two 15-min washes at 95°C in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% sodium dodecyl sulfate and rehybridized with a different probe. Occasionally, a trace amount of radioactivity in the main restriction fragment remained on the blot after this procedure. Thus, for each blot, probes hybridizing to the largest DNA fragment were used first, followed by probes recognizing successively smaller fragments. Any radioactivity remaining from a previous hybridization would not interfere with the results of the next.
Probes for each of the M26/CRE sites in this study were prepared by PCR with primer pairs specific to each of the M26/CRE sites as follows: 1-1, 5′-CGACGTCGACGACAAGAGAGATTT-3′ and 5′-GGCATTCTGAATTTCAAACTTTCGCA-3′; 1-2, 5′-TAGCAGCCTCAATCTGAACATCGG-3′ and 5′-GTGGGAAGACGATTTAGCGAATGC-3′; 1-3, 5′-TGTCTTGGCATCAGCATACAGTT-3′ and 5′-TATGAACTCGCTCAGGGAATGGAG-3′; 1-4, 5′-GAAGCTTCAAAGGATCGAATTGAGTTGTG-3′ and 5′-GATAACCTTTCCAAAGACCGCCTTC-3′; 2-1, 5′-AGGAGCTTACGCCGTCTCAAGTAT-3′ and 5′-TTCGTGATCTTCTACCATCGCGTC-3′; 2-2, 5′-CAACACGTTATTATGGTAAACCGAACG-3′ and 5′-ACGGTCGAGATGAGTACAAAGTCA-3′; 2-3, 5′-GCTCAAGTTCCACCGATTGTGAAG-3′ and 5′-GCAAGCGCAGTTTGAGGTCTGTTA-3′; 2-4, 5′-CCCTGCGAAAGTAAATGAAGTGCG-3′ and 5′-CCCTCTTCTTGTATGAAGTTGTGGATCA-3′; 2-5, 5′-CCAATAGAAGGGAACTTCATCATAGGC-3′ and 5′-GCTCTCATCGTCGTTATGTCGAGT-3′; 2-6, 5′-TTTGAGCCAACTATCGCTGGACG-3′ and 5′-GAGAGGAAATCCATTCAGTACCGTTAAA-3′; 2-7, 5′-GCCTAATCAGCAACAATTAGCCCA-3′ and 5′-CCGAATTATGGCATTTAGCAAGTAAAGCA-3′; 3-2, 5′-AACAACACCTACCATGTCAGGGTCTC-3′ and 5′-TCCGAACGGAAACGCTCAAGAGAA-3′; 3-3, 5′-CACACCAACTGAATATTGTATCCCATCC-3′ and 5′-AACGGCATGCTACCAGAAAGT-3′; 3-4, PacI, 5′-TGTGAAAGTGTCTAATGCCTTATCTGA-3′ and 5′-GGAAATTAAGACAAGCAAAGCGTGC-3′; 3-4, HaeII, 5′-CCAGCATACACAGCCATTAACCGA-3′ and 5′-ACGTCAACTTGGACGGAGGTTATG-3′ or 5′-GGCTTGTCCAGATATTACGGCATC-3′ and 5′-ACAGTCGACACAAACATCTAAATAAGC-3′; 3-5, 5′-GCTCAAGCGATCCTTCCATCAAAC-3′ and 5′-TGATTACTCGCCAATGCTTTCA-3′.
RESULTS AND DISCUSSION
Meiotic DSBs at natural M26/CRE sites.
The Atf1-Pcr1 consensus binding sequence M26CS, noted above, contains 15 nonrandom bases (32) (Fig. 1) and does not occur in the sequenced S. pombe genome (http://www.sanger.ac.uk/Projects/S_pombe/). However, the central 10-bp M26 palindrome appeared to be the most critical element for strong binding of Atf1-Pcr1 in vitro and therefore most likely for hot spot activity as well. This sequence occurs four times in S. pombe (Fig. 1, sites 2-4, 2-5, 2-6, and 2-7). In order to broaden our search for hot spots beyond just these four sites, we also included a shorter sequence, 5′-ATGACGTCA-3′, which appeared essentially as active as the full 10-bp palindrome (32). This extended M26 sequence, termed M26ES, occurs an additional 11 times in the genome (Fig. 1). We also included in our analysis one palindromic CRE sequence, 5′-CTGACGTCAG-3′ (site 1-4), that forms a strong hot spot in ade6 (13). Since engineered M26 hot spots in the ade6 gene are sites of meiotic DSBs and since DSB formation at the one site tested (ade6-M26) requires the Pcr1 protein (31), we tested whether the naturally occurring sites (M26ES or CRE) are also sites of Pcr1-dependent meiotic DNA breaks.
