Abstract
We show that histone-DNA interactions are disrupted across entire yeast heat shock genes upon their transcriptional activation. At HSP82, nucleosomal disassembly spans a domain of ∼3 kb, beginning upstream of the promoter and extending through the transcribed region. A kinetic analysis reveals that histone H4 loses contact with DNA within 45 s of thermal upshift. Nucleosomal reassembly, prompted by temperature downshift, is also rapid, detectable within 60 s. Prior to their eviction, promoter-associated histones are transiently hyperacetylated, while those in the coding region are not. An upstream activation sequence mutation that weakens the binding of heat shock factor obviates domain-wide remodeling, while deletion of the TATA box that nearly abolishes transcription is permissive to 5′-end remodeling. The Swi/Snf complex is rapidly recruited to HSP82 upon heat shock. Nonetheless, domain-wide remodeling occurs efficiently in Swi/Snf mutants despite a sixfold reduction in transcription; it is also seen in gcn5Δ, set1Δ, and paf1Δ mutants. Contrary to current models, we demonstrate that a high density of RNA polymerase (Pol) is insufficient to elicit histone displacement. This finding suggests that histone eviction is modulated by factors that are not linked to elongating Pol II. It further suggests that histone depletion plays a causal role in mediating vigorous transcription in vivo and is not merely a consequence of it.
The genomes of eukaryotic cells are packaged into a protein-DNA complex termed chromatin, whose basic structural unit is the nucleosome. Each nucleosome core, which consists of 147 bp of DNA wrapped in nearly two left-handed supercoils around an octamer of histones (55), is assembled from one H3/H4 tetramer and two H2A/H2B dimers (20). The tripartite nature of the nucleosome, in addition to facilitating its assembly onto newly replicated DNA (36), also facilitates its disassembly during such processes as transcription, replication, recombination, and repair (37, 41; for reviews, see references 7 and 78).
Chromatin is intimately involved in the regulation of transcription, and there is much evidence to suggest that its remodeling, particularly over regulatory DNA sequences, plays a key role in both the activation and repression of genes. For example, the regulatory regions of genes poised for or actively engaged in transcription are hypersensitive to digestion by nucleases such as DNase I (for reviews, see references 21 and 30). Such nuclease-hypersensitive sites tend to contain hyperacetylated histones and be depleted of nucleosomes (9, 47, 49, 67). In budding yeast, the promoters of heat shock genes are assembled into constitutive DNase I-hypersensitive sites (22, 83). These sites contain both acetylated and methylated histones under noninducing conditions (16, 76; this study), suggesting that they are comprised of remodeled nucleosomes.
Surprisingly little is known of the chromatin structure of the coding regions of actively transcribed genes, although the great weight of evidence suggests that core histones, particularly H3 and H4, are present. Early studies of chick chromatin revealed that the transcription units of active genes are highly sensitive to digestion by DNase I (27, 89). These observations suggested that active genes might be associated with structurally altered, unfolded nucleosomes. Indeed, chromatin enriched in transcriptionally active sequences exhibits enhanced accessibility of histone H3 sulfhydryl groups and is selectively retained on organomercurial agarose columns (66). In addition, active transcription units may be assembled into nucleosomes that are depleted of H2A and H2B. This is suggested by the fact that nucleosome core particles deficient in H2A and H2B are strongly enriched for transcribed DNA sequences and are preferentially bound by RNA polymerase II (Pol II) (6). More direct evidence for H2A/H2B depletion has come from the finding that transcription of an in vitro-reconstituted mononucleosome results in the loss of one H2A/H2B dimer (41) and recent in vivo evidence that Pol II transcription of coding sequences induces H2A/H2B exchange with free histone pools in Physarum (84).
Actively transcribed chromatin is also enriched in hyperacetylated core histones. This has been shown by nuclease digestion (75), biochemical fractionation (35, 88), and chromatin immunoprecipitation (ChIP) assays (72). One role for this acetylation may be to unfold the nucleosomal fiber, rendering regulatory sites more accessible and transcriptional units more pliable (for a review, see reference 19). Moreover, actively transcribed regions are hypermethylated at Lys4 and Lys79 of H3 (62, 68, 72). These marks may promote the association of coactivators (69) and/or suppress the binding of corepressors (86). Interestingly, transcription-associated histone acetylation is restricted to the promoter regions of two inducible yeast genes, HIS3 and HO (44, 48), suggesting that the correlation between coding region histone modification and transcriptional activation might not always exist.
Cells adapt to stress by rapidly and selectively inducing the transcription of specific genes. The broadly conserved heat shock factor (HSF) activates heat shock protein genes in eukaryotic cells in response to thermal, chemical, or oxidative stress; exposure to heavy metals; and glucose starvation (33, 60, 91). Yeast HSF, encoded by the essential HSF1 gene, contains two potent activation domains (63, 80) and possesses the unusual property of being able to activate transcription in the absence of a number of critical coactivators and general transcription factors. These include TFIIA, TAF9 (TAFII17, a subunit of both TFIID and SAGA), the TFIIH kinase Kin28, the Med17 and Med22 (Srb4 and Srb6) subunits of Mediator (5, 13, 50, 59), and even the carboxyl-terminal domain (CTD) of Pol II (57). Thus, the mechanism by which HSF activates its target genes is of unusual interest.
Here, we have used ChIP to investigate the occupancy and modification state of core histones at HSF-regulated genes in Saccharomyces cerevisiae. In response to heat shock, promoters and coding regions of heat shock genes rapidly lose contact with all histone isoforms. In response to attenuating conditions, histones promptly rebind DNA. At HSP82, heat shock-induced histone loss is strictly dependent upon HSF, yet histone depletion within the gene’s promoter region occurs independently of elevated transcription. Domain-wide histone loss also occurs independently of the Swi/Snf ATP-dependent chromatin remodeling complex, despite severely impaired transcription, and is unaffected by mutations in prominent histone modification and transcription elongation complexes. While a high density of Pol II is seen at the induced HSP82 gene (in both Swi/Snf+ and Swi/Snf − contexts), we provide evidence that elongating Pol II is not of itself sufficient to elicit histone displacement.
MATERIALS AND METHODS
Yeast strains.
All strains used in this study (Table 1) are derived from the HSP82+ strain SLY101 (53). In vitro mutagenesis, in combination with two-step gene transplacement, was performed as previously described (24, 56) to introduce a 19-bp substitution mutation into the HSP82 core promoter. The wild-type sequence, TAAAACATATAAATATGCA (−145 to −127, relative to AUG), was replaced by the mutant sequence, GATCTCCTTTAGCTTCTCG (found within the PET56 open reading frame [ORF]), creating an allele termed hsp82-ΔTATA (strain DSG118). The allele termed hsp82-P2, present in strain DSG101, bears a 2-bp substitution mutation in the high-affinity HSF site HSE1. The P2 mutation has been extensively characterized (24, 52, 56). The snf2Δ deletion was engineered into SLY101 via one-step transplacement with the KANr cassette (31), creating strain JHD204. The knockout was confirmed by genomic PCR. To create a swi1Δ chromosomal knockout, we crossed a swi1Δ::LEU2 allele into strain EAS2001, creating strain DGY101. Note that EAS2001 contains the hsp82-2001 allele, whose basal and induced transcription is indistinguishable from that of HSP82+ (77).
TABLE 1.
Yeast strains used
| Strain | Genotype | Reference or source |
|---|---|---|
| SLY101 | MATα ade−can1-100 his3-11,15 leu2-3,112, trp1-1 ura3 cyh2r | 53 |
| SLY102 | SLY101; hsp82Δ::CYH2S | 53 |
| DSG101 | SLY101; hsp82-P2 | 24 |
| DSG118 | SLY101; hsp82-ΔTATA | This study |
| JHD201 | SLY101; gcn5Δ | This study |
| JHD202 | SLY101; set1Δ | This study |
| JHD203 | SLY101; paf1Δ | This study |
| JHD204 | SLY101; snf2Δ | This study |
| EAS2001 | SLY101; sir4Δ2::HIS3 hsp82-2001 | 77 |
| DGY101 | EAS2001; swi1Δ | This study |
| JHD105 | EAS2001; snf2-9xMyc | This study |
| W303-1B | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. J. Rothstein |
| T2 | W303-1B; hsp82-T2 | 52; W. T. Garrard |
To C-terminally tag the chromosomal SNF2 gene with the 9-Myc epitope, we performed a one-step gene transplacement of strain EAS2001. The transforming DNA was PCR amplified using pWZV87 as a template (42). To permit homologous recombination, forward and reverse PCR primers were appended with 45 bp of SNF2 ORF and downstream flanking sequences, respectively. Proper targeting of the SNF2-9Myc-KlTRP1 fragment was confirmed by genomic PCR, creating strain JHD105. To permit expression of Myc-tagged histone H4, strains used in the pertinent ChIP assays (see Fig. 3, 4A, 4B, 5D, 6B, 7C, and 9A) were transformed with an episomal myc-HHF2 gene (borne on the TRP1-CEN6-ARS4 plasmid pNOY436; a gift of E. A. Sekinger and K. Struhl).
