Abstract
Hydrolysis of the mRNA cap plays a pivotal role in initiating and completing mRNA turnover. In nematodes, mRNA metabolism and cap-interacting proteins must deal with two populations of mRNAs, spliced leader trans-spliced mRNAs with a trimethylguanosine cap and non-trans-spliced mRNAs with a monomethylguanosine cap. We describe here the characterization of nematode Dcp1 and Dcp2 proteins. Dcp1 was inactive in vitro on both free cap and capped RNA and did not significantly enhance Dcp2 activity. Nematode Dcp2 is an RNA-decapping protein that does not bind cap and is not inhibited by cap analogs but is effectively inhibited by competing RNA irrespective of RNA sequence and cap. Nematode Dcp2 activity is influenced by both 5′ end sequence and its context. The trans-spliced leader sequence on mRNAs reduces Dcp2 activity ∼10-fold, suggesting that 5′-to-3′ turnover of trans-spliced RNAs may be regulated. Nematode Dcp2 decaps both m7GpppG- and m2,2,7GpppG-capped RNAs. Surprisingly, both budding yeast and human Dcp2 are also active on m2,2,7GpppG-capped RNAs. Overall, the data suggest that Dcp2 activity can be influenced by both sequence and context and that Dcp2 may contribute to gene regulation in multiple RNA pathways, including monomethyl- and trimethylguanosine-capped RNAs.
RNA turnover is an important part of mRNA metabolism and contributes to gene expression (7, 24, 41, 59, 60). A number of pathways of eukaryotic mRNA decay have been described (1, 7, 41, 56, 59, 60). Among these are two general pathways of mRNA turnover, 3′-to-5′ and 5′-to-3′ exonucleolytic decay. Specialized mRNA turnover pathways are also known and include nonsense-mediated decay, endonuclease cleavage, and nonstop decay. Enzymes that cleave the mRNA cap participate in all of these turnover pathways (7, 10, 16, 21, 41).
In both general pathways of mRNA turnover (3′-to-5′ and 5′-to-3′ decay), decay is initiated by deadenylation or shortening of the mRNA poly(A) tail by deadenylases. In the 3′-to-5′ pathway, deadenylation facilitates access of a complex of 3′-to-5′ exonucleases, the exosome, to the 3′ end of the RNA. The exosome degrades the RNA 3′ to 5′ until it reaches the RNA cap. The RNA cap consists of a guanine residue added cotranscriptionally to the first nucleotide of an mRNA transcript through an inverted 5′-5′ triphosphate bridge. This unusual triphosphate linkage is not a substrate for general exo- or endonucleases. Consequently, complete 3′-to-5′ exonucleolytic decay results in the RNA degraded to its constituent nucleotide monophosphates and the remaining dinucleotide, the mRNA cap. The remaining cap is a substrate for the DcpS scavenger decapping enzyme, which hydrolyzes the m7GpppN cap to m7Gp and ppN (6, 10, 20, 21, 31, 32, 40, 45, 54, 58). In the 5′-to-3′ pathway, deadenylation of the RNA triggers decapping of the RNA by the enzyme complex, Dcp1/Dcp2 (2, 7, 10, 14, 21, 23, 26, 41, 42, 49, 50, 53, 57). Decapping of the deadenylated RNA by Dcp1/Dcp2 results in m7Gpp and p-RNA products. The 5′ monophosphate RNA is a substrate for 5′-to-3′ exonucleolytic decay by an exonuclease such as Xrn1. Thus, hydrolysis of the cap plays important roles in both initiating and completing mRNA decay in the two general mRNA turnover pathways. These decapping enzymes also participate in the specialized mRNA turnover pathways (18, 28, 34).
Spliced leader (SL) RNA trans splicing generates the mature 5′ ends of mRNAs by addition of a spliced leader sequence to the pre-mRNA (4, 11, 29, 38, 39). This mechanism of gene expression is present in diverse eukaryotes including some flagellate protozoa, hydra, nematodes, rotifers, flatworms, and even primitive chordates (38, 43). In metazoa, addition of the spliced leader sequence also adds an atypical cap to the RNA, a trimethylguanosine (TMG) cap (m2,2,7GpppN), compared to the typical m7GpppN eukaryotic cap (30, 37, 52, 55). However, not all cellular mRNAs in trans-splicing metazoa undergo spliced leader addition. Therefore, two populations of mRNA are present in the cell, non-trans-spliced mRNAs with the typical m7GpppN cap and variable 5′ untranslated region sequence and trans-spliced mRNAs with the 5′ spliced leader sequence and an m2,2,7GpppN cap. mRNA metabolism and cap-interacting proteins must deal with these two distinctly capped mRNA populations. One of the interests of our laboratory is to understand the posttranscriptional role of spliced leader trans splicing on mRNA metabolism and to understand how mRNA metabolism and cap-interacting proteins have accommodated these two RNA populations. To address these questions, we have chosen to use two nematodes as model systems, Caenorhabditis elegans and Ascaris. Using Ascaris embryos, we have developed both in vitro translation and decay systems as well as biolistic methods to evaluate the role of the spliced leader sequence and TMG cap on mRNA translation and decay (6, 12, 27). Using the in vitro decay system, we recently demonstrated that the predominant general pathway of mRNA decay is 3′-to-5′ exonucleolytic decay followed by “scavenger” hydrolysis of the resulting mRNA cap (6). 5′-to-3′ decay also occurs in the Ascaris extracts but is ∼15-fold less active than the 3′-to-5′ decay pathway. Both pathways in vitro are capable of hydrolysis of both the m7GpppN cap and the m2,2,7GpppN cap derived from the spliced leader. We have also shown that the recombinant C. elegans “scavenger” enzyme DcpS can hydrolyze both the m7GpppN and m2,2,7GpppN caps. In contrast, human DcpS is not capable of hydrolyzing m2,2,7GpppN caps, and the C. elegans enzyme substrate requirements differ from those of the human enzyme (6).
To extend these studies on mRNA turnover and decapping in nematodes, we have now cloned, expressed, and analyzed recombinant C. elegans Dcp1 and Dcp2. Nematode Dcp2 requires an RNA substrate of at least 50 nucleotides (nt), is an RNA binding protein, does not directly bind the RNA cap, and is competitively inhibited by RNA irrespective of sequence and cap. Nematode Dcp2 can also hydrolyze m2,2,7GpppG caps. This is a general property of eukaryotic Dcp2, since we also demonstrate that budding yeast and human Dcp2 can also hydrolyze m2,2,7GpppG caps. In addition, nematode Dcp2 activity is affected by both the 5′ terminal sequence and the context. Overall, these data suggest that Dcp2 could be involved in both mRNA and snRNA or snoRNA turnover, that the Dcp2 activity is affected by RNA sequence, and that these properties have implications for RNA turnover and its regulation.
MATERIALS AND METHODS
Cloning.
Total C. elegans RNA was isolated from mixed-stage C. elegans cultures using Trizol (Invitrogen, Carlsbad, CA). First-strand cDNA was generated using SuperScript II reverse transcriptase and oligo(dT) primers (Invitrogen). The C. elegans Dcp1 and Dcp2 open reading frames were amplified from the cDNA using specific primers (see Fig. S4 in the supplemental material) and either Expand high-fidelity polymerase (Roche, Indianapolis, IN) or Taq DNA polymerase (Promega, Madison, WI). The Dcp1 coding region PCR product was cloned into pET16b (Novagen, Madison, WI) as an NdeI and BamHI fragment or the yeast vector pESP-1 (Stratagene, La Jolla, CA) as a BamHI fragment using Escherichia coli DH5α as a host. The Dcp2 coding-region PCR products were digested with BamHI and EcoRI (Dcp2ΔBoxB and Dcp2 1-659) or BamHI and NotI (full-length Dcp2) and cloned into pET32a (Novagen, Madison, WI) using DH5α as a host. Recombinants were identified and confirmed by DNA sequencing. Dcp2 1-479 was subcloned from the full-length Dcp2 as a BamHI and EcoRV fragment into the BamHI and Hind III (blunted) sites of pET32a. The single Dcp2 mutants I259T and I295T were Taq DNA polymerase mutants identified during sequencing of clones derived from the original cDNA PCR. Additional clones of Dcp2 were generated either by direct subcloning or PCR cloning into pET32a. Clones were then transformed into Rosetta DE3 (Novagen) for protein expression. The single E275Q mutation in the nudix motif of Dcp2 1-479 was generated using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Recombinant protein expression and purification.