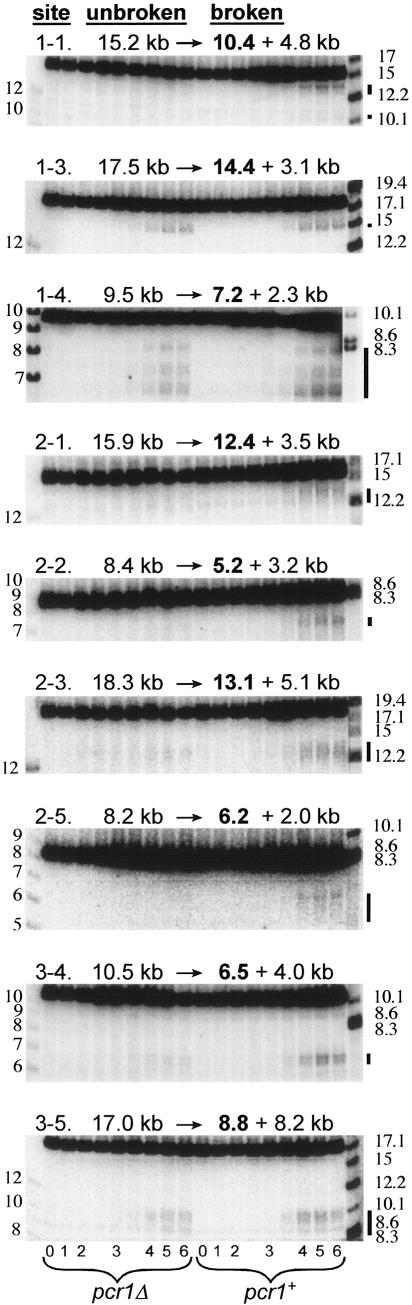
By Southern blot hybridization, we tested the M26- or CRE-containing genome fragments for meiotic DSBs (Fig. 2; the complete set of uncropped images is in Fig. S1 of the supplemental material). DSBs were observed in 13 of the 15 fragments probed. Nine of the 13 observed break sites were prominent (≥1% of the total DNA was broken; Table 2) and often appeared as clusters of breaks spread over regions of up to 2 kb (e.g., site 1-4, Fig. 2). Since approximately 200 kb of the genome was surveyed in our analysis, this corresponds to an average of one prominent break site per ∼22 kb. This density of breaks appears to be somewhat greater than that observed in previous analyses, where prominent break sites were estimated to occur ∼25 to 200 kb apart (40). If so, this implies that the presence of M26/CRE is positively correlated with DNA breaks. Of course, a more important consideration is the positions of the break sites relative to the M26/CRE sequence in each fragment. Fifteen DSB sites were found in 15 DNA fragments averaging ∼14 kb in length (two fragments, 1-1 and 2-4, had two clusters of breaks each; Fig. 2; see Fig. S1 of the supplemental material). Ten of those breaks were ≤1 kb from the M26/CRE sequence (Table 2; see Fig. S1 of the supplemental material). In any fragment, the probability of a break occurring ≤1 kb from M26/CRE by chance is 2 kb/14 kb = 0.14, and the probability of ≥10 of 15 breaks falling at least that close to M26/CRE can be calculated from the binomial distribution to be 4.4 × 10−6. Thus, most, and perhaps all, of the breaks ≤1 kb from the M26/CRE sequence likely result from the presence of that motif at those sites.
FIG. 2.