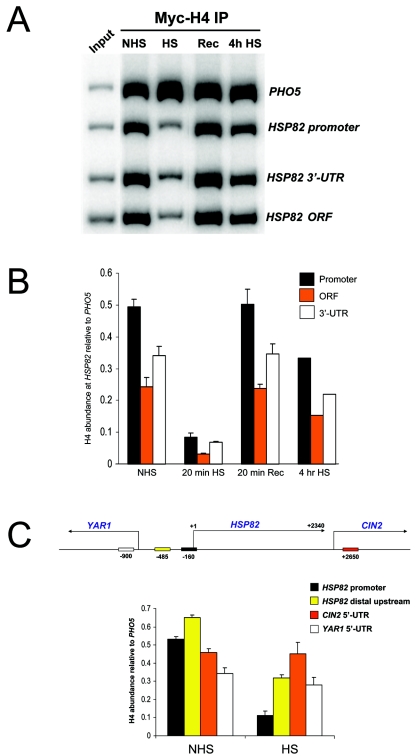
FIG. 3.
Myc-H4 is depleted over the entire HSP82 domain in response to a 20-min heat shock but reassociates with DNA following either brief recovery (Rec) or during extended heat shock (4h HS). ChIPs were performed with Myc MAb 9E10 on chromatin isolated from SLY101 cells containing an episomal copy of myc-HHF2. (A) Electrophoretic analysis of quantitative multiplex PCRs of input (lane 1, reading from left to right) and IPs of the chromatin samples indicated (lanes 2 to 5). (B) Summary of three to four independent experiments (means ± SD; 4-h heat shock was analyzed only once). (C) In vivo cross-linking of Myc-H4 to the HSP82 promoter, HSP82-YAR1 intergenic region, and the 5′ ORF regions of YAR1 and CIN2 under control and 20-min heat shock conditions (depicted are means ± SD for two experiments). Also illustrated is a physical map of the heat shock gene locus, with the PCR amplicons (rectangles) as well as the mapped 3′ end of the HSP82 transcript (25) indicated. Midpoint coordinates of each amplicon are relative to the principal transcription start site of HSP82 (+1).
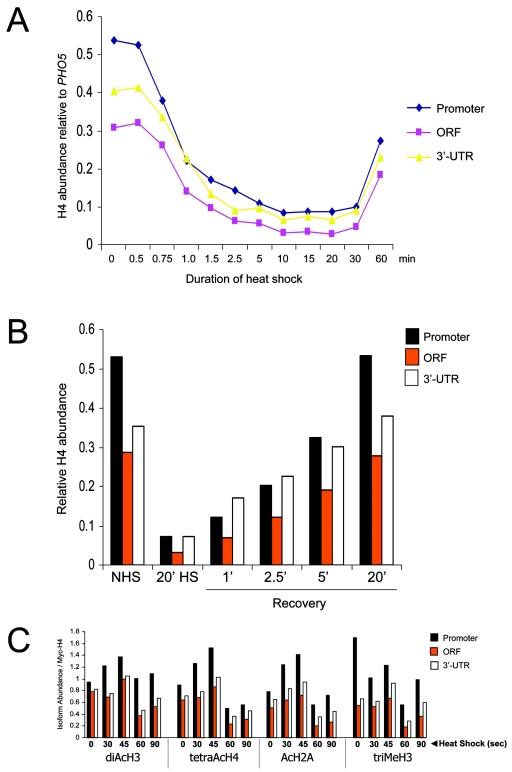
FIG. 4.
Histone abundance and modification state at HSP82 rapidly changes in response to inducing conditions. (A) Kinetics of Myc-H4 depletion over the promoter, ORF, and 3′ UTR at the indicated times following an instantaneous 30 to 39°C temperature shift. ChIP analysis was conducted as described in the legend to Fig. 3. Values represent the means of two independent experiments. (B) Myc-H4 association with DNA under noninducing (NHS) and acutely inducing (HS) conditions, as well as at 1, 2.5, 5, or 20 min following a 39 to 30°C downshift. (C) Histones H2A, H3, and H4 are transiently hyperacetylated at the HSP82 promoter prior to their eviction from all regions of the gene. The relative abundance of the indicated isoforms was measured in cells subjected to an instantaneous 39°C heat shock. Illustrated are quotients of isoform abundance (data not shown) divided by Myc-H4 abundance for each time point (obtained from the means of two independent sets of experiments).
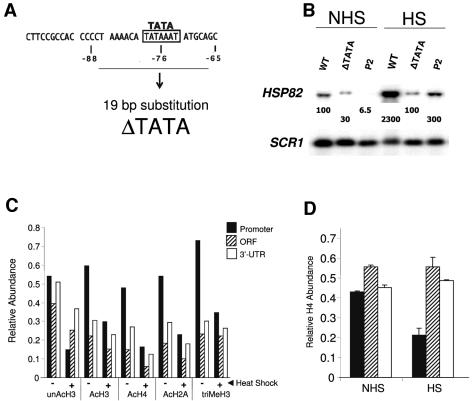
FIG. 5.
Histone displacement takes place within the promoter of an activated hsp82 TATA mutant. (A) The ΔTATA mutation (coordinates as in Fig. 1). (B) Northern analysis of non-heat-shocked (NHS) and 20-min heat-shocked (HS) isogenic strains bearing the HSP82+ (WT), hsp82-ΔTATA, and hsp82-P2 alleles as indicated. HSP82 transcript levels, normalized to those of the Pol III transcript SCR1, are provided within each lane (values represent the means of ≥3 independent assays). (C) In vivo cross-linking analysis of hsp82-ΔTATA. Abundance of histone isoforms at the indicated hsp82 regions under control (−) and 20-min heat-shocked (+) states was determined by ChIP as described in the legend to Fig. 1. (D) As in panel B, except Myc-H4 abundance was assayed (means ± SD for two experiments).
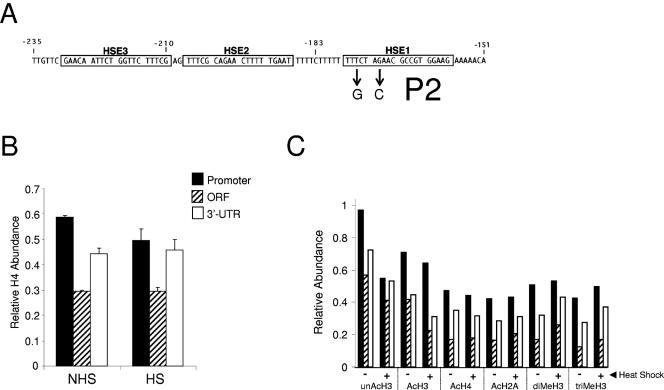
FIG. 6.
Histone displacement is obviated at the UASHS mutant hsp82-P2. (A) The P2 mutation (numbering as in Fig. 1) (56). (B) In vivo cross-linking of Myc-H4 to the hsp82-P2 promoter and coding region ± heat shock (means ± SD for two experiments). (C) Isoform abundance at the indicated hsp82 regions (as in panel B) under control (−) and 20-min heat-shocked (+) states was determined by ChIP as in Fig. 1.
FIG. 7.
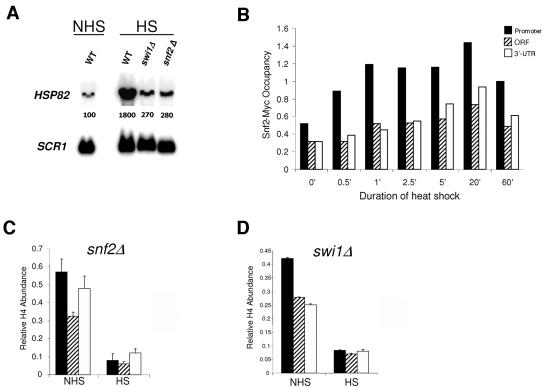
Snf2-Myc is rapidly recruited to HSP82 upon heat shock, yet efficient histone H4 displacement occurs in strains bearing functionally inactivated Swi/Snf. (A) Northern analysis of non-heat-shocked and 20-min-heat-shocked Swi/Snf+ (WT), swi1Δ, and snf2Δ strains bearing the HSP82+ gene. Normalized HSP82 transcript levels are provided within each lane as in Fig. 5B (values represent means of three independent experiments). (B) Snf2-Myc ChIP time course analysis at HSP82. Illustrated is abundance of Snf2-Myc at the promoter, ORF, and 3′ UTR of HSP82 at the indicated times, following instantaneous shift from 30 to 39°C. (C and D) In vivo cross-linking of Myc-H4 to HSP82 in snf2Δ and swi1Δ cells maintained at 30°C (NHS) or heat shocked for 20 min (HS) (depicted are means ± SD for two experiments).
