Abstract
The mammalian SWI/SNF chromatin remodeling complex, whose function is of critical importance in transcriptional regulation, contains approximately 10 protein components. The expression levels of the core SWI/SNF subunits, including BRG1/Brm, BAF155, BAF170, BAF60, hSNF/Ini1, and BAF57, are stoichiometric, with few to no unbound molecules in the cell. Here we report that exogenous expression of the wild type or certain deletion mutants of BAF57, a key subunit that mediates the interaction between the remodeling complex and transcription factors, results in diminished expression of endogenous BAF57. This down-regulation process is mediated by an increase in proteasome-dependent degradation of the BAF57 protein. Furthermore, the protein levels of BAF155/170 dictate the maximum cellular amount of BAF57. We mapped the domains responsible for the interaction between BAF57 and BAF155 and demonstrated that protein-protein interactions between them play an important role in this regulatory process. These findings provide insights into the physiological mechanisms responsible for maintaining the proper stoichiometric levels of the protein components comprising multimeric enzyme complexes.
The SWI/SNF chromatin remodeling complexes are evolutionarily conserved multimeric enzymatic machines that alter the nucleosomal structure using energy derived from ATP hydrolysis (34). Ample experimental evidence suggests that the SWI/SNF complexes play important roles in fundamental cellular processes such as transcription, replication, and the repair of chromatin (24, 28). As a result, mammalian SWI/SNF complexes have been implicated in diverse physiological and pathological processes, including cell proliferation and differentiation, retrovirus infection, and carcinogenesis (17, 21, 25).
The human SWI/SNF complexes contain either BRG1 or Brm as the catalytic ATPase subunit and approximately 10 BRG1-associated factors (BAFs) (36, 37). The BAF170 and/or BAF155, BAF60, BAF57, BAF53, and BAF47 (hSNF5/Ini1) subunits are present in all mammalian SWI/SNF complexes and conserved from yeast to humans, except for BAF57 (36). The BAF155 and BAF170 proteins are highly homologous and likely exist either as heterodimers (BAF155/BAF170) or as homodimers (BAF155/155 or BAF170/170) through a leucine zipper motif in the cell (37). In addition, BAF155 or BAF170 contains two highly conserved motifs that are commonly found in chromatin-associated proteins. One is the SANT (Swi3, Ada2, N-coR, and TFIIIB) domain (1), a motif believed to function as a histone tail binding module (5). The other motif, termed the SWIRM (Swi3, Rsc8, and Moira) domain, is predicted to adopt an α-helical structure and mediate specific protein-protein interactions (2). Human BAF57 is a modular protein containing a proline-rich region at the amino terminus, a high-mobility-group (HMG) domain, a conserved region termed the NHRLI domain (named after a group of conserved amino acids in this domain), a putative coiled-coil domain, and a charged C-terminal region (Fig. 1A) (27, 35). Studies using transgenic mice overexpressing a dominant-negative mutant form of mouse BAF57 in T-cell precursors revealed an essential role of BAF57 in CD4 silencing during T-cell lineage commitment (7). A role for BAF57 in gene silencing is also supported by the finding that BAF57 interacts with the methyl cytosine binding protein MeCP2 and participates in MeCP2-dependent transcriptional repression (16). Recently, BAF57 was shown to interact directly with estrogen receptor alpha and the androgen receptor (AR) and to help recruit the SWI/SNF complex to estrogen- and androgen-responsive promoters for hormone-dependent transcriptional activation (4, 23).
FIG. 1.
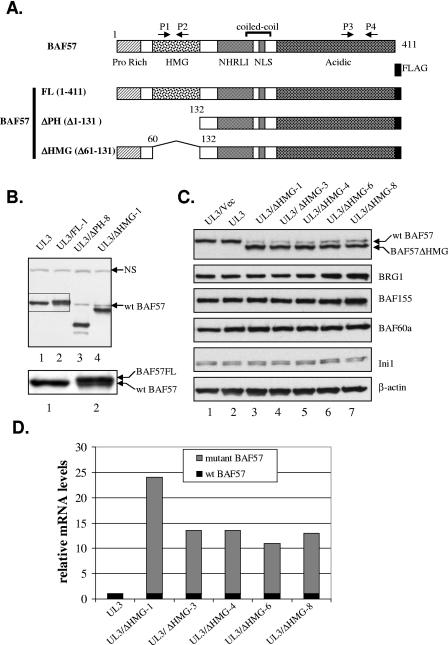
Exogenous expression of BAF57 down-regulates endogenous BAF57. (A) Schematic representation of human BAF57 domain structure and the FLAG-tagged constructs used in this study. Arrows mark the positions of real-time PCR primer pairs. HMG, high mobility group; NLS, nuclear localization signal. (B) Whole-cell extracts (WCEs) from parental UL3 cells (lane 1) and UL3-derived cell lines expressing either full-length BAF57 (lane 2) or the BAF57ΔPH (lane 3) or BAF57ΔHMG (lane 4) mutant were analyzed by Western blotting with anti-BAF57 antibody. The boxed area was enlarged and is shown under the main figure to illustrate the separation of wt BAF57 and BAF57FL proteins. “NS” denotes a nonspecific band recognized by the BAF57 antibody and indicates equivalent loading. (C) WCEs from parental UL3 cells (lane 2) and UL3-derived cell lines expressing either an empty vector (lane 1) or the BAF57ΔHMG mutant (lanes 3 to 7) were analyzed by Western blotting for the expression of BAF57, other SWI/SNF subunits, and β-actin. (D) Relative mRNA levels of wt BAF57 and BAF57ΔHMG in UL3 cells and various UL3/BAF57ΔHMG cell lines, as determined by quantitative RT-PCR. The relative wt BAF57 mRNA level was determined with the P1/P2 primer set. The relative amount of wt BAF57 mRNA in UL3 cells as well as the amount of combined wt and ΔHMG BAF57 mRNAs in various UL3/BAF57ΔHMG cell lines was first determined with the primer set P3/P4 and then adjusted according to the amplification efficiency difference between P1/P2 and P3/P4, as estimated by comparing CT values of P1/P2- and P3/P4-directed amplification of equal amounts of mRNA from UL3 cells. An arbitrary value of 1 was assigned to the wt BAF57 mRNA level in UL3 cells determined with the P1/P2 primers. All other values for a given cell line are presented relative to this value in the graph.