Meiosis-induced DSBs are produced at the M26/CRE motif at several sites in the genome. pcr1+ or pcr1Δ strains were induced for meiosis, and genomic DNA was purified at the indicated time points. The DNA was digested and analyzed by Southern blot hybridization with probes specific to each of the indicated sites. The predicted sizes in kilobase pairs of the unbroken restriction fragment and of fragments generated by a break at the M26/CRE motif are indicated above each blot. The size of the broken fragment recognized by the probe is shown in boldface type. The positions of broken DNA fragments are indicated by black bars to the right of each gel. DNA size markers in kilobases are shown to the left and right of the gels. The exposure of these markers was sometimes adjusted separately from the rest of the blot for clarity.
TABLE 2.
Meiotic DSBs at M26/CRE sites in wild-type S. pombe
| Site | Enzyme used for DNA digestion | Unbroken fragment sizea(kb) | Predicted size of broken fragmentb(kb) | Break ≤1 kb from predicted positionc | Break ≥1 kb from predicted positionc | % breakage pcr1+d | % breakage pcr1Δd |
|---|---|---|---|---|---|---|---|
| 1-1 | PacI | 15.2 | 10.4 | + | 0.5 | 0.5 | |
| 1-1 | PacI | 15.2 | 10.4 | + | 2.3 | 1.1 | |
| 1-2 | NruI | 15.0 | 10.8 | ||||
| 1-3 | NruI | 17.5 | 14.4 | + | 3.2, 2.7 | 2.3, 2.2 | |
| 1-4 | NcoI | 9.5 | 7.2 | + | 7.0 | 5.0 | |
| 2-1 | PacI | 15.9 | 12.4 | + | 3, 4.3 | 1.5, 2.8 | |
| 2-2 | PacI | 8.4 | 5.2 | + | 1.9 | 0.6 | |
| 2-3 | PacI | 18.3 | 13.1 | + | 3.3, 5.0 | 1.8, 2.9 | |
| 2-4 | NruI | 7.9 | 5.1 | + | <0.5 | <0.5 | |
| 2-4 | NruI | 7.9 | 5.1 | + | <0.5 | <0.5 | |
| 2-5 | NruI | 8.2 | 6.2 | + | 0.8, 0.5 | <0.5, <0.5 | |
| 2-6 | NruI | 14.0 | 10.7 | ||||
| 2-7 | NruI | 18.7 | 12.8 | + | <0.5, <0.5 | <0.5, <0.5 | |
| 3-2 | NruI | 10.3 | 4.4 | + | <0.5, <0.5 | <0.5, <0.5 | |
| 3-3 | NruI | 28.7 | 16.0 | + | 2.0 | 1.0 | |
| 3-4 | PacI | 10.5 | 6.5 | + | 1.8, 1.6 | 0.9, <0.5 | |
| 3-5 | NruI | 17.0 | 8.8 | + | 3.4 | 2.1 |
Predicted size of unbroken fragment from S. pombe genomic sequence (http://www.sanger.ac.uk/Projects/S_pombe/).
Size of hybridizing fragment predicted from a break within the M26/CRE sequence.
Fig. 2; see Fig. S1 of the supplemental material.
Percentage of total DNA in broken fragments. Pairs of values are from separate blots of one induction. pcr1+ and pcr1Δ strains were compared side by side on the same blot. The first value of each pair (pcr1+ or pcr1Δ) is from one blot, and the second value is from the second blot.
We looked for a common feature among the M26/CRE sites with detectable DSBs. Of the 10 M26/CRE sites with DSBs, 9 were located in intergenic regions and 1 (site 3-4) occurred within a gene intron. Seven of the 10 DSB-associated sites were located <750 bp upstream of open reading frames (potential gene promoter regions), 5 of which show Atf1-dependent induction in response to stress (sites 1-1, 1-3, 1-4, 2-2, and 2-7; http://www.sanger.ac.uk/PostGenomics/S_pombe/projects/stress/). These results are comparable to observations on S. cerevisiae, in which meiotic DSBs are usually found in intergenic regions, often in gene promoters (5, 15).