FIG. 9.
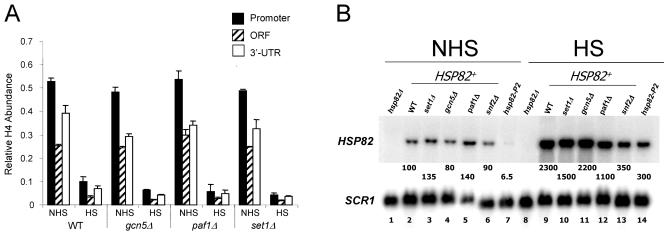
Myc-H4 is efficiently displaced from HSP82 in cells bearing deletions of histone modification or transcription elongation factors. (A) In vivo cross-linking of Myc-H4 to HSP82 in isogenic wild-type (WT), gcn5Δ, paf1Δ, and set1Δ strains under NHS and 20-min HS states (depicted are means ± SD for two experiments). (B) Northern analysis of noninduced and 20-min heat-shocked isogenic HSP82+ strains bearing knockouts of the indicated coactivator genes (lanes 2 to 6 and 9 to 13). Lanes 7 and 14, hsp82-P2 in a WT coactivator background; lanes 1 and 8, hsp82Δ strain SLY102 (negative hybridization control). HSP82 transcript levels are provided as in Fig. 4B (values represent means of ≥3 independent assays).
Cultivation, heat shock, and recovery conditions.
Saccharomyces cerevisiae strains were cultivated at 30°C to early log phase (A600 = 0.3 to 0.7) in rich yeast extract-peptone-glucose broth supplemented with 0.03 mg/ml adenine. Those transformed with pNOY436 were grown similarly but in synthetic complete medium lacking tryptophan. Heat shock induction was achieved by transferring the culture (typically 50 to 100 ml) to a vigorously shaking 39°C water bath; once the temperature reached 39°C, incubation was allowed to continue for an additional 20 min or 4 h before being terminated through the addition of either sodium azide to a final concentration of 10 mM (Northern) or formaldehyde to a 1% concentration (ChIP). For recovery, cultures were first heat shocked for 20 min, then immersed in an ice water bath for a few seconds with rapid swirling, and transferred to a 30°C shaking incubator for 20 min prior to being terminated as above. For time course assays (see Fig. 4A, 4C, and 7B), instantaneous 30 to 39°C upshift was achieved by rapidly mixing equal volumes of 30°C culture and prewarmed medium (55°C) and then incubating with rapid shaking at 39°C for the times indicated. To achieve an instantaneous 39 to 30°C downshift (see Fig. 4B), 20-min heat-shocked cultures were mixed with a 0.75 volume of prechilled (0°C) medium and then transferred to a shaking 30°C water bath for the times indicated.
ChIP analysis.
ChIP was performed essentially as previously described (76). Cells were cross-linked with 1% formaldehyde, then converted to spheroplasts with lyticase, and lysed using 1 volume of 0.5-mm glass beads for 30 min at 4°C on an Eppendorf 5432 mixer. Chromatin was sheared to a mean length of ∼0.4 kb with a Branson 250 sonifier equipped with a microtip (three 25-s pulses at constant power and an output setting of 22 W). Antibodies specific for the following epitopes were used: histone H3 (unacetylated isoform), diacetyl histone H3 (K9 and K14), tetra-acetyl histone H4 (K5, K8, K12, and K16), monoacetyl histone H2A (K7), and dimethyl histone H3 (K4) (all obtained from Upstate Biotechnology); trimethyl histone H3 (K4) (Abcam); Myc (monoclonal antibody [MAb] 9E10; Santa Cruz Biotechnology); and mouse Pol II CTD (a gift of David Bentley, University of Colorado Health Sciences Center) (71). IPs were achieved by adding generally either 3 μl of polyclonal antiserum or 5 μl of MAb to 300 μl of chromatin lysate. They were mixed on a nutator at 4°C overnight. Pansorbin cells (40 μl; Calbiochem) were then added, and the incubation was continued for an additional 3 h. For Pol II ChIPs, the 300-μl chromatin lysate was preincubated with 50 μl blocked protein A-Sepharose beads for 3 h at 4°C, and then 6 to 8 μl of Pol II antibody and 3 μl of 30% Sarkosyl were added to the clarified supernatant and permitted to incubate overnight at 4°C. A total of 50 μl of fresh protein A beads was then added, and the mixture was incubated at 4°C for 2 h. Beads were then washed as previously described (76).
Following washing and reversal of formaldehyde-induced cross-links, DNA was ethanol precipitated and dissolved in 30 μl Tris-EDTA (TE). Two microliters was then used as a template in a PCR. For input, 200 μl of soluble chromatin was ethanol precipitated and dissolved in TE, and cross-links were reversed. The chromatin was reprecipitated, and DNA was purified and dissolved in 150 μl TE. One microliter of this was used as template in a PCR. In addition to template DNA, the 50-μl reaction mixtures contained 2.5 mM MgCl2; 400 μM (each) dCTP, dGTP, dTTP, and dATP; and 1 μCi of [α-32P]dATP (250 Ci/mmol). After 2 min of denaturation at 93°C and addition of 1.25 U Taq DNA polymerase, the temperature was lowered to 60°C for 1 min, followed by 32 s at 72°C. Samples were then subjected to a program of 22 to 24 cycles, each consisting of 1 min at 93°C, 1 min at 60°C, and 32 s at 72°C. PCR products were precipitated, electrophoresed on 8% Tris-borate-EDTA polyacrylamide gels, dried, exposed to a Phosphor Screen, and quantified on a Storm 860 PhosphorImager (Molecular Dynamics) utilizing ImageQuant 5.2 software. Linearity of the PCR was tested by assaying threefold serial dilutions of 15 individual templates. The mean increase in multiplex PCR signal for the heavier loaded sample of each pair was 2.2 (see Fig. 8A).
FIG. 8.
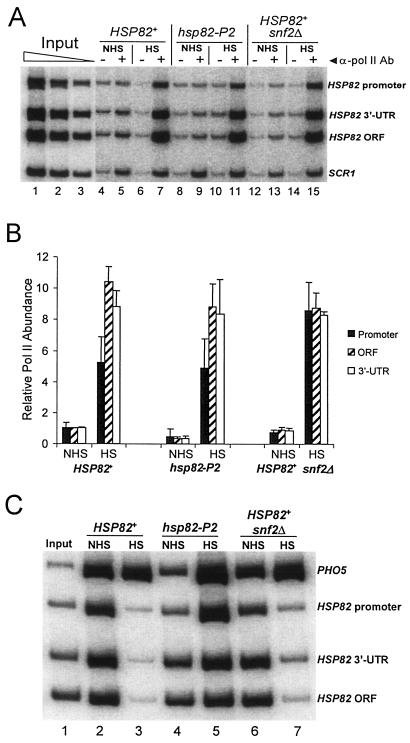
Pol II density within the promoter, ORF, and 3′ UTR of activated HSP82 is not diminished by mutations that markedly impair its transcription. (A) Electrophoretic multiplex PCR analysis of Pol II ChIPs of noninduced and 20-min heat shock-induced SLY101 (HSP82+), DGY101 (hsp82-P2), and JHD204 (snf2Δ) cells. SCR1 serves as an internal load control (see Materials and Methods). +, Pol II (mouse CTD) antibody; −, no antibody. Input lanes, threefold serial dilutions of DNA template purified from sonicated soluble chromatin (SLY101). (B) Relative abundance of Pol II at the HSP82 promoter, ORF, and 3′ UTR under NHS and HS states in the indicated strains (depicted are means ± SD for two experiments). All values are normalized to Pol II abundance within the noninduced HSP82+ ORF (SLY101), arbitrarily set to 1.0. (C) Electrophoretic multiplex PCR analysis of input chromatin (lane 1) and tetra-acetylated H4 ChIPs (lanes 2 to 7) of the indicated samples (identical to those used in panel A). As in all other histone ChIPs, the PHO5 promoter served as an internal load control.