The subunit stoichiometry of mammalian SWI/SNF complexes has yet to be determined but is probably similar to that recently determined for yeast, considering that most of the core subunits are conserved between those two organisms (31). For BAF57, it has been shown previously that each mammalian SWI/SNF complex contains only one copy (35). Importantly, the biochemical purification process of the mammalian SWI/SNF complex revealed that no free subunits are present within the cell, suggesting that most, if not all, subunit proteins are assembled into the complex (10, 36). Thus, cells must coordinate the expression/degradation of multiple SWI/SNF subunits in order to maintain the correct stoichiometric protein level for each subunit. How cells accomplish this is largely unknown. Previous observations have suggested that a cellular mechanism(s) may exist to monitor the quantitative amount of at least some SWI/SNF subunits in vivo. For example, the overexpression of Brm protein in HeLa cells by transient transfection induces a drastic decrease in the level of endogenous BRG1 (29). In addition, the stable expression of exogenous wild-type or ATPase-deficient BRG1 in mammalian cells results in no or only a modest increase in the overall cellular BRG1 level (9, 11, 30). Furthermore, the expression of an N-terminally truncated form of BAF57 leads to a diminished expression of endogenous BAF57 in mouse T-cell precursors (7). Finally, mouse embryonic stem cells containing a targeted deletion of one genomic copy of the SNF5/Ini1 gene produce the same amount of Ini1 protein as wild-type cells (15).
In this study, we present evidence to support a critical role for BAF155/BAF170 in regulating the steady-state protein level of BAF57 and the overall stoichiometry of the SWI/SNF complex. We demonstrate that protein-protein interactions among those subunits and proteasome-mediated protein degradation are involved in this regulatory process. Our results provide a mechanistic explanation for the use of potential protein quality control systems to maintain the subunit stoichiometry of multimeric enzymes such as the SWI/SNF complex.
MATERIALS AND METHODS
Plasmids.
The mammalian expression vector for FLAG-tagged human BAF57 was constructed by inserting a PCR fragment containing the entire coding region of BAF57 and a C-terminal FLAG epitope into pcDNA3.1(−)Zeo (Invitrogen). The same fragment was also inserted into the vectors pGEX5T1 and pET16 to generate plasmids for the expression of glutathione S-transferase (GST)- and histidine-tagged BAF57 protein, respectively, in Escherichia coli. The vectors for the expression of various BAF57 deletion mutants were generated using a standard PCR-based mutagenesis procedure. The mammalian expression vector pBJ5-BRG1 has been described previously (37). The expression vectors for human SNF5/Ini1, BAF155, and BAF170 were generated by inserting appropriate PCR fragments into the vector pcDNA3.1D/V5-His-TOPO (Invitrogen). The vectors for the expression of various BAF155 deletion mutants were generated by inserting appropriate PCR fragments into the vector pCMV/myc/nuc (Invitrogen). The authenticity of PCR-generated sequences was verified by DNA sequencing.
Antibodies and chemicals.
A recombinant histidine-tagged BAF57 protein was expressed in E. coli BL21-codon plus (DE3)-RP cells (Stratagene) and isolated under denaturing conditions as described previously (6). The recombinant protein was used as an antigen to immunize rabbits. The anti-BAF57 antibody was affinity purified from rabbit serum. Antibodies to BRG1 (G7), BAF155 (H76), BAF170 (H116), and Brm (N20) were obtained from Santa Cruz Biotech. Antibodies to β-actin (clone AC15), SNF5/Ini1, and FLAG (M2) were purchased from Sigma. Antibodies to Myc and BAF60a were purchased from Invitrogen and BD Bioscience, respectively. Cycloheximide, epoxomycin, and MG132 were purchased from Calbiochem.
Establishment of BAF57 stable cell lines and drug treatment.
UL3 cells, a derivative of the U2OS osteosarcoma cell line, were previously described (12). To establish UL3-BAF57 cell lines, cells were transfected with 1 μg wild-type or truncated BAF57 expression vector using the FuGene 6 reagent (Roche). Forty-eight hours after transfection, cells were selected in medium containing 100 μg/ml Zeocin (Invitrogen). The medium was changed every 3 days for approximately 2 weeks. Drug-resistant clones were expanded and screened by Western blot analysis using a FLAG antibody. For drug treatment, cells were treated with either 10 μM MG132, 5 μM epoxomycin, or 50 μg/ml cycloheximide for 2 h, 4 h, 6 h, and 8 h and then collected in urea buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl [pH 7.5]). Cells receiving no treatment were considered treated for 0 h.
Fluorescence-activated cell sorting (FACS).
UL3 cells stably expressing the BAF57ΔHMG mutant were cotransfected with 2 μg pEGFP-C1 plasmid (BD Clontech) and 6 μg of a specific subunit expression vector. Forty-eight hours after transfection, cells were trypsinized and suspended in medium at a density of approximately 5 × 106 cells/ml. Transfected cells were sorted on a Becton Dickinson FACSVantage SE flow cytometer, and green fluorescent protein (GFP)-positive and -negative cells were collected.
Real-time RT-PCR.
Total RNA was extracted from cells and treated with amplification-grade DNase I (Invitrogen) before reverse transcription (RT). Quantitative RT-PCR was performed with a Stratagene Mx3000P machine and Brilliant SYRB green QPCR master mix (Stratagene). PCRs were carried out with the primers 5′-AAAGCCACCAGATAAGCCG-3′ (P1), 5′-ATCAGTGAGATCTCGCCACA-3′ (P2), 5′-ACGAGAACATTCCGATGGAG-3′ (P3), and 5′-GCTCTCCGAGCCAGTGTTAC-3′ (P4) and with β-actin and glyceraldehyde-3-phosphate dehydrogenase primers. Average cycle threshold (CT) values for P1/P2 and P3/P4 amplifications were calculated and normalized to CT values for either β-actin or glyceraldehyde-3-phosphate dehydrogenase.
Western blot and IP assays.
Whole-cell extracts, except for those used for immunoprecipitation (IP), were prepared by lysing cells in urea buffer. Protein concentrations were determined using Bradford reagent (Bio-Rad). Equal amounts of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a Hybond-P membrane (Amersham), and subjected to Western blot assays with appropriate antibodies. Proteins on the membranes were detected with a chemiluminescence kit (Perkin-Elmer).
For IP, cells were lysed in high-salt buffer (20 mM Tris-HCl [pH 7.5], 400 mM NaCl, 1 mM EDTA, 0.5% NP-40) containing a protease inhibitor cocktail (Sigma). A whole-cell extract (500 μg) was first diluted with an equal volume of dilution buffer (lysis buffer without NaCl), brought to 500 μl by adding IP buffer (lysis buffer with 200 mM NaCl), and incubated with approximately 3 to 5 μg of antibody at 4°C overnight. The incubation was continued for an additional 2 h after the addition of 20 μl of protein A/G-plus-agarose bead slurry (Santa Cruz Biotech). Agarose beads were washed four times with IP buffer and eluted into 40 μl 2× SDS-PAGE gel loading buffer by heating at 95°C for 5 min. The bound proteins were separated by SDS-PAGE.