As noted in the introduction, Gerton et al. (15) also reported a correlation between DSBs in S. cerevisiae and the G+C content in 5-kb windows surrounding the break site. In our more limited survey of break sites, we found a similar correlation, but only in a much smaller window. Viewed in 100-bp windows, 9 of the 10 M26/CRE sites showing breaks were in regions where the G+C content exceeded the genomic average, and 4 of 5 sites showing no detectable breaks were in regions of lower-than-average G+C content. There was also a positive, but weak, quantitative correlation between G+C content and break intensity (linear regression analysis, R2 = 0.30; unpublished observations). These correlations declined as larger windows were considered. Thus, the pattern of nucleotide composition around hot spots in S. cerevisiae noted by Gerton et al. (15) may be similar to but more extensive than that around the active M26/CRE sites in S. pombe.
Partial Pcr1 independence of M26CS hot spot activity.
As previously mentioned, the M26 hot spot was first identified by the ade6-M26 mutation (17). The activity of that hot spot is completely dependent on the Atf1-Pcr1 transcription factor. Mutation of pcr1 eliminates detectable breakage at the hot spot (31), and mutation of either atf1 or pcr1 reduces recombination to non-hot spot levels (22). A CRE hot spot created at the same position as the ade6-M26 mutation also showed complete dependence on both Atf1 and Pcr1 (13). Thus, we expected that other M26/CRE sites would have similar requirements for hot spot activity, and DSBs, if they occurred at these sites, would not be observed in strains lacking Pcr1. We were therefore surprised to see substantial Pcr1 independence of breakage at some of the M26/CRE sites whose positions coincided with DSBs (e.g., Fig. 2, sites 1-3, 1-4, and 3-5). In our DSB analysis (Fig. 2), an atf1 mutant was not tested because it cannot be induced for meiosis in pat1-114 (ts) strains (31). Consequently, we used genetic tests for Atf1 and Pcr1 dependence of the M26/CRE sites examined here.
Previous studies have shown that although mutations in atf1 or pcr1 have similar phenotypes, they are not identical. Both mutations result in poor mating and sporulation efficiency, but an atf1 mutation is more deficient in these processes (23; our unpublished observations). Furthermore, Atf1, but not Pcr1, is required for growth at high osmolarity (22, 23). And mutation of pcr1, but not atf1, results in cold-sensitive growth on rich medium (37; our unpublished observations). Thus, both of these polypeptides may also act alone or in conjunction with other protein partners in vivo.
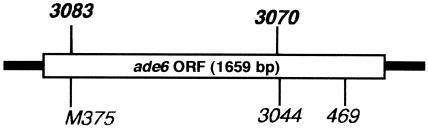
Since many of the DSBs at or near M26/CRE sites were not eliminated in the pcr1 mutant, we tested whether Atf1 and Pcr1 were required for hot spot activity of the M26 consensus sequence (M26CS) at two sites in the ade6 gene (Fig. 3, ade6-3083 and ade6-3070) (32). Both of these hot spots showed substantial activity in the absence of Pcr1: hot spot activity in the pcr1 mutant was significantly reduced, but less than threefold relative to the corresponding pcr1+ strains, and was 10- to 20-fold greater than that of the non-hot spot control alleles (Table 3). The substantial hot spot activity of these sites in the absence of Pcr1 is consistent with the Pcr1-independent breakage observed at or near several of the M26/CRE sites in the genome, as noted above.
FIG. 3.
Positions of ade6 alleles used in crosses (Tables 3 and 4). The two M26CS hot spot alleles, ade6-3083 and -3070 (32), are shown in boldface type above the gene; non-hot spot alleles appear below. ORF, open reading frame.
Given these results, we also tested whether the M26CS sites in ade6 showed atf1-dependent hot spot activity. In these experiments, the atf1 mutations decreased recombination >12-fold relative to the corresponding atf1+ strains for both hot spot alleles (Table 4). For one of the alleles (ade6-3070), recombination was reduced to the same level as the non-hot spot control (ade6-3044), suggesting that its activity is completely dependent on Atf1; for the other hot spot allele (ade6-3083), recombination remained slightly, but significantly, elevated relative to the non-hot spot control (P < 0.02, one-tailed t test). The residual hot spot activity observed in the absence of either atf1 or pcr1 may reflect the ability of these polypeptides to bind weakly as homodimers to M26 in the absence of its partner protein (36). Homodimer binding to M26CS, particularly the Atf1 homodimer, may be substantially greater than to the original ade6-M26 heptamer, accounting for its activity in the absence of Pcr1.
