Abstract
River microbial communities play an important role in global nutrient cycles, and aggregated bacteria such as those in epilithic biofilms may be major contributors. In this study the bacterial diversity of River Taff epilithon in South Wales was investigated. A 16S ribosomal DNA (rDNA) clone library was constructed and analyzed by partial sequencing of 76 of 347 clones and hybridization with taxon-specific probes. The epilithon was found to be very diverse, with an estimated 59.6% of the bacterial populations not accounted for by these clones. Members of the Cytophaga-Flexibacter-Bacteroides division (CFBs) were most abundant in the library, representing 25% of clones, followed by members of the α subdivision of the division Proteobacteria (α-Proteobacteria), γ-Proteobacteria, gram-positive bacteria, Cyanobacteria, β-Proteobacteria, δ-Proteobacteria, and the Prosthecobacter group. This study concentrated on the epilithic CFB populations, and a new set of degenerate 16S rDNA probes was developed to enhance their detection, namely, CFB560, CFB562, and CFB376. The commonly used probe CF319a/b may frequently lead to the underestimation of CFB populations in environmental studies, because it does not fully detect members of the division. CFB560 had exact matches to 95.6% of CFBs listed in the Ribosomal Database Project (release 8.0) small-subunit phylogenetic trees, compared to 60% for CF319a/b. The CFB probes detected 66 of 347 epilithon TAF clones, and 60 of these were partially sequenced. They affiliated with the RDP-designated groups Cytophaga, Sphingobacterium, Lewinella, and Cytophaga aurantiaca. CFB560 and CF319a/b detected 94% (62 of 66) and 48.5% (32 of 66) of clones, respectively, and therefore CFB560 is recommended for future use. Probe design in this study illustrated that multiple degenerate positions can greatly increase target range without adversely effecting specificity or experimental performance.
Epilithon is a biofilm which covers stones in aqueous environments such as river water. Nutrients are concentrated in the polysaccharide matrix of the biofilm, providing a carbon and energy reserve, and there is also physical protection from external hazards such as pollutants (16). This indicates that epilithic populations of bacteria are different from the free-living river water communities, and extensive differences have been noted between particle-attached and free-living bacteria in rivers (13). River microbial communities play an important role in global nutrient cycles (1), and aggregated bacteria such as those found in epilithon may be major contributors. Indeed, attached and particle-associated bacteria are 100 times more active than free-living bacteria and can account for 90% of heterotrophic bacterial activity (12, 13). Therefore, the relatively unknown and potentially important bacterial populations of river epilithon need to be investigated.
Until recently, the molecular analysis of bacterial diversity in freshwater ecosystems has received less attention than other environments (8, 22, 23, 36, 37, 39, 40, 46, 51, 52). In view of the estimate that <1% of bacteria in nature can be cultured (4, 17, 19), the analysis of 16S ribosomal DNA (rDNA) clone libraries represents the best method for examining epilithon communities, especially as most sequences retrieved from environmental samples represent previously undescribed species (24).
A clone library of natural freshwater biofilm has not yet been investigated, although one study of a marine biofilm has been carried out (14). Fluorescence in situ hybridization (FISH) has revealed the division level composition in river (9, 33), glacial stream (5), and drinking water biofilms (26, 27). River biofilms had diverse populations dominated by members of the α subdivision of the division Proteobacteria (α-Proteobacteria) and β-Proteobacteria, and the Cytophaga group of the Cytophaga-Flexibacter-Bacteroides (CFB) division. Conversely, stream and drinking water biofilms were dominated by β-Proteobacteria, with lower proportions of CFBs, and α- and γ-Proteobacteria.
The aim of this study was to estimate the bacterial composition and diversity of River Taff epilithon, via partial sequencing and taxon-specific oligonucleotide probing of a 16S rDNA clone library. We chose to concentrate on the CFB component of the epilithon clone library for the following reasons. First, CFB populations are frequently detected in diverse environments ranging from Antarctic sea ice (35) to terrestrial soils (21). Second, however, the number of representative CFB 16S rDNA sequences is less than that of other major divisions, such as the Proteobacteria. The Ribosomal Database Project (RDP) (31) release 8.0 phylogenetic trees divide the CFB division into 12 groups; Bacteroides and Cytophaga, Sphingobacterium, Lewinella, Persicobacter, Cytophaga aurantiaca, Flexibacter litoralis, Thermonema lapsum, strain SBR 2085, Rhodothermus, and three groups represented solely by environmental clone sequences. The Bacteroides and Cytophaga group has previously characterized the entire division and contains by far the most cultures and sequences. The former are obligately anaerobic rods, which are almost exclusively animal associated, while the latter include a phenotypically and physiologically diverse range of pigmented bacteria, some of which exhibit gliding motility and produce extracellular enzymes which degrade biomacromolecules.
Finally, CFB populations are potentially underestimated in the natural environment. This is because the commonly used CFB-specific probe CF319a/b (32) does not fully detect the whole CFB division (50). Therefore, we developed a new set of degenerate CFB-specific 16S rDNA-targeted oligonucleotide probes to obtain better coverage of CFB diversity.
MATERIALS AND METHODS
Organisms and culture conditions.
Reference strains used in this study are described fully in Results. They were cultured as indicated in the corresponding catalogues of the American Type Culture Collection (ATCC), the National Collections of Industrial and Marine Bacteria (NCIMB) (Scotland, United Kingdom), the Pasteur Culture Collection of Cyanobacteria (PCC) (Paris, France), and the National Collection of Type Cultures (NCTC) (London, United Kingdom).
Sampling.
Five stones were collected from each of three unshaded sites (15 stones in total), roughly 15 m apart on the River Taff in Cardiff (Ordnance Survey map reference, SO 783165) in October 1998. The stones were transported to the laboratory in individual sterile plastic bags and processed within 1 h of collection. Epilithon samples were obtained by scrubbing each stone in 100 ml of sterile deionized water with a sterile toothbrush (10). Aggregates in the samples were then dispersed by a 5-min treatment in a stomacher (Colworth Stomacher; A.J. Seward & Co. Ltd., London, United Kingdom), which works by beating the sterile plastic bag containing the samples with paddles, as described by Burton et al. (10). Epilithon samples were stored at −20°C in sterile containers.
DNA extraction.
Total community DNA was extracted according to the guanidinium thiocyanate method (41) modified by Björkroth and Korkeala (6). Briefly, 3-ml epilithon samples were centrifuged for 10 min at 15,800 × g and resuspended in 200 μl of HTE (50 mM Tris-HCl, 20 mM EDTA disodium salt [pH 8.0]). Next, 50 μl of lysozyme (50 mg ml−1) and 10 μl of mutanolysin (1 U μl−1; Sigma catalog no. M9901) were added before incubation for 15 min at 37°C; followed by 10 μl of proteinase K (16 mg ml−1) for 15 min at 37°C. Cells were lysed by addition of 500 μl of GES reagent (5 M guanidinium thiocyanate [Sigma], 100 mM EDTA, and 0.5% [wt/vol] n-lauryl sarcosine) and mixed for 5 to 10 min until clear. Then, 385 μl of 7.5 M sodium acetate was added and the mixture was centrifuged at 15,800 × g for 5 min. DNA was precipitated from the supernatant on ice with 0.6 volume of isopropanol for 10 min, and the pellet was washed twice with 100 μl of 70% ice-cold ethanol and air dried, before being resuspended in 100 μl of 10 mM Tris-HCl, pH 8.0, and stored at 4°C. The gentle enzymatic rather than physical disruption of cells potentially limited DNA shearing and chimera formation, while sequencing indicated that gram-positive bacteria were successfully disrupted.