Yeast expression and glutathione S-transferase purification of Dcp1 in pESP-1 was carried out as described by the manufacturer (Stratagene). Bacterial protein expression was induced with 0.4 mM isopropyl β-d-thiogalactoside for 3 h at 30°C in cultures previously grown at 37°C to an optical density at 600 nm of 0.6. Frozen bacterial pellets were resuspended in ice-cold lysis buffer (50 mM Na2HPO4, pH 7.5, 600 mM NaCl, 600 mM urea, 10% glycerol, 1% Triton X-100), lysozyme was added to a final concentration of 1 mg/ml, and the suspension was incubated on ice for 30 min and then sonicated. His-tagged Dcp2 was bound to Ni2+-nitrilotriacetic acid (NTA)-agarose (QIAGEN Inc., Valencia, CA) for 60 min at 4°C, unbound proteins removed with washing buffer (50 mM Na2HPO4, pH 7.5, 600 mM NaCl), and the bound proteins eluted with wash buffer containing increasing concentrations of imidazole (20 to 300 mM imidazole). Fractions containing Dcp1 or Dcp2 were dialyzed against 20 mM Tris, pH 7.5, 50 mM KCl, 20% glycerol, 1 mM dithiothreitol (DTT), and 0.5 mM phenylmethylsulfonyl fluoride and stored at −80°C. Dcp2 was further purified following dialysis into 50 mM Na2HPO4 (pH 8.0) and 150 mM NaCl on a HiTrap heparin column using a gradient from 0.15 to 1 M NaCl in 50 mM Na2HPO4 (pH 8.0) (Pharmacia, Piscataway, NJ). The most-pure Dcp2 fractions with activity were dialyzed against 20 mM Tris, pH 7.5, 50 mM KCl, 20% glycerol, 1 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride and stored at −80°C. Human Dcp2 was expressed and purified as described previously (23). Budding yeast Dcp1 and Dcp2 (generously provided by Carolyn Decker and Roy Parker) were coexpressed and purified as described previously (47).
Substrate preparation.
Preparation of PCR templates for in vitro transcription and transcription reactions was carried out as previously described (6). Cap-labeled RNAs were prepared from uncapped RNA substrates using [α-32P]GTP (Perkin-Elmer, Boston, MA) and recombinant vaccinia RNA guanylyltransferase and (guanine-N7)-methyltransferase (generously provided by Stewart Shuman), human capping enzyme (generously provided by Aaron Shatkin), and cap-specific 2′-O-methyltransferase (generously provided by Paul Gershon) as described previously (6). Cap-labeled RNAs were typically gel purified by denaturing polyacrylamide gel electrophoresis (PAGE) prior to use or passed several times over Sephadex G-50. m32,2,7Gp*ppGm-SL snRNA was produced by hypermethylation of an m7Gp*ppGm-SL snRNA in Ascaris whole-cell embryo translation extracts followed by phenol-chloroform extraction and ethanol precipitation or gel purification as described previously (6). Enrichment for the hypermethylated SL RNA was carried out by immunoprecipitation with anti-TMG antibodies (5) using conditions recommended by the supplier (Synaptic Systems, Gottingen, Germany). Cap-labeled dinucleoside triphosphates were prepared by nuclease P1 digestion as described previously (6).
Decapping reactions.
Decapping reactions were carried out as previously described (6) in decapping buffer [50 mM Tris, pH 7.9, 30 mM (NH4)2SO4, 1 mM MgCl2, 1 mM DTT] for 30 to 60 min at 30°C (nematode and yeast) or 37°C (human) using 1 to 5 ng of 32P cap-labeled RNA (0.03 to 0.15 pmol) and 5 to 500 ng of purified Dcp1 or Dcp2. For human and yeast Dcp2 reactions and in some nematode experiments the buffer was supplemented with 2.5 mM MnCl2. Decapping reactions on 32P cap-labeled dinucleotides were carried out as described previously (6). Reactions were stopped either by addition of 50 mM EDTA or by freezing at −80°C. Reaction products were resolved by thin-layer chromatography (TLC) and 25% PAGE-5 M urea and subjected to autoradiography as described previously (6).
Substrate and product characterization.
Cap-labeled RNA substrates, the RNA cap, and decapping products were characterized using a variety of methods including comigration with known standards using TLC or denaturing PAGE analysis and several enzyme shift strategies as described previously (6). TLC was carried out using polyethyleneimine (PEI)-cellulose as the stationary phase and ammonium sulfate (0.45 M) or lithium chloride (0.3 to 1 M) as the mobile phase. PAGE analysis was done using 5 M urea 20% or 25% denaturing gels as described previously (3, 6). Reaction substrates and products were then quantified by phosphorimager analysis of the TLC or PAGE separations using Molecular Dynamics STORM 860 and ImageQuant software. Dinucleoside cap analogues and methylated nucleotides used as standards or competitors were prepared as described previously (6) and were generously provided by Edward Darzynkiewicz.
Nucleotide sequence accession number.
The C. elegans Dcp2 mRNA sequence has been deposited in GenBank under accession no. DQ143943.
RESULTS
Identification of C. elegans Dcp2 and Dcp1.
Sequence similarity was used to identify potential C. elegans database orthologs of the yeast decapping enzymes Dcp1 and Dcp2. A 332-amino-acid Dcp1 ortholog was identified that contains a Pfam Dcp1 domain and similarity with other divergent Dcp1 proteins (see Fig. S1 in the supplemental material), particularly in the amino-terminal ∼150 amino acids. An annotated Dcp2 ortholog was also identified in the C. elegans database (ndx-5), predicted to encode either 367- or 344-amino-acid proteins (CE23761 and CE17847) that differ in their amino termini as a function of alternative 5′ exon use. Sequence alignment of these proteins with other Dcp2 orthologs indicated that these predicted proteins lacked a conserved carboxy-terminal domain present in other Dcp2 proteins, the Box B domain, as well as a portion of the nudix fold (see Fig. 1A, Dcp2ΔBoxB). Our bioinformatic reevaluation of the genomic region encoding C. elegans Dcp2 identified a 5′ splice site within several amino acids of the database's predicted ndx-5 carboxy terminus that could lead to inclusion of additional 3′ exons.
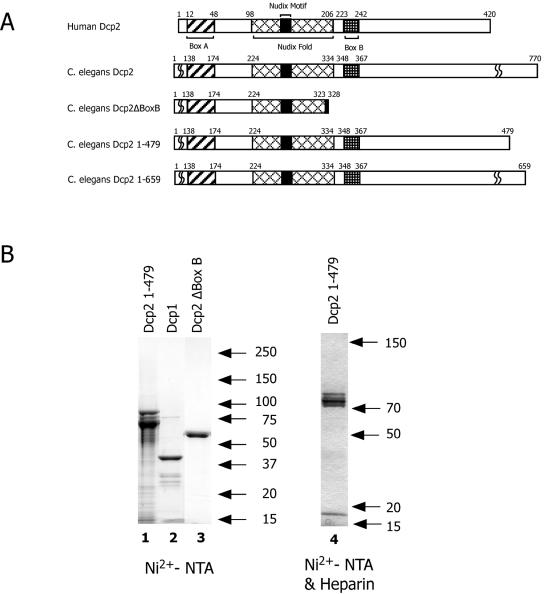
FIG. 1.
Schematic of Dcp2 proteins and purified recombinant Dcp1 and Dcp2 proteins. (A) Schematic of Dcp2 proteins. C. elegans proteins used in the current study are schematically shown and compared to human Dcp2. Note that the database annotation for C. elegans Dcp2, shown here as Dcp2ΔBoxB, lacks the COOH terminus of the nudix fold and Box B. The three other proteins used for decapping assays (Dcp2, Dcp2 1-479, and Dcp2 1-659) contain all conserved regions of known Dcp2 proteins. The COOH-terminal domains of Dcp2 proteins vary considerably and contain limited similarity. (B) Sodium dodecyl sulfate-PAGE illustration of purified recombinant Dcp1 and Dcp2 proteins. Proteins were purified by Ni2+-NTA chromatography or both Ni2+-NTA and heparin chromatography, resolved by sodium dodecyl sulfate-PAGE, and stained with Coomassie blue stain. Ni2+-NTA/heparin chromatography preparations were used for most of the analyses.
To experimentally determine the organization of the expressed open reading frame(s) from the putative Dcp2 locus, PCR primer pairs were designed to test several of our revised coding region predictions. Reverse transcription (RT)-PCR and sequence analysis confirmed the expression of C. elegans RNAs encoding the database-predicted 367- and 344-amino-acid ndx-5 proteins that lack the conserved Box B domain (Fig. 1A, Dcp2ΔBoxB). A longer open reading frame that used the 5′ spliced site we predicted and several 3′ exons was identified in C. elegans RNA that contained a 2,313-nucleotide open reading frame encoding a 770-amino-acid protein (Fig. 1A, C. elegans Dcp2). Importantly, the 770-amino-acid protein contains the complete nudix fold and a Dcp2 Box B region (Fig. 1A; also see Fig. S2 in the supplemental material). This protein also contains what was previously predicted as an independent C. elegans database open reading frame, CE18392. The fused 3′ CE18392 coding region has no significant similarity with characterized proteins. Notably, the C. briggsae gene and mRNA annotation for this locus support the longer protein-encoding transcript we predicted and identified (see Fig. S2 in the supplemental material). In addition, antibodies raised against C. elegans Dcp2 recognize a protein in C. elegans extracts of the predicted size for the 770-amino-acid protein (molecular weight, ∼85,000).