PCR primers were as follows (all coordinates are relative to ATG): PHO5 promoter (forward, −507 to −478; reverse, −8 to −33), HSP82 promoter (forward, −401 to −374; reverse, −34 to −70), HSP82 ORF (forward, +1248 to +1275; reverse, +1444 to +1416), HSP82 3′ untranscribed region (UTR) (forward, +1883 to +1911; reverse, +2155 to +2125), HSP82 core promoter (forward, −227 to −201; reverse, −140 to −163), HSP82 5′ intergenic region (forward, −643 to −620; reverse, −431 to −456), CIN2 5′ ORF (forward, +149 to +179; reverse, +427 to +398), CIN2 3′ UTR (forward, +523 to +550; reverse, +797 to +767), YAR1 5′ ORF (forward, +3 to +31; reverse, +200 to +172), YAR1 3′ UTR (forward, +379 to +405; reverse, +579 to +551), SSA4 ORF (forward, +1109 to +1136; reverse, +1297 to +1269), HSP12 ORF (forward, +160 to +187; reverse, +359 to +333), HSP26 ORF (forward, +222 to +250; reverse, +515 to +488) and SCR1 (forward, +343 to +366; reverse, +467 to +444).
To calculate the relative abundance of a given gene promoter or coding sequence present in an IP, we used the following formula: Qgene = IPgene/inputgene. For histone ChIPs, the abundance of each test promoter or coding region is expressed relative to that of the PHO5 promoter, which served as an internal recovery control (76). For the Snf2-Myc ChIP assay (see Fig. 7B), nonspecific background (the PCR signal arising from immunoprecipitation of chromatin isolated from a non-Myc-tagged strain) was subtracted from each specific IP signal prior to calculating Qgene. For Pol II ChIPs, abundance at each hsp82 region is expressed relative to its abundance at a neutral locus on chromosome V (SCR1, a 521-bp Pol III gene closely flanked by two non-heat-inducible Pol II genes, YERWdelta17 and YER137W-A). To accurately quantify Pol II abundance, we subtracted the signal arising from a mock ChIP (−Ab) from the corresponding ChIP signal (+Ab) (see Fig. 8A) prior to calculating Qgene.
Northern analysis.
RNA was isolated, purified, electrophoresed, and blotted to Gene Screen as previously described (77). Blots were sequentially hybridized to HSP82- and SCR1-specific probes at 45°C and 55°C, respectively. The HSP82-specific probe was generated by 25 cycles of linear PCR using as a template an oligonucleotide corresponding to the transcript's unique 3′ UTR (spanning +2167 to +2228 relative to ATG). The SCR1 probe was generated by 25 cycles of linear PCR amplification of the gene's region spanning +343 to +467 with the SCR1 PCR fragment as a template.
RESULTS
Histone-DNA interactions are disrupted across the entire HSP82 domain in response to heat shock.
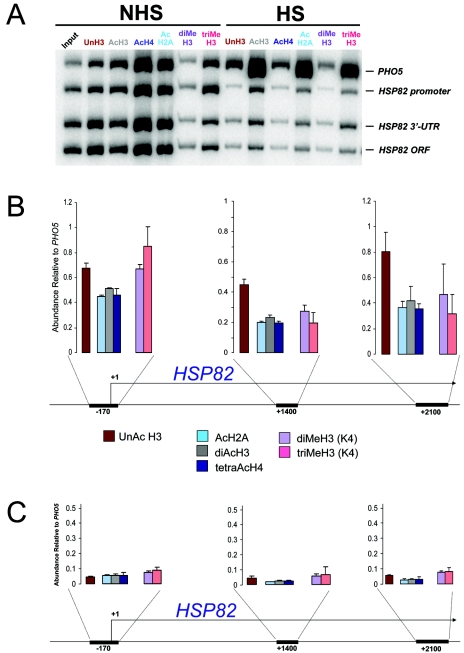
To investigate the fate of histones within the HSP82 coding region, we conducted ChIP assays of nonacetylated, acetylated, and methylated isoforms. Since the gene is robustly transcribed in response to heat shock, we anticipated that there would be an increase in the abundance of both acetylated and methylated histone isoforms, as has been reported for other actively transcribed eukaryotic genes (54, 62, 68, 72). Indeed, histone acetylation has been shown to facilitate HSF-induced transcription of an in vitro-reconstituted heat shock gene (64). However, as shown by the multiplex PCR analysis of Fig. 1, the abundance of acetylated H2A, H3, and H4, as well as that of methylated H3 (both di- and trimethyl K4), was diminished 70 to 95% following a 20-min heat shock. This drastic loss of covalently modified isoforms occurred over the gene's upstream region, ORF, and 3′ UTR (hereafter referred to as the HSP82 domain) and mirrored the fate of unacetylated H3 (compare HSP82 signals with those of the coamplified PHO5 promoter) (Fig. 1).
FIG. 1.
Heat shock results in displacement of unacetylated, acetylated, and methylated histones throughout the HSP82 domain. (A) In vivo cross-linking analysis of the HSP82+ strain SLY101 under non-heat-shock inducing (30°C) (NHS) and 20-min heat shock-inducing (39°C) (HS) conditions. Depicted is a polyacrylamide gel electrophoretic analysis of multiplex PCRs of ChIPs obtained using isoform-specific antibodies (lanes 2 to 13, reading from left to right). Lanes: Input, input DNA; UnH3, unacetylated H3; AcH3, diacetylated H3; AcH4, tetra-acetylated H4; AcH2A; monoacetylated H2A; diMeH3, dimethylated H3; triMeH3, trimethylated H3. Note that the lane-to-lane variation in the PHO5 signal reflects differences in IP efficiency and DNA recovery; it has not proven reproducible. (B) Summary of three independent ChIP experiments. Depicted is the mean abundance (± standard deviation [SD]) of six histone isoforms at the indicated HSP82 regions relative to their abundance at the PHO5 promoter. Coordinates represent the midpoint of each amplicon; numbering is relative to the principal transcription start site (76). (C) Same as the results shown in panel B, except that the mean abundance of the six isoforms following a 20-min heat shock is shown.
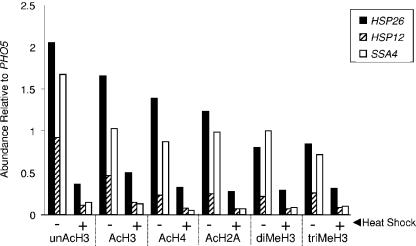
To determine whether loss of histone DNA contacts was unique to HSP82 or also occurred at other heat shock genes, we extended our analysis to HSP12, HSP26, and SSA4. The promoters of these genes contain heat shock elements and, by the criterion of ChIP, are regulated by HSF (32; J. Iqbal and D. S. Gross, unpublished data). HSP12 and HSP26 are also regulated by Msn2/Msn4 (4, 70). At each ORF, drastic reductions in all histone isoforms (up to 90%) were seen upon heat shock (Fig. 2), closely paralleling previous findings of histone loss at heat shock gene promoters (16, 23, 76). Thus, domain-wide loss of histone-DNA contacts seems to be characteristic of HSF-regulated genes.
FIG. 2.
Histone abundance is drastically reduced within the coding regions of HSP12, HSP26, and SSA4 following heat shock. ChIPs were performed as described in the legend to Fig. 1, and isoform abundance within each gene's ORF (relative to that at the PHO5 promoter) under control (−) and 20-min heat-shocked (+) states was determined.
The foregoing results suggest the possibility that histones lose physical contact with the promoters and transcribed regions of activated heat shock genes. Alternatively, heat shock may induce epitope masking. To distinguish between these possibilities, we employed a strategy that permitted detection of histone H4 irrespective of its modification state. To allow pan-detection of H4, we transformed strains with an episomal myc-HHF2 gene and assayed H4 abundance using a Myc-specific antibody. When expressed from an episomal construct, Myc-tagged H4 is incorporated into nucleosomes and causes no obvious growth defects; indeed, strains harboring myc-HHF2 as the sole source of H4 are viable (40). Using this strategy, we found that the abundance of Myc-H4 was reduced 75 to 85% across the HSP82 domain, following 20 min of acute heat shock (Fig. 3A and B), closely paralleling reductions observed with specific histone isoforms (Fig. 1) and consistent with histone displacement.
Two further observations suggest that the apparent loss of histone occupancy at HSP genes reflects their physical displacement from DNA and not, for example, inefficient cross-linking caused by heat shock. First, the effect is highly specific to strongly activated heat shock genes. There was no decrease in histone cross-linking at the coamplified PHO5 promoter, nor was there any diminishment of histones within either the promoter or coding sequence of a less vigorously transcribed HSP gene (see Fig. 6B). Second, nonhistone proteins such as HSF, TATA-binding protein (TBP), and Pol II are readily detected within the promoters and coding regions of activated heat shock genes (see Fig. 8A) (76). Together with previous observations (16, 23), the results indicate that chromatin spanning entire heat shock gene domains is grossly altered upon heat shock, an alteration that likely involves the loss of histone octamers.