PCR amplification of 16S rRNA genes.
Directly extracted epilithon DNA template was used in PCRs with no requirement for further purification. The primers 63F (5′-CAGGCCTAACACATGCAAGTC-3′) and 1387R (5′-GGGCGGWGTGTACAAGGC-3′) were used to amplify bacterial 16S rDNA (34), and primers 27F (5′-AGAGTTTGATCMTGGCTCAG 3′) and 1392R (5′-ACGGGCGGTGTGTRC-3′) were used to amplify bacterial and archeal 16S rDNA (29). Two different primer sets were used to reduce amplification bias associated with any one primer set. A typical 100-μl PCR mixture contained 20 pmol of each forward and reverse primer, 200 μM (each) deoxynucleoside triphosphates, 1 U of Taq polymerase (Bioline), Taq polymerase PCR buffer (Bioline), 1.5 mM MgCl2, and approximately 250 ng of template DNA. PCR was performed using an MJ Research PTC-100 machine. The cycles used were as follows: 1 cycle at 94°C for 10 min; 30 cycles at 92°C for 30 s, 55°C for 30 s, and 75°C for 1 min; and 1 cycle at 75°C for 5 min.
Clone library construction.
Two epilithon 16S rDNA clone libraries were constructed using DNA extracted from mixed epilithon samples from 15 stones collected at the three sampling sites. Library A (TAF-A) was made with primer pair 63F and 1387R in six cloning reactions to reduce potential random biases associated with individual reactions. Library B (TAF-B) was made with primer pair 27F and 1392R in a single cloning reaction, to compare the effects of different primer pairs on the composition of the clone library. Five replicate PCRs were mixed together for each cloning reaction to reduce random biases within individual PCRs. Clone libraries were constructed with the TOPO TA Cloning System (Invitrogen), according to the manufacturer’s instructions. Recombinant transformants were selected by blue-white screening and were checked for correct insert size by colony PCR using primers M13F (5′-GTAAAACGACGGCCAGT-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′). Clones were stored at −70°C as glycerol stocks in Luria-Bertani liquid medium containing 50 μg of ampicillin liter−1.
Sequencing and phylogenetic analysis.
The clone library was initially analyzed by partial sequencing of 76 randomly chosen clones. Of these, 44 clones were randomly selected from library TAF-A and 32 clones were separately selected from library TAF-B. In order to sequence a comparable region at the start of the 16S rRNA gene, the insert orientation of clones was determined via PCR using primers M13F and 1387R. Insert orientation screening was found to be successful in 71 of 76 cases; the clones sequenced in the reverse direction were not included in phylogenetic tree construction. Plasmid DNA was prepared for sequencing using Wizard plus SV Minipreps (Promega), and sequencing was done by a Licor DNA 4000L machine (MWG Ltd. Biotech, Milton Keynes, United Kingdom) using fluorescent primer labeling with near-infrared IRD800 according to the manufacturer’s instructions. Sequences were analyzed for chimera formation with the Check_Chimera software of the RDP. Nonchimeric sequences were compared to the RDP SSU_Prok data set (release 8.0) using the Sequence_Match analysis service (http://rdp.life.uiuc.edu/) and were also submitted to the BLAST search program (2) of the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). Clone 16S rDNA sequences, their closest relatives identified from database searches, and appropriate type strain sequences were aligned with CLUSTAL W (47). Reference sequences were obtained from the RDP Select_Sequence function and the GenBank database. Evolutionary distances were calculated by the method of Jukes and Cantor (25) and phylogenetic trees were constructed by the neighbor-joining method with TREECON for Windows (48), including bootstrap analysis (15).
Coverage (C) is a percentage term used to quantify how much of the environmental diversity is described by a clone library. It was estimated for the epilithon clone library by the equation C = [1 − (n1/N)] × 100, where n1 represents the number of clone types occurring only once (identical clones having >97% sequence similarity) and N is the number of clones being examined (18).
Slot blot hybridizations.
Clone libraries were screened with appropriate positive and negative controls by slot blot hybridization with taxon-specific 16S rDNA-targeted oligonucleotide probes, to determine the division and subdivision level composition of the clone libraries. Details of all probes used in this study are shown in Table 1. Approximately 0.5 μg of amplified clone insert was put in 100 μl of sterile polished water. This was denatured for 10 min at 95°C and cooled immediately on ice. Positively charged nylon membrane (Boehringer Mannheim) was soaked in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and placed above two pieces of Whatman 3MM filter paper on a 3-by-16 array slot blot manifold (Flowgen). Wells were loaded with the cooled sample and the vacuum was applied until wells were empty. All traces of sample were finally washed through by 100 μl of 2× SSC. Membranes were air dried, and the DNA was covalently fixed to the membrane by UV cross-linking for 2 to 3 min. Oligonucleotide probes were manufactured and 3′ end labeled with fluorescein by MWG Ltd. Biotech. Hybridization procedures were carried out using reagents and stringency washes as described previously (3, 32, 38, 43, 44). The chemiluminescence detection method with anti-fluorescein-AP Fab fragments (Boehringer Mannheim) was used according to manufacturer’s instructions. Membranes were exposed to photographic film (Kodak) for 2 to 10 min and developed according to manufacturer’s instructions.
TABLE 1.
16S rRNA-targeted oligonucleotide probes used in this study
| Probe | Specificity cited by original authors | Sequence (5′-3′) | Target sitea | Reference |
|---|---|---|---|---|
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 338–355 | 3 |
| ALF968 | α- and several δ-Proteobacteria | GGTAAGGTTCTGCGCGTT | 968–985 | 38 |
| CYA361 | Cyanobacteria | CCCATTGCGGAAAATTCC | 361–378 | 44 |
| G+VE | Gram-positive bacteria | TCATCATGCCCCTTATG | 43 | |
| CF319a/b | Cytophaga-Flavobacterium cluster | TGGTCCGTRTCTCAGTAC | 319–336 | 32 |
| CFB560 | CFB division | WCCCTTTAAACCCART | 560–575 | This study |
| CFB562 | CFBs excluding Cytophaga group | TACGYWCCCTTTAAACCCA | 562–580 | This study |
| CFB376 | CFBs excluding Cytophaga and Bacteroides groups | TGMCCAATATTCCYTACTG | 376–394 | This study |
Position in the 16S rRNA gene of Escherichia coli.
CFB probe design and specificity testing.
A CLUSTAL W alignment of 16S rDNA sequences from CFBs and representatives of other major lineages of Bacteria was manually screened for signatures characteristic of the CFB division. Potential probes were checked using the RDP Probe_Match software to determine theoretical range and specificity of the probe. To estimate the proportion of CFBs detected by the proposed probes, a complete list of CFB sequences in the RDP was obtained and compared with the Probe_Match results. In both cases the number of sequences with a catalogue number for a culture collection was determined. Environmental clone sequences were excluded because they were often quite short. The probes identified as having good theoretical specificity and range were then tested experimentally against a panel of target and nontarget 16S rDNAs amplified from reference organisms. One of the main concerns in oligonucleotide probe design is that one mismatch will not be discriminated. Bacteria showing one mismatch to the CFB probes were grouped according to the specific mismatch they had, and some examples were tested under optimized hybridization conditions (data not shown).