Cloning, expression, and purification of Dcp1 and Dcp2.
The predicted C. elegans Dcp1 332-amino-acid coding region was amplified by RT-PCR from mixed-stage C. elegans cDNA and cloned into the bacterial expression vector pET16b and into the yeast expression vector pESP-1. Recombinant proteins derived from the two hosts were purified by Ni2+-NTA agarose chromatography and GST-affinity or anti-Flag chromatography, respectively. C. elegans Dcp2 proteins lacking Box B (Dcp2Δ BoxB) and the full 770-amino-acid open reading frame were amplified by RT-PCR from mixed-stage C. elegans cDNA and cloned into pET32a, and the bacterially expressed recombinant proteins were purified by Ni2+-NTA agarose chromatography.
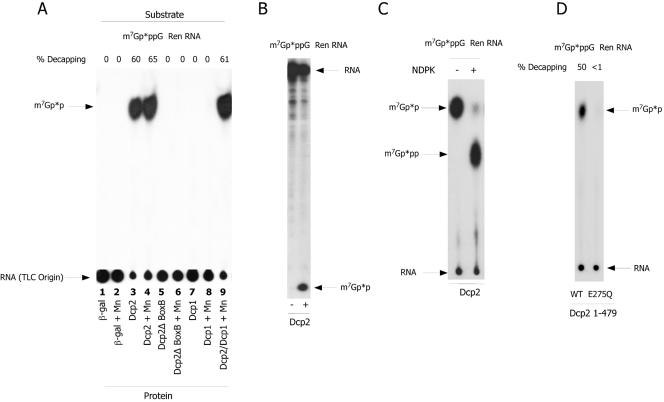
To determine if the recombinant Dcp1 or Dcp2 proteins were functional decapping enzymes, decapping assays were carried out using purified Dcp1 and Dcp2 proteins. The decapping assay determines whether a protein is able to hydrolyze the cap of a 250-nucleotide cap-labeled m7Gp*ppG RNA. As shown in Fig. 2A (lane 3), protein derived from the long Dcp2 open reading frame had significant decapping activity, producing a decapping product that comigrated on TLC analysis with m7Gp*p, the nucleoside diphosphate product characteristic of Dcp1/Dcp2-type decapping enzymes. The decapping product was independently confirmed to be m7Gp*p based on comigration of the product with standards using independent TLC solvent separations, 25% PAGE analysis, and demonstration of the conversion of the nucleoside diphosphate product to its corresponding nucleoside triphosphate with nucleoside diphosphate kinase (Fig. 2B and C). In contrast, recombinant C. elegans Dcp1 protein purified from either bacteria (see Fig. 1B, lane 2) or yeast showed no decapping activity under a variety of assay conditions including reaction buffers with Mn2+ (Fig. 2A, lanes 7 and 8). The nematode Dcp2 protein lacking a portion of the nudix fold and Dcp2 Box B (Dcp2Δ BoxB) (see Fig. 1B, lane 3) was inactive in decapping (Fig. 2A, lanes 5 and 6). Consistent with the essential nature of the Box B regions, the N-terminal fragment of Dcp2 (1-479) that includes Box B was as active as the full-length protein (Fig. 2A). Single point mutations in Dcp2 1-479 within the nudix motif (I259T or E275Q) lead to almost complete loss of decapping activity, whereas the single mutation I295T leads only to reduced activity (Fig. 2D and data not shown). Overall, these data indicate that the RNA decapping we observe is derived from expression of the C. elegans Dcp2 protein. For the majority of the experiments described below, the enzyme activity illustrated is derived from the Dcp2 protein (1-479) expressed and purified as a fusion protein from pET32a by Ni2+-NTA agarose and then heparin chromatography (Fig. 1B, lane 4).
FIG. 2.
C. elegans Dcp2 is active in RNA decapping, but Dcp1 is not. (A) Decapping activity of C. elegans Dcp1 and Dcp2. Ni2+-NTA-purified Dcp1 and Dcp2 were assayed for RNA-decapping activity using an m7Gp*ppG cap-labeled 250-nucleotide test RNA (p* designates which phosphate is 32P) as described in Materials and Methods. Following the reaction, samples were spotted onto PEI-cellulose TLC and the plates resolved in 0.45 M ammonium sulfate and subjected to autoradiography. The arrows designate the TLC origin (where RNA remains following chromatography), and m7Gp*p illustrates where an m7Gpp marker runs. The β-gal lane represents a decapping assay carried out with similarly purified recombinant β-galactosidase protein. Dcp1 was inactive in the assay with or without Mn2+ and did not significantly enhance Dcp2 activity. % Decapping denotes the percentage of input RNA decapped, as determined by ImageQuant analysis of both the labeled RNA at the origin and the m7Gpp spot. (B) Dcp2 reactions analyzed by PAGE. An aliquot of the Dcp2 reaction was also resolved by 25% PAGE/5 M urea analysis to confirm comigration of the Dcp2 product with m7Gpp markers. (C) Conversion of the m7Gp*p product to m7Gp*pp by nucleoside diphosphokinase. An aliquot of the Dcp2 reaction was extracted with phenol-chloroform and treated with nucleoside disphosphokinase (NDPK) in the presence of ATP. NDPK will convert nucleoside diphosphates to their corresponding triphosphates. This assay in concert with other TLC and PAGE assays indicates the product of the C. elegans Dcp2 reaction is a diphosphate. (D) Mutation within the Nudix motif eliminates Dcp2 decapping activity. A mutation (E275Q) (see Fig. S2 in the supplemental material) was introduced into the Nudix motif of C. elegans Dcp2 1-479. Simultaneous purification of this expressed protein with the wild-type protein was carried out and the proteins assayed for Dcp2 activity as in part A. This mutation results in almost complete loss of Dcp2 activity.
Dcp1 activity.
Dcp2 was originally identified as an enhancer of Dcp1 decapping activity (14). More recently it has been shown that Dcp2 is the catalytic subunit of the Dcp1/Dcp2 complex (31, 33, 53) and that in budding yeast Dcp1 can enhance the decapping activity of Dcp2 (47, 49). In addition, the decapping activity of the yeast or human proteins was reported to be significantly higher in the presence of the Mn2+ ion (42, 49). Our initial assays for decapping activity of Dcp1 and Dcp2 were conducted with Mg2+ as the sole cation. We reevaluated Dcp1 activity in the presence of both the Mg2+ ion and the Mn2+ ion and also examined the effect of Dcp1 on Dcp2 activity. As illustrated in Fig. 2A (lanes 7 to 9), Dcp1 was also inactive for decapping in the presence or absence of the Mn2+ ion and had only a small effect on the decapping activity of Dcp2. Furthermore, Dcp1 was shown to be inactive on several different test RNA substrates (different RNA sequences and different caps including GpppG-, m7GpppGm-, and m2,2,7GpppGm-RNA) and also had no decapping activity directly on dinucleotide caps (GpppG, m7GpppGm, and m2,2,7GpppGm cap) (data not shown). We also tested whether the addition of Dcp1 to Dcp2 differentially enhanced the decapping of different RNA substrates and different cap structures (e.g., test RNAs with or without SL, SL RNA, and U1 RNA with GpppG, m7GpppG, and hypermethylated caps). Enhancement of nematode Dcp2 decapping on different caps or RNA substrates with the addition of Dcp1 was limited (observed less than twofold) and in most cases did not enhance Dcp2 activity (data not shown). Although Dcp1 was purified using the same conditions and simultaneously with Dcp2, it remains possible that bacterially expressed nematode Dcp1 may not be functional.
Substrate length specificity of C. elegans Dcp2.