RNA interference.
Small interfering RNA (siRNA) duplexes targeting BAF57 (5′-AAGGAGAACCGTACATGAGCA-3′) and BAF60a (5′-AAGACACATAAGCTCCAGGAC-3′) were synthesized by QIAGEN. siRNA duplexes targeting BRG1, Brm, BAF155, BAF170, and hSNF5/Ini1 were purchased from Santa Cruz Biotech. siRNA targeting lamin A/C was purchased from Dharmacon. The Oligofectamine reagent (Invitrogen) was used to transfect siRNA oligonucleotides into cells according to the manufacturer's protocol.
GST pull-down assay.
GST or GST-BAF57 fusion protein was expressed in E. coli strain BL21-codon plus-RP cells (Stratagene) and isolated with a GST fusion protein purification kit (Pierce). One microgram of GST fusion protein was incubated with [35S]methionine-labeled BRG1 or BAF protein produced by in vitro translation using the TNT T7 Quick reticulocyte system (Promega). The binding reactions were performed in 400 μl of binding buffer (20 mM Tris-HCl [pH 7.5], 5 mM MgCl, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol) for 2 h at 4°C, 20 μl of glutathione-Sepharose 4B beads (Amersham Biosciences) was added to the reaction, and the incubation was continued for 1 h. The beads were washed five times with binding buffer. Bound proteins were eluted into SDS-PAGE gel loading buffer, separated by SDS-PAGE, and analyzed on a PhosphorImager machine (Molecular Dynamics).
RESULTS
Down-regulation of endogenous BAF57 by exogenous BAF57.
During the course of studying BAF57, we stably transfected mammalian expression vectors expressing either full-length BAF57 or one of two deletion mutants of human BAF57 into UL3 cells, a U2OS osteosarcoma cell line derivative (Fig. 1A) (12). The expectation was that the exogenous BAF57 mutants would function as dominant-negative inhibitors of the endogenous protein, based on previous observations suggesting that similar BAF57 deletion mutants can be assembled into the SWI/SNF chromatin remodeling complex (7, 35). Stable UL3 cell lines were established which express highlevels of each transgene, i.e., BAF57FL, BAF57ΔPH, or BAF57ΔHMG (as judged by immunoblot analysis using FLAG antibody [data not shown]), after antibiotic selection. In order to determine the relative expression level of the transgene compared to endogenous BAF57 in the established lines, whole-cell extracts were prepared from one representative cell clone for each transgene and analyzed by Western blotting using a BAF57-specific antibody. Surprisingly, exogenous expression of the BAF57ΔPH or BAF57ΔHMG transgene leads to a drastic down-regulation of endogenous BAF57 compared to that in parental UL3 cells (Fig. 1B, upper panel, compare BAF57 levels in lanes 1, 3, and 4). Since the BAF57FL transgene was C-terminally tagged with a FLAG epitope, it migrated slightly slower by SDS-PAGE than the endogenous protein due to the highly acidic nature of the tag. Thus, a distinct visual comparison can be made between the BAF57FL transgene and endogenous BAF57 in our cell model. Careful examination of the protein banding pattern indicates that endogenous BAF57 is also down-regulated in the cell line expressing BAF57FL (Fig. 1B, lower panel, which is an enlarged version of the boxed area in the upper panel). These observations suggest that down-regulation of the endogenous protein is not specifically due to expression of the deletion mutants. Interestingly, a marked decrease in endogenous BAF57 expression was also seen when a BAF57 deletion mutant similar to BAF57ΔPH was introduced into mouse T lymphocytes (7), suggesting that the down-regulation effect is neither cell nor species specific. Therefore, we speculated that there is a universal cellular mechanism involved to monitor, control, and/or maintain the expression of BAF57.
As a first step to investigate this mechanism, we utilized the UL3/BAF57ΔHMG cell line as a model system due to the ability to easily separate BAF57ΔHMG from endogenous BAF57 protein by SDS-PAGE and the availability of several optimal BAF57ΔHMG expression clones with which to work. In order to rule out the possibility that the antibiotic selection process might have exerted a negative effect on BAF57 expression, we established an additional UL3 cell line using empty vector (denoted UL3/Vec) via the same selection procedure. The expression of endogenous BAF57 was determined in the parental UL3, UL/Vec, and five independently derived UL3/ΔHMG cell lines (lines 1, 3, 4, 6, and 8). While endogenous BAF57 expression was similar in both parental UL3 and UL/Vec cells (Fig. 1C, top panel, lanes 1 and 2), all five UL3/ΔHMG clones tested displayed a marked reduction in wild-type (wt) BAF57 protein (Fig. 1C, top panel, lanes 3 to 7). However, the total amount of BAF57, including wt BAF57 and the BAF57ΔHMG protein, remained relatively constant in those cells and similar to the levels found in UL3 or UL3/Vec cells (compare lane 1 or 2 with lanes 3 to 7 in Fig. 1C, top panel). The expression levels of other SWI/SNF subunits, including BRG1, BAF155, BAF60a, and hSNF5/Ini1, did not vary much among those cell lines, as shown by Western blot analysis using subunit-specific antibodies (Fig. 1C, middle and bottom panels).
To determine whether the decreased expression of BAF57 occurs at the transcriptional or posttranscriptional level, we carried out quantitative real-time RT-PCR analysis on cDNAs prepared from total RNA isolated from UL3 or UL3/BAF57ΔHMG cells using either the P1/P2 or P3/P4 primer set, whose locations are shown in Fig. 1A. These primer sets were designed to allow detection and comparison between the two BAF57 species within our cell model. Primers P1 and P2, located within the HMG domain, only amplify cDNA fragments derived from endogenous wt BAF57, while the P3/P4 set, located in a common region of wt BAF57 and BAF57ΔHMG, will amplify cDNAs from both species. The results from real-time PCR analysis showed that the mRNA levels of wt BAF57 remained constant among the cell lines, while message levels for BAF57ΔHMG varied but were generally much higher than those of wt BAF57 (Fig. 1D). The high level of transgene messenger expression could be due to the use of a strong vector-derived cytomegalovirus promoter and/or to multiple integration copies of the transgene. Amplification using cDNA templates produced in the absence of reverse transcriptase did not generate any products, indicating that genomic DNA was not present in the reaction (data not shown). Based on these results, we concluded that the down-regulation of endogenous BAF57 by BAF57ΔHMG expression occurs at the posttranscriptional level.
Effects of proteasome and translation inhibition on BAF57 protein level.