Hybridization conditions for the CFB probes were determined empirically by increasing the percentage of formamide in the hybridization solution, while decreasing the NaCl concentration in the washing solution according to the formula of Lathe (30). The temperature was held constant at 35, 46, and 46°C for probes CFB560, CFB562, and CFB376, respectively. Prehybridization was for 2 h in 5× 0, 40, and 30% formamide in the hybridization solution, respectively, and at 0.037, 0.037, and 0.074 M NaCl in the stringency wash, respectively.
Nucleotide sequence accession numbers.
River Taff epilithon 16S rDNA non-CFB TAF clones were assigned GenBank accession numbers as follows: α-Proteobacteria, AY038695 to AY038709 β-Proteobacteria, AY038710 to AY038714 γ-Proteobacteria, AY038715 to AY038724 δ-Proteobacteria, AY038725 Cyanobacteria, AY038726 to AY038733 gram-positive bacteria, AY038734 to AY038742; and Prosthecobacter group, AY038743. For the following CFB TAF clones and additional CFB reference sequences used in phylogenetic analysis, GenBank accession numbers are indicated in parentheses: TAF-A168 (AY038744), TAF-A35 (AY038745), TAF-A84 (AY038746), TAF-A174 (AY038747), TAF-A67 (AY038748), TAF-A86 (AY038749), TAF-A187 (AY038750), TAF-A38 (AY038751), TAF-A191 (AY038752), TAF-A188 (AY038753), TAF-A120 (AY038754), TAF-A14 (AY038755), TAF-B87 (AY038756), TAF-A133 (AY038757), TAF-A228 (AY038758), TAF-A144 (AY038759), TAF-A70 (AY038760), TAF-A87 (AY038761), TAF-A199 (AY038762), TAF-A115 (AY038763), TAF-A147 (AY038764), TAF-A194 (AY038765), TAF-A198 (AY038766), TAF-A242 (AY038767), TAF-B41 (AY038768), TAF-B64 (AY038769), TAF-A131 (AY038770), TAF-B8 (AY038771), TAF-B2 (AY038772), TAF-B76 (AY038773), TAF-B80 (AY038774), TAF-A131 (AY038770), TAF-B56 (AY038775), TAF-B81 (AY038776), TAF-B90 (AY038777), TAF-B75 (AY038778), TAF-B40 (AY038779), TAF-B66 (AY038780), TAF-B77 (AY038781), TAF-A28 (AY038782), TAF-A44 (AY038783), TAF-A119 (AY038784), TAF-A125 (AY038785), TAF-A195 (AY038786), TAF-B100 (AY038787), TAF-A4 (AY038788), TAF-B9 (AY038789), TAF-A11 (AY038790), TAF-A130 (AY038791), TAF-A173 (AY038792), TAF-A186 (AY038793), TAF-A231 (AY038794), TAF-A237 (AY038795), TAF-B84 (AY038796), TAF-A99 (AY038797), Flavobacterium sp. strain A265 (U85888), CFB group bacterium A1002 (AF236015), metal-contaminated soil clone K20-54 (AF145849), Cytophaga sp. strain P1 (AF260716), Flavobacterium hibernum ATCC 51468 (L39067), Flavobacterium hydratis ATCC 29551 (M58764), Flavobacterium columnare strain RR-19 (M58781), uncultured bacterium BS3 (AF087055), Flavobacterium xylanivorum (AF162266), Flavobacterium aquatile ATCC 11947 (M62797), Flexibacter maritimus strain R2 ATCC 43398 (M64629), Cytophaga latercula LEWIN SIO-1 ATCC 23177 (M58769), Cytophaga marinoflava ATCC 19326 (M58770), Cytophaga uliginosa NCIMB1863 (D12674), Microscilla aggregans subsp. Catalatica strain HI-3 ATCC 23190 (M58791), Riemerella columbina LMG11607 (AF181448), Riemerella anatipestifer ATCC 11845 (U60101), unidentified (X85208), Haloanella gallinarum (AB035150), Chryseobacterium proteolyticum (AB039830), Chryseobacterium indoltheticum ATCC 27950 (M58772), Chryseobacterium balustinum ATCC 33487 (D14016), Cytophaga fermentans NCIMB2218 (D12661), Bacteroides distansonsis ATCC 8503 (M86695), Bacteroides fragilis ATCC 25285 (M61006), Bacteroides eggerthii NCTC11185 (L16485), Flexibacter canadensis ATCC 29591 (M62793/M28055), soil bacterium SC-I-70 (AJ252650), Sphingobacterium sp. strain OM-E81(AB020206), Pedobacter heparinus strain HIM762-3 ATCC 13125 (M11657), clone Sva1038 (AJ240979), Cytophaga sp. strain JTB251 (AB015265), soil bacterium clone SC-I-12 (AJ252615), Flexibacter elegans (M58782), Saprospira grandis ATCC 23119 (M58795), Flexibacter filiformis Fxe1 ATCC 29495 (M58782), Flavobacterium ferrugineum ATCC 13524 (M62798), Flexibacter polymorphus ATCC 27820 (M58786), Flexibacter roseolus strain Lewin CR-155 ATCC 23088 (M58787), Flexibacter litoralis Lewin SIO-4 ATCC 23117 (M58784), Persicobacter diffluens strain LEWIN LIM-1 ATCC 23140 (M58765), Microscilla arenaria strain HJ-1 ATCC 23161 (M60455), Flectobacillus sp. GWF23A (AJ011696), Flectobacillus major ATCC 29496 (M62787/M27800), Spirosoma linguale strain Mcl ATCC 23276 (M62789/M27802), Spirosoma-like sp. strain PC5. 1A (X89911), Cytophaga aurantiaca ATCC 12208 (D12658), Cytophaga hutchinsonii D465 ATCC 33406 (D12663), Flexibacter flexilis CR-63 ATCC 23079 (M62794/M28056), unidentified bacterium IFAM3211 (X90701), soil bacterium clone C113 (AF013535), Microscilla sericea strain SIO-7 ATCC 23182 (M58794), Cytophagales strain S23328 (D84607), Taxeobacter strain Txcl (Y18837), Hymenobacter actinosclerus (Y17356), Thermonema lapsum ATCC 43542 (L11708), Rhodothermus obamensis (X77140), and green sulfur bacterium (root) Chlorobium limicola strain UdG6940 (Y10642).
RESULTS
Clone library construction.
Two epilithon 16S rDNA clone libraries, containing a total of 347 clones, were constructed. Library A (TAF-A) contained 243 clones and was made with primers 63F and 1387R, while library B (TAF-B) contained 104 clones and was constructed with 27F and 1392R.
Clone library analysis. (i) Random partial sequencing.
A sample of 76 clones was initially selected for partial sequencing and phylogenetic analysis. Partial sequence lengths ranged from 393 to 1,327 bp (median, 836 bp; 80% > 700 bp). Forty-five clones had unique sequences, and 31 fell into 10 sequence similarity groups (≥97% similar), giving 55 different sequences altogether. Coverage of the epilithon clone library based on the initial random sequencing was estimated at 40.7%.