As mentioned above, five of the sites tested for meiotic DSBs showed breaks >1 kb from the M26/CRE motif without an accompanying prominent break at the predicted location (sites 1-1, 2-2, 2-4, 2-7, and 3-3, Table 2; Fig. 2; Fig. S1 of the supplemental material). In one case (site 3-3), a cluster of breaks was observed >6 kb from the predicted site, but at none of the five sites was a second M26 or CRE sequence found near the actual break position. Since most meiotic recombination in S. pombe apparently does not require the Atf1 or Pcr1 protein (22; our unpublished observations), the simplest explanation for these breaks is that they are unrelated to the presence of the nearby M26 sequence. However, most of these breaks appeared to be strongly dependent on the Pcr1 protein, for example, sites 1-1, 2-2, and 3-3 (Fig. 2; Fig. S1 of the supplemental material). Since many other meiosis-specific DSBs were seen in the same pcr1 mutant meiosis (for example, at sites 1-3, 1-4, and 3-5, Fig. 2), the lack of DSBs at some sites must, therefore, reflect Pcr1 dependence.
Might M26 stimulate DNA breaks distant from it? Previously, we showed that one M26 hot spot (ade6-3011) is able to stimulate visible breaks up to 1.3 kb distant from it (31). In that case, there were also breaks much closer to M26. Nevertheless, this result demonstrated that M26 can stimulate breaks a considerable distance from it. A similar phenomenon might explain breaks distant from M26 at some of the sites mentioned above, but it does not explain the absence of breakage proximal to M26 at those same sites. Perhaps the DNA near M26 at these sites is protected from cleavage, which occurs instead at the nearest available position. This admittedly unorthodox model would be one means of explaining how hot spots of recombination are preserved through evolutionary time (27): if hot spots of recombination are determined by the DNA sequence at or near the site of breakage, and break repair involves copying from a homolog without the hot spot, they should be lost at high frequency. If, however, the break determinant is located distant from the actual site of breakage, it would be lost at a much lower frequency.
M26 is a recombination hot spot in cds1.
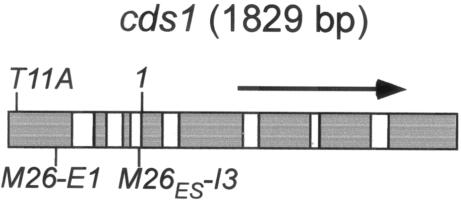
Meiotic DSBs are generally assumed to be hot spots of recombination. In the two organisms in which meiotic DSBs have been observed (S. cerevisiae and S. pombe), strong hot spots of recombination are sites of DNA breakage and vice versa, where tested (e.g., 8, 11, 26, 31, 33). Thus, the DSBs we found in this study are very likely also hot spots of recombination. In order to confirm this assumption, we analyzed one site (3-4, Fig. 1 and 2) more thoroughly because it lies within a well-characterized gene, cds1, encoding a checkpoint protein kinase (14, 25, 35). The cds1 gene contains two M26 motifs; M26ES, which we identified originally in our genome survey, occurs in the third intron (M26ES-I3), and a second site containing all but the last nucleotide of the M26ES motif occurs 343 bp upstream, in the first exon of the gene (M26-E1; Fig. 4). Since either or both of these sites could contribute to the DSBs observed in Fig. 2, we mapped the break sites more precisely in order to determine their positions relative to the two M26 motifs. Figure 5 shows a 6-kb DNA fragment containing cds1 probed from each side for meiotic DSBs. In both analyses, M26ES-I3 fell between two prominent break sites ∼200 bp apart, making it the likely feature responsible for most, if not all, of the observed breakage. Twin break sites bracketing the M26 sequence were also observed at the ade6-M26 hot spot (31). This pattern of breakage may reflect exclusion of Rec12 over the narrow region of DNA to which Atf1-Pcr1 is actually bound. Thus, while stimulating the cleavage of neighboring DNA, the hot spot sequence itself is protected.
FIG. 4.