The maintenance of a constant protein level in the presence of overexpressed mRNA can be achieved either by inhibition of mRNA translation or by an increased degradation of newly synthesized protein. Given that the translational control of specific mRNAs usually occurs at 5′ and 3′ untranslated regions, we consider translational inhibition an unlikely cause since any 5′ and 3′ untranslated region sequences derived from the transgene are products of the expression vector (32). Therefore, we carried out experiments to test whether the degradation of wt BAF57 and BAF57ΔHMG is enhanced in UL3/BAF57ΔHMG-1 cells compared to that in UL3 cells, given that this stable clone expresses the highest level of BAF57ΔHMG mRNA (Fig. 1D). Since most cellular protein degradation is mediated by the proteasome pathway, we treated UL3/BAF57ΔHMG-1 cells with the potent proteasome inhibitor MG132 for 0, 2, 4, 6, and 8 h and determined the expression levels of wt BAF57 and BAF57ΔHMG by Western blotting at each time point. As shown in Fig. 2A, inhibition of the proteasome by MG132 leads to a rapid buildup of both wt and ΔHMG BAF57 proteins (the increase is obvious as early as 2 h after treatment), suggesting that rapid degradation of both proteins takes place in those cells. Since MG132 also inhibits nonproteasomal enzymes, we repeated the experiment using another proteasome inhibitor, epoxomycin, whose only known targets are proteasomal enzymes (20), and obtained similar results (Fig. 2B). To estimate the half-lives of wt and ΔHMG BAF57 proteins, we blocked protein translation in UL3/BAF57ΔHMG-1 cells with cycloheximide and determined the protein levels of wt BAF57 and BAF57ΔHMG at the same time points after treatment. Surprisingly, blocking translation only led to a slight decrease in wt and ΔHMG BAF57 levels during the length of the treatment, suggesting that both wt and ΔHMG BAF57 proteins are very stable (Fig. 2C) in UL3/BAF57ΔHMG-1 cells. In contrast to the case with UL3/BAF57ΔHMG-1 cells, an increase in BAF57 protein was not obvious in the parental UL3 line treated with MG132, even after 8 h of treatment (Fig. 2D), indicating a much slower degradation rate. However, as observed in UL3/BAF57ΔHMG-1 cells, the treatment of UL3 cells with cycloheximide only led to a marginal decrease in BAF57 protein, again suggesting that BAF57 is very stable in UL3 cells (Fig. 2E).
FIG. 2.
Inhibition of proteasome and translation has distinct effects on the BAF57 protein level in UL3 and UL3/BAF57ΔHMG-1 cells. UL3/BAF57ΔHMG-1 cells were treated with either MG132 (A), epoxomycin (B), or cycloheximide (C) for 0, 2, 4, 6, and 8 h. Whole-cell extracts were prepared at each time point and resolved by 12% SDS-PAGE. The relative protein levels of wt BAF57 and BAF57ΔHMG in the extracts were determined by Western blotting. UL3 cells were also treated with either MG132 (D) or cycloheximide (E) for 0, 2, 4, 6, and 8 h. The relative protein level of wt BAF57 present at each time point was again determined by Western blotting. “NS” denotes a nonspecific band recognized by the BAF57 antibody and indicates equivalent loading.
BAF155/170 dictates the steady-state level of BAF57 protein.
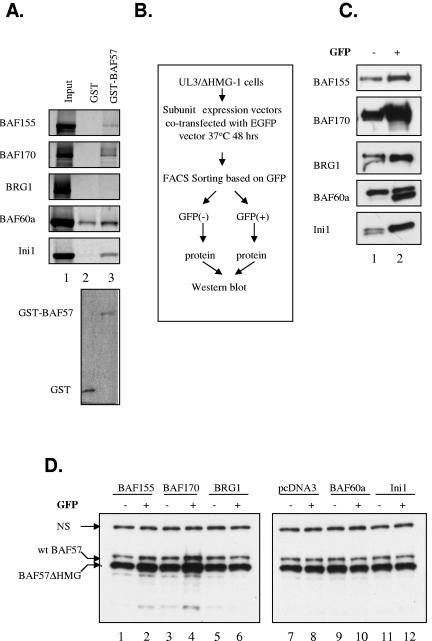
The rapid proteasome-mediated degradation, along with the long half-life of wt BAF57 and BAF57ΔHMG proteins, observed in UL3/BAF57ΔHMG-1 cells seems to present a paradox. In order to resolve this contradiction, we reasoned that there may be two pools of wt and ΔHMG BAF57 proteins present in these cells, one that is highly stable (as demonstrated by the long half-life) and one which is highly unstable (as revealed by the rapid protein accumulation after proteasome inhibition). Previous studies have shown that protein-protein interactions can facilitate the stability of a protein. For example, the tumor suppressor p14ARF is stabilized when bound to NPM/B23 in a higher-molecular-weight complex (22). In addition, the yeast mating type switch factors MATa1 and MATα2 are mutually stabilized by dimerization (18). Since BAF57 is a component of the SWI/SNF complex and since the BAF57ΔHMG protein can be efficiently incorporated into the remodeling complex (7, 35; data not shown), we speculated that other SWI/SNF subunits may aid in the regulation and distribution of newly synthesized wt BAF57 or BAF57ΔHMG protein into the two theoretical pools, possibly via protein-protein interactions. To test this, we first determined which subunits can directly interact with BAF57 in a GST pull-down assay. A GST-BAF57 fusion protein was able to pull down BAF155, BAF170, hSNF5/Ini1, and possibly BAF60a, but not BRG1, in this assay, suggesting that BAF57 can interact with multiple SWI/SNF subunits directly and independently (Fig. 3A, lane 3). Next, expression vectors for several common SWI/SNF subunits, including BRG1, BAF155, BAF170, BAF60a, and hSNF5/Ini1, along with a GFP expression plasmid, were transiently transfected into UL3/BAF57ΔHMG-1 cells to achieve enforced overexpression of those proteins in transfected cells. To eliminate the effect of variations in transfection efficiency, cells were sorted based on their expression of GFP by FACS in a scheme described in Fig. 3B. Western blot analysis of GFP-expressing cells showed an increase in expression of individual subunits versus GFP-negative cells, indicating a successful enrichment of transfected cells (Fig. 3C). Lysates from FACS were then used to determine the steady-state amounts of wt BAF57 and BAF57ΔHMG present in these two cell populations. Interestingly, the enforced overexpression of BAF155 and BAF170, but not other subunits, was able to augment the steady-state levels of wt and ΔHMG BAF57 in these cells (Fig. 3D, compare lanes 1 and 2 or lanes 3 and 4), suggesting a unique role of BAF155/BAF170 in enlarging the stabilized pool of wt and ΔHMG BAF57.
FIG. 3.