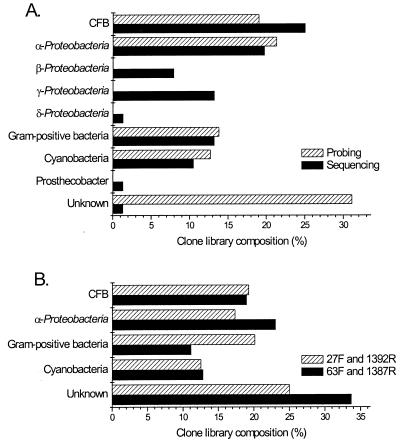
Clone sequences analyzed fell into nine major lineages of the domain Bacteria (24): CFB phylotypes constituted the most abundant division with 25.0% (19 of 76) of the sequenced clones, followed by 19.7% (15 of 76) α-Proteobacteria, 13.2% (10 of 76) γ-Proteobacteria, 13.2% (10 of 76) gram-positive bacteria, 10.5% (8 of 76) Cyanobacteria, 8% (6/76) β-Proteobacteria, 1.3% (1 of 76) δ-Proteobacteria, and 1.3% (1 of 76) Prosthecobacter group; 1.3% (1 of 76) of the clone library could not be assigned to any known lineage. The RDP analysis service Check_Chimera identified 6.6% (5 of 76) of the putatively chimeric clones; these were between distinctly different phylogenetic groups.
Clone library analysis. (ii) Hybridization with taxon-specific oligonucleotide probes.
PCR-amplified inserts from all 347 epilithon 16S rDNA clones were slot blotted onto positively charged nylon membrane and hybridized with a range of probes (Table 1). EUB338 detected 98.3% (341 of 347) of the clones; the undetected clones represented one α-Proteobacteria clone, two γ-Proteobacteria clones, two chimeras, and a Prosthecobacter-affiliated clone. The α-Proteobacteria probe ALF968 detected 21.3% (74 of 347) of the clone library, making this the most abundant group detected by probing, followed by 19.0% (66 of 347) of CFB clones detected by at least one of the probes CF319a/b, CFB560, CFB562, and CFB376; 13.8% (48 of 347) of gram-positive bacteria clones detected by G+VE; and 12.7% (44 of 347) of Cyanobacteria clones detected by CYA361. Only 2.0% (7 of 347) of clones gave positive signals with more than one probe and so were perhaps chimeric molecules; this may be an underestimation since chimeras can sometimes be detected by zero or one probe depending upon their nature. Altogether, 31.1% (108 of 347) of clones were not detected by any of the group-specific probes, and it is probable that these were affiliated with the β-, γ-, or δ-Proteobacteria or Prosthecobacter groups which were not detected by hybridization. No 16S rDNA probes are currently available for the entire β- and γ-Proteobacteria subdivisions, while δ-Proteobacteria and Prosthecobacter group probes were not used due to the low frequency of these groups found by sequencing.
Sequencing and hybridization revealed similar levels of CFBs, α-Proteobacteria, gram-positive bacteria, and Cyanobacteria (Fig. 1A); however, the hybridization approach was limited by lack of suitable probes for β- and γ-Proteobacteria. A comparison of the proportions of different divisions found in the TAF-A and TAF-B libraries is shown in Fig. 1B. Similar levels of CFBs and Cyanobacteria were found in both libraries, while TAF-A contained more α-Proteobacteria but fewer gram-positive bacteria than TAF-B.
FIG. 1.
(A) Comparison of composition of the epilithon TAF 16S rDNA clone library estimated by random partial sequencing of 76 of 347 clones and hybridization of the library with taxon-specific oligonucleotide probes ALF968 (38), CYA361 (44), G+VE (43), CF319a/b (32), and CFB560, CFB562, and CFB376 (this study). (B) Comparison of the effects of primers (for TAF-A, 67F and 1387R [n = 243]; for TAF-B, 27F and 1392R [n = 104]) used on the composition of epilithon 16S rDNA clone libraries estimated by hybridization with taxon-specific oligonucleotide probes.
CFB probe design and specificity testing.
Our studies with the extensively used probe CF319a/b showed that it did not fully detect the CFB division, and neither was it specific for the Cytophaga group. A theoretical estimate of the target range of CF319a/b was obtained by comparing the number of 16S rDNA sequences for culture collection CFBs in the RDP database with the number from the RDP Probe_Match results. CF319a/b had exact target matches for only 60% of CFBs and was particularly poor at detecting the Lewinella, F. litoralis, and T. lapsum groups (Table 2).
TABLE 2.
Theoretical target ranges of various CFB 16S rDNA probesa
| CFB group | No. of sequences with zero mismatches/no. tested (probe specificity)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CFB560 (all CFBs) | CFB562 (most CFBs excluding Cytophaga group) | CFB376 (most CFBs excluding Bacteroides and Cytophaga groups) | CF319a/b (Cytophaga-Flavobacterium cluster) | BAC303 (Bacteroides group) | CFB286 (mainly freshwater, but some marine, soil, and food CFB isolates) | CFB563 (marine and animal-associated CFB isolates) | CFB719 (mainly animal-associated CFB isolates) | CFB972 (mainly animal-associated CFB isolates) | CFB1082 (human-associated CFB isolates) | FC (Cytophaga-Flavobacterium cluster) | |
| Bacteroides | 65/67 | 52/67 | 0/67 | 17/67 | 37/67 | 30/67 | 0/67 | 41/67 | 0/67 | 50/67 | 0/67 |
| Cytophaga | 91/96 | 0/96 | 0/96 | 77/96 | 0/96 | 0/96 | 73/96 | 1/96 | 60/96 | 0/96 | 26/96 |
| Sphingobacterium | 10/10 | 10/10 | 7/10 | 8/10 | 0/10 | 0/10 | 0/10 | 6/10 | 0/10 | 0/10 | 0/10 |
| Lewinella | 10/10 | 10/10 | 8/10 | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| Persicobacter | 5/6 | 3/6 | 5/6 | 5/6 | 0/6 | 3/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
| C. aurantiaca | 15/16 | 15/16 | 11/16 | 9/16 | 0/16 | 4/16 | 0/16 | 1/16 | 0/16 | 0/16 | 0/16 |
| F. litoralis | 3/3 | 2/3 | 2/3 | 0/3 | 0/3 | 1/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| T. lapsum | 1/1 | 1/1 | 1/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Overall CFBs | 200/209 | 93/209 | 34/209 | 117/209 | 37/209 | 38/209 | 73/209 | 49/209 | 60/209 | 50/209 | 25/209 |
Probes CFB560, CFB562, and CFB376 (this study); CF319a/b and BAC303 (32); CFB286, CFB563, CFB719, CFB972, and CFB1082 (50); and FC (28). Target range was calculated as the proportion of culture collection CFBs in the RDP small-subunit prokaryotic data set (release 8.0) which contained the probe target sequence with zero mismatches. The Rhodothermus, strain SBR 2085, and the three environmental clone groups were not included in the table because they contain no culture collection isolates.