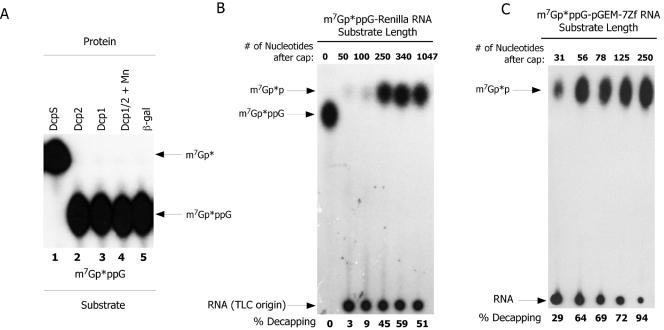
Decapping proteins differ in their substrate length requirements as a function of both the type of decapping enzyme and the organism source. We therefore examined the substrate length requirement of nematode Dcp2. Dcp2 was not capable of hydrolyzing the dinucleotide cap m7GpppG lacking an RNA (Fig. 3A, lane 2) or other dinucleotide caps (GpppG, m7GpppGm, and m2,2,7GpppGm) (data not shown). These dinucleotide cap substrates are readily cleaved by nematode DcpS (Fig. 3A, lane 1) (6). Dcp2 exhibited minimal activity on m7GpppG cap-labeled Renilla substrate up to ∼25 nucleotides (5, 10, 20, and 25 nucleotide substrates tested) (data not shown). Optimal activity was observed with a ∼250-nucleotide Renilla substrate (Fig. 3B). However, using a second series of RNA substrates derived from the pGEM-7Zf polylinker, a small amount of Dcp2 activity was observed on a 31-nucleotide substrate and optimal activity was observed on capped substrates greater than 50 nucleotides (Fig. 3C). Therefore, minimal substrate length demonstrated some variation with the two substrates examined. This indicates that optimal activity on a substrate is likely a function of length as well as sequence and/or structure.
FIG. 3.
C. elegans Dcp1 and Dcp2 are inactive on cap dinucleotides, and Dcp2 activity is optimal on RNA of at least 50 nucleotides. (A) Dcp1 and Dcp2 are inactive on cap dinucleotide substrate. m7Gp*ppG dinucleotide cap prepared as described previously (6) was used as a substrate for Dcp1, Dcp2, DcpS, and Dcp1/2 decapping. The reaction products were resolved using PEI-cellulose TLC and 0.45 M ammonium sulfate followed by autoradiography. No decapping products were observed following incubation of Dcp1 or Dcp2 with the substrate. The arrows illustrate the location of the input m7Gp*ppG substrate and the only products observed from the reactions, the m7Gp* derived from DcpS complete hydrolysis of the substrate. The β-gal lane represents a decapping assay carried out with a similarly purified recombinant β-galactosidase protein. (B) RNA length and substrate dependence of C. elegans Dcp2 on a Renilla RNA. m7Gp*pppG-capped Renilla RNAs of increasing length were evaluated as substrates for C. elegans Dcp2. In addition to the RNAs illustrated, Renilla RNAs of 5, 10, 20, and 25 nucleotides were also examined and were poor substrates for Dcp2 (data not shown). The percent conversion of the input RNA substrate to m7Gp*p is illustrated (% Decapping), determined as described in the legend to Fig. 2. Zero length represents dinucleotide cap, m7Gp*ppG, which as illustrated is not hydrolyzed by Dcp2. (C) RNA length and substrate dependence of C. elegans Dcp2 on pGEM-7Zf polylinker-derived RNAs. m7Gp*ppG-capped RNAs of increasing length derived from the pGEM-7ZF polylinker were evaluated as substrates for C. elegans Dcp2. Note that for this substrate in comparison with the Renilla RNA (see B above), decapping activity becomes optimal on RNA as small as ∼50 nucleotides.
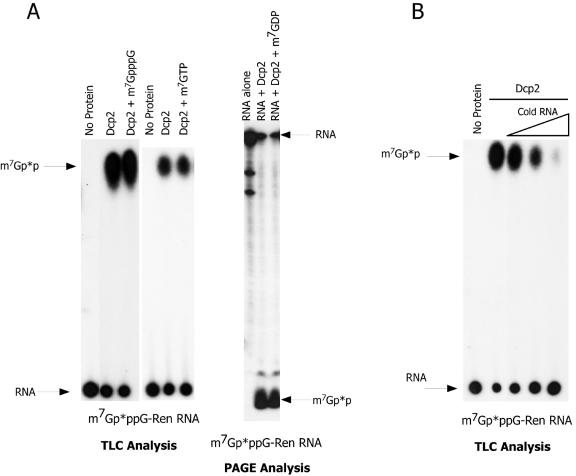
Consistent with a requirement for the RNA moiety attached to the cap, Dcp2 activity is not competitively inhibited by dinucleotide cap or methylated nucleotides added to reactions, as illustrated in the TLC and PAGE assays in Fig. 4A. In addition, Dcp2 does not bind to cap based on m7GTP-Sepharose affinity chromatography assays (data not shown). However, addition of capped or uncapped RNA as a competitor to Dcp2 decapping reactions effectively inhibited Dcp2 activity (Fig. 4B and 5B).
FIG. 4.
C. elegans Dcp2 is not inhibited by cap or cap nucleotide analogs but is effectively competed with 10- to 100-fold excess of cold RNA. (A) Effects of cap and cap analog on Dcp2 activity. C. elegans Dcp2 activity (50 ng of protein) was examined on a 250-nucleotide m7Gp*ppG cap-labeled RNA in the absence and presence of 200 μM m7GpppG or 200 μM of the cap analog m7GTP or m7GDP (often a more effective inhibitor of cap-binding proteins) at concentrations ∼50-fold higher than required to effectively inhibit other cap-binding proteins, such as eIF4E. Reaction products from the competition assays were resolved using both a TLC and PAGE assay. Note that these cap analogs (+ m7GpppG, + m7GDP, or + m7GTP) do not inhibit the decapping reactions (i.e., levels of m7GDP produced). (B) RNA is a competitor of Dcp2 activity. Dcp2 activity (50 ng of protein) on a 250-nucleotide m7Gp*pppG-capped Renilla RNA was evaluated in the absence and presence of increasing amounts of the same cold RNA. Two nanograms of labeled RNA was present in the reaction. Cold RNA competitor was thus present at equal, 10-fold, or 100-fold excess. Note that addition of competing RNA to the decapping reaction effectively inhibits Dcp2 decapping. Similar results were obtained using the Renilla RNA substrate and a nonhomologous firefly luciferase RNA as competitor RNA (data not shown).
FIG. 5.
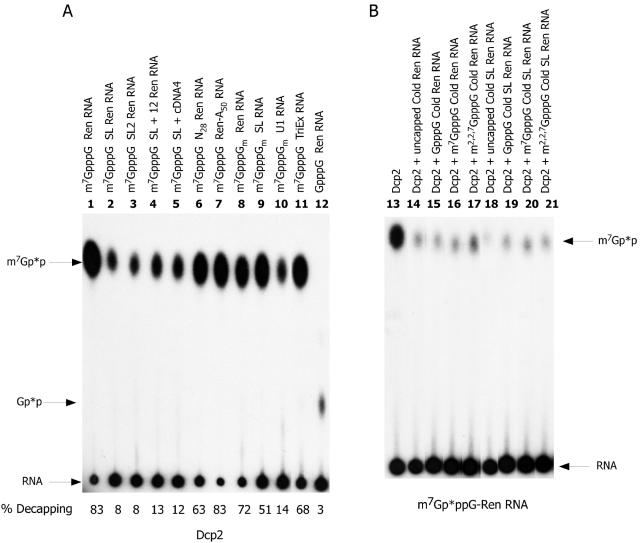
C. elegans Dcp2 activity is sequence and context dependent. (A) Sequence and context dependence of C. elegans Dcp2 activity. The illustrated RNAs were cap labeled, and equivalent amounts of RNA were used as substrates for C. elegans Dcp2 (50 ng of protein). Reactions were carried out in the linear range of both protein and RNA concentration. All RNAs are ∼250 nucleotides in length except for the SL RNA (108 nt), U1 RNA (170 nt), and TriEx RNA (1,900 nt). Reaction products were resolved on PEI-cellulose TLC with 0.45 M ammonium sulfate and visualized by autoradiography. Percent decapping of the substrate was determined as described in the legend to Fig. 2A. (B) RNA competition is independent of cap and SL sequence. Five nanograms of m7Gp*ppG cap-labeled 250-nucleotide Renilla RNA was incubated with 50 ng of Dcp2 protein in the presence or absence of 100 ng of the illustrated competitor RNAs. The competition assay was carried out in the linear range of both the assay and competitor RNA. RNA competition is generally equivalent regardless of the cap or presence of the SL sequence. Similar results were obtained using the Renilla RNA as the test substrate and a nonhomologous firefly luciferase RNA as the competitor RNA (data not shown).
RNA sequence and Dcp2 activity.