BAF155/170 interacts with BAF57 and increases steady-state levels of BAF57 protein in UL3/BAF57ΔHMG-1 cells. (A) Pull-down assay of core SWI/SNF subunits translated in vitro by GST-BAF57. The 35S-labeled subunit proteins were incubated with either GST (lane 2) or GST-BAF57 (lane 3), and bound proteins were detected with a phosphorimager (upper panel). A Coomassie-stained gel of GST and the GST-BAF57 fusion protein is shown in the lower panel. (B) Strategy for enriching UL3/BAF57ΔHMG-1 cells transfected with pcDNA3 or plasmids expressing various core SWI/SNF subunits. (C) WCEs from GFP-negative (lane 1) and -positive (lane 2) cells after transfection were analyzed by Western blotting with antibodies to core SWI/SNF subunits. (D) The same extracts used in panel C as well as WCEs from pcDNA3-transfected cells were analyzed by Western blotting with BAF57 antibody. “NS” denotes a nonspecific band and indicates equivalent loading.
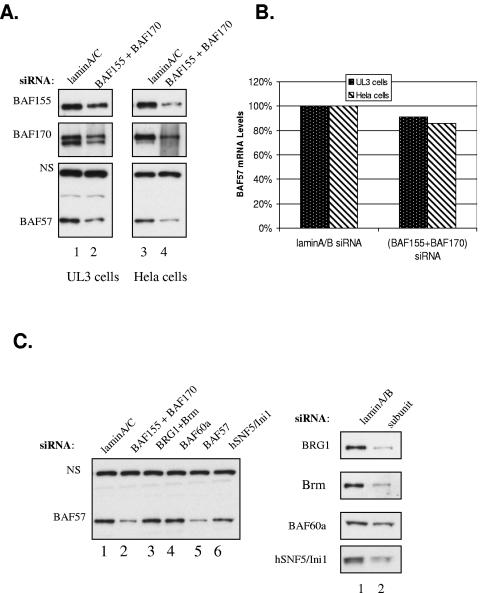
To further examine the role of BAF155/BAF170 in determining the steady-state level of BAF57 under physiological cellular conditions, we exploited BAF155- and BAF170-specific siRNAs to knock down BAF155 and BAF170 expression in UL3 cells. Whole-cell extracts were prepared from cells transfected with either a control lamin A/C siRNA or a mixture of BAF155 and BAF170 siRNAs and subjected to Western blot analysis using antibodies specific to BAF57, BAF155, and BAF170. As expected, the down-regulation of BAF155 and BAF170 led to a concomitant reduction in expression of endogenous BAF57 in these cells (Fig. 4A, left panel). To determine whether this coregulation effect was specific to UL3 cells, we repeated the experiment with HeLa cells and obtained the same result (Fig. 4A, right panel). In order to rule out any potential off-target effects by the siRNAs used in the study, we determined the mRNA levels of BAF57 in siRNA-transfected cells by real-time PCR analysis. Compared to the significant decrease in protein levels, there was a small, ∼10% decrease in BAF57 mRNA in cells treated with BAF155 and BAF170 siRNAs compared to those treated with control siRNA (Fig. 4B). The down-regulation of the protein levels of several common SWI/SNF components in HeLa cells by siRNA treatment demonstrated that only BAF155 and BAF170 siRNAs, besides the BAF57 siRNA, were able to significantly decrease the steady-state level of BAF57 protein in these cells (Fig. 4C, compare lanes 2 and 5 to the other lanes). The transfection of siRNAs targeting BRG1, Brm, BAF60a, and hSNF5/Ini1 into HeLa cells did not result in an evident decrease in the BAF57 protein level compared to that in control siRNA-transfected cells (Fig. 4C, compare lanes 3, 4, and 6 to lane 1), despite the fact that they could significantly knock down their intended targets, with the exception of BAF60a siRNA, which only modestly decreased BAF60a protein expression (Fig. 4D, compare lane 1 to lane 2). This result is completely consistent with the observation made with UL3/BAF57ΔHMG-1 cells overexpressing BAF155 or BAF170 (Fig. 4C). Together, these data indicate that the amounts of BAF155 and BAF170 determine the steady-state level of BAF57 protein in human cells, most likely through increasing the protein stability of BAF57.
FIG. 4.
Reduction of BAF155 and BAF170 expression decreases BAF57 protein level in UL3 and HeLa cells. (A) UL3 and HeLa cells were transfected with lamin A/C siRNA (lanes 1 and 3) or a mixture of BAF155 and BAF170 siRNAs (lanes 2 and 4). Whole-cell extracts were prepared 48 h after transfection and analyzed by Western blotting for the expression of BAF155, BAF170, and BAF57. (B) UL3 and HeLa cells were treated as described for panel A, but total RNA was prepared from those cells, and the relative mRNA level of wt BAF57 was determined with the P1/P2 primer set by real-time PCR. The BAF57 mRNA level in UL3 and HeLa cells transfected with the control siRNA was arbitrarily set at 100%. (C) HeLa cells were transfected with either lamin siRNA (lane 1) or siRNAs targeting BAF155/170, BRG1/Brm, BAF60a, BAF57, and hSNF5/Ini1, respectively (lanes 2 to 6). Whole-cell extracts were prepared 48 h after transfection and analyzed by Western blotting for the expression of BAF57 (left panel). The same extracts were also probed with antibodies to BRG1/Brm, BAF60a, and hSNF5/Ini1 to show the knockdown efficiency (right panel).
Specific BAF155 domains are needed to control the BAF57 protein level.