Therefore, three new probes were designed with various specificities for detection of all CFB clones in the epilithon 16S rDNA clone libraries: CFB560, CFB562, and CFB376. The theoretical specificities of these probes compared very favorably with others described in the literature (Table 2) and were experimentally supported in the majority of cases by slot blot hybridizations against a panel of target and nontarget 16S rDNAs from culture collection organisms (Table 3). Exceptions were that CFB562 and CFB376 did not detect P. heparinus, while CF319a/b detected M. aggregans, C. indoltheticum, and S. grandis, but not M. arenaria, which was in contrast to theoretical predictions. Although only culture collection bacteria were used to estimate the theoretical probe specificities, environmental clone sequences in the same groups were also detected (data not shown). However, no clone sequences were theoretically detected from the Rhodothermus sp., strain SBR 2085, or the three environmental clone groups.
TABLE 3.
Results of control hybridizations for CFB-specific 16S rDNA-targeted oligonucleotide probesa
| Species | Catalogue no. | Phylogeny | Hybridization with:
|
|||
|---|---|---|---|---|---|---|
| CFB560 | CFB562 | CFB376 | CF319a/b | |||
| CFBs | ||||||
| Cytophaga fermentans | NCIMB 2218 | Bacteroides | + | + | − | + |
| Bacteroides distasonis | ATCC 8503 | Bacteroides | + | + | − | + |
| Bacteroides fragilis | ATCC 25285 | Bacteroides | + | + | − | − |
| Bacteroides eggerthii | NCTC 11185 | Bacteroides | + | + | − | − |
| Microscilla aggregans | NCIMB 1418 | Cytophaga | + | − | − | + |
| Chryseobacterium indoltheticum | NCIMB 2220 | Cytophaga | + | − | − | + |
| Flavobacterium aquatile | NCIMB 8694 | Cytophaga | + | − | − | + |
| Flexibacter maritimus | NCIMB 2154 | Cytophaga | + | − | − | + |
| Cytophaga uliginosa | NCIMB 1863 | Cytophaga | + | − | − | + |
| Cytophaga latercula | NCIMB 1399 | Cytophaga | + | − | − | + |
| Cytophaga marinoflava | ATCC 19326 | Cytophaga | + | − | − | + |
| Pedobacter heparinus | NCIMB 9290 | Sphingobacterium | + | − | − | + |
| Saprospira grandis | NCIMB 1363 | Lewinella | + | + | + | + |
| Microscilla arenaria | NCIMB 1413 | Persicobacter | + | + | + | − |
| Microscilla sericea | NCIMB 1403 | Cytophaga aurantiaca | + | + | + | + |
| Non-CFBs | ||||||
| Agrobacterium tumefaciens | Putative isolate | α-Proteobacteria | − | − | − | − |
| Burkholderia cepacia | NCTC 10661 | β-Proteobacteria | − | − | − | − |
| Acinetobacter calcoaceticus | NCIMB 8250 | γ-Proteobacteria | − | − | − | − |
| Proteus vulgaris | NCTC 6213 | γ-Proteobacteria | − | − | − | − |
| Pseudomonas fragi | NCTC 10689 | γ-Proteobacteria | − | − | − | − |
| Pseudomonas aeruginosa | Putative isolate | γ-Proteobacteria | − | − | − | − |
| Myxococcus xanthus | NCIMB 9412 | δ-Proteobacteria | − | − | − | − |
| Synechococcus sp. | PCC 7942 | Cyanobacteria | − | − | − | − |
| Synechocystis sp. | PCC 6803 | Cyanobacteria | − | − | − | − |
| Nostoc sp. mac | PCC 8009 | Cyanobacteria | − | − | − | − |
| Nostoc sp. | PCC 7524 | Cyanobacteria | − | − | − | − |
| Anabaena sp. | PCC 7126 | Cyanobacteria | − | − | − | − |
| Bacillus megaterium | ATCC 12872 | Gram positive | − | − | − | − |
| Bacillus cereus | NCTC 7464 | Gram positive | − | − | − | − |
| Bacillus subtilis | NCTC 3610 | Gram positive | − | − | − | − |
| Staphylococcus aureus | NCTC 6571 | Gram positive | − | − | − | − |
| Brevibacter acetylicum | Putative isolate | Gram positive | − | − | − | − |
| Arthrobacter atrocyaneus | Putative isolate | Gram positive | − | − | − | − |
| Clone 83.8% related to Verrucomicrobiales clone CO19 | Verrucomicrobiales | − | − | − | − | |
| Clone 87.8% related to Pirellula marina | Planctomyces | − | − | − | − | |
Probes CFB560, CFB562, and CFB376 (this study) and probe CF319a/b (32). PCR-amplified 16S rDNAs from target and nontarget bacteria were fixed onto nylon membrane and hybridized against probes under optimized conditions. The phylogeny of test organisms stated here is as listed in the RDP (release 8.0) small-subunit phylogentic trees.
Probe CFB560 was a degenerate probe that had exact target matches in 95.6% (200 of 209) of culture collection CFBs; the small number of undetected sequences could be due to database sequencing errors. The four sequences that make up the degenerate probe CFB560 detect different members of the CFB division. Briefly, CFB560a (ACCCTTTAAACCCAAT; boldface indicates degenerate positions) detects all the Cytophaga, Sphingobacter, and Persicobacter groups, most of the C. aurantiaca group, and a third of the Bacteroides group. CFB560b (TCCCTTTAAACCCAAT) detects roughly two-thirds of the Bacteroides group. CFB560c (ACCCTTTAAACCCAGT) detects most of the Lewinella and Flexibacter litoralis groups and one sequence from the Cytophaga aurantiaca group. Lastly CFB560d (TCCCTTTAAACCCAGT) detects only F. ferrugineum ATCC 13534 from the Lewinella group. CFB562 was a degenerate probe made up of four separate sequences, which had exact target matches in 44.5% of culture collection CFBs in the RDP database. CFB562 theoretically detects most CFBs outside the Cytophaga group. CFB376 was also a degenerate probe which had exact target matches in only 16.3% of culture collection CFBs in the RDP database. It theoretically detects 74% of CFBs outside Bacteroides and the Cytophaga groups.
Sequencing of CFB clones detected by CF319a/b, CFB560, CFB562, and CFB376.