Approximately 70% of nematode mRNAs undergo maturation through trans splicing (27, 36, 61). This processing event forms the mature 5′ ends of mRNA by addition of a conserved 22-nucleotide sequence known as the spliced leader. To determine whether this sequence had any effect on C. elegans Dcp2 decapping in vitro, we compared Dcp2 activity on cap-labeled RNA that differed only by substitution of the first 22 nucleotides with the nematode SL1 sequence (GGUUUAAUUACCCAAGUUUGAG). Dcp2 decapping activity was reduced ∼10-fold on an RNA substrate with a 5′ terminal SL sequence (Fig. 5, compare lanes 1 and 2). This effect was also observed when a second C. elegans spliced leader (SL2, GGUUUUAACCCAGUUACUCAAG) that is 45% identical to SL1 (Fig. 5, compare lanes 1 and 3) was substituted for the 5′ end sequence of the Renilla RNA test substrate. Addition of the complete 5′ untranslated region (33 nt) of an SL1 trans-spliced mRNA (SL-12) to the test substrate also significantly reduced decapping activity (Fig. 5A, lane 4). Similar results were also observed with the 5′ 250 nucleotides of a native trans-spliced mRNA (lane 5). However, several control RNAs (e.g., actin and elongation factor) lacking an SL and the test Renilla RNA with either random nucleotides at the 5′ end or a 3′ 50-nucleotide poly(A) tail were efficiently decapped (lanes 6 to 7 and 11; also data not shown). At present it is not clear whether the sequence or a structural component of the SL is responsible for the decrease in decapping. However, M-Fold RNA analysis failed to detect a stable structure or extensive base pairing within the SL or between the SL and the 3′ RNA substrate. These data suggest that either the sequence or structure of the SL at the 5′ end of an RNA leads to a reduction in decapping.
We also tested two other small RNA substrates, the SL RNA and U1 RNA. The 108-nucleotide spliced leader RNA is the substrate utilized for the trans-splicing reaction. The first 22 nucleotides of this RNA are trans spliced to a pre-mRNA to form the 5′ end of a trans-spliced mRNA. Interestingly, the SL sequence at the 5′ end of SL RNA was cleaved with relatively high efficiency in comparison to other test RNAs with the same SL sequence at their 5′ ends (Fig. 5A, compare lanes 1, 2, and 9). This was unexpected, since the SL RNA substrate is predicted to have the spliced leader sequence in a relatively stable 5′ stem-loop structure (consisting of the first 30 nucleotides) including base pairing of the first two nucleotides (13). These attributes led us to predict that the SL RNA would be a poor Dcp2 substrate. However, the SL RNA is readily decapped in the assays. A second and somewhat larger small nuclear RNA (U1) whose 5′ end is predicted to not be base paired (the 5′ end of U1 is free to base pair with the 5′ spliced site in cis splicing) was also tested as a decapping substrate. In spite of the longer length (170 nt) and predicted absence of 5′-end base pairing or a stem-loop immediately adjacent to the cap, the U1 RNA was a relatively poor substrate for Dcp2 (Fig. 5A, compare lanes 1, 9, and 10). Overall, these data indicate that the decapping rate of different cellular RNAs is likely to vary.
Ten- to 25-fold molar excess cold competitor RNA added to a decapping reaction is an effective competitor and inhibits Dcp2 activity (Fig. 4B and 5B). However, neither the RNA sequence, type of cap, nor addition of the SL sequence to the RNA has a large effect in these competition experiments (Fig. 5B and data not shown). One interpretation of these data is that Dcp2 contains a general and independent RNA binding domain. Following RNA binding, subsequent nucleotide interactions and access to the cap within the cap-binding/cleavage pocket can influence catalysis.
Cap specificity for nematode decapping.
Addition of the spliced leader during trans splicing also brings a different cap to the 5′ ends of trans-spliced mRNAs, a trimethylguanosine cap (m2,2,7GpppN) compared to the typical m7GpppN cap (30, 37, 52). Seventy percent of nematode mRNAs have a m2,2,7GpppG cap, while 30% have the typical m7GpppN cap. To examine the cap specificity of nematode Dcp2, we prepared cap-labeled RNA substrates with Gp*ppG, m7Gp*ppG, m7Gp*ppGm, and m2,2,7Gp*ppGm caps. Illustration of these and other substrates (see below) and analysis of their RNA caps is provided in Fig. S3 in the supplemental material.
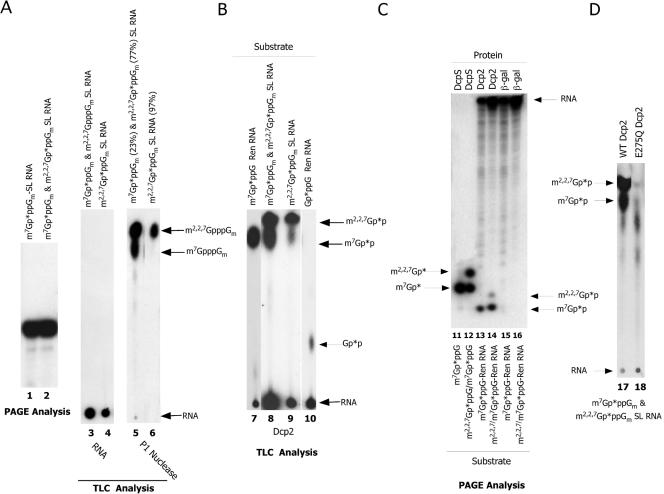
To produce cap-labeled trimethylguanosine substrates, m7GpppGm-capped SL RNA was incubated in Ascaris embryo whole-cell translation extracts (6, 37). Following this treatment, the RNA was intact (Fig. 6A, lanes 1 to 4), and ∼70% of the m7GpppGm-capped SL RNA was trimethylated (m2,2,7GpppGm-SL RNA), as deduced by nuclease P1 characterization of the resulting RNA cap structure (Fig. 6A, lane 5). To further enrich for TMG cap-labeled RNA, the RNA was immunoprecipitated using anti-TMG antibodies. Immunoprecipitation of the SL RNA produced SL RNA preparations that were typically 95% m2,2,7GpppGm-SL RNA based on P1 nuclease digestions and TLC analysis of the RNA cap (Fig. 6A, lane 6).
FIG. 6.
C. elegans Dcp2 hydrolyzes m2,2,7GpppGm-capped RNA. (A) RNA substrates used for decapping assays. PAGE analysis of m7Gp*ppGm-SL RNA (lane 1) prior to and after hypermethylation in whole-embryo nematode extract to the mixed products of m7Gp*ppGm- and m2,2,7Gp*ppGm-SL RNA (PAGE, lane 2; TLC, lane 3). The mixed m7Gp*ppGm- and m2,2,7Gp*ppGm-SL RNA was then immunoprecipitated with anti-TMG antibodies (see Materials and Methods) and evaluated by TLC analysis (lane 4). The mixed m7Gp*ppGm- and m2,2,7Gp*ppGm-SL RNA and the anti-TMG precipitated RNA were treated with P1 nuclease to remove the cap, and the cap products were characterized and identified by TLC analysis and autoradiography (lanes 5 and 6). ImageQuant was used to determine the percentages (see values in parentheses) of m7Gp*ppGm- versus m2,2,7Gp*ppGm-SL RNA before (lane 5) and after (lane 6) anti-TMG antibody precipitation. (B) C. elegans Dcp2 activity on different RNA caps. Dcp2 reactions were carried out using 50 ng of Dcp2 with m7Gp*ppG-Ren RNA, the mixed m7Gp*ppGm- and m2,2,7Gp*ppGm-SL RNA substrate, the anti-TMG precipitated m2,2,7Gp*ppGm-SL RNA, and Gp*pppG-Ren RNA. Reactions products were resolved and visualized as described above. (C) PAGE analysis of DcpS and Dcp2 decapping products to confirm comigration of the identified products with known standards. (D) The E275Q mutation in nematode Dcp2 1-479 eliminates most decapping of the m2,2,7Gp*ppGm-SL RNA as well as m7Gp*ppGm-SL RNA. Reactions were carried out and the products resolved and visualized as described above.
m7GpppG-capped RNAs are readily cleaved by nematode Dcp2. In higher eukaryotes, the RNA cap typically includes methyl groups on the 2′-ribose of the first and second encoded bases, cap 1 and cap 2, respectively. Addition of a 2′-O-ribose methyl group on the first encoded base did not have a significant effect on nematode Dcp2 activity (Fig. 5A, compare lanes 1 and 8). However, the presence of an N-7 methyl group is an important determinant for nematode decapping, since its absence leads to a significant reduction of decapping activity (evidenced by production of only small amounts of Gp*p product derived from Gp*ppG-capped RNA) (Fig. 5A, compare lanes 1 and 12; Fig. 6B, compare lanes 7 and 10). m2,2,7GpppGm on the SL RNA (and U1 RNA; data not shown) is also decapped, producing m2,2,7Gp*p (Fig. 6B, lanes 8 and 9). The diphosphate products produced by Dcp2 decapping of GpppG, m7GpppG-, m7GpppGm, and m2,2,7GpppG m RNAs were further characterized by comigration with markers on PAGE analysis (Fig. 6C, lanes 11 to 14) and the ability to convert the diphosphate products to triphosphates with nucleoside diphosphate kinase treatment (data not shown). These analyses confirm that the primary products derived from all these cap substrates are the diphosphates of their capped RNA substrates. Therefore, nematode Dcp2 cleaves m7GpppG-, m7GpppGm-, and m2,2,7GpppGm-capped RNAs. GpppG-capped RNAs are also cleaved, but at a much-reduced efficiency (Fig. 6B, compare lanes 7 and 10). Importantly, the mutation in the nudix motif (E275Q) of nematode Dcp2 also abolishes most of the m2,2,7GpppGm decapping activity (Fig. 6D). Quantitative comparison of decapping of m7GpppGm and m2,2,7GpppGm in the mixed substrate indicates that nematode Dcp2 can hydrolyze m7GpppGm- and m2,2,7GpppGm-capped RNA at the same efficiency.