Chromatin remodeling is confined to a circumscribed domain encompassing the 5′ flank, ORF, and 3′ UTR of HSP82.
To delimit the extent of heat shock-induced remodeling, we assayed the histone content of the two genes flanking HSP82. This was of interest, since it was unclear whether the dramatic reduction in histone abundance at HSP82 was locus-wide or circumscribed to the heat shock gene's domain. Using ChIP as above, we found that Myc-H4 was not significantly depleted within the coding region of either YAR1 or CIN2 upon heat shock (Fig. 3C). However, it was 50% reduced within the YAR1-HSP82 intergenic region (position −485 relative to HSP82). Therefore, while heat shock-induced histone displacement spanned a domain of nearly 3 kb, it did not spread into the closely flanking neighboring genes. This was true even though the coding region of one of them, CIN2, begins just 110 bp downstream of the 3′-mapped end of the HSP82 transcript (25). Indeed, histones were hypoacetylated within both YAR1 and CIN2 coding regions, concomitant with their heat shock-induced transcriptional repression (data not shown). This finding is consistent with previous observations correlating coding region histone hypoacetylation with transcriptional down-regulation (10, 46).
Kinetics of domain-wide remodeling are remarkably rapid.
We next sought to determine the kinetics with which histones are displaced. To do so, we formaldehyde-cross-linked cells at various times following a rapid 30 to 39°C upshift. This approach afforded instantaneous snapshots of protein-DNA interactions, since formaldehyde rapidly induces cross-linking and simultaneously freezes all metabolic processes (39), including those induced by heat shock (Iqbal and Gross, unpublished). As shown in Fig. 4A, Myc-H4 dissociation at HSP82 was detectable within 45 s (14 to 28% loss) and was essentially complete by 10 min (80 to 88% loss). Notably, displacement occurs over the three assayed regions with similar, although not necessarily identical, kinetics. Histone loss within the promoter is detectable as early as 30 s of postthermal upshift, whereas comparable changes in the ORF and 3′ UTR do not appear until 45 s. This difference might reflect distinct mechanisms and is further explored below. An additional and intriguing finding is that histone loss is transient. Histone-DNA contacts were reestablished 30 to 60 min into the heat shock (Fig. 4A), coincident with a decline in the rate of HSP82 transcription (data not shown).
To extend this analysis, we also examined the kinetics with which Myc-H4 reestablished contact with DNA upon a 39 to 30°C downshift (recovery). If histone loss reflects nucleosomal disassembly, its restoration may reflect de novo nucleosomal assembly, a process thought to be generally dependent on DNA replication. We found that Myc-H4 rapidly rebound to all three regions of HSP82 in response to temperature downshift, a condition that results in drastically reduced HSP82 transcription (77). This was detectable as early as 60 s and was essentially complete by 20 min (Fig. 4B; see also Fig. 3B). We conclude that histone H4 and, by extension, the H3-H4 tetramer itself reassociates with HSP82 with rapid kinetics over the entire length of the gene. Given the rapidity of the kinetics and the fact that the cell population is asynchronous, these striking findings argue against a replication-coupled process and instead suggest that nucleosome reassembly is replication independent (see Discussion).
A transient burst of histone acetylation at the HSP82 promoter precedes gene-wide histone displacement.
The histone code hypothesis (38, 81) postulates that covalent modifications within histone tails serve as essential recognition sites for the binding of factors including, for example, ATP-dependent remodeling enzymes (2, 34) and histone chaperones (1). Thus, covalent modifications of histones might precede their dissociation from DNA, as has been recently shown for the activated PHO5 promoter (67). We assayed the abundance of both acetylated and methylated histones throughout the HSP82 gene during the first 60 s of heat shock. Consistent with a role for lysine acetylation, we found that the relative abundance of acetylated histones increases 40 to 50% during the first 45 s of heat shock (Fig. 4C). This enrichment was short lived and was restricted to the promoter. At 60 s following heat shock, there was a marked reduction in acetylated H2A and H4. As above, this change took place relative to total histone (calculated as isoform occupancy/Myc-H4 occupancy), suggesting that nucleosomes acetylated at H2A and H4 are preferentially targeted for eviction. Notably, the net abundance of promoter-associated trimethylated H3 precipitously decreased during the first minute, arguing against a role for H3 K4 methylation in domain-wide remodeling. We conclude that heat shock-induced acetylation of promoter-associated nucleosomes precedes, by a matter of seconds, their disassembly.
Remodeling of the hsp82 promoter and 5′ UTR occurs independently of elevated transcription.
Demonstration of domain-wide histone displacement at HSF-regulated genes raises the question of what underlies this phenomenon. To test whether histone displacement at HSP82 is dependent upon transcription, we engineered a 19-bp substitution of its core promoter (Fig. 5A), encompassing the TATA box and coinciding with a region strongly protected from hydroxyl radical cleavage in chromatin (29). The mutant, termed ΔTATA, exhibited a 23-fold reduction in heat shock-induced transcription (Fig. 5B). Despite this, partial displacement of each histone isoform, as well as of Myc-H4, took place over the gene's promoter in response to acute heat shock (Fig. 5C and D). By comparison, there was little change in histone abundance over the ORF or 3′ UTR (Fig. 5D). Virtually identical results (data not shown) have been observed in a strain bearing a less crippling mutation in the hsp82 TATA box (a 2-bp substitution, termed T2, converting TATAAA to TAGCAA) and exhibiting a 12-fold reduction in induced transcription (52). We conclude that the dramatic remodeling which takes place at the 5′ end of HSP82 is at least partially independent of elevated transcription.
Mutation of HSE1 blocks histone displacement.
We next asked whether HSF itself is critical to domain-wide remodeling. To do so, we assayed histone abundance at an hsp82 promoter mutant bearing a 2-bp mutation in HSE1, the high-affinity HSF binding site (Fig. 6A). This mutation, termed P2, significantly weakens HSF binding to the hsp82 promoter both in vivo and in vitro (24, 56) and drastically reduces noninduced expression. hsp82-P2 nonetheless activates transcription to a level which exceeded that of hsp82-ΔTATA (Fig. 5B; see Fig. 9B). In contrast to the TATA box mutants, however, the upstream activation sequence (UAS) mutant showed little evidence of histone displacement upon heat shock (Fig. 6B). Moreover, despite the nearly 50-fold increase in transcription, neither the promoter nor the coding region of hsp82-P2 showed evidence of increased histone acetylation or methylation (Fig. 6C). Therefore, weakening of HSF's binding to the hsp82 promoter effectively eliminates heat shock-induced histone modification and displacement.
Swi/Snf is rapidly recruited to the activated HSP82 gene, is required for its vigorous transcription, yet is dispensable for domain-wide remodeling.
We next sought to test the contribution of Swi/Snf, given its critical role in the transcriptional activation of such inducible yeast genes as HO, SUC2, INO1, and PHO8 (for a review, see reference 90) and its demonstrated role in regulating transcription, particularly elongation, of the mammalian hsp70 gene (14, 17). As shown in Fig. 7A, heat shock-induced HSP82 expression was reduced >6 fold in cells bearing a deletion of either SNF2 or SWI1. Consistent with this, Myc-tagged Snf2 was rapidly recruited to the HSP82 promoter in response to heat shock (Fig. 7B), and its abundance positively correlates with that of HSF (32, 76). Swi/Snf was also detected within the HSP82 coding region, although with delayed kinetics and reduced occupancy. Recruitment of Swi/Snf suggests that the expression phenotypes associated with the snf2Δ and swi1Δ mutations are due to a direct role for Swi/Snf in regulating heat shock-induced transcription. Despite this, Myc-H4 was efficiently displaced from all three regions of the gene following acute heat shock of either mutant (Fig. 7C and D). This outcome is especially striking, given that histone depletion was not seen in the UAS mutant hsp82-P2 (Fig. 6B), whose rate of activated transcription is similar to that of the Swi/Snf mutants (see Fig. 9B, lane 13 versus lane 14). Therefore, even though HSP82 transcription is Swi/Snf dependent, remodeling of both promoter and coding region occurs efficiently in its absence.
Pol II density at HSP82 is unaffected by either a SNF2 gene deletion or a UAS promoter mutation.