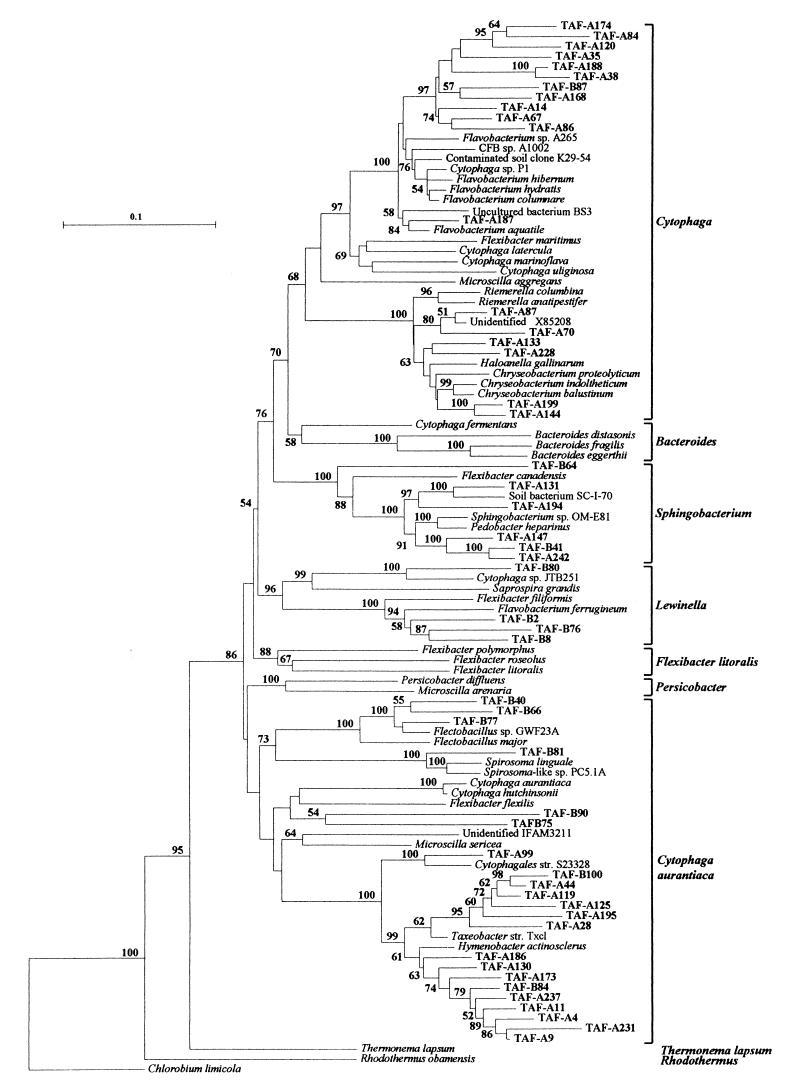
Altogether, 66 out of the 347 clones in libraries TAF-A and TAF-B were detected by one or more of the CFB probes used. Subsequently, 60 of these were partially sequenced and used in tree construction (Fig. 2). Partial sequence lengths ranged from 485 to 1,327 bp (median, 875 bp; 80% > 800 bp). The inclusion of shorter sequence lengths did not alter the topology of the phylogenetic tree shown in Fig. 2, compared to a tree constructed from type strain and clone sequences of more than 1,300 bp (results not shown). In addition, CFB TAF clones clustered in the same groups in the phylogenetic trees as predicted by the RDP and GenBank searches with Sequence_Match and BLAST, respectively. Unique sequences were found for 35 clones, while 25 were divided into six groups with >97% sequence similarity to each other. Overall, 41 unique taxa were found, and coverage for CFBs within the epilithon clone library was 37.5%. Twenty-two sequences affiliated most closely to the Cytophaga group; eight sequences belonged to the Sphingobacterium group; five clones belonged to the Lewinella group; and 24 sequences were affiliated with the Cytophaga aurantiaca group. A greater proportion of epilithon CFB clones, compared to non-CFB clones, represented novel 16S rDNA sequences, as 96.6% of CFBs but only 47.1% of non-CFBs were <97% similar to the closest match in the GenBank database.
FIG. 2.
Phylogenetic tree illustrating the relationship between the closest relatives in the RDP and GenBank databases and 50 CFB TAF clones (sequence lengths, 485 to 1,327 bp; median, 875 bp; 80% > 800 bp). This tree also illustrates the organization of groups within the CFB division. The strain SBR 2085 group and the three environmental clone groups are not included, because they contain only a few sequences in the RDP database. Epilithon clones from this study are indicated in boldface type and are designated TAF-A if they originated from library A constructed with primers 63F and 1387R (34) or designated TAF-B if they originated from library B constructed with primers 27F and 1392R (29). Phylogenetic groupings correspond to those given in the RDP (release 8.0) small-subunit phylogenetic trees. DNA sequences were aligned using CLUSTAL W, and the trees were constructed by the neighbor-joining program from a similarity matrix of pairwise comparisons made by the Jukes-Cantor algorithm in TREECON. The tree was rooted with the green sulfur bacterium C. limicola. Bootstrap values over 50, from 100 replicate trees, are shown at the nodes. The scale bar represents 10% difference in nucleotide sequence. The GenBank database accession numbers for all sequences used in the phylogenetic trees are listed in Materials and Methods.
Probe CF319a/b detected only 48.5% (32 of 66) of CFB clones in the epilithon clone library, which included all sequenced Cytophaga group clones, three of eight Sphingobacterium group clones, and 2 of 24 C. aurantiaca group clones. This reflected its theoretically inconsistent detection through the CFB division and bias toward the Cytophaga group. CF319a/b also detected three TAF clones not detected by any of the other probes, two of which were sequenced and found to represent a γ-Proteobacteria (TAF-B43) and a Cytophaga (TAF-A191) group sequence. Conversely, CFB560 detected 93.9% (62 of 66) of CFB clones, which was consistent with theoretical analysis showing its almost universal detection of CFBs. CFB562 detected 57.6% (38 of 66) of CFB clones, and, more specifically, no Cytophaga group sequences were detected, while all Sphingobacterium, Lewinella, and C. aurantiaca group clones were detected. CFB376 identified 45.5% (30 of 66) of CFB clones, matching exactly its theoretical specificity range.
DISCUSSION
Epilithon 16S rDNA clone library.
Sequencing and hybridization screening of TAF 16S rDNA clone libraries gave very similar results, suggesting that either method was acceptable for predicting division level abundances (Fig. 1A). The hybridization approach was limited by the lack of suitable probes available and difficulty in identifying chimeric clones: in this study 31% of the library was undetected mainly due to lack of probes for β- and γ-Proteobacteria, which together with chimeras constituted 28% of the sequenced clones. There appeared to be some bias associated with either primers 63F and 1387R or 27F and 1392R for α-Proteobacteria and gram-positive bacteria (Fig. 1B). The use of both primer sets in this study potentially limited the effect of this bias. Coverage of the whole clone library was estimated at 40.7%; thus, almost 60% of diversity in the epilithon sample was not accounted for. Coverage of the more extensively sequenced CFB component was a similar 37.5%, showing that epilithic bacteria were very diverse and that more complete sequencing of the whole library would not increase coverage. Therefore, a much larger library would be required to describe the complete community.
CFBs; α-, β-, γ-, and δ-Proteobacteria; gram-positive bacteria; Cyanobacteria; and Prosthecobacter group bacteria were all identified in the epilithon. These divisions were mostly consistent with those found in other river biofilms and aggregate populations; however, the proportions of these divisions appear to vary with both season and location. It is unlikely that the River Taff epilithon sampled in October 1998 would be representative of the Taff epilithon throughout the year or of epilithon in other rivers at the same time of year. Therefore, this study should be regarded as an investigation into a previously poorly described habitat rather than a comprehensive ecological study of river biofilms. It provides valuable phylogenetic information and guidance for future spatial and temporal investigations of structure-function relationships in river epilithon.
Comparisons can be made between the composition of River Taff epilithon assessed in this study and that of other freshwater and biofilm habitats. However, the evaluation of such comparisons is difficult because season, location, and sampling methodologies can all have a great influence on the described composition. CFBs and α-, β-, and γ-Proteobacteria have been found by FISH in two other river biofilms (9). β-Proteobacteria have been reported to dominate river biofilms during initial development, followed by a rapid increase in CFBs and α-Proteobacteria in the mature biofilm (33). Sediment biofilms of the Rotmoos glacial stream in the Austrian Alps were also dominated by β-Proteobacteria (5). The absence of members of the order Planctomycetales in epilithon was surprising given the widespread occurrence of these organisms in freshwater (49), but this may be explained by the observation that Planctomyces exhibits seasonal variation in river biofilms (9). Seasonal patterns have also been observed for CFBs in the same biofilm (9), and maximum aquatic CFB densities are frequently observed in winter (28, 42). FISH has shown that planktonic bacteria of two Japanese rivers was dominated by CFBs (28), but organic aggregates of the Elbe River were dominated by β- and γ-Proteobacteria (7). In the Columbia River α-, β-, and γ-Proteobacteria; gram-positive bacteria; CFBs; Planctomyces; and Verrucomicrobiales were observed as both particle-attached and free-living bacteria (13).