Human and budding yeast Dcp2 is active on m2,2,7GpppGm-capped RNAs.
We previously demonstrated that two nematode cap-interacting proteins (eIF4E and DcpS) can bind and function on both m7GpppG and m2,2,7GpppG caps, whereas their mammalian counterparts cannot (6, 27). In a comparative study we determined the abilities of human and yeast Dcp2 enzymes to hydrolyze m2,2,7GpppG-capped RNA. Unexpectedly, incubation of human Dcp2 with m2,2,7GpppG-capped RNA resulted in the hydrolysis of the TMG cap-containing RNA, producing m2,2,7Gpp, as illustrated by-product migration in TLC analysis (Fig. 7A). The identity of the diphosphate product was further confirmed by its conversion to its corresponding triphosphate with NDPK (Fig. 7B). In addition, budding yeast Dcp1/Dcp2 was also able to cleave m2,2,7GpppG-capped RNA, producing m2,2,7Gpp (Fig. 7C). These data indicate that the ability to cleave m2,2,7GpppG-capped RNA is phylogenetically conserved in diverse Dcp2 proteins. The human enzyme cleaves m7GpppGm- and m2,2,7GpppGm-capped RNA with equivalent efficiencies, whereas the yeast enzyme is ∼30% less efficient on m2,2,7GpppGm-capped RNA. snRNAs and some snoRNAs have trimethylguanosine caps. Thus, in addition to its function in decapping cellular mRNAs, Dcp2 may also function in decapping and the turnover of snRNAs and some snoRNAs.
FIG. 7.
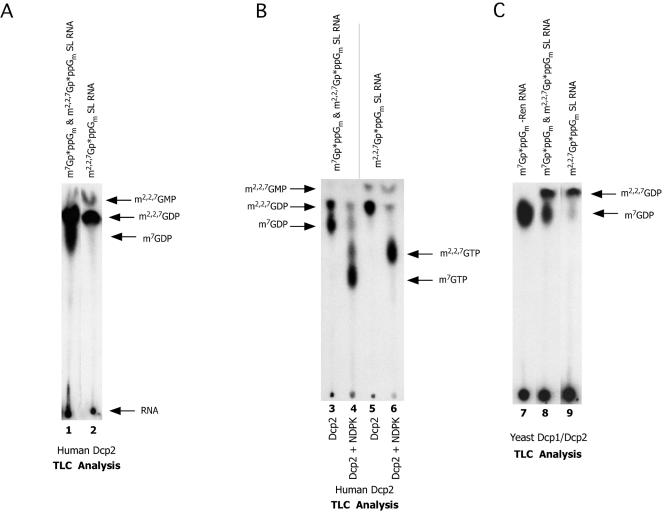
Human and yeast Dcp2 effectively hydrolyze m2,2,7GpppGm-capped RNA. (A) Human Dcp2 decaps m2,2,7GpppGm-capped RNA. Ni2+-NTA purified human Dcp2 (300 ng) reactions were carried out at 37°C in the presence of both Mg2+ and Mn2+ ions with the illustrated substrates as described in Materials and Methods. The reaction products were resolved by PEI-cellulose TLC and visualized by autoradiography. (B) Human Dcp2 decapping products are nucleotide diphosphates. Human Dcp2 decapping products (lanes 3 and 5) were treated with nucleoside diphosphate kinase (lanes 4 and 6) and the products of the reactions resolved on PEI-cellulose TLC and visualized with autoradiography. Note that the majority of the reaction products in lanes 3 and 5 are converted to their corresponding diphosphates in lanes 4 and 6. The reactions did not proceed to completion, leaving some diphosphates unconverted. (C) Yeast Dcp1/Dcp2 decaps m2,2,7GpppGm-capped RNA. Coexpressed and copurified budding yeast Dcp1/Dcp2 reactions were carried out at 30°C in the presence of both Mg2+ and Mn2+ ions with the illustrated substrates as described in Materials and Methods. The reaction products were resolved by PEI-cellulose TLC and visualized by autoradiography.
DISCUSSION
We have shown that nematode Dcp2 is an RNA-decapping enzyme and functions in the absence of C. elegans Dcp1. The optimal substrate length of C. elegans Dcp2 is ∼50 nucleotides or greater, but this length requirement may also be substrate dependent. C. elegans Dcp2 does not directly bind to cap and is not inhibited by cap or cap analogs. RNA, however, is an effective competitive inhibitor of Dcp2 activity. Nematode Dcp2 is therefore similar in several respects to human and yeast Dcp2 (42, 49). The effect of RNA competition on nematode Dcp2 activity is not significantly affected by the RNA sequence, cap, or addition of the spliced leader sequence. However, the 5′ terminal sequence of the RNA substrate can influence decapping activity. These data indicate that nematode Dcp2 is a general RNA binding protein, that initial RNA binding is independent of the cap, and that decapping activity can be influenced by 5′ terminal sequence.
Influence of the SL sequence on decapping.
Addition of a nematode spliced leader sequence to the 5′ end of a test RNA leads to an ∼10-fold reduction in Dcp2 decapping. The reduction in Dcp2 activity appears context dependent, since the spliced leader on the SL RNA is a relatively good substrate for decapping. Competing RNA with the SL sequence did not show any increased effectiveness as a competitor. These data suggest that C. elegans Dcp2 first binds RNA (perhaps in a nonspecific manner), and subsequent decapping can be influenced by the context of the RNA elements near the 5′ end of the RNA. This view suggests the protein has at least two independent domains, one for RNA binding and the second for cap cleavage. The catalytic activity of the latter domain can be influenced by both sequence and context. Steiger et al. found that annealing a DNA oligonucleotide to the 5′ end of an RNA leading to an RNA/DNA duplex within the first 25 nt reduced yeast decapping by ∼5-fold (17, 49). These data suggest that sequestering the 5′ end of the RNA can inhibit decapping. Yeast Dhhp1, an RNA helicase, has also been shown to be an enhancer of yeast decapping (8, 17), supporting the idea that access to the cap or 5′ end of the RNA can be an important determinant in decapping. Our data provide the first evidence that a specific sequence and its context at the 5′ end of an RNA can influence decapping.
Reduced decapping of mRNAs with a spliced leader sequence has implications for nematode mRNA metabolism. Since 70% of the mRNAs in C. elegans and Ascaris have a spliced leader sequence (27, 36, 61), trans-spliced mRNAs might typically be subject to limited 5′-to-3′ decay due to reduced decapping of the SL mRNAs. Our in vitro decay data in Ascaris whole-cell embryo extracts indicate that the 3′-to-5′ decay pathway is at least 15 times more active than the 5′-to-3′ decay pathway (6). Dcp2 decapping followed by 5′-to-3′ decay might be a more highly regulated pathway. Under this scenario, Dcp2 decapping of SL-containing mRNAs might be limited but could be influenced and regulated by trans-interacting proteins. Several decapping enhancing proteins have been identified and characterized in yeast (8, 15, 17, 25, 46).
The presence of the SL sequence at the 5′ end of an RNA appears to have differential effects on Dcp2 decapping. When present on the SL RNA, decapping does not appear significantly limited in spite of the known secondary structure of the SL RNA. These data suggest that both sequence and context are important determinants for decapping. Therefore, individual trans-spliced mRNAs could contain additional sequence elements that can influence decapping. Notably, both in vitro and in vivo we do not see significant differences in the mRNA half-lives of test transcripts comparing a test RNA with or without a spliced leader and with a trimethylguanosine cap (27) (L. S. Cohen et al., submitted for publication). This further suggests that under the conditions of the experiments, 3′-to-5′ decay may be predominant, and that our test RNA does not contain sequences that facilitate enhanced decapping in the presence of the SL.