The cds1 gene. The positions of two Cds1-inactivating mutations, T11A and 1 (see Materials and Methods) and the two M26 motifs, M26-E1 in exon 1 and M26ES-I3 in intron 3, are shown. Exons and introns are indicated by gray and white boxes, respectively.
FIG. 5.
M26ES lies between two prominent meiotic break sites in cds1. Genomic DNA from meiotically induced cells was digested with HaeII and probed from each end of the resulting 6.0-kb cds1 fragment, as indicated in the schematic diagram below each blot. The inferred positions of both M26 motifs are shown to the right of the blots. The values on the left are molecular sizes in kilobase pairs.
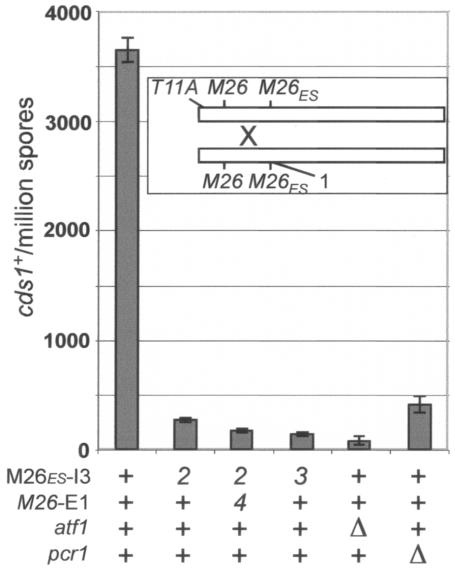
In order to test our prediction that M26ES-I3 was responsible for the DSBs in cds1, we also analyzed it genetically for hot spot activity. Mutations in cds1 result in sensitivity to the DNA synthesis inhibitor HU (25, 35), so cds1+ recombinants can be readily selected. Figure 6 shows results from crosses between cds1-T11A (35) and cds1-1, a nonsense mutation 512 bp downstream of the T11A mutation (Fig. 4). When no other mutations were present, we observed approximately 3,700 cds1+ recombinants per million viable spores. This frequency of recombination is comparable to that observed for the ade6-M26 hot spot (12, 17), which shows about the same frequency of meiotic DSBs (31). When M26ES-I3 was also mutated in both parent strains by a single base pair change (cds1-2), the frequency of recombinants dropped 13-fold. When the second M26 motif (M26-E1) was also abolished (cds1-4), recombination was reduced further, compared to mutation of only the first site (P < 0.004, one-tailed t test). Thus, while the majority of recombination can be attributed to only one site (M26ES-I3), there may be a small contribution from the second site as well. (The cds1-2 single mutant is wild type with respect to HU sensitivity [unpublished observations], and the cds1-4 mutation is translationally silent.) This observation supports our earlier conclusion that bases outside of the originally defined 7-bp M26 sequence can dramatically affect hot spot activity (32). It also suggests that some of the >800 M26/CRE sites scattered throughout the genome, which may be only weakly recognized by Atf1 or the Atf1-Pcr1 heterodimer, could contribute to low-level (non-hot spot) recombination in regions of the genome lacking prominent break sites (40). Consistent with this view, we noted that the single-base-pair mutation in the active hot spot of cds1 (cds1-2) may not have completely eliminated its activity, since a double-base-pair change at that hot spot (cds1-3) also decreased recombination significantly, albeit modestly, compared to the single-base-pair change (P < 0.0007, one-tailed t test; Fig. 6).
FIG. 6.
M26 is a hot spot of recombination in cds1. Bars show the mean frequency of cds1+ recombinants (± the standard error of the mean) obtained from crosses between strains with the cds1-T11A and cds1-1 mutations (inset) and M26, atf1, and pcr1 mutations as indicated at the bottom. Each cross was performed three times, except for the atf1 mutant, which was performed twice; error bars for that cross show the range of values obtained. M26ES-I3 and M26-E1 are the M26 sites in the third intron and first exon of cds1, respectively (Fig. 4). Mutations in these sites are indicated by allele numbers (2, 3, and 4) at the bottom (see Materials and Methods for allele descriptions).