To explore the basis for the contrasting chromatin phenotypes of the P2 and snf2Δ mutants, we investigated Pol II density within the hsp82 promoter, ORF, and 3′ UTR of the wild-type strain and of the two isogenic mutants. If histone displacement is a consequence of elongating Pol II, then Pol II abundance should be high in cases where domain-wide histone displacement is observed (HSP82+) and low in cases where it is not (hsp82-P2). Indeed, ChIP revealed that Pol II density was high within all three regions of HSP82+ in both SNF2+ and snf2Δ cells following a 20-min heat shock (Fig. 8A and B). As expected, abundance of histone H4 negatively correlated with that of Pol II in the same chromatin samples (compare Fig. 8C, lanes 3 and 7, with 8A, lanes 7 and 15). These results are consistent with the idea that elongating Pol II underlies histone displacement and are in agreement with results obtained with the hsp82-ΔTATA strain that showed a low level of induced transcription and no depletion of coding region histones. Interestingly, since Pol II density was not diminished in snf2Δ cells despite a sixfold reduction in transcription, this suggests that loss of Swi/Snf impairs not only the rate of initiation but also the rate of elongation (see Discussion).
Arguing against a correlation between Pol II density and histone loss, however, are results obtained with the P2 mutant. Despite the fact that activated transcription was significantly reduced (Fig. 5B and 9B), Pol II density at the heat shock gene was unaffected (Fig. 8A, compare lane 11 with lane 7; Fig. 8B). This suggests that similar to snf2Δ, the P2 mutation affects the rate of both transcription initiation and elongation. In contrast to the case with snf2Δ, however, the P2 mutation diminished Pol II density under non-heat-shocked conditions (Fig. 8B) and, more importantly, abrogated histone displacement following HS (Fig. 8C, lane 5; Fig. 6B). This latter observation indicates that a high density of Pol II within the hsp82 coding region is insufficient, by itself, to bring about histone displacement.
Nucleosomal disassembly is independent of the Gcn5 and Set1 histone-modifying enzymes as well as the Paf1 elongation complex.
To investigate additional coactivators that might underlie domain-wide histone displacement (and thereby be recruited to HSP82+ but not to hsp82-P2), we constructed isogenic strains bearing deletions of GCN5, SET1, or PAF1. Gcn5, the catalytic subunit of SAGA, was of interest, since the recruitment of SAGA to a variety of yeast promoters has been demonstrated. Indeed, it is a primary target of both Gal4 and Gcn4 (8, 12, 26, 82), and HSF-mediated trans-activation of target genes in response to stress is thought to occur principally through the SAGA (as opposed to the TFIID) pathway (92). Moreover, the finding that promoter-associated H3 was transiently hyperacetylated at Lys9 and Lys14 in response to heat shock (Fig. 4C) is consistent with a role for Gcn5, given its preference for acetylating these residues in vivo (48). However, as shown in Fig. 9, deletion of GCN5 had no detectable effect on either HSP82 expression (Fig. 9B) or histone displacement (Fig. 9A). This suggests that SAGA's role in the activation of HSP82 may derive more from its ability to facilitate TBP binding to core promoters (18) than from its ability to acetylate chromatin.
We next examined the role of the Set1 histone methyltransferase, given its suggested role in Pol II elongation at actively expressed yeast genes. While the absence of enhanced H3 K4 methylation at the induced HSP82 gene (Fig. 1 and 4C) would seem to argue against a role for Set1, it is possible that this modification occurs over a discrete region of HSP82 that eluded detection by the primer pairs we employed. In particular, H3 K4 trimethylation has been reported to be maximal within the 5′ coding regions of activated genes (62, 68); thus a comparable, albeit transient, modification may occur over the HSP82 coding region. However, as shown in Fig. 9, a set1Δ mutation had little impact on either the extent of histone displacement or transcriptional activation of HSP82, arguing against a role for the Set1 complex, COMPASS, in regulating domain-wide histone displacement.
Finally, we investigated whether the Paf1 complex played a role in domain-wide histone displacement at HSP82. Paf1 associates with Pol II at multiple stages during the transcription cycle (for a review, see reference 61) and has been detected within the coding regions of HSF-regulated genes, including HSP82 and SSA4 (65). Despite this, deletion of the PAF1 gene had no effect on histone displacement, although it did reduce activation approximately twofold (Fig. 9A and B).
DISCUSSION
Histone contacts with DNA are rapidly and reversibly lost from heat shock gene promoter and coding regions.
Through the use of ChIP assays, we have obtained evidence that the yeast HSP82 heat shock gene undergoes remarkably rapid and widespread alteration in chromatin structure upon its activation, and that this alteration, which is reflected by a drastic reduction in histone cross-linking to DNA, is fully reversible upon recovery. While it remains a formal possibility that the histones are rearranged into a conformation that precludes efficient cross-linking by formaldehyde, the fact that we see substantial reduction in the cross-linking of three different core histones and that such reduced cross-linking occurs irrespective of their modification state most likely indicates dissociation of histone octamers from DNA. This dramatic, domain-wide nucleosomal disassembly is not unique to HSP82 but is also seen at other heat shock-inducible genes, including HSP12, HSP26, and SSA4. Additional support for histone eviction comes from similar findings of histone removal at activated Drosophila heat shock genes (see below) (74).
It is noteworthy that an earlier study employing nuclease digestion of chromatin also showed transcription-dependent alterations in nucleosomal structure over the HSP82 coding region (51). However, these alterations, which consisted of enhanced DNase I sensitivity and half-nucleosomal cleavage periodicity, were present in nuclei obtained from both noninduced and heat-shocked cells bearing the wild-type HSP82 gene; full-nucleosome cleavage periodicity was seen at two transcriptionally quiescent hsp82 mutants (51). Therefore, structurally altered (and possibly unfolded) nucleosomes may exist within the coding region of basally transcribed HSP82. The ChIP experiments presented here suggest that such nucleosomes are rapidly displaced upon heat shock. In the earlier study, these displaced histones may have partially reassociated over the coding DNA during isolation of nuclei and subsequent DNase I digestion, thereby masking the dramatic alterations documented here.
There is considerable evidence that the coding regions of actively transcribed eukaryotic genes contain altered nucleosomes. These altered nucleosomes have variously been shown to be unfolded (66), to contain hyperacetylated and hypermethylated core histones (35, 62, 68, 88), and to be depleted in histones H2A and H2B (6, 41). It is thus possible that domain-wide loss of histone octamers is peculiar to yeast heat shock genes. However, this is unlikely. First, electron micrographs have been obtained that show loss of nucleosomes over the coding regions of yeast rRNA genes (15). Second, recently published ChIP analyses demonstrate partial loss of histones over the GAL1 and GAL10 coding regions, following galactose induction (45, 49, 73). Therefore, nucleosome depletion within gene coding regions may be a characteristic of vigorously transcribed genes. Indeed, as alluded to above, a recent analysis of green fluorescent protein-H3 distribution in Drosophila polytene chromosome spreads reveals that H3 is rapidly lost from heat shock gene loci upon heat shock. In contrast to yeast heat shock genes, however, H3 is efficiently replaced by a variant, H3.3, and such replacement occurs early during transcriptional induction (74).
Distinct mechanisms of histone displacement over promoter and coding region.
Evidence presented here suggests that domain-wide histone displacement at HSP82 occurs in two discrete steps: (i) 5′-end remodeling that is dependent on a stably bound activator and (ii) coding region remodeling that is dependent on elongating Pol II. A critical role for HSF in triggering domain-wide displacement is indicated by the phenotype of the hsp82-P2 allele, which bears a 2-bp mutation in HSE1 that significantly reduces HSF binding to the UAS (24, 56). At this allele, domain-wide histone loss is obviated. In addition, elongating RNA polymerase is required for disrupting coding region chromatin. This conclusion is based on the phenotype of the hsp82-ΔTATA allele, whose promoter is efficiently remodeled upon heat shock, while its coding region is not. (Polymerase's role in histone displacement is more extensively discussed below.) Since activated transcription is more severely affected in hsp82-ΔTATA than in hsp82-P2, disruption of promoter chromatin in the TATA mutant likely stems from activated HSF (and the factors it recruits). A similar distinction between proximal and distal chromatin remodeling has been previously shown for the human hsp70 gene (11). That HSF is in fact bound to the UAS of the hsp82-ΔTATA gene has not been formally shown. However, chromatin footprinting analysis of the closely related hsp82-T2 allele reveals that promoter-associated DNase I hypersensitivity, a signature of DNA-bound HSF (28), is fully retained (52). Thus, it is probable that HSF constitutively binds the promoter region of hsp82-T2, and by extension, to that of hsp82-ΔTATA.
An additional mechanistic difference between promoter versus coding region remodeling is that the former is characterized by a burst of histone acetylation that precedes nucleosomal disassembly by a matter of seconds. No comparable modification is seen over the coding region. It is tempting to speculate that these promoter-specific marks, which are especially prominent on H2A and H4, facilitate the recruitment of bromodomain-containing enzymatic complexes (2, 34) that ultimately catalyze domain-wide histone displacement. Given the in vitro preference of Esa1 to acetylate H2A and H4 (85), it is possible that this histone acetyltransferase marks promoter-associated nucleosomes for eviction. Acetylated octamers appear to be preferentially targeted, since acetylated H2A and H4 are disproportionately depleted (relative to that of Myc-H4) at time points subsequent to 45 s.