We have also observed a reduction of decapping efficiency on the nematode U1 RNA. The 5′-terminal 12 nucleotides of the RNA are predicted to not be base paired (this is a phylogenetically conserved property of U1 RNAs) and are thus not a likely explanation for reduced decapping on this substrate. Reduced decapping of U1 RNA could be due to sequence or structural elements that effect either initial RNA binding or catalysis or both. One possibility is that Dcp2 requires more than 12 unpaired nucleotides for access to or catalysis of the cap. Both U1 and the SL RNA are predicted to form significant secondary structures during the decapping incubation period. Little is known regarding the tertiary structure of these RNAs that might influence decapping.
Dcp2 is a TMG-decapping enzyme.
Nematode Dcp2 has limited activity on GpppG-capped RNA substrates. Dcp2 activity is significantly greater on m7GpppG-capped RNA, indicating that the N-7 methyl group is an important determinant for decapping activity. Similar observations have been made for both the budding yeast and human enzymes (26, 53, 57). C. elegans Dcp2 does not appear to be significantly influenced by a 2′-O-methyl (cap 1). This is consistent with the presence of at least a cap 1 in higher eukaryotes. Budding yeast, however, are thought to not have a cap 1 (35, 48). Yeast Dcp2 is also capable of hydrolyzing m7GpppGm-capped RNA (data not shown). This suggests that at least 2′-O-methyl ribose residues are not likely to influence decapping.
Nematode Dcp2 is also equally active in the hydrolysis of m2,2,7GpppGm-capped RNA. Given that a large percentage of nematode mRNAs have a trimethylguanosine cap, we predicted that the nematode enzyme was likely to be functional on this cap structure. We have observed that other cap-interacting proteins in nematodes (DcpS and eIF4E) are capable of interacting with both m7GpppG and m2,2,7GpppG caps (6, 27). In contrast, the human orthologs of these proteins have limited affinity and activity on m2,2,7GpppG caps, suggesting that the nematode enzymes have evolved to accommodate mRNAs with m2,2,7GpppG caps. Unexpectedly, we observed that both the human and budding yeast Dcp2 are very efficient in hydrolyzing m2,2,7GpppG-capped RNA. Cleavage of m2,2,7GpppG-capped RNA is thus a phylogenetically conserved property of Dcp2 proteins. Furthermore, since snRNA and some snoRNAs have hypermethylated caps, Dcp2 proteins may be involved in the decapping and turnover of sn- and snoRNAs.
Notably, Dcp2 activity is also substrate dependent, as illustrated by the much lower efficiency of decapping U1 snRNA than that for the SL RNA. Whether the greater activity on the SL RNA in vitro is reflected in higher turnover rates in vivo remains to be determined. During the trans-splicing reaction, the SL RNA is turned over as it is consumed in the reaction. SL RNA substrate levels might require tight regulation, since either excess or insufficient SL RNA substrate could be deleterious to the cells. Decapping of the SL RNA might be necessary under normal or particular conditions to maintain an appropriate level of the SL RNA substrate in cells. In addition, regulated turnover of the SL RNA substrate by Dcp2 could lead to global regulation of gene expression of trans-spliced genes. Nematode Dcp2 is active on the assembled SL RNP, since we previously demonstrated that SL RNAs assembled into snRNPs in cell extracts are substrates for Dcp2 decapping as well as 3′-to-5′ decay (6). Overall, our data suggest that Dcp2 proteins can efficiently decap TMG-capped RNAs and may be involved in the turnover of RNAs involved in several metabolic pathways.
Dcp1/Dcp2 have been primarily localized by several groups in yeast and mammalian cells throughout the cytoplasm and in discrete cytoplasmic foci (P-bodies or processing bodies), although nuclear staining has also been observed (9, 22, 32, 33, 51, 53). Dcp2 is primarily cytoplasmic and is present within discrete foci (both P-bodies and P-granules) in early embryos of C. elegans (S. Lall, F. Piano, and R. Davis, submitted for publication). C. elegans nuclear staining is not prominent, but it is detectable. Overall, these data suggest that Dcp2 is also present in the nuclei of cells and thus could participate in sn- and snoRNA turnover. Alternatively, as some snRNAs undergo maturation in the cytoplasm, Dcp2 could function on these RNAs during their cytoplasmic maturation.
Recently, a vertebrate U8 snoRNA binding protein with decapping activity was identified (19). This decapping protein is primarily nucleolar in localization and decaps GpppG-, m7GpppG-, and m2,2,7GpppG-capped RNAs, generating primarily cap-derived diphosphate products in the presence of the Mn2+ cation. Different decapping efficiencies were noted for different RNA substrates, including U8 snoRNA, U3 snoRNA, and 5S RNA. However, homologs of this protein are not readily identifiable outside of the vertebrate lineage. Thus, it seems likely that Dcp2 may be involved in the turnover of sn- and snoRNAs in nonvertebrate lineages.
Nematode Dcp1.
Several lines of evidence indicate that Dcp2 is the primary catalytic subunit of the Dcp1/Dcp2 complex (31, 33, 53). We have not been able to detect any significant decapping activity using a variety of substrates (several RNAs containing different caps or free caps) and assay conditions (including Mn2+) with either bacterially expressed or yeast-expressed C. elegans Dcp1. Using similar assay conditions, human Dcp1 is also inactive in vitro (33, 53). More recently it has been suggested that in yeast Dcp1 is an enhancer of Dcp2 activity (7, 44, 47, 49). Using several RNA substrates with different caps, we have not consistently observed significant enhancement of Dcp2 activity with Dcp1. Recently, overexpression of human Dcp1 in mammalian cells also was not found to significantly enhance decapping in vivo (34). It remains to be determined whether posttranslational modifications, specific assay conditions, or auxiliary factors are required to enable Dcp1 to enhance Dcp2 in higher eukaryotes.
Expression of truncated forms of Dcp2.
The C. elegans WormBase predicted two different RNAs and proteins that would lack the carboxy-terminal portion of the Nudix domain and Box B. We have identified RNAs by RT-PCR that would encode these proteins and have expressed and purified these proteins from bacteria. However, these proteins are catalytically inactive in our in vitro assays. What role these proteins might play or if they are catalytically active in vivo remains to be determined.
Conclusions.
Nematode Dcp2 is an RNA-decapping enzyme that does not bind or function directly on caps but is effectively competed against by RNA. Our data suggest Dcp2 has at least two domains: the first binds to the substrate RNA, and the second is involved in catalysis of the decapping reaction. Notably, nematode Dcp2 decapping is influenced by both RNA sequence and context that appears independent of the initial RNA binding step. Finally, phylogenetically diverse Dcp2 proteins are capable of decapping TMG-capped RNAs and may therefore be involved in sn- and snoRNA turnover. Consequently, Dcp2 activity may influence several metabolic pathways.
Supplementary Material
Acknowledgments
A dual expression plasmid containing recombinant vaccinia RNA guanylyltransferase and (guanine-N7)-methyltransferase was generously provided by Stewart Shuman, human RNA guanylyltransferase by Aaron Shatkin, mRNA cap-specific 2′-O-methyltransferase by Paul Gershon, and cap analogs and nucleotides by Edward Darzynkiewicz. Purified yeast Dcp1/Dcp2 was generously provided by Carolyn Decker and Roy Parker. We thank Jens Lykke-Andersen and members of the Davis lab for their comments on the manuscript.