The proportions of divisions identified in epilithon were very different from those found in drinking water and seawater biofilms. A drinking water biofilm study found β-Proteobacteria dominant during all stages of development, and CFBs were <1% of total bacterial populations (26). The single clone library investigation of a seawater biofilm revealed a limited selection of only Rhodobacter and Alteromonas species (14).
The epilithon CFB populations.
CFBs are known to play an important role in the turnover of organic matter (11), and they represented one of the most abundant divisions found in our epilithon clone library. These results further suggest that if aggregate populations are responsible for the majority of heterotrophic activity in river ecosystems, CFBs would be some of the major contributors. The abundance and diversity of CFBs in the epilithon may be due to some CFBs possessing attributes which make them ideal biofilm organisms. For example, some CFBs exhibit gliding motility, which could be advantageous on a surface habitat, and the filamentous nature of some CFBs may act as a defense mechanism against invertebrate grazing (20). Some CFBs also utilize cellulose, chitin, DNA, lipids, and proteins, which would be abundant where dead organisms were trapped in the epilithic matrix.
Epilithon CFB 16S rDNA clones belonged to the Cytophaga, Sphingobacterium, Lewinella, and C. aurantiaca groups. No Bacteroides species were identified, which was consistent with the fact that they are obligately anaerobic and typically associated with animal hosts (45). More surprisingly, no TAF clones were assigned to the F. litoralis, Persicobacter, T. lapsum, strain SBR 2085, or Rhodothermus group or to the three environmental clone groups. However, these groups are the smallest in the CFB division, and the few sequences reported so far come from soil, hot springs, and deep sea hydrothermal vents. The fact that a greater proportion of epilithon CFB clones than non-CFB clones represented novel 16S rDNA sequences indicates that there is currently a lack of both cultured CFBs and environmental CFB clones in the sequence databases. This suggests that further studies are needed to provide more-reliable information about the diversity of the CFB division in all natural environments.
CFB probe design.
Probes CFB560, CFB562, and CFB376 were designed to enhance and complement existing CFB probes which do not detect the entire division and therefore potentially underestimate the proportion of environmental CFBs (Table 2). The theoretical probe specificities calculated here suggest these probes would be more effective than CF319a/b, which is commonly used. This was confirmed in practice as CFB560 detected nearly twice as many epilithon environmental clones as CF319a/b. However, no sequences were theoretically detected from the Rhodothermus, strain SBR 2085, or the three environmental clone groups, which could limit our probes’ utility. However, it should be noted that the latter groups contain a very small number of sequences that mainly come from extreme environments such as saline hot springs and deep sea hydrothermal vents, so the probes should be used with caution in studies of such environments.
Each probe designed in this study was degenerate, consisting of four individual sequences, some of which theoretically have the potential to be used alone with greater specificity. For example, CFB560a and CFB560b, when used together, would theoretically detect most members of the Bacteroides, Cytophaga, Sphingobacter, Persicobacter, and C. aurantiaca groups. While CFB560c would detect most of the Lewinella, F. litoralis, and T. lapsum groups. However, CFB560d would only detect F. ferrugineum, an identical profile to probe FFE8b (32) which has a different target region. F. ferrugineum may somehow be unique within the CFBs, although its phylogenetic position within the Lewinella group does not suggest this. The new probes could also be used to complement existing probes; for example, CFB563 detects most of the Cytophaga group, while CFB562 detects most CFBs outside the Cytophaga group.
Probe design in this study has illustrated that the inclusion of degenerate positions can greatly increase target range while still retaining phylogenetic specificity. No CFB-specific sequence of sufficient length to be a probe exists within the 16S rDNA sequence, and this is likely to be the case for other major divisions. Therefore, if the current range of bacterial phylogenetic probes is to be expanded, increased use of degeneracies should be considered. In this study probes CFB560 and CFB562 were designed to target an overlapping region, both containing the short core sequence TGGGTTTAAAGGG, specific to all CFBs. Careful use of degeneracies and variation of probe start and end positions allowed discrimination of phylogenetic groups within the CFB division. The fact that this same core signature sequence is present in CFB563 (50) further illustrates that small changes in a probe sequence can result in very different target ranges. CFB563 was less effective as a CFB general probe than our CFB560 and was designed with the Probe_Design tool of the ARB software package (O. Strunk, O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckmann, B. Nonhoff, M. Lenke, A. Ginhart, A.Vilbig, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig, Department of Microbiology, Technische Universitat München, Munich, Germany [http://www.mikro.biologie.tu-muenchen.de]), while our probes were designed manually. Both methods identified the same core region, but the software package failed to introduce degeneracies, showing that there is still room for manual probe design in molecular microbial ecology. We recommend that probe CFB560 be used to screen for CFBs in clone libraries and be tested as a FISH probe for counting CFBs in nature.
Acknowledgments
This work was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) and Syngenta Industrial CASE Studentship awarded to Louise A. O’Sullivan (reference number 98/A4/P/04730).
REFERENCES
- 1.Allan, J. D. 1995. Stream ecology structure and function of running waters. Chapman and Hall, London, United Kingdom.
- 2.Altschul, S. F., T. L. Hadden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chrisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battin, T. J., A. Wille, B. Sattler, and R. Psenner. 2001. Phylogenetic and functional heterogeneity of sediment biofilms along environmental gradients in a glacial stream. Appl. Environ. Microbiol. 67:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Björkroth, J., and H. Korkeala. 1996. Evaluation of Lactobacillus sakei contamination in vacuum-packaged sliced cooked meat products by ribotyping. J. Food Prot. 59:398–401. [DOI] [PubMed] [Google Scholar]
- 7.Bockelmann, U., W. Manz, T. R. Neu, and U. Szewzyk. 2000. Characterization of the microbial community of lotic organic aggregates (“river snow”) in the Elbe River of Germany by cultivation and molecular methods. FEMS Microbiol. Ecol. 33:157–170. [DOI] [PubMed] [Google Scholar]
- 8.Bossard, P. P., Y. Santini, D. Gruter, R. Stettler, and R. Bachofen. 2000. Bacterial diversity and community composition in the chemocline of the meromictic alpine Lake Cadagno as revealed by 16S r DNA analysis. FEMS Microbiol. Ecol. 31:173–182. [DOI] [PubMed] [Google Scholar]
- 9.Brummer, I. H. M., W. Fehr, and I. Wagner-Dobler. 2000. Biofilm community structure in polluted rivers: abundance of dominant phylogenetic groups over a complete annual cycle. Appl. Environ. Microbiol. 66:3078–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton, N. F., M. J. Day, and A. T. Bull. 1982. Distribution of bacterial plasmids in clean and polluted sites in a South Wales river. Appl. Environ. Microbiol. 44:1026–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crump, B. C., and J. A. Baross. 1996. Particle-attached bacteria and heterotrophic plankton in the Columbia River estuary. Mar. Ecol. Prog. Ser. 138:265–273. [Google Scholar]
- 13.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang, H., and C. R. Lovell. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. [DOI] [PubMed] [Google Scholar]
- 16.Freeman, C., and M. A. Lock. 1995. The biofilm polysaccharide matrix: a buffer against changing organic substrate supply? Limnol. Oceanogr. 40:273–278. [Google Scholar]
- 17.Giovannoni, S., and M. Rappé. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p.47–84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, Inc., New York, N.Y.