Elongating Pol II is necessary but not sufficient to trigger nucleosomal disassembly.
The requirement of Pol II elongation for coding region disruption suggests the possibility that these chromatin alterations are simply a consequence of high rates of transcription. In support of this idea is the finding that even though heat shock-induced HSP82 transcript levels are drastically reduced in a snf2Δ mutant, Pol II density remains unchanged. A high density of Pol II within the gene's coding region provides a plausible explanation for the efficient displacement of histones that is seen in response to heat shock in Swi/Snf mutants. Furthermore, histone-DNA contacts are lost at the coding regions of galactose-induced GAL genes, and such loss is both correlated with high rates of transcription and dependent on the presence of elongating Pol II (45, 49, 73).
Nevertheless, we have obtained evidence that RNA polymerase is not, of itself, sufficient to elicit histone eviction. This is the case, since Pol II density is as high at the heat shock-induced hsp82-P2 gene (which exhibits both reduced transcription and no detectable loss of histones) as at wild-type HSP82. Therefore, activities in addition to elongating Pol II must contribute to the disruption of coding region chromatin. We suggest that activated HSF, tightly bound to the HSP82+ promoter, triggers the recruitment of remodeling activities that efficiently disassociate histones over the length of the entire gene. Such histone-displacing activities, by definition, would fail to be recruited to hsp82-P2, where HSF's occupancy of the UAS is significantly reduced (24). Importantly, the finding that high Pol II density can coexist with nucleosomes over the length of a gene is not simply an oddity of the hsp82-P2 mutant. We see a similar coupling of high Pol II density with high histone occupancy at the induced SSA3 gene, whose promoter appears to be weakly bound by HSF (J. Zhao, J. Iqbal, and D. S. Gross, unpublished data). Given the absence of a correlation between Pol II abundance and nucleosomal disassembly, it is probable that coding region histone displacement plays a causal role in the vigorous transcription seen in genes such as HSP82, HSP26, SSA4, and GAL1 and is not merely a consequence of it.
A role for Swi/Snf in governing Pol II elongation rate in vivo.
An additional implication of these experiments is that Swi/Snf affects the rate of Pol II elongation at HSP82. This is based on the finding that, as discussed above, Pol II density within the HSP82 coding region is unaffected by an snf2Δ mutation despite a sixfold reduction in transcription. Moreover, Pol II abundance within the promoter is actually higher in heat-shocked snf2Δ cells than in SNF2+ cells, consistent with Pol II having an increased dwell time and/or impairment in promoter escape. In this respect, the role of yeast Swi/Snf is reminiscent of that demonstrated for mouse Swi/Snf (BRG1), which has been shown to mediate release of paused Pol II within the 5′ UTR of the hsp70 heat shock gene (14). Given the striking similarity of the snf2Δ and P2 phenotypes, the apparent elongation defect of the snf2Δ mutant might be a consequence of impaired binding by HSF to the chromatinized UAS, whose default state is nucleosomal (28, 87). Thus, the role of Swi/Snf in governing the elongation rate could be indirect. It will be interesting to test whether other Swi/Snf-dependent genes similarly retain a high density of Pol II in the context of an inactivating Swi/Snf mutation.
Implications of domain-wide nucleosomal disassembly and reassembly.
Nucleosomes carry important epigenetic information (3, 79). This information exists in the pattern of covalent modifications of the constituent histones, as well as in the presence of histone variants. Normally, this information is preserved during such processes as replication, recombination, and transcription. An intriguing finding is that histone H4 rapidly reassociates with HSP82 in response to thermal downshift, detectable within 60 s and restoring its original level within 20 min; it also reassociates with DNA during continuous heat shocks of ≥30 min. The rapidity of Myc-H4 deposition, presumably accompanying its reassembly into nucleosomes, implies that such a process occurs independently of DNA replication. Replication-independent assembly of the histone variant H3.3 has been previously shown to mark transcriptionally active genes in Drosophila (3), and the Swr1 complex mediates ATP-dependent, replication-independent exchange of the H2AZ variant in S. cerevisiae (43, 58). Our results suggest that if epigenetic information is preserved at activated yeast heat shock genes, then retention of as little as 20% of preexisting H3-H4 tetramers may suffice to seed the domain-wide formation of nucleosomes with a similar epigenetic profile. This residual 20% may also be necessary for tethering Swi/Snf to HSP82's promoter and coding region during heat shock (Fig. 7B). We did not examine the composition of nucleosomes reassembling heat shock gene domains following recovery from heat shock. It might be anticipated that their modification profile and histone variant composition will resemble those seen in the pre-heat-shocked state, although the possibility of such nucleosomes bearing deposition-specific marks cannot be ruled out (93).
Role of other coactivators in domain-wide nucleosomal disassembly.
We have shown here that a number of prominent remodeling and modification enzymes, such as Swi/Snf, Gcn5, and Set1, as well as the Paf1 elongation complex, are dispensable for domain-wide histone displacement. In addition, in other experiments we have found that the histone H3/H4 binding and deposition protein, Asf1, is not involved in histone displacement at HSP82 (unpublished data). This contrasts with Asf1's pivotal role in histone displacement from the activated PHO5 and PHO8 promoters (1). Absence of a detectable role for any of these factors may due to their being functionally redundant or because other proteins (such as Esa1) mediate nucleosomal disassembly at yeast heat shock genes. Whatever their identity, our work suggests that such enzymatic activities are recruited by HSF to wild-type HSP82, yet fail to be recruited to the 2-bp HSE1 mutant hsp82-P2.
Acknowledgments
We thank Sri Balakrishnan for assistance with the Northern assays, Serena Magrogan for construction of the hsp82-ΔTATA strain, David Bentley for providing RNA Pol II antiserum, Bill Garrard for the gift of the hsp82-T2 strain, Ned Sekinger and Kevin Struhl for providing the myc-HHF2 expression vector, and Selena Kremer, Sri Balakrishnan, and Ricky DeBenedetti for helpful comments on the manuscript.
D.S.G. wishes to dedicate this paper in memory of his father.