This work was supported by NIH grant AI49558 and CUNY-CSI startup funds to R.E.D and by NIH grant GM67005 to M.K.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baker, K. E., and R. Parker. 2004. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr. Opin. Cell Biol. 16:293-299. [DOI] [PubMed] [Google Scholar]
- 2.Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield, D. M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382:642-646. [DOI] [PubMed] [Google Scholar]
- 3.Bergman, N., M. Opyrchal, E. J. Bates, and J. Wilusz. 2002. Analysis of the products of mRNA decapping and 3′-to-5′ decay by denaturing gel electrophoresis. RNA 8:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumenthal, T. 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11:132-136. [DOI] [PubMed] [Google Scholar]
- 5.Bringmann, P., J. Rinke, B. Appel, R. Reuter, and R. Luhrmann. 1983. Purification of snRNPs U1, U2, U4, U5 and U6 with 2,2,7-trimethylguanosine-specific antibody and definition of their constituent proteins reacting with anti-Sm and anti-(U1)RNP antisera. EMBO J. 2:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, L. S., C. Mikhli, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R. E. Davis. 2004. Nematode m7GpppG and m3(2,2,7)GpppG decapping: activities in Ascaris embryos and characterization of C. elegans scavenger DcpS. RNA 10:1609-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coller, J., and R. Parker. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861-890. [DOI] [PubMed] [Google Scholar]
- 8.Coller, J. M., M. Tucker, U. Sheth, M. A. Valencia-Sanchez, and R. Parker. 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7:1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cougot, N., S. Babajko, and B. Seraphin. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cougot, N., E. van Dijk, S. Babajko, and B. Seraphin. 2004. ‘Cap-tabolism.’ Trends Biochem. Sci. 29:436-444. [DOI] [PubMed] [Google Scholar]
- 11.Davis, R. E. 1996. Spliced leader RNA trans-splicing in metazoa. Parasitol. Today 12:33-40. [DOI] [PubMed] [Google Scholar]
- 12.Davis, R. E., A. Parra, P. T. LoVerde, E. Ribeiro, G. Glorioso, and S. Hodgson. 1999. Transient expression of DNA and RNA in parasitic helminths by using particle bombardment. Proc. Natl. Acad. Sci. USA 96:8687-8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denker, J. A., P. A. Maroney, Y. T. Yu, R. A. Kanost, and T. W. Nilsen. 1996. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA 2:746-755. [PMC free article] [PubMed] [Google Scholar]
- 14.Dunckley, T., and R. Parker. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunckley, T., M. Tucker, and R. Parker. 2001. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics 157:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fillman, C., and J. Lykke-Andersen. 2005. RNA decapping inside and outside of processing bodies. Curr. Opin. Cell Biol. 17:326-331. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, N., and K. Weis. 2002. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 21:2788-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, M., C. J. Wilusz, S. W. Peltz, and J. Wilusz. 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 20:1134-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh, T., B. Peterson, N. Tomasevic, and B. A. Peculis. 2004. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol. Cell 13:817-828. [DOI] [PubMed] [Google Scholar]
- 20.Gu, M., C. Fabrega, S. W. Liu, H. Liu, M. Kiledjian, and C. D. Lima. 2004. Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity. Mol. Cell 14:67-80. [DOI] [PubMed] [Google Scholar]
- 21.Gu, M., and C. D. Lima. 2005. Processing the message: structural insights into capping and decapping mRNA. Curr. Opin. Struct. Biol. 15:99-106. [DOI] [PubMed] [Google Scholar]
- 22.Ingelfinger, D., D. J. Arndt-Jovin, R. Luhrmann, and T. Achsel. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8:1489-1501. [PMC free article] [PubMed] [Google Scholar]
- 23.Khanna, R., and M. Kiledjian. 2004. Poly(A)-binding-protein-mediated regulation of hDcp2 decapping in vitro. EMBO J. 23:1968-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khodursky, A. B., and J. A. Bernstein. 2003. Life after transcription—-revisiting the fate of messenger RNA. Trends Genet. 19:113-115. [DOI] [PubMed] [Google Scholar]
- 25.Kshirsagar, M., and R. Parker. 2004. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166:729-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaGrandeur, T. E., and R. Parker. 1998. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 17:1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lall, S., C. C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R. E. Davis. 2004. Contribution of trans-splicing, 5′-leader length, cap-poly(A) synergism, and initiation factors to nematode translation in an Ascaris suum embryo cell-free system. J. Biol. Chem. 279:45573-45585. [DOI] [PubMed] [Google Scholar]
- 28.Lejeune, F., X. Li, and L. E. Maquat. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12:675-687. [DOI] [PubMed] [Google Scholar]
- 29.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell. 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liou, R. F., and T. Blumenthal. 1990. trans-spliced Caenorhabditis elegans mRNAs retain trimethylguanosine caps. Mol. Cell. Biol. 10:1764-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, H., N. D. Rodgers, X. Jiao, and M. Kiledjian. 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 21:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, S. W., X. Jiao, H. Liu, M. Gu, C. D. Lima, and M. Kiledjian. 2004. Functional analysis of mRNA scavenger decapping enzymes. RNA 10:1412-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22:8114-8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lykke-Andersen, J., and E. Wagner. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mager, W. H., J. Klootwijk, and I. Klein. 1976. Minimal methylation of yeast messenger RNA. Mol. Biol. Rep. 3:9-17. [DOI] [PubMed] [Google Scholar]
- 36.Maroney, P. A., J. A. Denker, E. Darzynkiewicz, R. Laneve, and T. W. Nilsen. 1995. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA 1:714-723. [PMC free article] [PubMed] [Google Scholar]
- 37.Maroney, P. A., G. J. Hannon, and T. W. Nilsen. 1990. Transcription and cap trimethylation of a nematode spliced leader RNA in a cell-free system. Proc. Natl. Acad. Sci. USA 87:709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilsen, T. W. 2001. Evolutionary origin of SL-addition trans-splicing: still an enigma. Trends Genet. 17:678-680. [DOI] [PubMed] [Google Scholar]
- 39.Nilsen, T. W. 1993. Trans-splicing of nematode premessenger RNA. Annu. Rev. Microbiol. 47:413-440. [DOI] [PubMed] [Google Scholar]
- 40.Nuss, D. L., Y. Furuichi, G. Koch, and A. J. Shatkin. 1975. Detection in HeLa cell extracts of a 7-methyl guanosine specific enzyme activity that cleaves m7GpppNm. Cell 6:21-27. [DOI] [PubMed] [Google Scholar]
- 41.Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121-127. [DOI] [PubMed] [Google Scholar]
- 42.Piccirillo, C., R. Khanna, and M. Kiledjian. 2003. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA 9:1138-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pouchkina-Stantcheva, N. N., and A. Tunnacliffe. 2005. Spliced leader RNA-mediated trans-splicing in phylum Rotifera. Mol. Biol. Evol. 22:1482-1489. [DOI] [PubMed] [Google Scholar]
- 44.Sakuno, T., Y. Araki, Y. Ohya, S. Kofuji, S. Takahashi, S. Hoshino, and T. Katada. 2004. Decapping reaction of mRNA requires Dcp1 in fission yeast: its characterization in different species from yeast to human. J. Biochem. (Tokyo) 136:805-812. [DOI] [PubMed] [Google Scholar]
- 45.Salehi, Z., L. Geffers, C. Vilela, R. Birkenhager, M. Ptushkina, K. Berthelot, M. Ferro, S. Gaskell, I. Hagan, B. Stapley, and J. E. McCarthy. 2002. A nuclear protein in Schizosaccharomyces pombe with homology to the human tumour suppressor Fhit has decapping activity. Mol. Microbiol. 46:49-62. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz, D., C. J. Decker, and R. Parker. 2003. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA 9:239-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.She, M., C. J. Decker, K. Sundramurthy, Y. Liu, N. Chen, R. Parker, and H. Song. 2004. Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat. Struct. Mol. Biol. 11:249-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sripati, C. E., Y. Groner, and J. R. Warner. 1976. Methylated, blocked 5′ termini of yeast mRNA. J. Biol. Chem. 251:2898-2904. [PubMed] [Google Scholar]
- 49.Steiger, M., A. Carr-Schmid, D. C. Schwartz, M. Kiledjian, and R. Parker. 2003. Analysis of recombinant yeast decapping enzyme. RNA 9:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevens, A. 1988. mRNA-decapping enzyme from Saccharomyces cerevisiae: purification and unique specificity for long RNA chains. Mol. Cell. Biol. 8:2005-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tharun, S., W. He, A. E. Mayes, P. Lennertz, J. D. Beggs, and R. Parker. 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404:515-518. [DOI] [PubMed] [Google Scholar]
- 52.Thomas, J. D., R. C. Conrad, and T. Blumenthal. 1988. The C. elegans trans-spliced leader RNA is bound to Sm and has a trimethylguanosine cap. Cell 54:533-539. [DOI] [PubMed] [Google Scholar]
- 53.van Dijk, E., N. Cougot, S. Meyer, S. Babajko, E. Wahle, and B. Seraphin. 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21:6915-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Dijk, E., H. Le Hir, and B. Seraphin. 2003. DcpS can act in the 5′-3′ mRNA decay pathway in addition to the 3′-5′ pathway. Proc. Natl. Acad. Sci. USA 100:12081-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Doren, K., and D. Hirsh. 1990. mRNAs that mature through trans-splicing in Caenorhabditis elegans have a trimethylguanosine cap at their 5′ termini. Mol. Cell. Biol. 10:1769-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasudevan, S., S. W. Peltz, and C. J. Wilusz. 2002. Non-stop decay—-a new mRNA surveillance pathway. Bioessays 24:785-788. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Z., X. Jiao, A. Carr-Schmid, and M. Kiledjian. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. USA 99:12663-12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, Z., and M. Kiledjian. 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107:751-762. [DOI] [PubMed] [Google Scholar]
- 59.Wilusz, C. J., and J. Wilusz. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 20:491-497. [DOI] [PubMed] [Google Scholar]
- 60.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 61.Zorio, D. A., N. N. Cheng, T. Blumenthal, and J. Spieth. 1994. Operons as a common form of chromosomal organization in C. elegans. Nature 372:270-272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.