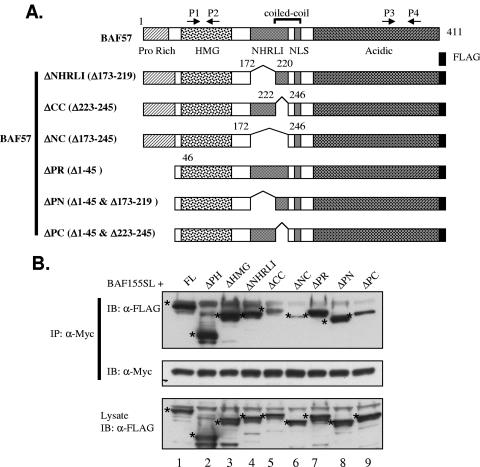
Since GST pull-down assays revealed an interaction between BAF155 or BAF170 and BAF57, we sought to examine the importance of this interaction in the regulation of the steady-state level of BAF57 by BAF155 or BAF170. Since BAF155 and BAF170 are highly homologous, we focused our study on BAF155. To map the minimal BAF155 domain(s) required for its stabilization effect in UL3/BAF57ΔHMG-1 cells, we constructed a battery of plasmid vectors for the expression of BAF155 mutants containing progressive deletions from both the amino and C termini (Fig. 5A, mutants BAF155Δ1 to Δ7). Since the functional nuclear localization signal (NLS) on the BAF155 molecule is not known, we fused all mutants at the C terminus with a 3× NLS sequence and the Myc epitope to aid in nuclear localization and detection (Fig. 5A). These mutants were then overexpressed in UL3/BAF57ΔHMG-1 cells by transient transfection. The stabilization effect by BAF155 in those cells is evident even without the separation of transfected cells from untransfected cells (Fig. 5B, compare lane b to lane a; data not shown), and the expression level of mutants can be assessed by one antibody (anti-Myc) in a Western blot. (Indeed, this represents a “lower limit” of the effect due to the presence of abundant nontransfected cells in the mixed population.) We then prepared extracts from UL3/BAF155ΔHMG-1 cells after transfection without sorting and determined the steady-state levels of wt BAF57 and BAF57ΔHMG by Western blot analysis of those cells. As shown in Fig. 5B, top left panel, the mutants BAF155Δ1 (lane c), Δ5 (lane g), and Δ6 (lane h), in addition to wild-type BAF155 (lane b), were able to elevate both wt BAF57 and BAF57ΔHMG expression, while the empty vector (lane a) and the rest of the mutants (lanes d, e, f, and i) could not. The expression levels of the mutants were comparable, with the possible exception of Δ1, as demonstrated by anti-Myc Western blotting (Fig. 5B, bottom left panel). This blot was reprobed with β-actin to show equal loading of the protein. Since all of the functional mutants, i.e., Δ1, Δ5, and Δ6, contain both the SANT and leucine zipper (LZ) domains while the nonfunctional ones, mutants Δ2, Δ3, Δ4, and Δ7, are devoid of at least one of them, we concluded that both domains are essential for the stabilization effect. To determine if those two domains are also sufficient, we constructed one additional mutant which only contains the SANT and LZ domains (Fig. 5A, mutant BAF155SL) and carried out the same transfection experiment. As shown in Fig. 5B, top right panel, BAF155SL was capable of augmenting wt BAF57 and BAF57ΔHMG expression as effectively as wild-type BAF155 (compare lane k or l to lane j). Thus, these data indicated that the SANT and LZ domains are essential and sufficient for the stabilization effect.
FIG. 5.
Specific BAF155 domains are required for increasing the BAF57 protein level and the interaction with BAF57 in vivo. (A) Schematic illustration of human BAF155 domain structure and the various deletion constructs used in this study. All mutants were fused to a nuclear localization signal (NLS) and Myc epitope at the C terminus. P/Q, proline- and glutamine-rich region; LZ, leucine zipper. (B) Whole-cell extracts from UL3/BAF57ΔHMG-1 cells transiently transfected with either an empty vector (lane a), wt BAF155 (lane b), or various deletion mutant expression vectors (lanes c to i) were analyzed by Western blotting with either BAF57 antibody (top panel) or Myc antibody (bottom panel). β-Actin was used as an internal loading control (middle panel). (C) Whole-cell extracts were prepared from UL3 cells transiently transfected with BAF57-FLAG and various BAF155 mutant expression vectors and immunoprecipitated with Myc antibody. The precipitated materials were analyzed by Western blotting with FLAG-horseradish peroxidase (FLAG-HRP) antibody (top panel). The same blot was stripped and reprobed with Myc-HRP antibody (middle panel) to demonstrate the IP efficiency. The bands corresponding to the expected BAF155 protein are marked with dots. About 5% of the total input lysates used for immunoprecipitation were directly analyzed by Western blotting with FLAG antibody (bottom panel).
Interaction between BAF155 and BAF57 is mediated by conserved protein domains.
To determine if these two domains are also required for the interaction between BAF155 and BAF57 in vivo, we transfected either an empty vector or the same battery of BAF155 mutant constructs, together with the BAF57FL plasmid, into UL3 cells. Whole-cell extracts were prepared after transfection and subjected to IP with the Myc antibody. The precipitated immunocomplexes were resolved by SDS-PAGE and probed with FLAG antibody to detect the presence of BAF57 protein. As shown in Fig. 5C, top panel, mutants Δ2 to Δ4 did not interact with BAF57, while the other mutants, including BAF155SL, retained the ability to interact with BAF57 (compare lanes c, d, and e to lanes b, f, g, h, and i). Reprobing the same blot with Myc antibody showed that comparable amounts of mutant protein were precipitated (Fig. 5C, middle panel) in each IP reaction. Given the domain structure of the mutants, this co-IP result suggested that the LZ domain is the major determinant for the interaction between BAF155 and BAF57 in vivo. Taken together with the data presented in Fig. 5B, we concluded that the interaction between BAF57 and BAF155 is essential, but probably not sufficient, for overexpressed BAF155 to increase the steady-state levels of wt BAF57 and BAF57ΔHMG in UL3/BAF57ΔHMG-1 cells.
In order to further evaluate the importance of the protein-protein interaction between BAF155 and BAF57 in regulating the cellular BAF57 level, we mapped the domain(s) of BAF57 that is important for the interaction with the minimal functional BAF155 molecule, the BAF155SL protein, in vivo by a similar co-IP strategy to that described for Fig. 5C. We employed the BAF155SL mutant instead of wt BAF155 in order to eradicate any protein interactions mediated by other domains of wt BAF155. In addition to the two BAF57 deletion mutants described in Fig. 1A, we constructed vectors for the expression of BAF57 mutants containing deletions of the NHRLI domain, the coiled coil (CC) domain, the proline-rich region, or a combination of domains (Fig. 6A). All of the mutants were tested for expression and found to localize to the nucleus in UL3 cells (data not shown). Those vectors were then cotransfected into UL3 cells together with the BAF155SL expression plasmid. Whole-cell extracts were then prepared and subjected to immunoprecipitation using the Myc antibody. As shown in Fig. 6B, top panel, all except mutants ΔCC, ΔNC, and ΔPC can be efficiently coprecipitated with BAF155SL, suggesting that the highly conserved CC domain is critical for efficient binding between BAF57 and BAF155SL. Deletion of the NHRLI domain, located just upstream of the CC domain, did not affect binding between BAF57 and BAF155SL to a significant extent (Fig. 6B, top panel, compare lane 4 to lanes 1 to 3). However, deletion of both the NHRLI and CC domains led to a weaker interaction than that after the loss of the CC domain alone (Fig. 6B, top panel, compare lanes 5 and 6), suggesting that the NHRLI domain might be required for optimal binding. The N-terminal proline-rich domain is not involved in this interaction (Fig. 6B, top panel, compare lane 7 to lanes 1 to 3 or lane 8 to lane 4). Reprobing the same blot with a Myc antibody demonstrated that comparable amounts of BAF155SL protein were pulled down by the Myc antibody in each IP reaction (Fig. 6B, middle panel, lanes 1 to 9). The expression levels of BAF57FL or mutants in the lysates used for IP were also similar (Fig. 6B, bottom panel). However, note that none of these deletions could entirely eliminate the interaction as detected in this co-IP assay, suggesting that there may be indirect interactions between BAF57 mutants and BAF155SL mediated through other SWI/SNF subunits present in the extract. This possibility is supported by the observation that BAF57 interacts with hSNF5/Ini1 in a GST pull-down assay, possibly through domains other than that critical for interaction with BAF155. We next attempted to establish stable UL3 cell lines overexpressing either the BAF57ΔNHRLI or BAF57ΔCC mutant protein, following the exact same procedure for the generation of UL3/BAF155ΔHMG cells. Despite multiple attempts and screening of many drug-resistant clones, we found that almost all of the mutants expressed very low levels of the mutant protein (data not shown). The failure to obtain high-level expression clones nevertheless suggests an intrinsic vulnerability of the BAF57ΔNHRLI and BAF57ΔCC proteins, which is consistent with the conclusion that the interaction between BAF57 and BAF155 is critical for the protein stability of BAF57 in vivo.