Since DSBs in cds1+ were largely, but not completely, eliminated by mutation of pcr1 (Fig. 2), we also tested how cds1 hot spot activity was affected by mutation of pcr1 or its partner atf1. Recombination was reduced 9-fold by a mutation in pcr1 but ∼40-fold by mutation of atf1 (Fig. 6). This large differential, along with the results in Tables 3 and 4, provides additional evidence that Atf1 can act independently of Pcr1 to promote recombination at M26 or related sites.
Conclusions.
Here we have shown that the M26/CRE sequence motif defines the locations of multiple meiotic DNA break sites in the wild-type S. pombe genome. At the one break site occurring within a gene, cds1, we also confirmed genetically that M26 acts as a recombination hot spot. Given this result and our current understanding of how recombination is initiated, it is very likely that M26/CRE is also a recombination hot spot at the other break sites we observed. To our knowledge, this is the first example in any eukaryote of a simple sequence motif accurately predicting the locations of recombination hot spots. However, since breaks were not observed at all of the sites examined and since there were large variations in break frequency at the different sites, there are clearly other factors involved in break site selection. These factors could include an influence of nucleotides adjacent to the M26ES sequence (32) or some broader aspect of the local chromatin structure.
Based on the results of our previous analysis of the ade6 gene (32), we restricted our present investigation to genomic sites containing the 9- to 10-bp M26ES and 10-bp CRE sequences, which we hypothesized were most likely to be sites of meiotic DSBs. The majority of these sites showed meiotic DNA breaks. However, this result does not imply that those longer sequences are essential for M26/CRE hot spot activity, and many of the hundreds of shorter versions of the motif could also be hot spots. M26/CRE may be just the first example among a diverse group of simple sequences determining the locations of recombination hot spots. Knowledge of such sequences would allow further predictions about meiotic recombination hot spots, similar to those presented here. This knowledge would facilitate the comparison of genetic and physical maps and aid, for example, the location of human disease genes by linkage studies.
Supplementary Material
Acknowledgments
We thank Paul Russell for strains and plasmids and Sue Amundsen, Gareth Cromie, Luther Davis, Joe Farah, Estelle Steiner, and Andrew Taylor for helpful comments on the manuscript.
This work was supported by National Institutes of Health grant GM31693 to G.R.S. and a New York State Office of Science, Technology and Academic Research Gen*NY*sis grant to Niagara University. W.W.S. was supported during part of this work by Special Fellowship 3230-05 from the Leukemia and Lymphoma Society.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Arcangioli, B., and A. J. S. Klar. 1991. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J. 10:3025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bähler, J., J.-Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 4.Baker, B. S., A. T. C. Carpenter, M. S. Esposito, R. E. Esposito, and L. Sandler. 1976. The genetic control of meiosis. Annu. Rev. Genet. 10:53-134. [DOI] [PubMed] [Google Scholar]
- 5.Baudat, F., and A. Nicolas. 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA 94:5213-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumental-Perry, A., D. Zenvirth, S. Klein, I. Onn, and G. Simchen. 2000. DNA motif associated with meiotic double-strand break regions in Saccharomyces cerevisiae. EMBO Rep. 1:232-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chedin, F., S. D. Ehrlich, and S. C. Kowalczykowski. 2000. The Bacillus subtilis AddAB helicase/nuclease is regulated by its cognate Chi sequence in vitro. J. Mol. Biol. 298:7-20. [DOI] [PubMed] [Google Scholar]
- 8.Cromie, G. A., C. A. Rubio, R. W. Hyppa, and G. R. Smith. 2005. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics 169:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Massy, B. 2003. Distribution of meiotic recombination sites. Trends Genet. 19:514-522. [DOI] [PubMed] [Google Scholar]
- 10.el Karoui, M., D. Ehrlich, and A. Gruss. 1998. Identification of the lactococcal exonuclease/recombinase and its modulation by the putative Chi sequence. Proc. Natl. Acad. Sci. USA 95:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, Q., F. Xu, and T. D. Petes. 1995. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15:1679-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, M. E., J. B. Virgin, J. Metzger, and G. R. Smith. 1997. Position- and orientation-independent activity of the Schizosaccharomyces pombe meiotic recombination hot spot M26. Proc. Natl. Acad. Sci. USA 94:7446-7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, M. F., T. Yamada, K. Ohta, and G. R. Smith. 2000. A family of CRE-related DNA sequences with meiotic recombination hotspot activity in Schizosaccharomyces pombe. Genetics 156:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furnari, B., A. Blasina, M. N. Boddy, C. H. McGowan, and P. Russell. 1999. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol. Biol. Cell 10:833-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown, and T. D. Petes. 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:11383-11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimm, C., P. Schaer, P. Munz, and J. Kohli. 1991. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol. Cell. Biol. 11:289-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutz, H. 1971. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics 69:317-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe, p. 395-446. In R. C. King (ed.), Handbook of genetics. Plenum Press, New York, N.Y.