- 18.Giovannoni, S. J., T. D. Mullins, and K. G. Field. 1995. Microbial diversity in oceanic systems: rRNA approaches to the study of unculturable microbes, p.217–248. In I. Joint (ed.), Molecular ecology of aquatic microbes. NATO ASI series, vol. G38. Springer-Verlag, Berlin, Germany.
- 19.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso sea bacterioplankton. Nature 345:60–63. [DOI] [PubMed] [Google Scholar]
- 20.Hahn, M. W., E. R. B. Moore, and M. G. Hofle. 1999. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl. Environ. Microbiol. 65:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hengstmann, U., K.-J. Chin, P. H. Janssen, and W. Liesack. 1999. Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil. Appl. Environ. Microbiol. 65:5050–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiorns, W. D., B. A. Methe, S. A. Nierzwicki-Bauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofle, M. G., H. Haas, and K. Dominik. 1999. Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as determined by 5S rRNA analysis. Appl. Environ. Microbiol. 65:3164–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p.21–132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 26.Kalmbach, S., W. Manz, and U. Szewzyk. 1997. Dynamics of biofilm formation in drinking water: phylogenetic and metabolic potential of single cells assessed by formazan reduction and in situ hybridisation. FEMS Microbiol. Ecol. 22:265–279. [Google Scholar]
- 27.Kalmbach, S., W. Manz, and U. Szewzyk. 1997. Isolation of new bacterial species from drinking water biofilms and proof of their in situ dominance with highly specific 16S rRNA probes. Appl. Environ. Microbiol. 63:4164–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenzaka, T., N. Yamaguchi, K. Tani, and M. Nasu. 1998. rRNA-targeted fluorescent in situ hybridisation analysis of bacterial community structure in river water. Microbiology 144:2085–2093. [DOI] [PubMed] [Google Scholar]
- 29.Lane, D. J. 1991. 16S/23S rRNA sequencing, p.115–175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom.
- 30.Lathe, R. 1985. Synthetic oligonucleotide probes deduced from amino acid sequence data, theoretical and practical considerations. J. Mol. Biol. 183:1–12. [DOI] [PubMed] [Google Scholar]
- 31.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manz, W., R. Amann, W. Ludwig, M. Vancanney, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteriodes in the natural environment. Microbiology 142:1097–1106. [DOI] [PubMed] [Google Scholar]
- 33.Manz, W., K. Wendt-Potthoff, T. R. Neu, U. Szewzyk, and J. R. Lawrence. 1999. Phylogenetic composition, spatial structure, and dynamics of lotic bacterial biofilms investigated by fluorescent in situ hybridisation and confocal laser scanning microscopy. Microb. Ecol. 37:225–237. [DOI] [PubMed] [Google Scholar]
- 34.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCammon, S. A., and J. P. Bowman. 2000. Taxonomy of Antarctic Flavobacterium species: description of Flavobacterium gillisiae sp. nov., Flavobacterium tegetincola sp. nov. and Flavobacterium xanthum sp. nov., nom. rev. and reclassification of [Flavobacterium] salengens as Salegentibacter salegens gen. nov., comb. nov. Int. J. Syst. Bacteriol. 50:1055–1063. [DOI] [PubMed] [Google Scholar]
- 36.Methe, B. A., W. D. Hiorns, and J. P. Zehr. 1998. Contrasts between marine and freshwater bacterial community composition: analysis of communities in Lake George and six other Adirondack lakes. Limnol. Oceanogr. 43:368–374. [Google Scholar]
- 37.Miskin, I. P., P. Farrimond, and I. M. Head. 1999. Identification of novel bacterial lineages as active members of microbial populations in a freshwater sediment using a rapid RNA extraction procedure and RT-PCR. Microbiology 145:1977–1987. [DOI] [PubMed] [Google Scholar]
- 38.Neef, A. 1997. Anwendung der insitu-einzelzell Identifizierung von Backterien zur Population Analyse in komplexen microbiellen Biozonozen. Ph.D. thesis. Technische Universität München, Munich, Germany.
- 39.Pedersen, K., J. Arlinger, L. Hallbeck, and C. Petterson. 1996. Diversity and distribution of subterranean bacteria in groundwater at Oklo in Gabon, Africa, as determined by 16S rRNA gene sequencing. Mol. Ecol. 5:427–436. [DOI] [PubMed] [Google Scholar]
- 40.Pernthaler, J., F.-O. Glockner, S. Unterholzner, A. Alfreider, R. Psenner, and R. Amann. 1998. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacteria genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151–156. [Google Scholar]
- 42.Reichenbach, H. 1992. The order Cytophagales, p.3631–3687. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 4. Springer-Verlag, Berlin, Germany.
- 43.Rheims, H., C. Sproer, F. A. Rainey, and E. Stackebrandt. 1996. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology 142:2863–2870. [DOI] [PubMed] [Google Scholar]
- 44.Schonhuber, W., B. Zarda, S. Eix, R. Rippka, M. Herdman, W. Ludwig, and R. Amann. 1999. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 65:1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah, H. H. 1992. The genus Bacteroides and related taxa, p.3593–3607. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 4. Springer-Verlag, Berlin, Germany.
- 46.Speksnijder, A. G. C. L., G. A. Kowalchuk, K. Roest, and H. J. Laanboek. 1998. Recovery of a Nitrosomonas-like 16S rDNA sequence group from freshwater habitats. Syst. Appl. Microbiol. 21:321–330. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Peer, Y., and R. De. Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569–570. [DOI] [PubMed] [Google Scholar]
- 49.Ward, N., F. A. Rainey, E. Stackebrandt, and H. Schlesner. 1995. Unraveling the extent of diversity within the order Planctomycetales. Appl. Environ. Microbiol. 61:2270–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weller, R., F. O. Glockner, and R. Amann. 2000. 16S rRNA-targeted oligonucleotide probes for the in situ detection of members of the phylum Cytophaga-Flavobacterium-Bacteroides. Syst. Appl. Microbiol. 23:107–114. [DOI] [PubMed] [Google Scholar]
- 51.Zwart, G., W. D. Hiorns, B. A. Methe, M. P. van Agterveld, R. Huismans, S. C. Nold, J. P. Zehr, and H. J. Laanbroek. 1998. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst. Appl. Microbiol. 21:546–556. [DOI] [PubMed] [Google Scholar]
- 52.Zwart, G., R. Huismans, M. P. van Agterveld, Y. V. de Peer, P. De Rijk, H. Eenhoorn, G. Muyzer, E. J. van Hannen, H. J. Gons, and H. J. Laanbroek. 1998. Divergent members of the bacterial division Verrucomicrobiales in a temperate freshwater lake. FEMS Microbiol. Ecol. 25:159–169. [Google Scholar]