FIG. 6.
Conserved BAF57 domains are required for interaction with BAF155 in vivo. (A) Schematic description of BAF57 deletion mutants used in this study. (B) Whole-cell extracts were prepared from UL3 cells transiently transfected with the BAF155SL expression plasmid (Fig. 5A) and various BAF57 deletion constructs and then were immunoprecipitated with Myc antibody. The precipitated materials were analyzed by Western blotting with FLAG-HRP antibody (top panel). The same blot was stripped and reprobed with Myc-HRP antibody (middle panel) to demonstrate the IP efficiency. About 5% of the total input lysates used for immunoprecipitation were directly analyzed by Western blotting with FLAG antibody (bottom panel). The band corresponding to the expected protein in each lane is marked with an asterisk.
DISCUSSION
It may appear axiomatic that the faithful function of multisubunit molecular machines would require mechanisms that maintain requisite levels of individual subunits for an active complex. However, the specific mechanisms by which such stoichiometry is maintained are not well established. In this study, we demonstrated that the levels of SWI/SNF subunits BAF155 and BAF170 dictate the maximum expression level of another subunit, BAF57. We further demonstrated that protein-protein interactions between BAF155 and BAF57, as well as protein degradation mediated by the proteasome, participate in this regulatory process. We believe this is the first report describing that cross talk between two subunits of the mammalian SWI/SNF complex helps maintain a constant protein level of one subunit.
Although the interaction between BAF57 and BAF155 is important in this regulatory process, note that the physical interaction, as revealed by co-IP assay, did not perfectly correlate with functional consequences. For example, both the SANT and LZ domains are required to augment the steady-state BAF57 protein level in UL3/BAF57ΔHMG-1 cells, yet the LZ domain alone is sufficient for binding to BAF57 in vitro. In addition, the BAF57ΔNHRLI mutant can bind to BAF155 very efficiently, but the stable overexpression of it in UL3 cells is difficult (data not shown). One possible resolution of these data is that the SANT domain of BAF155 and the NHRLI domain of BAF57 may promote the interaction in an optimal conformation, which cannot be revealed by co-IP analysis. Alternatively, both the SANT domain and the NHRLI domain may indeed be important for the interaction yet may become dispensable when BAF155 and BAF57 are overexpressed in cells.
Human BAF57 has been shown to interact directly with several specific transcription factors and cofactors and may play an important role in the recruitment of the SWI/SNF complex to gene promoters (3, 4, 16, 26). Under these circumstances, the non-complex-bound BAF57 could potentially interfere with the recruitment process and elicit a dominant-negative effect. Consistent with this idea, ectopic expression of wt BAF57 has previously been shown to impair estrogen receptor transactivation in cells containing endogenous BAF57, but not in those devoid of endogenous BAF57 (4). Therefore, maintenance of a constant physiological level of BAF57 is critical for SWI/SNF-dependent activation of certain estrogen-responsive genes and possibly other SWI/SNF-dependent gene regulation events.
Indeed, one could propose a model where the cell must distinguish between free and complex-bound BAF57 in order for targeted degradation to occur. Given that protein-protein interactions are involved in this process, one potential mechanism cells can exploit to separate those two different pools of BAF57 is that BAF57 may adopt a destabilized conformation without binding to BAF155/170. In other words, BAF155 and BAF170 may serve as molecular chaperones for the correct folding and maturation of BAF57 in vivo.
An imbalanced subunit synthesis of multisubunit protein complexes has been known to generate aberrant proteins and is remedied by protein quality control (PQC) systems (14). Therefore, it is likely that a PQC system checks on the endogenous BAF57 level and triggers proteasome-mediated degradation of BAF57 when it is not bound to BAF155/BAF170. Since BAF57 is a nuclear protein and the interaction between BAF57 and BAF155/170 most likely occurs in the nucleus, our data suggest the presence of a nucleus-specific PQC system in mammalian cells. Interestingly, a PQC system operating solely in the nucleus was recently discovered in yeast (13). However, given that this SAN1 system appears to target only mutated nuclear proteins, it would appear unlikely that it is involved in the degradation of free BAF57 in mammalian cells. It will be important to identify the potential PQC system involved in maintaining the stoichiometry of multimeric enzyme complexes such as SWI/SNF.
The expression profiles of various SWI/SNF subunits in a large collection of human tumor cell lines have been reported previously (8). Interestingly, no BAF155 or BAF170 null expression cell line was discovered. In fact, it appears that the total expression level of BAF155 and BAF170 is relatively constant among most cell lines, regardless of the expression status of the other SWI/SNF subunits, including BAF57, BRG1/Brm, and Ini1. However, significantly less expression of BAF155 and BAF170 was detected in two cell lines that also express less BAF57 than other cell lines evaluated, consistent with the conclusion from our study.
It is possible that BAF155/170 functions as a scaffold for the assembly of the SWI/SNF remodeling complex; therefore, its expression may be regulated by alternative mechanisms. This concept is supported by the fact that the loss of either BRG1/Brm, BAF57, or hSNF5/Ini1 expression does not affect the interaction of the remaining subunits (10, 33, 36; data not shown). Thus, the particular attention that the cell apparently focuses on tightly regulating free BAF57 may reflect the range of its protein partners outside the SWI/SNF complex and speak to its unique expression in higher eukaryotes (35). In the future, additional experiments will help to further define the detailed protein-protein interaction map among SWI/SNF complex subunits and establish an essential quality control role for BAF155/170 in the assembly of a functional human SWI/SNF complex for transcription (19).