- 19.Haring, S. J., L. J. Lautner, J. M. Comeron, and R. E. Malone. 2004. A test of the CoHR motif associated with meiotic double-strand breaks in Saccharomyces cerevisiae. EMBO Rep. 5:41-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keeney, S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1-53. [DOI] [PubMed] [Google Scholar]
- 21.Keeney, S., C. N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375-384. [DOI] [PubMed] [Google Scholar]
- 22.Kon, N., M. D. Krawchuk, B. G. Warren, G. R. Smith, and W. P. Wahls. 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 94:13756-13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kon, N., S. C. Schroeder, M. D. Krawchuk, and W. P. Wahls. 1998. Regulation of the Mts1-Mts2-dependent ade6-M26 meiotic recombination hot spot and developmental decisions by the Spc1 mitogen-activated protein kinase of fission yeast. Mol. Cell. Biol. 18:7575-7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKittrick, N. H., and G. R. Smith. 1989. Activation of Chi recombinational hotspots by RecBCD-like enzymes from enteric bacteria. J. Mol. Biol. 210:485-495. [DOI] [PubMed] [Google Scholar]
- 25.Murakami, H., and H. Okayama. 1995. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature 374:817-819. [DOI] [PubMed] [Google Scholar]
- 26.Petes, T. D. 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2:360-370. [DOI] [PubMed] [Google Scholar]
- 27.Pineda-Krch, M., and R. Redfield. 2005. Persistence and loss of meiotic recombination hotspots. Genetics 169:2319-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuchert, P., M. Langsford, E. Käslin, and J. Kohli. 1991. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 10:2157-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiozaki, K., and P. Russell. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10:2276-2288. [DOI] [PubMed] [Google Scholar]
- 30.Smith, G. R. 2001. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35:243-274. [DOI] [PubMed] [Google Scholar]
- 31.Steiner, W. W., R. W. Schreckhise, and G. R. Smith. 2002. Meiotic DNA breaks at the S. pombe recombination hotspot M26. Mol. Cell 9:847-855. [DOI] [PubMed] [Google Scholar]
- 32.Steiner, W. W., and G. R. Smith. 2005. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics 169:1973-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, H., D. Treco, N. P. Schultes, and J. W. Szostak. 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338:87-90. [DOI] [PubMed] [Google Scholar]
- 34.Szankasi, P., W. D. Heyer, P. Schuchert, and J. Kohli. 1988. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe: wild-type and mutant alleles including the recombination hotspot allele ade6-M26. J. Mol. Biol. 204:917-925. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka, K., M. N. Boddy, X. B. Chen, C. H. McGowan, and P. Russell. 2001. Threonine-11, phosphorylated by Rad3 and ATM in vitro, is required for activation of fission yeast checkpoint kinase Cds1. Mol. Cell. Biol. 21:3398-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wahls, W. P., and G. R. Smith. 1994. A heteromeric protein that binds to a meiotic homologous recombination hotspot: correlation of binding and hotspot activity. Genes Dev. 8:1693-1702. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe, Y., and M. Yamamoto. 1996. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol. Cell. Biol. 16:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson, M. G., M. Samuels, T. Takeda, W. M. Toone, J.-C. Shieh, T. Toda, J. B. A. Millar, and N. Jones. 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10:2289-2301. [DOI] [PubMed] [Google Scholar]
- 39.Wood, V., R. G. William, M.-A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, et al. 2002. The genome sequence of the eukaryote fission yeast Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 40.Young, J. A., R. W. Schreckhise, W. W. Steiner, and G. R. Smith. 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9:253-263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.