This work was supported by grants to D.S.G. from the National Science Foundation (MCB-0350190 and MCB-0450419), the National Institutes of Health (GM45842), and the Center for Excellence in Cancer Research at the Louisiana State University Health Sciences Center.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 4.Amoros, M., and F. Estruch. 2001. Hsf1p and Msn2/4p cooperate in the expression of the Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol. Microbiol. 39:1523-1532. [DOI] [PubMed] [Google Scholar]
- 5.Apone, L. M., C. A. Virbasius, F. C. P. Holstege, J. Wang, R. A. Young, and M. R. Green. 1998. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell 2:653-661. [DOI] [PubMed] [Google Scholar]
- 6.Baer, B. W., and D. Rhodes. 1983. Eukaryotic RNA polymerase II binds to the nucleosome cores of transcribed genes. Nature 301:482-488. [DOI] [PubMed] [Google Scholar]
- 7.Belotserkovskaya, R., A. Saunders, J. T. Lis, and D. Reinberg. 2004. Transcription through chromatin: understanding a complex FACT. Biochim. Biophys. Acta 1677:87-99. [DOI] [PubMed] [Google Scholar]
- 8.Bhaumik, S. R., T. Raha, D. P. Aiello, and M. R. Green. 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 10.Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592-604. [DOI] [PubMed] [Google Scholar]
- 11.Brown, S. A., and R. E. Kingston. 1997. Disruption of downstream chromatin directed by a transcriptional activator. Genes Dev. 11:3116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant, G. O., and M. Ptashne. 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Cell 11:1301-1309. [DOI] [PubMed] [Google Scholar]
- 13.Chou, S., S. Chatterjee, M. Lee, and K. Struhl. 1999. Transcriptional activation in yeast cells lacking transcription factor IIA. Genetics 153:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corey, L. L., C. S. Weirich, I. J. Benjamin, and R. E. Kingston. 2003. Localized recruitment of a chromatin remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 17:1392-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deckert, J., and K. Struhl. 2001. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol. 21:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Serna, I. L., K. A. Carlson, D. A. Hill, C. J. Guidi, R. O. Stephenson, S. Sif, R. E. Kingston, and A. N. Imbalzano. 2000. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 20:2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberharter, A., and P. B. Becker. 2002. Histone acetylation: a switch between repressive and permissive chromatin. EMBO Rep. 3:224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eickbush, T. H., and E. N. Moudrianakis. 1978. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry 17:4955-4964. [DOI] [PubMed] [Google Scholar]
- 21.Elgin, S. C. R. 1988. The formation and function of DNase hypersensitive sites in the process of gene activation. J. Biol. Chem. 263:19259-19262. [PubMed] [Google Scholar]
- 22.Erkine, A. M., C. C. Adams, M. Gao, and D. S. Gross. 1995. Multiple protein-DNA interactions over the yeast HSC82 heat shock gene promoter. Nucleic Acids Res. 23:1822-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erkine, A. M., and D. S. Gross. 2003. Dynamic chromatin alterations triggered by natural and synthetic activation domains. J. Biol. Chem. 278:7755-7764. [DOI] [PubMed] [Google Scholar]
- 24.Erkine, A. M., S. F. Magrogan, E. A. Sekinger, and D. S. Gross. 1999. Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol. Cell. Biol. 19:1627-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farrelly, F. W., and D. B. Finkelstein. 1984. Complete sequence of heat shock-inducible HSP90 gene of Saccharomyces cerevisiae. J. Biol. Chem. 259:5745-5751. [PubMed] [Google Scholar]
- 26.Fishburn, J., N. Mohibullah, and S. Hahn. 2005. Function of a eukaryotic transcription activator during the transcription cycle. Mol. Cell 18:369-378. [DOI] [PubMed] [Google Scholar]
- 27.Garel, A., and R. Axel. 1976. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc. Natl. Acad. Sci. USA 73:3966-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross, D. S., C. C. Adams, S. Lee, and B. Stentz. 1993. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 12:3931-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gross, D. S., K. E. English, K. W. Collins, and S. Lee. 1990. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J. Mol. Biol. 216:611-631. [DOI] [PubMed] [Google Scholar]
- 30.Gross, D. S., and W. T. Garrard. 1988. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57:159-197. [DOI] [PubMed] [Google Scholar]
- 31.Guldener, U., S. Heck, T. Fiedler, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hahn, J.-S., Z. Hu, D. J. Thiele, and V. R. Iyer. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24:5249-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hahn, J.-S., and D. J. Thiele. 2004. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J. Biol. Chem. 279:5169-5176. [DOI] [PubMed] [Google Scholar]
- 34.Hassan, A., P. Prochasson, K. Neely, S. Galasinski, M. Chandy, M. Carrozza, and J. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 35.Hebbes, T. R., A. W. Thorne, and C. Crane-Robinson. 1988. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson, V. 1988. Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry 27:2109-2120. [DOI] [PubMed] [Google Scholar]
- 37.Jackson, V. 1990. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry 29:719-731. [DOI] [PubMed] [Google Scholar]
- 38.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 39.Katan-Khaykovich, Y., and K. Struhl. 2002. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 16:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keener, J., J. A. Dodd, D. Lalo, and M. Nomura. 1997. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl. Acad. Sci. USA 94:13458-13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kireeva, M. L., W. Walter, V. Tchernajenko, V. Bondarenko, M. Kashlev, and V. M. Studitsky. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of H2A/H2B dimer during transcription. Mol. Cell 9:541-552. [DOI] [PubMed] [Google Scholar]
- 42.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Windsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 43.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krebs, J. E., M.-H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13:1412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kristjuhan, A., J. Walker, N. Suka, M. Grunstein, D. Roberts, B. R. Cairns, and J. Q. Svejstrup. 2002. Transcriptional inhibition of genes with severe histone H3 hypoacetylation in the coding region. Mol. Cell 10:925-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuo, M.-H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269-272. [DOI] [PubMed] [Google Scholar]
- 48.Kuo, M.-H., J. Zhou, P. Jambeck, M. E. A. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 50.Lee, D., and J. T. Lis. 1998. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature 393:389-392. [DOI] [PubMed] [Google Scholar]
- 51.Lee, M. S., and W. T. Garrard. 1991. Transcription-induced nucleosome “splitting”: an underlying structure of DNase I sensitive chromatin. EMBO J. 10:607-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee, M. S., and W. T. Garrard. 1992. Uncoupling gene activity from chromatin structure: promoter mutations can inactivate transcription of the yeast HSP82 gene without eliminating nucleosome-free regions. Proc. Natl. Acad. Sci. USA 89:9166-9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee, S., and D. S. Gross. 1993. Conditional silencing: The HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol. Cell. Biol. 13:727-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 56.McDaniel, D., A. J. Caplan, M. S. Lee, C. C. Adams, B. R. Fishel, D. S. Gross, and W. T. Garrard. 1989. Basal-level expression of the yeast HSP82 gene requires a heat shock regulatory element. Mol. Cell. Biol. 9:4789-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McNeil, J. B., H. Agah, and D. Bentley. 1998. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 12:2510-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 59.Moqtaderi, Z., M. Keaveney, and K. Struhl. 1998. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2:675-682. [DOI] [PubMed] [Google Scholar]
- 60.Morimoto, R. I. 1993. Cells in stress: transcriptional activation of heat shock genes. Science 259:1409-1410. [DOI] [PubMed] [Google Scholar]
- 61.Mueller, C. L., S. E. Porter, M. G. Hoffman, and J. A. Jaehning. 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14:447-456. [DOI] [PubMed] [Google Scholar]
- 62.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 63.Nieto-Sotelo, J., G. Wiederrecht, A. Okuda, and C. S. Parker. 1990. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell 62:807-817. [DOI] [PubMed] [Google Scholar]
- 64.Nightingale, K. P., R. E. Wellinger, J. M. Sogo, and P. B. Becker. 1998. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 17:2865-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 66.Prior, C. P., C. R. Cantor, E. M. Johnson, V. C. Littau, and V. G. Allfrey. 1983. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell 34:1033-1042. [DOI] [PubMed] [Google Scholar]
- 67.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 68.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 69.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12:1325-1332. [DOI] [PubMed] [Google Scholar]
- 70.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schubeler, D., D. M. MacAlpine, D. Scalzo, C. Wirbelauer, C. Kooperberg, F. van Leeuwen, D. E. Gottschling, L. P. O'Neill, B. M. Turner, J. Delrow, S. P. Bell, and M. Groudine. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18:1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwartz, B. E., and K. Ahmad. 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sealy, L., and R. Chalkley. 1978. DNA associated with hyperacetylated histone is preferentially digested by DNase I. Nucleic Acids Res. 5:1863-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sekinger, E. A., and D. S. Gross. 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105:403-414. [DOI] [PubMed] [Google Scholar]
- 77.Sekinger, E. A., and D. S. Gross. 1999. SIR repression of a yeast heat shock gene: UAS and TATA footprints persist within heterochromatin. EMBO J. 18:7041-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 79.Smith, M. M. 2002. Histone variants and nucleosome deposition pathways. Mol. Cell 9:1158-1160. [DOI] [PubMed] [Google Scholar]
- 80.Sorger, P. K. 1990. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 62:793-805. [DOI] [PubMed] [Google Scholar]
- 81.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 82.Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23:2800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szent-Gyorgyi, C., D. B. Finkelstein, and W. T. Garrard. 1987. Sharp boundaries demarcate the chromatin structure of a yeast heat-shock gene. J. Mol. Biol. 193:71-80. [DOI] [PubMed] [Google Scholar]
- 84.Thiriet, C., and J. J. Hayes. 2005. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 19:677-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 86.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745-756. [DOI] [PubMed] [Google Scholar]
- 87.Venturi, C. B., A. M. Erkine, and D. S. Gross. 2000. Cell cycle-dependent binding of yeast heat shock factor to nucleosomes. Mol. Cell. Biol. 20:6435-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walker, J., T. A. Chen, R. Sterner, M. Berger, F. Winston, and V. G. Allfrey. 1990. Affinity chromatography of mammalian and yeast nucleosomes. Two modes of binding of transcriptionally active mammalian nucleosomes to organomercurial-agarose columns, and contrasting behavior of the active nucleosomes of yeast. J. Biol. Chem. 265:5736-5746. [PubMed] [Google Scholar]
- 89.Weintraub, H., and M. Groudine. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193:848-856. [DOI] [PubMed] [Google Scholar]
- 90.Winston, F., and M. Carlson. 1992. Yeast Snf/Swi transcriptional activators and the Spt/Sin chromatin connection. Trends Genet. 8:387-391. [DOI] [PubMed] [Google Scholar]
- 91.Yamamoto, A., Y. Mizukami, and H. Sakurai. 2005. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 280:11911-11919. [DOI] [PubMed] [Google Scholar]
- 92.Zanton, S. J., and B. F. Pugh. 2004. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc. Natl. Acad. Sci. USA 101:16843-16848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang, L., S. Schroeder, N. Fong, and D. L. Bentley. 2005. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress.’ EMBO J. 24:2379-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]