Acknowledgments
We thank Paul Wade (NIEHS), Alex Merrick (NIEHS), and Harriet Kinyamu (NIEHS) for critical reviews of the manuscript, Kevin Trotter (NIEHS) for helpful suggestions, Carl Bortner and the NIEHS FACS core facility for support, and Weidong Wang (NIA) for BAF plasmids.
This research was supported by the Intramural Research Program of the NIH and NIEHS.
REFERENCES
- 1.Aasland, R., A. F. Stewart, and T. Gibson. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87-88. [PubMed] [Google Scholar]
- 2.Aravind, L., and L. M. Iyer. 24July2002, posting date. The SWIRM domain: a conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol. 3:RESEARCH 0039. [Online.] http://genomebiology.com/2002/3/8/research0039. [DOI] [PMC free article] [PubMed]
- 3.Baker, K. M., G. Wei, A. E. Schaffner, and M. C. Ostrowski. 2003. Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. J. Biol. Chem. 278:17876-17884. [DOI] [PubMed] [Google Scholar]
- 4.Belandia, B., R. L. Orford, H. C. Hurst, and M. G. Parker. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 21:4094-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, L. A., R. R. Latek, and C. L. Peterson. 2004. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5:158-163. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., and J. R. Patton. 1999. Cloning and characterization of a mammalian pseudouridine synthase. RNA 5:409-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi, T. H., M. Wan, K. Zhao, I. Taniuchi, L. Chen, D. R. Littman, and G. R. Crabtree. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418:195-199. [DOI] [PubMed] [Google Scholar]
- 8.Decristofaro, M. F., B. L. Betz, C. J. Rorie, D. N. Reisman, W. Wang, and B. E. Weissman. 2001. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J. Cell Physiol. 186:136-145. [DOI] [PubMed] [Google Scholar]
- 9.de la Serna, I. L., Y. Ohkawa, C. A. Berkes, D. A. Bergstrom, C. S. Dacwag, S. J. Tapscott, and A. N. Imbalzano. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25:3997-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doan, D. N., T. M. Veal, Z. Yan, W. Wang, S. N. Jones, and A. N. Imbalzano. 2004. Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene 23:3462-3473. [DOI] [PubMed] [Google Scholar]
- 11.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 12.Fryer, C. J., H. K. Kinyamu, I. Rogatsky, M. J. Garabedian, and T. K. Archer. 2000. Selective activation of the glucocorticoid receptor by steroid antagonists in human breast cancer and osteosarcoma cells. J. Biol. Chem. 275:17771-17777. [DOI] [PubMed] [Google Scholar]
- 13.Gardner, R. G., Z. W. Nelson, and D. E. Gottschling. 2005. Degradation-mediated protein quality control in the nucleus. Cell 120:804-815. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg, A. L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature 426:895-899. [DOI] [PubMed] [Google Scholar]
- 15.Guidi, C. J., T. M. Veal, S. N. Jones, and A. N. Imbalzano. 2004. Transcriptional compensation for loss of an allele of the Ini1 tumor suppressor. J. Biol. Chem. 279:4180-4185. [DOI] [PubMed] [Google Scholar]
- 16.Harikrishnan, K. N., M. Z. Chow, E. K. Baker, S. Pal, S. Bassal, D. Brasacchio, L. Wang, J. M. Craig, P. L. Jones, S. Sif, and A. El-Osta. 2005. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 37:254-264. [DOI] [PubMed] [Google Scholar]
- 17.Iba, H., T. Mizutani, and T. Ito. 2003. SWI/SNF chromatin remodelling complex and retroviral gene silencing. Rev. Med. Virol. 13:99-110. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, P. R., R. Swanson, L. Rakhilina, and M. Hochstrasser. 1998. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 94:217-227. [DOI] [PubMed] [Google Scholar]
- 19.Kinyamu, H. K., J. Chen, and T. K. Archer. 2005. Linking the ubiquitin-proteasome pathway to chromatin remodeling/modification by nuclear receptors. J. Mol. Endocrinol. 34:281-297. [DOI] [PubMed] [Google Scholar]
- 20.Kisselev, A. F., and A. L. Goldberg. 2001. Proteasome inhibitors: from research tools to drug candidates. Chem. Biol. 8:739-758. [DOI] [PubMed] [Google Scholar]
- 21.Klochendler-Yeivin, A., C. Muchardt, and M. Yaniv. 2002. SWI/SNF chromatin remodeling and cancer. Curr. Opin. Genet. Dev. 12:73-79. [DOI] [PubMed] [Google Scholar]
- 22.Kuo, M. L., W. den Besten, D. Bertwistle, M. F. Roussel, and C. J. Sherr. 2004. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 18:1862-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Link, K. A., C. J. Burd, E. Williams, T. Marshall, G. Rosson, E. Henry, B. Weissman, and K. E. Knudsen. 2005. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol. Cell. Biol. 25:2200-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 25.Muchardt, C., and M. Yaniv. 2001. When the SWI/SNF complex remodels the cell cycle. Oncogene 20:3067-3075. [DOI] [PubMed] [Google Scholar]
- 26.Pal, S., R. Yun, A. Datta, L. Lacomis, H. Erdjument-Bromage, J. Kumar, P. Tempst, and S. Sif. 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 23:7475-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papoulas, O., G. Daubresse, J. A. Armstrong, J. Jin, M. P. Scott, and J. W. Tamkun. 2001. The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc. Natl. Acad. Sci. USA 98:5728-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson, C. L., and J. Cote. 2004. Cellular machineries for chromosomal DNA repair. Genes Dev. 18:602-616. [DOI] [PubMed] [Google Scholar]
- 29.Reyes, J. C., J. Barra, C. Muchardt, A. Camus, C. Babinet, and M. Yaniv. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J. 17:6979-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salma, N., H. Xiao, E. Mueller, and A. N. Imbalzano. 2004. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Mol. Cell. Biol. 24:4651-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, C. L., R. Horowitz-Scherer, J. F. Flanagan, C. L. Woodcock, and C. L. Peterson. 2003. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 10:141-145. [DOI] [PubMed] [Google Scholar]
- 32.Standart, N., and R. J. Jackson. 1994. Regulation of translation by specific protein/mRNA interactions. Biochimie 76:867-879. [DOI] [PubMed] [Google Scholar]
- 33.Trotter, K. W., and T. K. Archer. 2004. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol. Cell. Biol. 24:3347-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, W., T. Chi, Y. Xue, S. Zhou, A. Kuo, and G. R. Crabtree. 1998. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. USA 95:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]